Abstract
Bicistronic expression vectors have been widely used for co-expression studies since the initial discovery of the internal ribosome entry site (IRES) about 25 years ago. IRES sequences allow the 5’ cap-independent initiation of translation of multiple genes on a single messenger RNA strand. Using a commercially available mammalian expression vector containing an IRES sequence with a 3’ green fluorescent protein fluorescent marker, we found that sequence length of the gene of interest expressed 5’ of the IRES site influences both expression of the 3’ fluorescent marker and overall transfection efficiency of the vector construct. Furthermore, we generated a novel construct expressing two distinct fluorescent markers and found that high expression of one gene can lower expression of the other. Observations from this study indicate that caution is warranted in the design of experiments utilizing an IRES system with a short 5’ gene of interest sequence (<300 bp), selection of single cells based on the expression profile of the 3’ optogenetic fluorescent marker, and assumptions made during data analysis.
Keywords: Bicistronic expression vectors, internal ribosome entry site, IRES sequences, transfection efficiency
Introduction
Internal ribosome entry site (IRES) elements provide the basis for 5’ cap-independent initiation of translation in the middle of a messenger ribonucleic acid (mRNA) molecule and were originally identified in both poliovirus RNA [1] and encephalomyocarditis virus RNA [2]. Viruses utilize IRES elements for cap and end-independent mRNA translation to maximize invasion and replication in the host [3]. In contrast, translation of eukaryotic messenger ribonucleic acid (mRNA) is critically dependent on the modification of the nucleotide located at the 5’ end of precursor mRNA [4]. The resulting 5’-7-methylguanosine cap structure defines the 5’-end of the message allowing the 43S complex of the ribosome to be transferred onto the 5’-end of the mRNA via the cap [4] and to then commence the scanning process to locate the initiation codon [5]. Recent studies have identified IRESs in eukaryotic cells supporting the notion that physiological processes that repress mRNA translation utilize IRES-mediated translation with numerous organisms characterized to date [6-8].
More recently, IRES elements from several RNA viruses [9] have been incorporated into bicistronic expression vectors widely used for co-expression studies including eukaryotic, retroviral, and lentiviral constructs [10,11]. Bicistronic vectors containing genetically-encoded fluorescent proteins, such as green fluorescent protein (GFP), have greatly simplified cell sorting and analysis in transfection studies by eliminating the intrinsic problems associated with co-transfection approaches [12].
Despite their widespread use there is very limited knowledge regarding the exact expression patterns of eukaryotic mammalian expression vectors, with most studies being carried out on retroviral constructs [12,13]. The issue is further complicated by the fact that translational efficiency is critically dependent upon IRES sequence and gene location [10]. Mizuguchi and colleagues reported that expression of the IRES-dependent gene was significantly lower than that of the cap-dependently translated one, both in vitro and in vivo [14].
In our own research studies, we observed large inter-experimental variability when working with IRES element-containing mammalian expression vectors, which could not be accounted for by experimental differences. More specifically, transfection efficiency of the plasmid and the expression level of the marker fluorescent protein, whose sequence was located 3’ to the IRES element, appeared to be significantly affected by the length of the gene sequence inserted 5’ to the IRES element. Previously, insert sequences of various lengths have been shown to affect expression levels of the second cistron, when inserted between the first cistron and the IRES sequence [15]. Similarly, hairpin loops interfered with gene expression in experimental systems [16]. This prompted us to conduct the present study in which we tested the effect of sequence length of the gene located 5’ of the IRES.
Using a commercially available mammalian expression vector containing an IRES element controlling the translation of GFP as a fluorescent marker, we found that sequence length of the gene of interest expressed 5’ of the IRES site influences both expression of GFP and overall transfection efficiency.
Furthermore, we generated a novel construct expressing a second fluorescent marker (tdTomato) [17] and found that high expression of one fluorophore was accompanied by significantly attenuated expression levels of the other, a finding with potentially profound consequences for studies using fluorescent markers in IRES element-containing mammalian expression vectors used to identify co-expression of unlabeled proteins.
We conclude that caution is warranted when using short (<200 bp) gene of interest sequences at the cap-dependent translation site and when selecting single cells based on the expression profile of a fluorescent marker under control of an IRES element.
Materials and methods
Vector design
Plasmid constructs were all based on the pIRES2.EGFP vector (Clontech, Mountain View, CA) that expresses enhanced green fluorescent protein (EGFP) [18] under the control of an IRES sequence. All vectors were assembled by standard PCR, restriction digest, ligation, and bacterial expansion with standard molecular biology methods, in all cases by the manufacturer’s protocol (Fermentas, Thermo, Glen Burnie, MD). Plasmid constructs were designed with Vector NTI v11 (Invitrogen, Carlsbad, CA) and DNA primers obtained from Integrated DNA Technologies (IDT, Coralville, IA). The 275 base pair (bp) insert was cloned from the N-terminus of mouse presenilin-2 (Accession# NM_011183; primers are listed in Supplementary Table 1) and inserted into pIRES2.EGFP inverted to create a space filling, non-coding sequence at the multiple cloning site (MCS) on the 5’ portion to the IRES with BamHI and EcoRI. The 1,400 bp insert was the fluorescent protein tdTomato, a generous donation by Roger Y. Tsien [17], a bright, red fluorescent protein construct. tdTomato was cloned into pIRES2.EGFP vector MCS at the BamHI and EcoRI sites 5’ to the IRES sequence via the primers listed in Supplementary Table 1. All plasmids were verified by sequencing at the DNA Analysis Facility (Yale, New Haven, CT). The DNA sequences of tdTomato and the inverted PS2 insert are given in Supplementary Figure 1.
Cell transfection
HEK293 cells (ATCC, Manassas, VA) were grown under standard cell culture conditions (5% CO2, 37°C) in Dulbecco’s Modified Eagles Medium (DMEM; Lonza, Walkersville, MD) supplemented to 10% with heat inactivated fetal bovine serum (FBS; PAA Laboratories, Etobicoke, ON). HEK293 cells were trypsinized (Mediatech, Manassas, VA), centrifuged at 500 g, three million cells were counted with a Cellometer T4 cell counter (Nexcelom Bioscience, Lawrence, MA), supplemented with 2.5 μg of plasmid DNA, and electroporated with the Nucleofector 4D with X-unit protocol CM-130 by the manufacturer’s protocol (Lonza, Walkersville, MD). Transfected cells were resuspended in DMEM with 10% FBS and seeded on 12 mm round, glass Poly-D-lysine/laminin Cellware coverslips (BD Biocoat, Bedford, MA) and incubated 24 hours prior to fixation.
Coverslips were fixed for 20 minutes with 4% paraformaldehyde (Acros Organics, Thermo Fisher Scientific, Glen Burnie, MD), and 0.2% Triton-X-100 (Sigma, St. Louis, MO) in phosphate buffered saline (PBS; Lonza, Walkersville, MD), then incubated with Hoechst-33342 (0.12 μg/ml; Life Technologies, Carlsbad, CA) in PBS for 1 hour in the dark at room temperature, washed three times with PBS for 10 minutes each, dipped in water, mounted on glass slides with Aqua-Polymount (Polysciences, Inc., Warrington, PA), and left to dry overnight at 4°C.
Image acquisition and statistical analysis
Fixed coverslips were imaged on a Leica SP5 white-light laser scanning confocal microscope (Leica Microsystems, Bannockburn, IL) with the Leica Application Suite Advanced Fluorescence v2.6 software and appropriate spectral separation. Random fields of cells were selected by DIC to prevent bias in the selection of especially bright green or red cells. Regions of Interest (ROIs) were traced around the perimeter of cells, the fluorescence quantified by mean grey intensity, and exported to Excel (v2010, Microsoft, Redmond, WA). EGFP fluorescence intensity values per cell were pooled by coverslip (n=5) and normalized to the average intensity of the positive control transfection (pIRES2.EGFP vector). Transfection efficiency was determined by dividing the number of fluorescent ROIs by the total cell number in the field. Values for coverslips were collected by treatment group and compared by one-way ANOVA with Newman-Keuls multiple comparisons test. Statistical analysis was conducted with Graphpad Prism (v5.04; Graphpad Software, La Jolla, CA). Coverslip mean fluorescence intensities were compared by 1way ANOVA with a Newman-Keuls multiple comparison test. Matched pairs of non-normalized EGFP and tdTomato fluorescence intensities were analyzed with a two-tailed Pearson’s correlation test.
Results
Length of the 5’ insert determines marker fluorophore expression and overall vector transfection efficiency
The length of the insert located at the 5’ portion of the IRES sequence increased both expression and transfection efficiency. Figure 1A and 1B display the increased expression of EGFP in HEK293 cells electroporated with pIRES2.EGFP vector (empty vector), 275 bpInsert.pIRES2.EGFP (275 bp insert), or tdTomato.pIRES2.EGFP (1400 bp insert). As indicated by the green fluorescence level, the longer 1400 bp insert 5’ of the IRES site increased expression of the EGFP marker (Figure 1A and 1B). Quantitative comparison of the EGFP fluorescence intensity between the three treatment groups indicated a significant 3-fold increase in EGFP fluorescence (P<0.001, n=5 coverslips per treatment group) by the 1400 bp insert (tdTomato.pIRES2.EGFP; 93.1 ± 6.2 Relative Fluorescent Units (RFU)) by 1way ANOVA with the Newman-Keuls multiple comparisons test (Figure 1C). There was no significant difference detected between the empty vector (pIRES2.EGFP; 31.7 ± 2.62 RFU) and the 275 bp Insert (275 bpInsert.pIRES2.EGFP; 45.9 ± 6.44 RFU). The apparent transfection efficiency was also significantly increased (P<0.001, one-way ANOVA with Newman-Keuls multiple comparisons test; n=5) by the larger insert located at the 5’ portion of the IRES element, as seen in Figure 1D. Apparent transfection efficiency was 50.2% ± 3.93% for HEK293 cells transfected with 1400 bpInsert.pIRES2.EGFP, almost twice the number of transfected cells. The other two treatment groups were found to have 29.3% ± 2.73% cells transfected with 275 bpInsert.pIRES2.EGFP, and 29% ± 2.25% cells transfected with Empty vector (pIRES2.EGFP vector).
Figure 1.
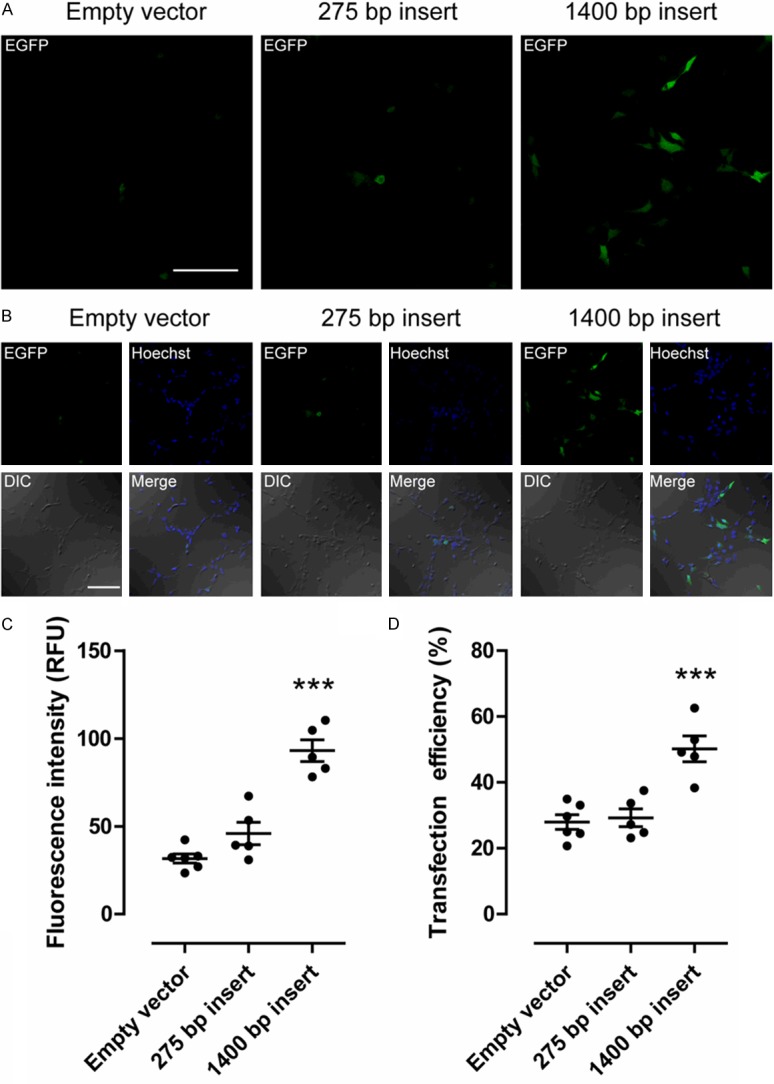
Length of inserts located at the 5’ portion of the IRES increases transfection efficiency of the marker gene EGFP located at the 3’ portion of the IRES. A. Representative confocal images of HEK293 cells transfected with pIRES2.EGFP constructs, EGFP fluorescence only. pIRES2.EGFP vector (Empty), 275 bpInsert.pIRES2.EGFP (275 bp), or tdTomato.pIRES2.EGFP (1400 bp). Scale bar: 100 μm (for all panels). B. Representative confocal images of EGFP fluorescence (EGFP), nuclei (Hoechst), DIC, and the three-color merge. Scale bar: 100 μm (for all panels). C. EGFP fluorescence intensity of HEK293 cells transfected with pIRES2.EGFP plasmids with inserts of various lengths at the 5’ portion of the IRES indicated. EGFP fluorescence per cell was normalized to the mean green fluorescence in the pIRES2.EGFP treatment group (positive control). Cells were then pooled and averaged by coverslip (n=5 coverslips per treatment group) and compared by one-way ANOVA with Newman-Keuls multiple comparisons test. Data is shown as the mean ± SEM. D. Apparent transfection efficiency of HEK293 cells transfected by electroporation with pIRES2.EGFP constructs was compared by one-way ANOVA with Newman-Keuls multiple comparisons test. Data is shown as the mean ± SEM [***P<0.001].
High expression of one gene is indicative of decreased expression of the other gene
Figure 2A displays tdTomato.pIRES2.EGFP transfected cells double-labeled with both the green (EGFP) and red (tdTomato) fluorescence. Figure 2B plots the mean intensity of the EGFP and tdTomato fluorescence signals of analyzed cells (n=362). The boxes represent 25th-75th quartiles, the bisecting line the median, and the whiskers for the 5th-95th percentile range with points outside the 5th-95th percentile indicated by dots. Note the EGFP marker intensity is prone to greater cell-to-cell variance than the tdTomato insert (62.7% vs. 40.6% coefficient of variance, respectively). This could be indicative of the competition for ribosomal entry between the 5’ cap and the IRES sequence (Figure 2B). Figure 2C is a Pearson’s correlation test of matched green and red fluorescence values on a cell by cell basis showing strong correlation (P<0.001, n=362 cells) but a poor adherence to a linear trend (solid line) of the intracellular green to red fluorescence ratio (R2=0.236).
Figure 2.
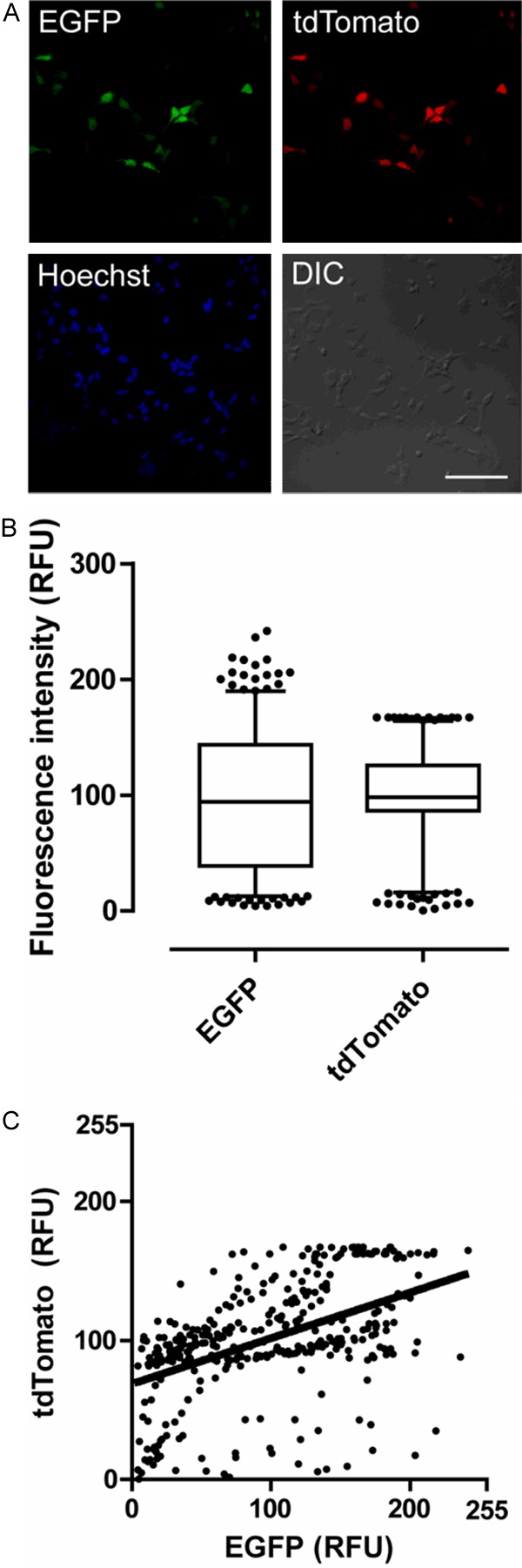
Expression of the two inserts of an IRES construct is correlated at median expression levels but not at high expression levels. A. HEK293 cells transfected with tdTomato.pIRES2.EGFP express both green (EGFP) and red (tdTomato) fluorescence signals. Scale bar: 100 μm (for all panels). B. The expression level of the EGFP marker 3’ to the IRES site has an increased coefficient of variance (62.7%) compared to the tdTomato gene 5’ to the IRES site (40.6%). Boxes represent quartiles (25th-75th percentile), the line is the median, and whiskers for the 5th-95th percentile with the indicated dots as values outside the 5th-95th percentile. C. Pearson’s correlation test for matched pairs of EGFP and tdTomato fluorescence signals (P<0.001, R2=0.236, n=362 cells). High relative fluorescence/expression of the product of one of the two genes flanking the IRES site is predictive of low relative fluorescence/expression of the product of the other gene.
Discussion
This study was conducted to test two hypotheses originating from observations made with past use of mammalian expression vectors reliant on an IRES sequence for co-transfection experiments. First, we hypothesized that expression of the IRES-dependent marker gene was regulated by the length of the insert 5’ to the IRES sequence. The 5’ transgene is often the gene of interest in experimental usage of IRES vectors with the gene 3’ to the IRES sequence serving as a marker for successful transfection or delivery of the plasmid, with fluorescent markers like EGFP being the label of choice for fluorescence imaging. Data presented in Figure 1 confirms the dependence of the 3’ marker gene expression on the length of the 5’ insert. The 275 bp insert formed no functional protein and only served to increase the space between the 5’ cap and the IRES sequence but the 275 bp insert was not sufficient to significantly increase expression of the 3’ marker gene. However, the longer tdTomato insert (1,400 bp) significantly increased EGFP marker gene expression. The diagram in Supplementary Figure 2 depicts these experimental findings. The increased apparent transfection efficiency is due either to increase plasmid uptake by the cells or increased expression of the marker EGFP though the latter is likely the case as the known transfection variables (amount of DNA, electroporation media, cell number and type) were held constant in all treatment groups.
We propose potential mechanisms that could underlie the observations reported above. While our data sets characterize an observed effect particular to the IRES system additional experiments beyond the scope of the present study are needed to discern a detailed molecular mechanism. Yet, two potential mechanisms can be easily proposed from the data presented here: First, the IRES bearing mRNA transcript may simply be stabilized by a transgene inserted 5’ of the IRES site. If one were to assume that the instability of the mRNA transcript is causative of the observed effect on 3’ marker expression, the empty vector transcript must be very unstable since transfection efficiency and expression were found to be significantly lowered. The IRES and EGFP portion of the mRNA transcript were held constant throughout all of our experiments but the differences in 3’ fluorescent marker and transfection efficiency were noticeable even at the crude level of the common bioassay on a simple tissue culture microscope. The increased expression observed in the tdTomato group would necessarily be caused by a stabilizing effect innate to either the tdTomato primary sequence or the shear presence of an insert of appropriate length to stabilize the IRES-EGFP portion of the transcript. If the former is true, then the great potential variability of the 5’ sequence and its ability to stabilize the IRES-EGFP portion of the mRNA would invalidate the use of the IRES system as a near universal experimental tool. The controls required to sufficiently test the large variety of 5’ insert sequence combinations and subsequent formulation of a workable theory for a specific primary sequence with the ability to stabilize the transcript is an effort beyond the scope of the present study. Indeed, if the IRES system were so sensitive to the nucleotide composition of the 5’ insert, but not to the length of the insert, where a dramatic effect such as we describe above were apparent, the IRES system would cease to be a useful experimental asset. Second, a small insert 5’ of the IRES sequence may be causative of the decreased 3’ fluorescent marker expression. Binding and assembly of the 5’ cap translation machinery in the case of an IRES construct with a small 5’ insert may involve sufficient steric hindrance as to interfere with the binding of the nearby IRES sequence and assembly of its translation machinery [3] within the active ribosome. This would also manifest as a false negative experimental error, where the plasmid has been successfully transfected but the transgene and marker are expressed below the detection threshold.
The second hypothesis we tested was that the expression of one gene in the IRES plasmid was favored by the cell at the expense of the other gene sharing the IRES site. This hypothesis was verified by the data and correlation given in Figure 2. The correlation is strong for cells with median expression of either fluorophore as can be seen by the near parity of the median fluorescent signal in Figure 2B. Thus, if one gene is expressed at a median level then the other is also expressed at a median level. This holds true for a wide range of expression levels as can be seen by the 5-95% confidence interval shown in Figure 2B. The wide distribution about the linear trend in Figure 2C also indicates that expression of the 5’ and 3’ genes will vary cell to cell but the level of expression of both genes is relatively similar over a broad range of intermediate values. However, when expression of one gene is extremely high such as to make it an outlier from the correlation this leads to the second gene being expressed at very low levels.
The mechanisms causative of the differential expression of the two genes sharing the IRES site cannot be fully described from the data above but these observations clearly point toward practical strategies for using expression vectors with IRES sequences. The 5’ cap translation is more efficient than IRES expression in experimental plasmids [14]. It is not known if the greater efficiency of 5’ cap translation compared to a standard IRES sequence (a Group 3 IRES sequence in the data above [3]) is achieved through increased ribosomal binding efficiency, enhanced assembly of the translational protein complex, or even increased processivity of the translational machinery [3,5,13].
Concluding remarks
We undertook this study to follow up on our previous observations of unequal gene expression in experiments utilizing an IRES system. The goal of the present study was to not identify molecular biology concepts beyond the scope of using fluorescent markers in bicistronic expression vectors as a means of evaluating expression levels of transgenic systems, but rather to clearly describe an effect particular to the IRES vector system. As such, our data provide guidance for future use of the IRES sequence and bicistronic expression vectors in experimental design. The experimenter should be aware that the expression of the marker gene is unreliable in plasmids with small or no inserts between the 5’ cap and the IRES sequence (Supplementary Figure 2). Our data also strongly contradicts an assumption often made when using an IRES system, where the investigator normalizes expression data by use of the empty IRES vector as a positive control or baseline. Such an assumption in data analysis is not unreasonable but it does turn out to be false since the population of cells expressing the 5’ insert is significantly different from the population transfected with the empty vector. Both of these equate to warnings over the decreased apparent transfection efficiency or the decreased expression of the IRES dependent marker in cases of small or no transgene inserted at the 5’ of the IRES sequence.
Finally, the experimenter should select cells in the median range of marker intensity, avoiding the especially bright or particularly dim cells, as preferential expression of one of the two genes flanking the IRES site is apparent in cells prominently overexpressing one of the genes. Overexpression of one gene results in the decreased expression of the other gene, invalidating the correlation between the marker and transgene and thus the assumed expression level of either gene within the cell. The IRES sequence remains an excellent choice in experimental design where expression of a transgene and marker are desirable but proper awareness of the limits of the experimental system are required for rational experimental planning and execution.
Acknowledgements
Research reported in this publication was supported by grants from the National Eye Institute (EY014227 and EY022774), the Institute on Aging (AG010485, AG022550 and AG027956), the National Center for Research Resources and National Institute of General Medical Sciences (RR022570 and RR027093) of the National Institutes of Health (PK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support by the Felix and Carmen Sabates Missouri Endowed Chair in Vision Research, a Challenge Grant from Research to Prevent Blindness and the Vision Research Foundation of Kansas City is gratefully acknowledged. The authors thank Dr. Roger Y. Tsien for the generous gift of the tdTomato construct. We thank Margaret, Richard and Sara Koulen for generous support and encouragement.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 2.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 5.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KH, Kim SH, Kim DY, Kim S, Kim KT. Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PLoS One. 2012;7:e37936. doi: 10.1371/journal.pone.0037936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 8.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 9.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim Biophys Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Bochkov YA, Palmenberg AC. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques. 2006;41:283–284. 286, 288 passim. doi: 10.2144/000112243. [DOI] [PubMed] [Google Scholar]
- 11.Pfutzner W. Retroviral bicistronic vectors. Drug News Perspect. 2008;21:473–480. doi: 10.1358/dnp.2008.21.9.1290817. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Constantinescu SN, Sun Y, Bogan JS, Hirsch D, Weinberg RA, Lodish HF. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal Biochem. 2000;280:20–28. doi: 10.1006/abio.2000.4478. [DOI] [PubMed] [Google Scholar]
- 13.Harries M, Phillipps N, Anderson R, Prentice G, Collins M. Comparison of bicistronic retroviral vectors containing internal ribosome entry sites (IRES) using expression of human interleukin-12 (IL-12) as a readout. J Gene Med. 2000;2:243–249. doi: 10.1002/1521-2254(200007/08)2:4<243::AID-JGM115>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 14.Mizuguchi H, Xu Z, Ishii-Watabe A, Uchida E, Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- 15.Payne AJ, Gerdes BC, Naumchuk Y, McCalley AE, Kaja S, Koulen P. Presenilins regulate the cellular activity of ryanodine receptors differentially through isotype-specific N-terminal cysteines. Exp Neurol. 2013;250:143–150. doi: 10.1016/j.expneurol.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 17.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 18.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


