Abstract
Microsatellites are abundant in the human genome and may acquire context-dependent functions. A highly polymorphic GT microsatellite is located downstream of the poly(A) signal of the human argininosuccinate synthetase (ASS1) gene. The ASS1 participates in urea and nitric oxide production and is a rate-limiting enzyme in arginine biosynthesis. To examine possible involvement of the GT microsatellite in ASS1 mRNA 3’-end formation, ASS1 minigene constructs were used in transient transfection for assessment of poly(A) site usage by S1 nuclease mapping. Synthesis of the major human ASS1 mRNA is found to be controlled by two consecutive non-canonical poly(A) signals, UAUAAA and AUUAAA, located 7 nucleotides apart where a U-rich sequence and the GU microsatellite serve as their respective downstream GU/U-rich elements. Moreover, AUUAAA utilization is affected by the GU-repeat number possibly leading to differential regulation of ASS1 polyadenylation in individuals with different repeat numbers. Interestingly, the less efficient UAUAAA motif is noted to be the major ASS1 poly(A) signal possibly as a result of an indispensable downstream U-rich element and restricted utilization of the AUUAAA motif by the presence of extended GU-repeats. The UAUAAA motif and the GT microsatellite are conserved only in primates whereas AUUAAA motif is present in all mammals analyzed. The suboptimal UAUAAA motif and the utilization of the polymorphic GT microsatellite as polyadenylation signal of the ASS1 gene may be used as a strategy in primates to modulate ASS1 level in response to interactions of genetic and environmental factors.
Keywords: Argininosuccinate synthetase 1 (ASS1), GT-repeat polymorphism, microsatellite, polyadenylation signal, poly(A) downstream element, gene regulation
Introduction
In eukaryotic cells, polyadenylation at the 3’-end of mRNA confers RNA stability and translatability and acts to signal transcription termination [1]. A crucial polyadenylation signal is the highly conserved AAUAAA hexamer located 10-30 bases upstream of the site of cleavage of the pre-mRNA and poly(A) addition. In addition, a much less conserved downstream GU- or U-rich sequence is also required for efficient polyadenylation. The AAUAAA hexamer and the GU- or U-rich region are known as the core of poly(A) signal [1]. The position of the GU- or U-rich element relative to the AAUAAA sequence is variable but usually lies 10-30 bases downstream of the poly(A) addition site [2]. The hexamer sequence and the GU- or U-rich elements are recognized by a number of trans-acting factors necessary for the first step of cleavage and polyadenylation. The hexamer element interacts with the cleavage and polyadenylation specificity factor (CPSF) while the cleavage stimulation factor (CstF) interacts with the GU- or U-rich elements [3].
Argininosuccinate synthetase 1 (ASS1; EC 6.3.4.5) participates in urea formation and is a rate-limiting enzyme in arginine biosynthesis. Arginine, one of the most versatile amino acids in animal cells, plays an important role in the synthesis of nitric oxide (NO) which is a messenger molecule with functions in a diverse range of processes including inflammation, immunity, vascular regulation and neurotransmission [4]. Since steps affecting arginine availability are potential control points for regulating NO synthesis, there have been growing interests in elucidating the mechanism of ASS1 gene regulation [5,6]. Moreover, tumours including hepatocellular carcinoma (HCC), melanoma, some mesotheliomas and renal cell cancers have been found not to express the ASS1 gene [7]. As a result, arginine deprivation employing the pegylated form of arginine deiminase (ADI-PEG20) as a targeted therapy is currently in clinical trials for patients with HCC and melanoma [7].
The human ASS1 gene encodes two mRNA species of 1.7 kb and 2.7 kb in size [8,9]. These transcripts are variable at the 3’-untranslated regions and the 1.7-kb species is about 20-fold more abundant than the 2.7-kb transcript. The canonical AAUAAA hexamer (Figure 1A, pA2) has been assigned as the poly(A) signal for the 2.7-kb transcript [9]. However, the poly(A) signal for the 1.7-kb mRNA, i.e. pA1, is still controversial. Two pre-mRNA cleavage/polyadenylation sites are located 17 nt apart and are designated in this study as the pA1prox cleavage site and pA1dist cleavage site (Figure 1A); these sites were identified based on cDNA sequences [10,11]. In the upstream sequence of these sites, there are two potential but non-canonical poly(A) hexamers, UAUAAA (pA1prox) and AUUAAA (pA1dist) (Figure 1A). The AUUAAA hexamer, the most common natural variant, was 77% efficient in in vitro polyadenylation analysis relative to the canonical AAUAAA signal whereas the UAUAAA motif that was found only in 4.37% of human mRNA poly(A) sites and was shown to be 17% efficiency in vitro [12,13]. The fact that the AUUAAA hexamer lies merely 7 nt upstream of the pA1prox cleavage site has led to the conclusion that AUUAAA might not function as a poly(A) signal in the generation of ASS1 mRNA [10]. On the other hand, we have identified a poly(GT) tract located 41 nt downstream of the pA1prox hexamer [9] (Figure 1A). GT-repeat is an abundant interspersed repetitive DNA widely distributed throughout the eukaryote genomes and the number of GT dinucleotides at each locus is highly polymorphic [14]. The GT-repeats in the genome are largely considered to be neutral in functions. Nevertheless, GT microsatellites have been suggested to act as transcriptional enhancers or silencers and to contribute to the recombination process [15]. Therefore, depending on its context, GT-repeat sequence may play some important roles in biological processes. Hence, we suspect the GT-repeat identified in the human ASS1 gene may serve as a downstream element for polyadenylation because of its location and sequence similarity to the GU-rich element.
Figure 1.

Effects of poly(A) hexamer sequences and the downstream GU/U-rich elements on pA1 usage. (A) Structure of the minigene plasmid pAS-2EBst. The bi-exonic minigene was transcriptionally driven by the SV40 early promoter (SV). The first exon (shaded box) included the human ASS1 cDNA exons 1 through 15 where a 30-nt foreign sequence (dark box) was inserted. The 3’-portion of the minigene included the complete intron 15 (Int15) and the terminal exon 16 (Ex16) of the gene. The sequence surrounding the pA1 signal is displayed where the proximal (pA1prox) and distal (pA1dist) poly(A) hexamers are underlined and their respective cleavage sites are indicated by arrows. The downstream U/GU-rich elements are indicated as follows: the U-rich (U1) element is double-underlined and the GU-repeat is designated as (GU)n where n referred to repeat number. (B) S1 nuclease mapping analysis of RNAs derived from the ASS1 minigene constructs. Total RNAs were prepared from minigene-transfected HuH-7 cells and were analyzed with the depicted probe. The structure of the 3’-end-labeled probe and the expected sizes of fragments protected by the respective RNA are shown. A probe was specifically tailored for each minigene. Asterisks indicate [32P] labels at the 3’-end of the probe. The minigene pAS-2EBst and its modification derivatives are listed. The region highlighted in (A) are displayed where “-“ indicates identical nucleotides to the prototype and nucleotides that are different from the prototype are shown. The intensity of the protected band was quantified by the use of a phosphorimager. The relative amount of pA1 RNA of the minigene is represented as a ratio of pA1 signal to total signals of pA1, pA2 and pA3 combined. Lane M is a 5’-end-labeled DNA marker where the fragment sizes are shown on the left. Lane Cont. was a control RNA prepared from cells transfected with the parental plasmid pSVpoly that was used in the minigene construction. The endogenous ASS1 mRNA-protected fragment (endo ASS1) is also indicated. The gel displayed is typical of two different experiments using independently prepared RNAs.
The goal of this work was to investigate such a possibility and to study which of the UAUAAA and AUUAAA hexamer serves as the major poly(A) signal for the 1.7-kb ASS1 mRNA production.
Materials and methods
DNA isolation and assessment of GT repeat genotype
Normal control DNAs used in this study included 25 leukocytes DNAs collected in the period 1984-1990 according to regulation of the Taipei Veterans General Hospital at the time of collection and 183 DNAs of unrelated Han Chinese individuals from the Taiwanese Cell and Genome Bank [16] kindly provided by the National Genotyping Center, Academia Sinica, Taiwan. The GT-repeat number was determined by PCR-based microsatellite genotyping using the following primer pair with the fluorescent dye FAM attached to the 5’-end of the forward primer: forward primer, 5’-FAM-GAGACACTAGTCTTTTATTTCTAG-3’; reverse primer, 5’-TTCGGCAGCACTTAGGTC-3’. PCR products were analyzed using an ABI PRISM 377 automated DNA sequencer or an ABI 3770 capillary sequencer (Applied Biosystems). Genotypes were defined and edited using the Applied Biosystems GeneScan and Genotyper software and validated by cloned DNAs with known GT-repeat number.
Plasmid construction
Plasmid pAS-2E was a human ASS1 expression plasmid with two exons under transcriptional control of the SV40 early promoter. This minigene plasmid originated from plasmid AS-2E-SAAC/123 [17] but carried the wild-type splice acceptor site and the polyadenylation signals pA1 and pA2. The plasmid pAS-2E served as a prototype for subsequent modifications. Plasmid pAS-2EBst (Figure 1A) carried a 30 nt insertion sequence (5’-GTGACCGGATCCACTCTGGCTACCTGGGTC-3’) at the unique BstEII site in the first exon of pAS-2E. The minigenes with the pA1 hexamer changed to the canonical signal AATAAA or the deleterious signal AAGAAA were constructed using the QuikChange site-directed mutagenesis protocol (Stratagene). The constructs with the pA1 downstream T-rich or GT- sequence modified to include sequence inversion, deletion or variation in the GT repeat number were generated by the common recombinant DNA techniques [18]. To create a DNA fragment as probe for S1 nuclease mapping, an intronless plasmid was constructed by replacing the Bsu36I-SpeI fragment covering the entire intron and part of the flanking exons of plasmid pAS-2EBst with the Bsu36I-SpeI fragment of the ASS1 cDNA. The homologous probe was used to examine RNA produced from each designated minigene in S1 nuclease mapping analysis.
Transient transfection and RNA analysis
The human HCC cell line, HuH-7, seeded at 1×107 cells per 150-mm culture dish, was transfected with 30 μg plasmid DNA by the calcium-phosphate co-precipitation method [17]. After transfection (72 h), RNA was isolated by the guanidium/caesium chloride method [18]. S1 nuclease mapping was performed using restriction fragments isolated from appropriate plasmids with the 3’-end labelled with [α-32P]dCTP by fill-in using DNA polymerase I, Klenow fragment [17]. The protected products were electrophorized through a 4% polyacrylamide gel containing 7 M urea. Gel was dried and analyzed either by autoradiography or by a phosphorimager for quantification with the ImageQuant software by Molecular Dynamics (Sunnyvale, CA).
Results
Effect of the pA1 hexamer sequence on the processing of the ASS1 pre-mRNA
To study the functional significance of the putative poly(A) signals and the associated downstream U-rich and GU-repeat elements in the synthesis of the human ASS1 mRNA, a transient transfection system using minigene construct was used. In order to extrapolate data obtained by in vitro studies to the in vivo situation, efforts were taken to generate a minigene construct with the 3’-terminus identical in structure to that of the endogenous gene. Thus, the minigene was designed such that the entire terminal intron (intron 15), the terminal exon (exon 16) and a genomic sequence located at about 980 nt downstream of the pA2 site of the human ASS1 gene were included (Figure 1A). Inclusion of intron 15 should provide not only the proper context for polyadenylation but also a splicing signal to facilitate mRNA export [17,19]. Moreover, since polyadenylation is also required for mRNA stabilization and accumulation, processing at the downstream pA2 site of exon 16 would stabilize transcripts that had bypassed processing at the pA1 site. The first exon of the minigene was derived from an ASS1 cDNA sequence and, thus, comprised of the sum of exons 1 to 15 of the ASS1 gene free of intron. To differentiate mRNA species expressed from the minigene and the endogenous ASS1 gene, the minigene was further modified by inserting a 30-nucleotide sequence (Figure 1A, dark bar) into the first cDNA-derived exon in the minigene construct. In S1 nuclease mapping analysis, the 30-nt insertion mismatch between the probe and the endogenous mRNA was digested by the S1 nuclease. The S1 probe was designed in such that mRNA employing the pA1 or pA2 signal would result in a protected fragment which was either 1330 nt or 2430 nt in size, respectively (Figure 1B). RNA generated without the intron being removed would be 1036 nt long which included the sequence from the 3’-end of the first exon of the minigene to the end-labelled site. For endogenous mRNA lacking the 30-nt insertion in the minigene, the protected region would be disrupted at the insertion site to generate a protected 552-nt fragment (Figure 1B).
A representative autoradiograph of an S1 mapping analysis performed on RNAs from transfected minigenes is shown in Figure 1B. When RNA from a wild-type minigene associated with 16 GT-repeats was examined, a major protected signal at 1330-nt position derived from the 1.7-kb RNA of the minigene was observed as would be expected (Figure 1B, construct 3). In addition, a protected fragment of 2430-nt from the 2.7-kb RNA was noted. A minor 1640-nt band was also detected that was likely derived from a mRNA species using a canonical poly(A) signal AAUAAA, designated as pA3, that resided at about 300 nt downstream of the proximal pA1 signal. The usage of pA3 was not particular to the minigene and could also be detected in the endogenous ASS1 mRNA in a canavanine-resistant epithelial cell line that overproduced the ASS1 mRNA [8] (data not shown). In this study, the fraction of RNA processed at pA1 was estimated by calculating the percentage of mRNA generated using the pA1 signal to that produced by the sum of pA1, pA2 and pA3 recognition. It is worth noting that both the 1.7-kb and 2.7-kb ASS1 mRNA species are highly stable with a half-life of approximately 15-20 h [20]. Thus, in the native sequence context, the efficiency of the use of pA1 was determined to be 95% (Figure 1B, construct 3), in good agreement with the in vivo situation in which the 1.7-kb RNA was found to be about 20-fold more abundant than that of the 2.7-kb RNA [8,9]. As a result, the minigene constructs employed in this study should have contained sufficient cis-elements to regulate mRNA 3’-end formation as in the case of the endogenous ASS1 gene. To study the effects of the variant poly(A) hexamer and the downstream U-rich and GU-repeat elements on pA1 usage, different modifications were introduced to obtain minigene constructs as shown in Figure 1B. When the UAUAAA motif (pA1prox) in the minigene was changed to the canonical AAUAAA sequence, a negligible amount of the mRNA was found that was processed at the pA2 site (Figure 1B, construct 1) suggesting that the appearance of the 2.7-kb mRNA is mainly due to recognition of the non-canonical pA1 hexamer. When the pA1prox hexamer was changed to AAGAAA that severely impaired the efficiency of polyadenylation in vitro [12], 87% of mRNA was still processed at the pA1 site (Figure 1B, construct 2). The mild effect of modifying the pA1prox AAGAAA on mRNA production suggests that the distal pA1 hexamer, i.e., AUUAAA, would also be functional (see below).
The GT microsatellite in the ASS1 gene is highly polymorphic
To investigate whether variations in the GT-repeat numbers would affect the efficiency of the preceding poly(A) signal, the extent of GT polymorphism in exon 16 of the human ASS1 gene was first examined. The repeat number was found to range from 14 to 25 by way of a bimodal distribution with peaks at 16 and 22 when genotypes of 208 unrelated Han Chinese individuals in Taiwan were determined (Figure 2). Furthermore, among the 208 individuals analysed, 164 cases are heterozygous at this locus. Thus, the ASS1 GT-repeat is highly polymorphic with 78.8% heterozygosity.
Figure 2.
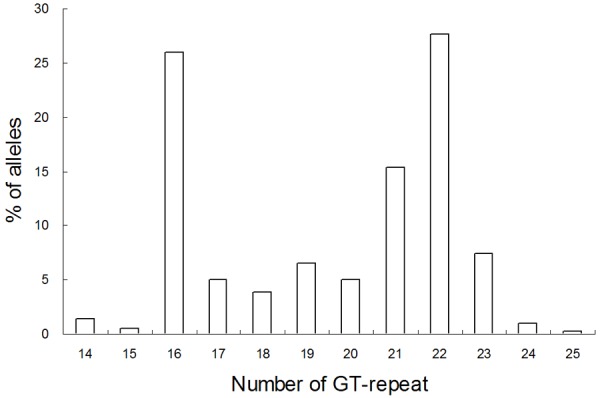
Frequency and distribution of GT-repeat number of the human ASS1 gene. The analysis was performed using the DNAs of 208 unrelated individuals.
Both the GU-repeat and the U-rich elements are required for efficient pA1 usage
Minigenes carrying 16 or 22 GT-repeats, the major alleles in the population under examination, were used to study the pA1 usage. When the number of GT-repeats in the minigene was increased from 16 to 22, the fractions of mRNA processed at the pA1 site were found to decrease from 0.95 to 0.93 with respect to the total amount of ASS1 mRNA (Figure 1B, constructs 3 and 4, respectively), or from 0.956 ± 0.009 to 0.926 ± 0.007 (P < 0.002 as determined by Student’s t test) when data from four independent analyses were compiled and statistically tested. Thus, a subtle but reproducible effect of GT-repeat polymorphism on ASS1 expression was, indeed, observed. Moreover, when the GT element was deleted entirely, a reduction of the 1.7-kb RNA to 66% occurred with an apparent concordant increase of the 2.7-kb RNA level (Figure 1B, construct 5). When the GT-repeat sequence was replaced by an AC-repeat of identical length, the pA1 RNA was 88% (Figure 1B, construct 6) indicating contribution of the GT microsatellite to the formation of the 3’-end of the ASS1 mRNA. On the other hand, when the U-rich element, designated as U1 in Figure 1A, was inverted, a reduction of relative pA1 RNA level to 78% was observed (Figure 1B, construct 7). It is conceivable that in the human ASS1 gene, even in the presence of the non-canonical poly(A) hexamer, efficient mRNA processing is achieved by cooperative interactions of the polyadenylation factors. Weak interactions of the CPSF protein with variant hexamer motifs may be compensated by stronger interactions of the CstF protein to downstream elements. In this regard, through in silico analysis of 3’-EST sequences, Graber et al. [21] have proposed that a complete polyadenylation signal is comprised of multiple cis-elements, and that no single exact sequence element is universally required for the processing of mRNA 3’-ends. Our findings are a demonstration that the overall polyadenylation efficiency is, indeed, a combined function and concerted effects of multiple elements.
Formation of the 3’-end of the 1.7-kb ASS1 mRNA is regulated by two consecutive poly(A) signals
Two poly(A) signals, UAUAAA and AUUAAA, separated by 7 nt, have been assigned to the generation of the 1.7-kb ASS1 mRNA [10,11]. To determine which of the hexamers is the major poly(A) signal, the cleavage/polyadenylation sites of the ASS1 mRNA were determined. To differentiate these sites in a close proximity, the S1 probe was end-labelled at the EagI site located 956 nt downstream of the HindIII site (Figure 3A). If the poly(A) addition site was at the pA1prox cleavage site as assigned by Bock et al. [10], a protected 392-nt probe fragment would be generated; otherwise, a pA1dist-derived 409-nt fragment was expected (Figure 3A). When RNAs prepared from HuH-7 cells transfected with the wild-type minigene were analyzed, three prominent bands of sizes 398 nt, 392 nt and 378 nt and a minor band at 409 nt were detected (Figure 3B, lane 1). It is worth noting that one of the termini of the RNA/DNA duplex formed between RNA processed at the pA1dist cleavage site and the DNA probe fell within the U/A-rich sequence and was prone to over-digestion by S1 nuclease [22]. The spurious 398-nt signal could, therefore, be a product derived from RNA generated using the pA1dist cleavage site due to S1 nuclease nibbling. Indeed, the signal of the 398-nt fragment was found to correlate with the strength of the pA1dist hexamer where the intensity of 398-nt fragment was about 29% of the total pA1 signal when pA1dist was associated with authentic AUUAAA hexamer (Figure 3B, lane 1) and was enhanced to 67% if pA1dist was associated with the canonical AAUAAA hexamer and reduced to 9% when associated with the deleterious AAGAAA hexamer (Figure 3C, lanes 1 and 2, respectively). Likewise, the 378-nt signal could have been derived from the 392-nt protected fragment as was evidenced by concurrent enhancement or reduction of signal intensities of both the 392-nt and 378-nt bands when the pA1prox hexamer was changed to the canonical AAUAAA or to a deleterious AAGAAA sequence, respectively (Figure 3B, lanes 2 and 3). For quantification, signal intensities of the 398-nt and 409-nt were taken together to represent activity of the pA1dist and that of 392-nt and 378-nt were taken as activity of pA1prox. It is noted in this study that mRNAs derived from the minigene construct and from the endogenous gene were indistinguishable by the S1 probe used. However, signal derived from the endogenous mRNAs could be disregarded due to their low level (Figure 3B, lane 7). Taken together, the data indicate that both the pA1prox and pA1dist hexamers are in use; in particular, the pA1prox is responsible for the production of about 70% to 80% of the amount of the 1.7-kb RNA (Figure 3B, lanes 1 and 4). A similar conclusion could be made for the endogenous ASS1 gene as shown in the analysis of mRNA derived from mock-transfected cells (Figure 3B, lane 7).
Figure 3.

Effects of the pA1 hexamer sequences and the downstream GU/U-rich elements on the usage of the proximal and distal pA1 signals. (A) Structure of the 3’-end-labeled probe and the expected sizes of fragments protected by RNA polyadenylated at the pA1prox or pA1dist cleavage site. The probe was 3’-end-labeled (asterisks) at the EagI site. (B) & (C) Polyacrylamide gel electrophoresis of the protected products derived from S1 nuclease analysis using total RNAs prepared from minigene-transfected HuH-7 cells. Structure features of the minigenes are indicated at the top of the gel where canon denotes the canonical hexamer, AAUAAA; mut denotes the AAGAAA hexamer and inv denotes an inverted sequence as indicated in Figure 1B. The U2 and U3 motifs are as shown in Figure 4. The intensity of the protected band was quantified by a phosphorimager. The pA1dist usage was calculated as a fraction of the pA1dist signal intensity to the combined signal intensities of pA1dist and pA1prox. The fragments of 409-nt along with 398-nt and those of 392-nt plus 378-nt are taken to represent pA1dist and pA1prox usage, respectively. In lane 6 in (B), the asterisked band was likely derived from the use of the pA1dist signal. And in lane 2 in (C), the band with open circle was likely derived from the use of pA1prox signal and the generation of such a band is probably due to stabilization of the DNA/RNA duplex terminus at the position of the G residue in the AAGAAA mutant. Open arrow on the left indicates non-specific band. The image of the left panel (lanes 1-8) was the result of phosphorimager and that of the right (lanes 9-12) was from autoradiography.
Differential regulation of the UAUAAA and AUUAAA motifs by the U-rich and the GU-repeat elements, respectively
In in vitro polyadenylation analysis, the UAUAAA and AUUAAA motifs are reported to be 17% and 77% efficient, respectively, relative to the canonical AAUAAA signal [12]. It is interesting that the less efficient UAUAAA pA1prox motif, instead of the AUUAAA pA1dist motif, is found in the above section to be the major poly(A) signal responsible for the synthesis of the human ASS1 mRNA. To delineate the role of the associated U-rich and the GU-repeat elements, poly(A) cleavage sites were examined in relation to these elements. Results show that when the number of the GT-repeats was increased from 16 to 22 in the minigene construct, a decrease in the signal intensity of the 398-nt and 409-nt bands together had led to a reduction of its contribution to total pA1 usage from 29% to 19% (Figure 3B, compare lanes 1 and 4). Thus, the reduction in pA1 usage observed in the minigene carrying 22 GT-repeats compared to that of 16 GT-repeats was due mainly to effect on the usage of the pA1dist signal (Figure 1B, constructs 3 & 4). Moreover, when the GT-repeat sequence was mutated to an AC-repeat sequence, the 398-nt band was found to reduce substantially (Figure 3B, lane 5) suggesting that the GU-repeat sequence is an important downstream element that facilitates the utilization of the pA1dist signal.
On the other hand, the U-rich element appears to affect mainly the pA1prox usage: when the U-rich element was modified by inversion, the 392-nt band was abolished and was replaced by a prominent 409-nt band (Figure 3B, lane 6, asterisked band). The 409-nt band was likely derived from an mRNA generated using the pA1dist signal in which case the shift of the protected band might be a result of disturbance of the distal cleavage sequence when the U-rich element was inverted. It is also possible that such inversion had led to the stabilization of the DNA/RNA duplex during S1 mapping analysis; the expected 409-nt band derived from pA1dist usage was, therefore, observed. It is noted that the probes used in the S1 nuclease mapping were tailored to match the RNA sequence derived from the particular minigene used in the study. Due to variations in the labelling efficiency of the S1 probes, it was difficult to compare the ASS1 mRNA levels derived from the various minigene constructs analyzed.
Extended GT-repeat sequence suppresses the efficiency of the distal pA1 signal
As demonstrated above, the GT microsatellite in the ASS1 gene acts as a GU-rich downstream element that facilitates pA1dist utilization. Moreover, efficiency of this motif is influenced by the number of the GU-repeats (Figure 3B, compare lanes 1 and 4). A survey has shown that the GU-rich downstream element is normally a short sequence like GUGUU in most of the mRNA species analyzed [23]. To study the optimal length of the GU-repeat in ASS1 transcripts, minigenes carrying different lengths of GT-repeats were tested (Figure 3C). The results show that when the number of GU-repeats was short (5 or 10 repeats), the pA1dist was the major signal in use and the usage of this signal was reduced as the length increased up to 26 GU-repeats (Figure 3C, lanes 4-8). Thus, to achieve efficient polyadenylation, the GU-repeat is maintained at an optimal length presumably to achieve ideal molecular spacing or an optimal RNA structure to allow stable interaction between the CPSF and CstF proteins which bind to the poly(A) hexamer and the downstream GU-rich element, respectively [3]. However, when the GU-repeat of the ASS1 gene was deleted entirely, major usage of the pA1dist hexamer was still observed (Figure 3C, lane 3). It is likely that under such a context, other U/GU-rich sequences residing further downstream participate in polyadenylation reaction. Two evolutionarily conserved stretches of T-rich sequences, designated as U2 and U3 (Figure 4), resided downstream of the GT-repeat of the ASS1 gene were found when sequences of placental mammals were aligned using the University of California Santa Cruz Genome Brower (http://genome.ucsc.edu/). We reasoned that if the U2 or U3 sequence could function as poly(A) downstream element in minigene when the GT-repeat was deleted, usage of the pA1dist motif would be affected by inversion of the U2/U3 elements. The data show that no matter if the U2 and U3 elements were altered singularly or in combination, recognition of pA1dist was unaffected (Figure 3C, compare lanes 9-11 to lane 12). Apparently, the U2 and U3 sequences did not function as a U-rich downstream element to promote pA1dist usage in the absence of the GU-repeat. It is likely that the pA1dist motif functioned adequately even in the absence of the downstream element although the presence of such element could enhance pA1dist activity substantially (Figure 3C, lane 4). Since CstF-64 has been shown to recognize UU dinucleotide (24), it is also possible that the GUU stretch resided between (GU)n and U2 (Figure 4) may serve as a suboptimal downstream element in combination with a relatively strong AUUAAA hexamer. However, when number of GU-repeat was increased, negative effects due either to the GU-length or to unfavourable RNA structure begin to appear (Figure 3C, lanes 6-8). And if the GU sequence was modified to poly(AC), strong deleterious effects resulted in low usage of pA1dist site (Figure 3B, lane 5).
Figure 4.
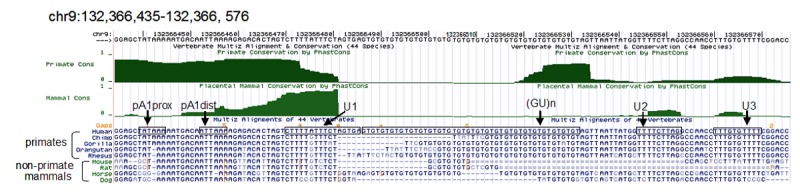
Analysis of conservation of sequences surrounding the polyadenylation site of the human ASS1 gene. Sequences equivalent to the human reference sequence hg18, chr9: 132,366,435-132,366,576 are displayed. Two measurements of evolutionary conservation are shown where Primate Conservation is derived from 9 primate species; Mammals Conservation is restricted to the placental mammals including 24 species. Shown below the conservation measurements are sequence alignments of human and the specified species. Double dashes indicate that the aligned species has one or more unalignable bases in the gap region. The pA1prox and pA1dist hexamer sequences and the downstream U-rich elements (U1, U2 and U3) and GU-repeats are boxed. The images are taken from the UCSC Genome Browser website (http:genome.ucsc.edu/).
Discussion
This study shows that the GT microsatellite in the human ASS1 gene serves as the poly(A) downstream GU-rich element to modulate ASS1 mRNA 3’-end formation. Such a finding grants microsatellite a new role in gene expression. On the other hand, studies have shown that dinucleotide repeat sequences may form Z-DNA to affect promoter activity when they reside at the 5’-end of a gene [25]. In the same token, one may envision that the particular DNA/RNA structure resulted from dinucleotide repeat sequences at the 3’-end of a gene can act as a signal to trigger transcription termination. Particularly, transcription termination is known to somehow couple to polyadenylation [1]. More work is needed to explore such a possibility.
Processing of the major 1.7-kb RNA of the human ASS1 gene is found in this work to use two non-canonical consecutive poly(A) signals, UAUAAA and AUUAAA, located 7 nt apart. Interestingly, the less efficient UAUAAA is shown to be the major signal employed in the ASS1 mRNA processing (Figure 3B, lanes 1, 4 & 7). This effect may be attributable to two factors. Firstly, the UAUAAA activity is facilitated by the indispensable downstream U-rich element. When the U-rich sequence was inverted, the function of this hexamer was severely hampered (Figure 3B, lane 6). Secondly, despite our demonstration that the GT-repeat served as a GU-rich downstream element for AUUAAA, extended GU-repeat lengths in the pre-mRNA in fact suppressed the functioning of AUUAAA as a poly(A) signal (Figure 3C). Thus, under the native context, the UAUAAA motif is selected. It is of interest to note that the pA1prox UAUAAA hexamer is found only in the ASS1 gene of primates (Figure 4). Similarly, longer GT-repeats are observed mainly in primates. For example, the GT-repeat numbers recorded in the reference sequences of human, chimp, gorilla, orangutan and rhesus are 23, 10, 15, 15 and 19, respectively, while it is only 3 in the mouse, rat and dog with one exception in the horse which is 18 (Figure 4). On the other hand, the pA1dist AUUAAA hexamer is highly conserved in mammalian ASS1 genes including those of the primate (Figure 4). One would envisage that in species, such as the mouse, that employ a single AUUAAA signal and a short GU downstream element, ASS1 mRNA levels would not be constrained by the poly(A) downstream element as found in the primate. In contrast, adaptation of the non-canonical UAUAAA hexamer in primates could be used as a strategy to modulate ASS1 expression at the polyadenylation step in response to physiological changes. In this regard, ASSI catalyzes the rate-limiting step in arginine biosynthesis. Its activity is critical for NO production since arginine is the only known substrate for all forms of NO synthase. Indeed, findings show that tumour necrosis factor-alpha (TNF-α) can affect ASS1 gene expression to deregulate NO production resulting in diseases [26,27]. Moreover, ASS1 was found to be seized by HSCARG, a redox sensor protein, to convey the cellular redox status to NO signalling [28]. Apparently, ASS1 participates in fine-tuning NO production to maintain cellular homeostasis in response to cellular and environmental stimuli. Moreover, argininosuccinate, the product of ASS1, has been proposed to function as a neuromodulator [29]. It is conceivable that the delicate control of ASS1 production is of pivotal importance for proper functioning of the brain and the nervous system. It would be, therefore, of advantage for primates to adapt the strategy of fine-tuning ASS1 expression at the polyadenylation level. However, the possibility that the appearance of the upstream UAUAAA hexamer in primates was evolved simply to compensate for the effect of a prior GT-repeat expansion cannot be ruled out.
In view of the critical roles of ASS1 in arginine synthesis and NO production, one would anticipate that the GT-repeat polymorphism that modulates the 3’-end formation of the ASS1 mRNAs is a likely candidate genetic determinant relevant to complex traits, including susceptibility to common disorders. Although the influence of this polymorphism on the baseline ASS1 gene expression levels may seem moderate, phenotypic outcome is likely to be amplified as a result of complex signal interactions within the genetic network. It is equally conceivable that such small quantitative variations observed in ASS1 may, by themselves, exert little effects. However, by a combination of multiple genetic variations from different genes, the consequence of such minute effects can have profound impact on disease susceptibility. Our preliminary study shows that the number of GT-repeats in some HCC patients skewed toward the longer size (T Su, unpublished data). Further studies on a much larger population are required to validate this initial finding.
Acknowledgements
We thank Dr. K.-B. Choo for critical reading and editing of the manuscript and the National Genotyping Center, Academia Sinica, Taiwan, for providing DNA samples. This work was supported in part by grant NSC 93-2311-B-075-002 from the National Science Council and by grants V90C-409 and V98C1-188 from the Taipei Veterans General Hospital, Taiwan.
References
- 1.Proudfoot N. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen JM, Férec C, Cooper DN. A systematic analysis of disease-associated variants in the 3’ regulatory regions of human protein-coding gene I: general principles and overview. Hum Genet. 2006;120:1–21. doi: 10.1007/s00439-006-0180-7. [DOI] [PubMed] [Google Scholar]
- 3.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.Morris SM Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev Nutr. 2002;22:87–105. doi: 10.1146/annurev.nutr.22.110801.140547. [DOI] [PubMed] [Google Scholar]
- 5.Tsai WB, Aiba I, Long Y, Lin HK, Feun L, Savaraj N, Kuo MT. Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res. 2012;72:2622–2633. doi: 10.1158/0008-5472.CAN-11-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haines RJ, Pendleton LC, Eichler DC. Argininosuccinate synthase: at the center of arginine metabolism. Int J Biochem Mol Biol. 2011;2:8–23. [PMC free article] [PubMed] [Google Scholar]
- 7.Feun L, You M, Wu CJ, Kuo MT, Wangpaichitr M, Spector S, Savaraj N. Arginine deprivation as a targeted therapy for cancer. Curr Pharm Des. 2008;14:1049–1057. doi: 10.2174/138161208784246199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su TS, Bock HGO, O’Brien WE, Beaudet AL. Cloning of cDNA for argininosuccinate synthetase mRNA and study of enzyme overproduction in a human cell line. J Biol Chem. 1981;256:11826–11831. [PubMed] [Google Scholar]
- 9.Su TS, Lin LH. Analysis of a splice acceptor site mutation which produces multiple splicing abnormalities in the human argininosuccinate synthetase locus. J Biol Chem. 1990;265:19716–19720. [PubMed] [Google Scholar]
- 10.Bock HG, Su TS, O’Brien WE, Beaudet AL. Sequence for human argininosuccinate synthetase cDNA. Nucleic Acids Res. 1983;11:6505–6512. doi: 10.1093/nar/11.18.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA Mammalian Gene Collection (MGC) Program Team. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber JL. Informativeness of human (dC-dA)n. (dG-dT)n polymorphisms. Genomics. 1990;7:524–530. doi: 10.1016/0888-7543(90)90195-z. [DOI] [PubMed] [Google Scholar]
- 15.Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- 16.Pan WH, Fann CS, Wu JY, Hung YT, Ho MS, Tai TH, Chen YJ, Liao CJ, Yang ML, Cheng AT, Chen YT. Han Chinese cell and genome bank in Taiwan: purpose, design and ethical considerations. Hum Hered. 2006;61:27–30. doi: 10.1159/000091834. [DOI] [PubMed] [Google Scholar]
- 17.Tsai TF, Wu MJ, Su TS. Usage of cryptic splice sites in citrullinemia fibroblasts suggests role of polyadenylation in splice-site selection during terminal exon definition. DNA Cell Biol. 1998;17:717–725. doi: 10.1089/dna.1998.17.717. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning - A Laboratory Manual. 3rd edn. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 19.Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci U S A. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai TF. Ph.D. dissertation. Taiwan: National Yang-Ming University; 1995. Regulation of human argininosuccinate synthetase gene expression. [Google Scholar]
- 21.Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3’-end-processing sequences in diverse species. Proc Natl Acad Sci U S A. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao HK, Hsiao KJ, Su TS. A silent mutation induces exon skipping in the phenylalanine hydroxylase gene in phenylketonuria. Hum Genet. 2001;108:14–19. doi: 10.1007/s004390000435. [DOI] [PubMed] [Google Scholar]
- 23.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenburg S, Koch-Nolte F, Rich A, Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc Natl Acad Sci U S A. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szlosarek PW, Grimshaw MJ, Wilbanks GD, Hagemann T, Wilson JL, Burke F, Stamp G, Balkwill FR. Aberrant regulation of argininosuccinate synthetase by TNF-α in human epithelial ovarian cancer. Int J Cancer. 2007;121:6–11. doi: 10.1002/ijc.22666. [DOI] [PubMed] [Google Scholar]
- 27.Goodwin BL, Pendleton LC, Levy MM, Solomonson LP, Eichler DC. Tumour necrosis factor-alpha reduced argininosuccinate synthetase expression and nitric oxide production in aortic endothelial cells. Am J Physiol Heart Circ Physiol. 2007;293:H1115–1121. doi: 10.1152/ajpheart.01100.2006. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Zhang J, Li H, Li Y, Ren J, Luo M, Zheng X. An NADPH sensor protein (HSCARG) down-regulates nitric oxide synthesis by association with argininosuccinate synthetase and is essential for epithelial cell viability. J Biol Chem. 2008;16:11004–11013. doi: 10.1074/jbc.M708697200. [DOI] [PubMed] [Google Scholar]
- 29.Wiesinger H. Arginine metabolism and the synthesis of nitric oxide in the nervous system. Prog Neurobiol. 2001;64:365–391. doi: 10.1016/s0301-0082(00)00056-3. [DOI] [PubMed] [Google Scholar]


