Abstract
Protein prenylation is a post translational modification that is indispensable for Ras–Rho mediated tumorigenesis. In mammals, three enzymes namely protein farnesyltransferase (FTase), geranylgeranyl transferase1 (GGTase1), and geranylgeranyl transferase2 (GGTase2) were found to be involved in this process. Usually proteins of Ras family will be farnesylated by FTase, Rho family will be geranylgeranylated by GGTase1. GGTase2 is exclusive for geranylgeranylating Rab protein family. FTase inhibitors such as FTI- 277 are potent anti-cancer agents in vitro. In vivo, mutated Ras proteins can either improve their affinity for FTase active site or undergo geranylgeranylation which confers resistance and no activity of FTase inhibitors. This led to the development of GGTase1 inhibitors. A well-defined 3-D structure of human GGTase1 protein is lacking which impairs its in silico and rational designing of inhibitors. A 3-D structure of human GGTase1 was constructed based on primary sequence available and homology modeling to which pubchem molecules library was virtually screened through AutoDock Vina. Our studies show that natural compounds Camptothecin (-8.2 Kcal/mol), Curcumin (-7.3 Kcal/mol) have higher binding affinities to GGTase-1 than that of established peptidomimetic GGTase-1 inhibitors such as GGTI-297 (-7.5 Kcal/mol), GGTI-298 (-7.5 Kcal/mol), CHEMBL525185 (-7.2 Kcal/mol).
Keywords: Prenylation, GGTase1, Rho, Auto dock, Camptothecin
Background
Protein prenylation is an important post translational modification through which naïve protein molecules are targeted to membranes. It also helps in protein-protein interactions and reversible binding of some transport proteins to membranes [1]. Prenylation is the addition of either Farnesyl or geranylgeranyl moieties to proteins. Many proteins including Ras superfamily of proteins require prenylation for their proper function [2]. Three independent prenylating enzymes namely protein farnesyl transferase (FTase) and two protein geranylgeranyl transferases (GGTase1 and 2) are responsible for addition of respective isoprenoids. The subtle change in amino acid recognition sequence by these three enzymes makes sure that any given protein is prenylated with only one of them. GGTase1 transfers geranylgeranyl diphosphate (GGPP) to proteins containing CAAX domain where C is cysteine, A can be any aliphatic amino acid, and X is always leucine. In a stark contrast GGTase2 recognize proteins with C-C or CXC domain and prenylate them. To date Rab family of proteins were the only known candidates to possess C-C or CXC domain [3, 4].
Rho family of GTPases (about eight members) belongs to Ras superfamily of protein that is geranylgeranylated by GGTase1. Members of Rho family especially RhoA and Rac1 play a vital role in Ras mediated transformation of NIH 3T3 cells [5]. Furthermore Ras prenylation, particularly farnesylation was targeted to prevent transformation of cells even today [6, 7]. Farnesylation was effectively stopped by inhibiting FTase through peptidomimetic compounds. Though proven to be very good drugs with amazingly nil side effects, FTase inhibitors (FTI) could not completely prevent tumor proliferation as some Ras isoforms like K-Ras-4B are resistant to FTIs like L-744,832, and FTI-277 or undergo alternative prenylation i.e. geranylgeranylation [8]. On the other hand most of the GGTase inhibitors found to date shows consistent results by arresting cells in G0/G1 phase and induce apoptosis [9].
FTase and GGTase1 are structurally very similar with a common alpha subunit [10]. The beta subunit gives specificity for the enzymes towards the protein substrate depends not only on the C- terminal tetrapeptide but also to the peptide sequence adjacent to it [11]. These two proteins also show high homology towards yeast mating factor proteins RAM1 and RAM2 which have farnesylation and geranylgeranylation transferase properties respectively. Unfortunately not much is known about the sequence of mammalian, especially human GGTase1 beta subunit but the gene for GGTase1 is homologous to yeast Saccharomyces cerevisiae CDC43 [2]. 3-D structures of human GGTase1 are also not available which severely impedes the usage of rapid techniques like high throughput screening etc. to find inhibitors for it. Computational modeling of protein is a handy technique through which structures of highly homologous proteins can be easily generated. One such structure for RabGGTase (GGTase2) has already been generated in order to understand the mechanism of prenylation [12]. Here we present a computation based 3-D model for human GGTase1 to which pubchem small molecule library was docked using AutoDock Vina run on PyRx0.8 graphic user interface (GUI).
Methodology
Sequence analysis:
Human GGTase1 protein amino acid sequence (P53609) was retrieved from UniProt website http://www.uniprot.org/. BLAST was performed from the blast tab available in UniProt website using default settings.
Homology modeling: the human GGTase1 protein sequence showed more than 90% homology with same protein from other organisms. Hence automatic modeling mode of SWISS MODEL was used without any optional template. The validated structure was then downloaded from the output file and used for further experiments [13, 14].
Docking:
Compounds with less than 1000 atoms and 1000 bonds were downloaded from pubchem small molecule library. Autogrid was constructed with default values around the active site predicted by CASTp [15]. Molecular docking was done using AutoDock Vina tool in a PyRx graphic user interface (GUI) version 0.8 [16] that run on a PC containing Windows7. These dockings were compared with those of well-known inhibitors like GGTI-297, GGTI-298 and CHEMBL525185. Chimera [17] was used for visualization and rendition of high resolution images.
Results & Discussion
Local alignment of P53609 showed that the protein sequence is about 98% similar to the sequence of Rat GGTase1 for which Xray structure is resolved with a resolution of 2.85Å. The high degree of sequence similarity provided us the way to use automatic modeling mode in SWISS MODEL and the output was generated (Figure 1A). GGTase1 model obtained from SWISS MODEL was superimposed with that of template protein 1N4Q (Figure 1B). Docking interactions of modeled protein with various putative inhibitors are shown in (Figure 2). Most of the inhibitors were docked to Thr49, Tyr272, Trp275. Docking scores of compounds with high dock score were tabulated Table 1 (see supplementary material). These results show that natural compounds camptothecin and curcumin with dock scores -8.2 and -7.3 respectively may potently inhibit GGTase1 better than the known inhibitors GGTI-287,297,298 and CHEMBL525185 with dock scores of -6.5, -7.5, -7.5, -7.2 respectively.
Figure 1.
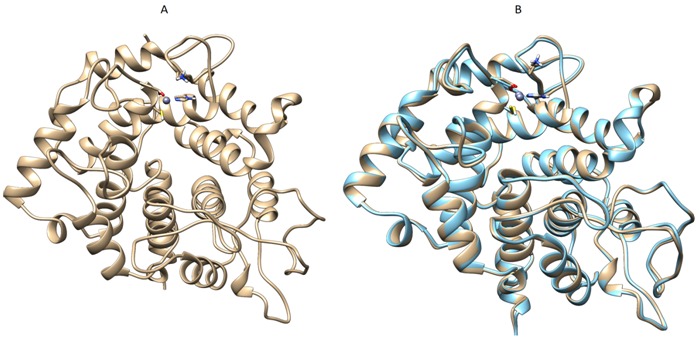
A Homology modeled of human GGTase1 downloaded from SWISS-MODEL workspace. B. Superimposition of modeled protein (gold) with its template 1N4Q (cyan).A blast of mouse GGTase1 protein against that of PDB database yielded 3-D model of 1N4Q with >90% sequence similarity. Using 1N4Q as template human GGTase1 is modeled (A) and sumperimposed with its template (B) for clarity.
Figure 2.
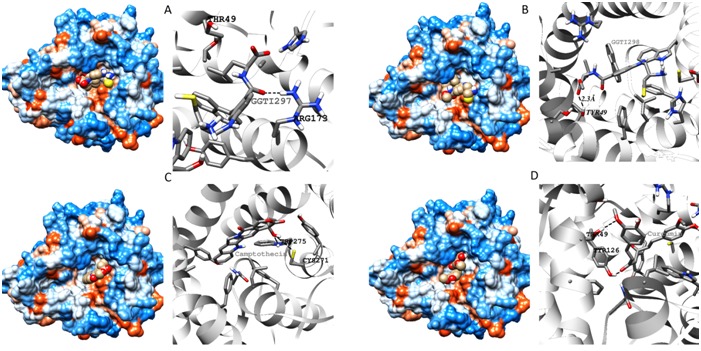
Docking simulation diagrams of GGTase1 with A. GGTI297 B. GGTI298 C. Camptothecin and D. Curcumin. Docking simulations using Autodock yielded structures which were saved as PDB files and later imaged using Chimera v1.8. Protein surfaces with ligand spheres are shown in left. In right side ribbon structures with hydrogen bonds (dashed lines) and bond length are shown.
Ras mediated transformation is manifested in many cancers. As mentioned earlier, a combined administration of FTase and GGTase1 inhibitors was proven to better cancers facilitated by Ras. To date the only proven drugs that act against GGTase1 were the peptidomimic compounds that are structurally similar to CAAX tetrapeptide. in silico docking is a fast track method for screening drugs with low cost but high efficiency and rapidity. Many compounds are already available in markets that were initially screened by in silico methods. But there are no inhibitors for GGTase1 other than peptidomimic compounds reported. This is mainly due to the lack of a 3-D structure of GGTase1. Here we present for the first time a homology modeled structure of GGTase1 and docked a library of natural compounds against it using AutoDock Vina. Our findings such that camptothecin and curcumin as putative inhibitors may go a long way in dealing with Ras mediated tumors. Camptothecin, a well-known topoisomerase I inhibitor was an anti-cancer drug [18]. It was discontinued as a drug due to its side effects. Curcumin, a pleiotropic molecule is shown to ameliorate cancer progression by a number of ways [19]. We hypothesize that anti-cancer activity of these compounds are in part may be due to GGTase1 inhibition. We have to further study and elucidate the complete mechanism of action of camptothecin and curcumin on GGTase1 inhibition.
Supplementary material
Acknowledgments
The financial support provided by the University Grants Commission, New Delhi [UGC F. No: 42-176/2013(SR)], India.
Footnotes
Citation:Thippanna et al, Bioinformation 9(19): 973-977 (2013)
References
- 1.Zhang FL, et al. Annu Rev Biochem. 1996;65:241. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 2.Casey PJ. J Lipid Res. 1992;33:1731. [PubMed] [Google Scholar]
- 3.Casey PJ, et al. Proc Natl Acad Sci U S A. 1991;88:8631. doi: 10.1073/pnas.88.19.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox AD, Der CJ. Small GTPases. 2010;1:2. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khosravi-Far R, et al. Mol Cell Biol. 1995;15:6443. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebti SM, et al. Oncogene. 2000;19:6584. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann G, et al. Nature. 2013;497:638. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 8.Fiordalisi JJ, et al. J Biol Chem. 2003;278:41718. doi: 10.1074/jbc.M305733200. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, et al. Oncogene. 1998;16:1467. doi: 10.1038/sj.onc.1201656. [DOI] [PubMed] [Google Scholar]
- 10.Seabra MC, et al. Cell. 1991;65:429. doi: 10.1016/0092-8674(91)90460-g. [DOI] [PubMed] [Google Scholar]
- 11.James GL, et al. J Biol Chem. 1995;270:6221. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 12.Wu YW, et al. J Biol Chem. 2009;284:13185. doi: 10.1074/jbc.M900579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordoli L, et al. Nat Protoc. 2009;4:1. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 14.Arnold K, et al. Bioinformatics. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 15.Dundas J, et al. Nucleic Acids Res. 2006;34:W116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf LK, et al. Chemical & Engineering. 2009;87:48. News Archive. [Google Scholar]
- 17.Pettersen EF. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.Gallo RC, et al. Journal of the National Cancer Institute. 1971;46:789. [PubMed] [Google Scholar]
- 19.Subramani PA, et al. Nat Prod Commun. 2013;8:121. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


