Abstract
The Arenaviridae family includes several hemorrhagic fever viruses which are important emerging pathogens. Junín virus, a member of this family, is the etiological agent of Argentine Hemorrhagic Fever (AHF). A collaboration between the Governments of Argentina and the USA rendered the attenuated Junín virus vaccine strain Candid#1. Arenaviruses are enveloped viruses with genomes consisting of two single-stranded RNA species (L and S), each carrying two coding regions separated by a stably structured, non-coding intergenic region. Molecular characterization of the vaccine strain and of its more virulent ancestors, XJ13 (prototype) and XJ#44, allows a systematic approach for the discovery of key elements in virulence attenuation. We show comparisons of sequence information for the S RNA of the strains XJ13, XJ#44 and Candid#1 of Junín virus, along with other strains from the vaccine lineage and a set of Junín virus field strains collected at the AHF endemic area. Comparisons of nucleotide and amino acid sequences revealed different point mutations which might be linked to the attenuated phenotype. The majority of changes are consistent with a progressive attenuation of virulence between XJ13, XJ#44 and Candid#1. We propose that changes found in genomic regions with low natural variation frequencies are more likely to be associated with the virulence attenuation process. We partially sequenced field strains to analyze the genomic variability naturally occurring for Junín virus. This information, together with the sequence analysis of strains with intermediate virulence, will serve as a starting point to study the molecular bases for viral attenuation.
Keywords: Arenaviridae, Junín virus, Natural variability, Point mutations, Vaccine lineage, Viral attenuation.
INTRODUCTION
The Arenaviridae family comprises only one genus formed of 25 species of arenaviruses, as categorized by the ICTV [1], and listed in (Table 1). A further classification is based on the geography of origin and distribution of the viruses and their hosts: Old World Arenaviruses (OWA) and New World Arenaviruses (NWA). The NWA group harbors all arenaviruses from the American continent and the OWA group includes the only arenaviral species present in all continents (Lymphocytic Choriomeningitis Virus – LCMV) as well as the African arenaviruses. To date, there are no reports of any native viral species from Europe or Australia. The precise geographic distribution pattern has its origin in a very narrow host range that is characteristic of all arenaviruses. The principal hosts for the NWA are several specific members of the rodent family Cricetidae, subfamilies Sigmodontinae and Neotominae [2]. In the case of LCMV, the rodent reservoir is the worldwide distributed Mus musculus, which explains the unusual ubiquity of this virus. One NWA, the Tacaribe Virus (TACV), was isolated from bats [3], in what was thought to be an atypical occurrence for an arenavirus. However, a recent discovery of new arenaviral species in boas [4] opens up new avenues of inquiry regarding the viral host range and biodiversity.
Table 1.
Arenaviridae family. NR: not reported; OWA: Old World Arenavirus; NWA: New World Arenavirus; the lineages are listed as: A, B, C and Rec A/B. The distribution is based on the country of virus isolation. Table modified from [6, 7]
| Virus | Acronym | Evolutionary Lineage | Distribution | Reservoir | Human Patogenicity |
|---|---|---|---|---|---|
| Flexal | FLEV | NWA-A | Brazil | Oryzomys spp. | Yes |
| Pichinde | PICV | NWA-A | Colombia | O. albigularis | NR |
| Paraná | PARV | NWA-A | Paraguay | O. buccinatus | NR |
| Allpahuayo | ALLV | NWA-A | Peru | Oecomys bicolor | NR |
| Pirital | PIRV | NWA-A | Venezuela | Sigmodon alstoni | NR |
| Junín | JUNV | NWA-B | Argentina | C. musculinus | Yes |
| Machupo | MACV | NWA-B | Bolivia | C. callosus, C. laucha | Yes |
| Guanarito | GTOV | NWA-B | Venezuela | Z. brevicauda | Yes |
| Sabia | SABV | NWA-B | Brazil | Unknown | Yes |
| Chapare | - | NWA-B | Bolivia | Unknown | Yes |
| Latino | LATV | NWA-C | Bolivia | Calomys callosus | NR |
| Tacaribe | TCRV | NWA-B | Trinidad | Artibeus spp. (bat) | Yes |
| Cupixi | CPXV | NWA-B | Brazil | O. capita | NR |
| Amapari | AMAV | NWA-B | Brazil | O. capita-Neacomys guianae | NR |
| Oliveros | OLVV | NWA-C | Argentina | Bolomys obscurus | NR |
| Whitewater Arroyo | WWAV | NWA-Rec A/B | USA Southwest (NM,TX,UT,OK) | Neotoma albigula, N. mexicana, N. micropus, N. sinerea | Yes |
| Lassa | LASV | OWA | Nigeria, Ivory Coast, Guinea, Sierra Leone | Mastomys sp. | Yes |
| Tamiami | TAMV | NWA-Rec A/B | Florida, USA | Sigmodon hispidus | NR |
| Bear Canyon | BCNV | NWA-Rec A/B | California, USA | Peromyscus sp. | NR |
| Lymphocytic Choriomeningitis | LCMV | OWA | Worldwide | Mus musculus | Yes |
| Mobala | MOBV | OWA | Central African Republic | Praomys sp. | NR |
| Mopeia | MOPV | OWA | Mozambique | Mastomys natalensis | NR |
| Ippy | IPPYV | OWA | Central African Republic | Arvicanthis sp. | NR |
| Lujo | LUJV | OWA | South Africa | Unknown | Yes |
| Luna | LUNV | OWA | Zambia | Mastomys natalensis | NR |
This viral family is of medical interest because 6 of the 25 members have been consistently associated with clinical symptoms and described as the etiological agents of human diseases. All pathogenic NWA produce hemorrhagic fevers, with very similar physiopathology (Table 2). The phases of the illness are well characterized and progress from prodromal to neurologic-hemorrhagic to convalescent [5]. There is a short incubation period before the onset of symptoms. In the case of Argentine Hemorrhagic Fever (AHF), the syndrome is characterized by renal, neurological, vascular, immunological and hematological alterations. If it remains untreated, the mortality rate can be as high as 15-30%.
Table 2.
Arenaviral hemorrhagic fevers pathological characteristics. The name of the disease (when existing), along with the prin-cipal clinical symptoms caused by Old World Arenaviruses and New World Arenaviruses are briefly explained. Modified from [13]
| Agent (Disease) | Pathological Characteristics |
|---|---|
| Old World Lassa (Lassa Fever) |
Incubation: 3 to 21 days.
Fever, headache, myalgia, backache, trembling and sickness. Generalized infection: hemorrhagic dissemination of virus to several organs and systems via bloodstream, lymphatic system, respiratory and digestive tract. Black vomit, aqueous diarrhea (dehydration), quantity decrease of lymphocytes and platelets, mild thrombocytopenia, abdominal, pleuritic and hepatic pain. Extensive reticulo-endothelial compromise: capillary injuries causing stomach, small intestine, kidney, lung and brain bleeding. Multifocal hepatocellular necrosis with Councilman-like bodies, hepatocytes cytoplasmic degeneration and minimal inflammatory response. Adrenal focal necrosis and cytoplasmic inclusions. Respiratory system: interstitial pneumonia, cough, dyspnea, bronchitis, pneumonia and pleurisy. Cardiovascular system: pericarditis, tachycardia, bradycardia, hypertension, hypotension, thrombocytopenia, leukopenia and hiperuricemia, lymphadenopathy, elevated aminotransferases, decreased prothrombin level, disorder of blood circulation and bleeding through the skin, lungs, gastrointestinal tract and other mucosa membranes. Nervous system: encephalitis, meningitis, uni- or bilateral hearing decrease, convulsions. |
| New World Junín (Argentine Hemorrhagic Fever) Machupo and Chapare (Bolivian Hemorrhagic Fever) Guanarito (Venezuelan Hemorrhagic Fever) Sabiá (Brazilian Hemorrhagic Fever) Whitewater Arroyo Flexal Lujo |
Incubation: 6 to 14 days. First
4 days: decaying, fever, anorexia, nausea and vomiting, headache and myalgia.
Second stage: acute hemorrhagic syndrome (epistaxis and hematemesis), or
acute neurologic syndrome. General: malaise, high fever, severe myalgia,
anorexia, back pain, abdominal tenderness, conjunctivitis, retro-orbital
pain, photophobia, and usually constipation. Acute phase of infection: peripheral blood viral active replication. Oropharyngeal enanthem. Gums swollen, congested and bleeding (gingival border). Proteinuria high dehydration and hemoconcentration. In women, early menorrhagia. Multifocal hepatocellular necrosis with formation of Councilman-like bodies, nuclear pyknosis, cytoplasmic eosinophilia, cytolysis, inflammation and a mild cellular infiltration composed of mononuclear cells and neutrophils. Kidney damage: distal tubular cells and collecting ducts. Glomeruli or proximal tubules. In a few cases, renal failure was reported. Cardiovascular system: postural hypotension and relative bradycardia, arrhythmias are transient and benign. Different degrees of dehydration, uremia, proteinuria, hematuria and oliguria. Respiratory: dry cough, without sore throat. Pharyngeal enanthem. Interstitial or bronchial pneumonia, pulmonary edema and hemorrhage. |
Since it was described, AHF has been reported to cause annual outbreaks during the autumn-winter season [6]. However, the number of actual cases in each outbreak has significantly diminished since the application of the vaccine to the risk population within the endemic area. For the 1992-2000 period the efficacy of the vaccine was estimated at 98% [7].
The development of the vaccine against AHF was the product of a collaboration between the US and Argentine governments. A greatly attenuated strain of Junín virus (JUNV), Candid#1, was extensively tested in rhesus monkeys and human volunteers with good results. Afterwards, a comprehensive clinical trial was conducted in the AHF endemic area [8, 9]. A diagram with the passage history that led to the attenuated JUNV Candid#1 strain is shown in (Fig. 1). The intermediate strains were named XJ#, in reference to the original strain and the number of passages in mouse brain. The vaccine was originally produced in the USAMRIID (United States Army Medical Research Institute for Infectious Diseases) facilities. In 2005, the National Institute of Human Viral Disease (INEVH) located in Pergamino, Argentina, finished the certification process and took over the vaccine production for local use.
Fig. (1).
History of passages for the Junín virus vaccine strain Candid#. Adapted from [11].
Other vaccines against Arenaviruses have not been released. Significant advances were made to develop an effective protection against Lassa fever, which is a disease endemic to the West-African region with 3-5x105 cases reported annually [10]. Its apparently expanding territory, together with the imported cases reported yearly all over the world, make this virus the biggest health threat of the Arenavirus family [11]. Several different approaches were used to try to achieve protective coverage with a Lassa virus vaccine. However, the necessity of a single-dose treatment and the great diversity among the many viral strains converge to increase the difficulty of this task [12].
Arenaviruses are enveloped viruses with a bipartite, single stranded RNA genome [14, 15]. The virion contains three structural proteins (L, N and Z) while the lipid envelope carries the three-piece glycoprotein (SSP, G1 and G2). The larger genomic segment (L RNA, ~7kb) encodes the RNA dependent RNA polymerase (L protein), which is the minor component of the virion, as well as Z, a Zinc finger-protein of 11 kDa that could be the arenaviral counterpart of a matrix protein [16]. The other genomic segment (S RNA, ~3,5 kb) codes for the nucleocapsid protein N and the glycoprotein precursor (GPC), which is processed by cellular proteases into three parts. The arrangement of the two open reading frames in opposite orientations, separated by the stably structured, non-coding intergenic region, gave rise to the term “ambisense coding” [17]. The first 19 nucleotides at each end of one segment are complementary, which allows a base-pairing that forms a panhandle structure highly conserved among arenaviruses [18].
SEQUENCE ANALYSIS OF THE VACCINE-RELATED JUNV STRAINS
In order to identify and characterize changes directly related with the attenuated phenotype of the JUNV vaccine strain, the complete nucleotide sequence of all available intermediate strains (XJ13, XJ17, XJ34, XJ39, XJ#44, XJ48 and Candid#1) from the vaccine genealogy were analyzed, along with a small number of field strains [19, 20].
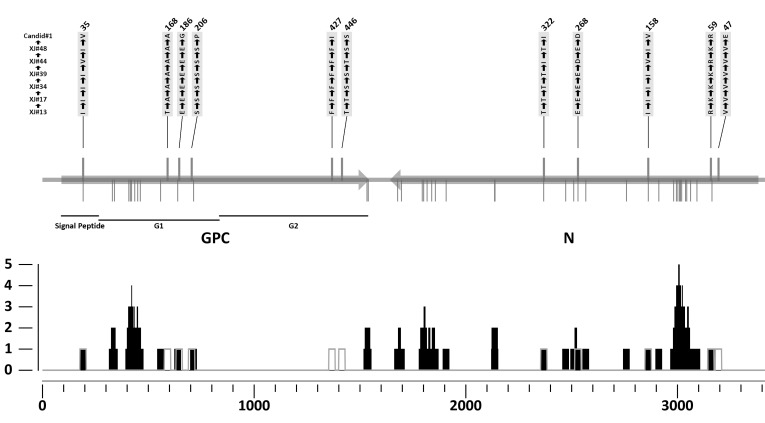
A comparison of the sequence of the vaccine genealogy strains revealed a set of differences that might be associated with the decrease in virulence (Fig. 2). An alignment of the S RNA-derived coding sequences disclosed eleven individual changes which translated into amino acid substitutions. The first of these changes was located in the stable signal peptide (XJ13 I35→ XJ17 I→XJ34 I→XJ39 I→XJ#44 V35→ XJ48 I→Candid#1 V), the following three were in the mid-part of G1 (T168→A→A→A→A→A→A; E186→E→E→E→E→E→G and S206→S→S→S→S→S→P), two more were found at the carboxyl terminus of G2 (F427→F→F→F→F→F→I and T446→T→S→S→T→S→S), and the last five were at the amino half of N (V47→V→V→V→V→V→E; R59→K→K→ K→R→K→R; I158→I→I→I→V→I→V; E268→E→E→E→E →E→D and T322→T→T→T→I→T→I). The same analysis, performed on the coding sequences derived from the L gene, revealed only nine nucleotide changes implied in amino acid substitutions. Two of these are reversions (XJ13 R881→XJ#44 G→Candid#1 R; S921→G→S), while the other seven might be associated with the attenuated phenotype (H76→Y→Y; V415→V→A; D462→N→N; L936→L→P; R1156→K→K; S1698→S→F and I1883→I→V). The Z gene showed no nucleotide changes among these three JUNV strains.
Fig. (2).
Summary of the changes found in Junín virus proteins. A. S RNA; the N and GPC ORFs are shown as arrows, indicating the translation sense. The strains used for the comparison are shown on the left side. The parts of GPC are marked below the diagram. Amino acid changes among the strains (XJ#13, XJ#17, XJ#34, XJ#39, XJ#44, XJ#48 and Candid#1) are drawn as lines above the gene, followed by the identity of the change and its position. Variations among the field strains (XJ#13, Romero, IV4454 and MC2) are drawn as lines below the gene. B. Relative frequency of mutation for the different types: black is for type-1and grey for type-2.
This analysis was extended to include two other completely sequenced JUNV strains, Romero and MC2, as well as a partial sequence from the Cba-IV4454 strain (S RNA and Z gene). The first strain is classified as of high virulence and was originally isolated from an AHF patient [21, 22]. In the Genebank its name is mistakenly spelled “Rumero”. The second strain (MC2) is of intermediate virulence and was isolated from a rodent captured in the AHF endemic area [23-25]. The Cba-IV4454 strain was also obtained from a patient and is considered of intermediate virulence. The analysis was performed on deduced amino acid sequences for the six aforementioned JUNV strains. Two distinct types of mutations were identified, according to a specific definition. Type 1 mutations are those where only one of the JUNV field strains (XJ13, Romero, IV4454, and MC2) differs from all others. Type 2 mutations refer to the positions with changes among the vaccine-related strains (XJ13, XJ#44 and Candid#1). This last type allows identifying key positions that might be involved in the attenuation process, while the Type 1 mutations take into account the naturally occurring variations. An overlap of these two types of changes uncovered several mutations with probably little significance for the viral virulence. Positions GPC35, N158, N268 and N322 all had changes among the six sequences that varied between two particular amino acids, regardless of the strain’s origin. Something similar happened with positions L881 and L921, where the only difference was found in XJ#44.
A mutation frequency index was calculated, defined as the number of mutations per amino acid in an overlapping-window strategy, with the resulting value assigned to the central residue. The analysis was made for both types of mutations for the complete JUNV genome sequence, using in-house software. This global plotting approach gives an overview of the distribution of the mutations along the genome. Several possible hot-spots were identified, that is, regions of the sequence with a marked tendency towards natural variation. Also, a small number of type 2 mutations were found outside the hot-spots, namely GPC168, GPC427, GPC446 and N47 for the S RNA and positions L76, L936, L1156 for the L RNA. These changes could more confidently be assigned to the virulence attenuation process.
CODING REGIONS
A further search for Type 2 mutations was performed on a set of field samples, isolated from human cases or rodents, captured in the endemic area during the period of 1963-91. Out of the genome, four regions were selected for this analysis: i) G fragment, comprising positions 303 to 941 within the GPC coding sequence; ii) N fragment, spanning nucleotides 1632 to 2095 of the nucleoprotein coding sequence and two regions within the open reading frame of the RNA dependent RNA polymerase [13]. The fragments were amplified by PCR and sequenced for 13 different JUNV field strains. Afterwards, the sequences were translated in silico, aligned and scanned for positions with differences between strains. A part of the Type 2 mutations obtained for the G fragment are shown in (Table 3). The selected region comprises most of G1, the consensus site for the arenavirus glycoprotein processing [26] and the beginning of G2. Some of the variants present among the field samples (from both humans and rodents) were also found among the attenuated JUNV strains. Consequently, the importance of these positions in the attenuation process should be re-evaluated.
Table 3.
G fragment positions with variations in amino acid sequence. The changes with regard to the XJ13 strain are annotated in bold letters. Type 2 mutations among vaccine related strains are shadowed in gray. Adapted from [13]
| Residue Position | XJ13 | XJ17 | XJ34 | XJ39 | XJ#44 | XJ48 | Candid#1 | MC2 | IV4454 | Romero | AN_8640 | AN_5185 | AN_13365 | AN_16501 | AN_17058 | AN_17246 | AN_17116 | AN_17064 | H_Lye63 | H_FHA5069 | H_p1879 | H_FHA5054 | H_8027 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 107 | S | S | S | S | S | S | S | S | T | S | S | S | S | S | S | S | S | S | S | T | T | T | S |
| 109 | Q | Q | Q | Q | Q | Q | Q | K | M | M | Q | Q | Q | M | Q | Q | M | M | Q | M | M | M | Q |
| 111 | S | S | S | S | S | S | S | S | T | S | S | S | S | S | T | S | S | S | S | T | T | T | S |
| 116 | A | A | A | A | A | A | A | A | A | E | A | A | A | A | A | A | A | A | A | A | A | A | A |
| 121 | Q | Q | Q | Q | Q | Q | Q | E | Q | Q | Q | Q | E | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q |
| 125 | I | I | I | I | I | I | I | I | V | I | I | I | I | I | V | I | I | I | I | V | V | V | I |
| 133 | S | S | S | S | S | S | S | S | S | S | S | S | S | N | S | S | N | N | S | S | S | S | S |
| 143 | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | W | R | R | R | W |
| 151 | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | A | V | V | V | A |
| 157 | H | H | H | H | H | H | H | Y | Y | H | H | H | H | H | H | H | H | H | H | H | H | H | H |
| 168 | T | A | A | A | A | A | A | T | T | T | A | A | T | T | A | A | T | T | A | T | T | T | A |
| 184 | V | V | V | V | V | V | V | I | V | V | V | V | V | V | V | V | V | V | V | V | V | V | V |
| 186 | E | E | E | E | E | E | G | E | E | E | G | G | E | E | G | G | E | E | G | E | E | E | G |
| 206 | S | S | S | S | S | S | P | S | S | S | P | P | S | S | P | P | S | S | P | S | S | S | P |
| 208 | P | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L |
| 209 | N | N | N | N | N | N | N | D | S | N | N | N | N | N | N | N | N | N | N | S | S | S | N |
| 244 | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | Q | H | Q |
| 245 | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | L | F | L |
The development of a reverse genetics system made it possible to directly test the role of the different variable positions in viral fitness and virulence. Albariño et al. used chimeric genomes, baring a combination of XJ13 and Candid#1-derived sequences, to assess the importance of each portion of GPC. They found that only the Candid#1-derived G2 part was critical for the attenuated phenotype. A further characterization mapped the significant points to the transmembrane domain within G2 [27]. However, a number of variable positions remain to be evaluated by this experimental method.
Previous studies described four conserved regions in the L protein of all arenaviruses [28]. The polymerase domain was mapped to the region III, with the presence of four conserved motifs: A, B, C and D [29]. The variation analyses showed only one change in this region, at position 1156 (R→K→K). Albeit the mutation has a conservative character, it is probably related to the attenuation process because there are no natural variants in this region. The other changes in the L ORF are located between regions II and III (L936, L→L→P) and in region I (L76, H→Y→Y). The modification of a Leucine for a Proline brings about a mayor structural change and is present only in the Candid#1 strain, in spite of being immersed in a region of high type 2 mutations index. The change at position L76 is near a predicted ATP/GTP binding site (www.expasy.org). Even though these results are preliminary, they suggest an important role for the polymerase in the virulence attenuation, in accord with reports for other viruses [29-31]. Also, some of the mutations in the structural proteins (nucleoprotein and the two glycoproteins) could be associated with the attenuation of virulence. The N protein has a highly conserved carboxyl-terminus, containing a zinc binding domain [32]. The mutation at N47 (V→V→E) lies outside that region, but could be related to other characteristics of the protein, such as the oligomerization capability [33], possibly interfering with one or both of N’s functions in the viral life cycle. In the genome replication, N promotes the synthesis of antigenomic, full-length copies of the S RNA. In the assembly of the virion, the protein binds the genomic RNA, forming the nucleocapsid. The other structural protein, GPC, sported a mutation at the carboxyl end of G1 (GPC168, T→A→A), directly affecting the conserved sequence (N166R167T168K169) for a predicted N-glycosylation site (NETGLYC 1.0). For the G2 portion, three domains have been described. First, the outer domain, located at the carboxyl end, which is outside the virion and interacts with G1; second, the transmembrane domain, which interacts with the stable signal peptide, and last, the inner domain, situated at the amino end, which could interact with the nucleocapsid protein or the Z protein, on the inside of the virion [26, 34]. The changes within G2 are in the transmembrane domain (GPC427, F→F→I) or in the cytoplasmic tail (GPC446, T→T→S) and, in both cases, could affect these important interactions.
The potential attenuation markers were found to be unevenly distributed in the genomic segments, with a higher degree of conservation in the L RNA, which indicated a faster evolution rate for the polypeptides encoded in the S RNA. In a comparison of genomes of virulent and avirulent strains of Pichinde arenavirus, Lan and collaborators [29] found that attenuation related mutations mapped to comparable genomic regions, but were fewer in number. If the comparison is limited to only the field strains of Junín virus (XJ13, Romero, MC2 and IV4454) the mutation distribution pattern became markedly different for the large and small genomic segments. In a protein sequence analysis of the field strains, 45 divergent sites were found for the S RNA derived ORFs and 48 sites for the L RNA derived ORFs. Interestingly, for the L RNA, 46 out of the 48 sites were XJ13-specific changes, while for the S RNA, this was only the case in 4 of the 45 sites.
As stated before, the G fragment included the sequence for the cleavage site between G1 and G2. The recognition site sequence has been published as QLPRRSLK251AFF [35] with the cleavage occurring between K251↓A or L↓K251 [36]. Most of the analyzed samples didn’t have mutations in that region. Only isolate H_FHA5054 presented changes at positions 244 and 245 (Q→H and L→F respectively), but it is not certain if these modifications would have any detectable outcome. A portion of the variation observed for the G fragment, namely the part corresponding to G1, could be associated with the host jump of a rodent strain to humans (receptor affinity differences). They could therefore be directly linked with changes in the strains virulence (increase or decrease) through a cellular tropism change. Another key feature of this region was the N-glycosylation target sequence N166R167T168K169, as determined by NETGLYC 1.0. This precise sequence is only present in the 65% of the field strains, while in the remaining 35% there is an alanine residue (A168). Complementary bioinformatics studies performed on this region predicted only slight changes to the protein properties.
On the other hand, the importance of two positions, strongly related to the N-glycosylation of JUNV and MACV was established. Also a high conservation degree of the residues involved, along with their environ was found to be necessary [37]. In recent years, an analysis of LCMV proved the implication of each N-linked glycan of the glycoprotein with GPC expression, fusion activity and infectivity [38]. Interestingly, all the attenuated strains derived from XJ13 have an A168 mutation. The hypothesis was that mutations present in field strains (natural variants) would be of minor importance in virulence attenuation. In fact, none of the type two mutation analyzed here were absent in field samples. Therefore, the data presented in (Table 3) could signify that the attenuated phenotype is not influenced by mutations in the G1 region. That result concurs with Albariño and collaborators [39], who mapped the essential attenuation-related mutations to G2.
NON CODING REGIONS
Previous studies suggested that sequence changes within the intergenic region could play a role in the attenuation processes of arenaviruses [40]. Conversely, our analysis of the intergenic regions from Candid#1, XJ48, XJ#44, XJ39, XJ34, XJ17 and XJ13 revealed a total conservation among the strains for both genomic RNAs. If there is a low tolerance for nucleotide changes in the region, it would suggest an evolutionary constraint, probably related to the calculated secondary structure and the associated transcription regulation function [32, 41]. Also, there was a high degree of conservation of the non-coding regions at the genomic ends of the analyzed strains, with nucleotide homology values ranging from 93% to 97%.
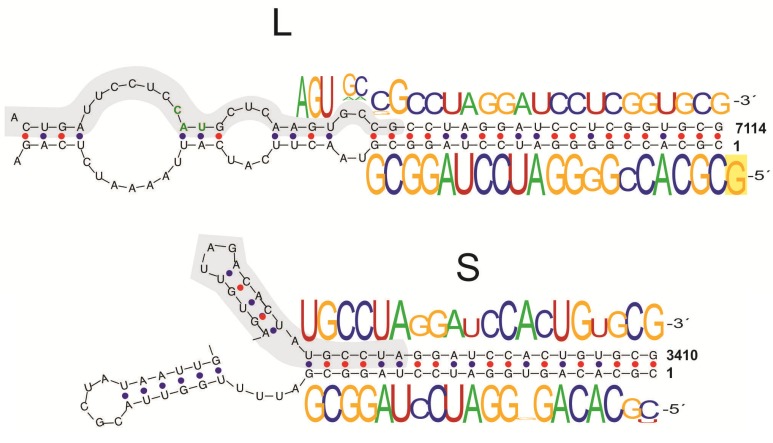
However, when viral RNAs obtained from infected cells were analyzed, a great variability could be observed at the 3’ and 5’ non-coding regions of RNAs derived from the same parental strain. The analysis was designed to evaluate the genomic and antigenomic forms separately. A number of 5’ non-templated bases were found for both Candid#1 genomic segments. When comparing the 5’ end of the genomic RNAs with the 3’ end of the antigenomic RNAs (which are the template for the former), at least one additional guanine was present at all 5’ ends of genomic clones with additional bases, as has been described for other arenaviruses [42]. On the contrary, the 3’ RACE of genomic forms yielded several clones with short deletions. A calculation of RNA secondary structure from Candid#1 S and L RNAs predicted the formation of a panhandle between the 5’ and 3’ ends of each genomic segment. The observed 3’ deletions all mapped to regions within the panhandle (Fig. 3).
Fig. (3).
Junín virus Candid#1 genomic end sequences. Predicted panhandle structures for the L and S RNAs. The region found in all 3´end sequences is shadowed. The sequence-logos show the nucleotide frequency determined by a RACE assey. A non-templated G is highlighted at the end of the L segment. Taken from [13]
Taken together, these results support the employ of cell-derived RNAs as primers for the synthesis of viral RNA, as well as 5’ terminal sequences from virus-derived RNAs into incomplete panhandle structures as templates for the completion of the 3’ terminal sequences. The resulting heterogeneity may well be a consequence of the action of different RNA editing mechanisms, as reported for other arenaviruses [43]. Still, the role of these regions in the process of attenuation remains to be established.
Interestingly, a comparison of the sequence of the ends (3’ and 5’) of both genomic segments revealed several positions that were highly conserved and could be part of the minimal promoter. The untranslated sequences on the extremes of the S RNA, as well as the 5’ end of the L RNA, have an approximate length of 80 nucleotides. This is not the case for the 3’ end of L RNA, which has only 30 nucleotides. Considering that the genomic forms serve as templates for the antigenomic forms and vice versa, all non-coding terminal regions have to include the promoter sequence that is the recognition site of the viral polymerase, in order to achieve the completion of the replicative cycle. Since one of these regions only has a size of 30 nucleotides, it is reasonable to conclude that the minimal promoter sequence is contained in that range. In support of this, if a sequence comparison is made of the 80 nucleotides of the ends of S and L RNAs, the homology value is 60% for the 5’ ends and only 50% for the 3’ ends. But, if the analysis is reduced to only 30 nucleotides, the value of the 3’ region ascends to 71%. In fact, there is a 19 nucleotide sequence motif highly conserved among arenaviruses (arena region) that can be found in all JUNV genomic ends. The remaining 11 bases of the 3’ end of the L RNA (GCTCAAGTGCC) have an elevated homology degree with two regions within the S RNA 3’ end. These repeated elements could be part of the translation or transcription processes. They have been found in other arenavirus species, in similar distributions. A comprehensive analysis of the genomic sequence of sublcade B1 New World arenaviruses [44] showed that the element that mapped to positions 38-46 (GCUCAAGUG for JUNV L RNA and GCUCAGUG for S RNA) was conserved within the group [20]. Therefore, it could be possible to identify a sequence motif at the 3’ end of both genomic RNAs of most arenaviruses. Also, in the particular cases of JUNV, Machupo and Tacaribe RNA S, the motif described as GSYC(A)1-2GUR, has a relatively high positional conservation within the RNA secondary structure when calculated by bioinformatic tools.
CONCLUSIONS AND SUMMARY
This works aim was to present a condensed report of the studies about the molecular determinants of viral virulence in JUNV. A set of putative attenuation markers have been identified within the GPC, Z and L ORFs of JUNV vaccine strain, Candid#1. Some of these positions were more confidently linked to the attenuated phenotype. The main reason for this is that these changes were found inside regions with a low natural mutation frequency (GPC168, GPC427, GPC446, N47, L76, L936 and L1156). They were analyzed with phylogenetic and bioinformatic tools.
Increasing complete genome sequence information availability, particularly from JUNV strains, has been a significant contribution for the identification of further putative virulence attenuation markers. In recent years several sequences were published for members of the vaccine strain lineage [20, 27]. They were included in previously done informatic analyses, confirming the results observed before, that is, only a few changes are necessary to obtain an attenuated phenotype. To further study the role in the virulence attenuation process of different point mutations, Albariño and collaborators developed a JUNV reverse genetics system [39]. Their results showed a direct involvement in the attenuated phenotype of certain point mutations within the GPC ORF of Candid#1.
Moreover, in order to analyze the natural mutation distribution, Goñi and collaborators designed a RT-Nested-PCR based technique and applied it to human and rodent derived samples from the Argentine Hemorrhagic Fever endemic area [45]. The sequences thus obtained were translated in silico and analyzed with different tools. A comparison was instituted between the GPC derived sequence data (G fragment) and the results published by Albariño and collaborators [25]. (Table 3) summarized the findings of that analysis. As is expected, the greater variability appears mainly in the portion of the glycoprotein complex that is exposed to the exterior and therefore comes in contact with the immune system. Changes to the sequence and conformation of epitopes could explain an increase in virulence due to escape from the natural host defenses. Also, slight variations within the receptor–binding region have been shown to confer the capability of infecting human cells as well as the animal host [46]. However, there are certain constraints acting on the outer part of the glycoprotein, related to its capacity of efficiently performing the original function within the viral cycle.
Studies addressing similar questions for other arenaviruses produced interesting results. Zhang and collaborators [47] created reassortants using an attenuated and a virulent PICV parental strain. They found that the virulence in guinea pigs could mainly be attributed to the S segment. Nevertheless, the virulence level of the parental strain was not attained, pointing to determinants being present in the L segment. Similar studies made with LCMV strains mapped the attenuation markers of this virus principally to its L segment [31]. Although both studies determine the virulence in an animal model of guinea pigs, the results obtained for each virus are different. This suggests that each attenuation process produced a distinct set of mutations in different regions. It would be interesting to assay what these mutations are for JUNV, principally taking into account the available intermediate strains of the vaccine genealogy, and the immense combination probabilities.
The sequence information accumulated through the analysis of a number of strains with varying virulence degrees will be the springboard for natural biodiversity studies. The mutations identified as potentially central to the attenuation process are ideal candidates for mutagenesis and reverse genetics assays. However, more complex studies will have to be made to determine the minimal changes that render an attenuated phenotype, since it is likely that many variations act synergically to decrease the observed virulence. Also, compensatory changes might arise to make up for structure or activity losses that hinder viral fitness [48, 49]. Therefore, even if a particular change is found in both field and vaccine strains, it does not necessarily negate its importance in virulence attenuation. Conversely, some variants present in natural and vaccine strains might have a cumulative effect upon the activity of a single viral protein or on the capacity of interaction with other viral or cellular proteins. The Z protein shows a low number of variations. This protein is responsible for the budding of the virus and the correct virion assembly. It also has several functions related to the modulation of the host immune response [50]. All of the roles of Z within the viral cycle are performed through interaction with proteins. The apparently low tolerance of changes within the Z ORF suggests that its functions greatly depend on the correct structure/sequence. Possibly, the regions of interaction of its partners are also submitted to a similar constraint. It will be interesting to analyze the precise regions once they have been positively identified. A great step towards identifying possible co-variation sites will come with the elucidation of the crystallographic structure of the viral polypeptides and their interaction partners. The beginning has been made with the first glycoprotein and partial nucleoprotein and L [51-53] structures and we are hoping for a steady advance in this area.
Summing up, this work presented a set of mutations that are probably related with an attenuated virulence phenotype. Also, most of the mutations analyzed here (Type 1 and 2) were mapped to a few discrete genomic regions. The development of a RT-PCR based method followed by sequencing, was the basis for a broad analysis of JUNV strains in search of natural genomic variability. This method could furthermore be useful if a program for the monitoring of vaccinated individuals is implemented.
ACKNOWLEDGEMENTS
We would like to acknowledge the current and previous members of the LIGBCM-AVEZ laboratory at National University of Quilmes for their contributions. The work is supported by CONICET and Agencia Nacional de Promoción Científica y Tecnológica.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Adams MJ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch Virol. 2012;157:1411–22. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cajimat MB, Milazzo ML, Hess BD, Rood MP, Fulhorst C F. Principal host relationships and evolutionary history of the North American arenaviruses. Virology. 2007;367:235–43. doi: 10.1016/j.virol.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downs W, Anderson C. Tacaribe virus a new agent isolated from Artibeus bats and mosquitoes in Trinidad. West Indies. Am. J .Trop. Med.: 1963;12:640–646. doi: 10.4269/ajtmh.1963.12.640. [DOI] [PubMed] [Google Scholar]
- 4.Hetzel U, Sironen T, Laurinmäki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher S. Isolation, identification and characterization of novel Arenaviruses, the etiological agent of Boid Inclusion Body Disease. . J.Virol. 2013;87(20) doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enria DA, Mills JN, Bausch D, Shieh W-J, Peters CJ, editors. 3rd ed. Philadelphia PA Elsevier : Pathogens and Practice; 2011. Arenavirus infections.Tropical Infectious Diseases Principles. pp. 449–461. [Google Scholar]
- 6.Ambrosio A M, Saavedra C, Riera L M, Fassio R M. producción nacional de vacuna a virus Jun in vivo atenuado (Candid #1) anti-fiebre hemorrágica argentina. Acta. Bioqu m. Cl n. Latinoam. 2006;40:5–17. [Google Scholar]
- 7.Ministery of Health National AHF Control Program Fourth Edition Azu. Buenos Aires Argetina. 2007;5:12. [Google Scholar]
- 8.Barrera Oro JG, Eddy GA, editors. In Fourth International Conference on Comparative. Canada: Virology Banff Alerta; 1982. Characteristics of candidate live attenuated Junín virus vaccine. pp. 17–22. [Google Scholar]
- 9.Maiztegui JI, Feinsod F, Briggiler AM, Peters CJ, Enría DA, Lupton HW, Ambrosio AM, Tiano E, Feuillade MR, Gamboa G, Conti O, Vallejos D, MacDonald C, Barrera Oro J G. In VII International Congress of Virology. Edmonton Canada:: 1987. Inoculation of Argentine volunteers with a live-attenuated Junín virus vaccine. p. 69. [Google Scholar]
- 10.Ogbu O, Ajuluchukwu E, Uneke CJ. Lassa fever in West African sub-region: an overview. J.Vector.Borne. Dis. 2007;44:1–11. [PubMed] [Google Scholar]
- 11.Sogoba N, Feldmann H, Safronetz D. Lassa Fever in west Africa: evidence for an expanded region of endemicity. Zoonoses and public health. 2012;59 Suppl 2:43–7. doi: 10.1111/j.1863-2378.2012.01469.x. [DOI] [PubMed] [Google Scholar]
- 12.Lukashevich I S. Advanced vaccine candidates for Lassa fever. Viruses. 2012;4:2514–57. doi: 10.3390/v4112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goñi S, Lozano M, Logie C. Molecular Attenuation Process in Live Vaccine Generation for Arenaviruses. In Point Mutations. Intech . 012 ;40(3 ):1–25. [Google Scholar]
- 14.Martínez Segovia Z , Grazioli F. The nucleic acid of Junín virus. Acta .Virologica. 1969;13:264–268. [Google Scholar]
- 15.Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Dutko FJ, Oldstone M BA. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J Virol. 1985;53:966–8. doi: 10.1128/jvi.53.3.966-968.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12978–83. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auperin D D, Romanowski V, Galinski M, Bishop D H. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy an ambisense viral. S RNA. J.Virol. 1984;52:897–904. doi: 10.1128/jvi.52.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young PR, Howard CR. Fine structure analysis of Pichinde virus nucleocapsids. J Virol . 1983;64 (Pt 4):833–42. doi: 10.1099/0022-1317-64-4-833. [DOI] [PubMed] [Google Scholar]
- 19.Goñi S E, Iserte J A, Ambrosio A M, Romanowski V, Ghiringhelli PD, Lozano ME. Genomic features of attenuated Junín virus vaccine strain candidate. Virus genes. 2006;32:37–41. doi: 10.1007/s11262-005-5843-2. [DOI] [PubMed] [Google Scholar]
- 20.Goñi SE, Iserte J A, Stephan BI, Borio CS, Ghiringhelli P D, Lozano M E. Molecular analysis of the virulence attenuation process in Junín virus vaccine genealogy. Virus genes. 2010;40:320–8. doi: 10.1007/s11262-010-0450-2. [DOI] [PubMed] [Google Scholar]
- 21.McKee K T, Mahlandt B G, Maiztegui J I, Eddy G A, Peters C J. Experimental Argentine hemorrhagic fever in rhesus macaques: viral strain-dependent clinical response. J. Infect. Dis. 1985;152:218–21. doi: 10.1093/infdis/152.1.218. [DOI] [PubMed] [Google Scholar]
- 22.Yun NE, Linde NS, Dziuba N, Zacks Ma, Smith JN, Smith J K, Aronson J F, Chumakova O V, Lander H M, Peters C J, Paessler S. Pathogenesis of XJ and Romero strains of Junin virus in two strains of guinea pigs. Am. J. Trop. Med. Hyg. 2008;79:275–82. [PMC free article] [PubMed] [Google Scholar]
- 23.Berria MI, Gutman Frugone LF, Girola R, Barrera Oro J G. [Immunological studies with Junin virus.I. Formation of antibodies in guinea pigs inoculated with live viruses]. Medicina: 1967;27:93–8. [PubMed] [Google Scholar]
- 24.Candurra NA, Damonte B, Coto CE. Antigenic relationships between attenuated and pathogenic strains of Junin virus. J Virol. 1989;27:145–50. doi: 10.1002/jmv.1890270215. [DOI] [PubMed] [Google Scholar]
- 25.Weissenbacher MC, Laguens R P, Coto CE. Argentine hemorrhagic fever. CTMI. 1987;134:79–116. doi: 10.1007/978-3-642-71726-0_4. [DOI] [PubMed] [Google Scholar]
- 26.York J, Romanowski V, Lu M, Nunberg JH. The signal peptide of the Junín arenavirus envelope glycoprotein is myristoylated and forms an essential subunit of the mature G1-G2 complex. J Virol. 2004;78:10783–92. doi: 10.1128/JVI.78.19.10783-10792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albariño CG, Bird BH, Chakrabarti A K, Dodd K . The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol. 2011; 85:10404–8. doi: 10.1128/JVI.00856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vieth S, Torda AE, Asper M, Schmitz H, Günther S. Sequence analysis of L RNA of Lassa virus. Virology. 2004;318:153–68. doi: 10.1016/j.virol.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Lan S, McLay L, Aronson J, Ly H, Liang Y. Genome comparison of virulent and avirulent strains of the Pichinde arenavirus. Arch.Virol. 2008;153:1241–50. doi: 10.1007/s00705-008-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Endres MJ, Griot C, Gonzalez-Scarano F, Nathanson N. Neuroattenuation of an avirulent bunyavirus variant maps to the L RNA segment. J.Virol. 1991;65:5465–70. doi: 10.1128/jvi.65.10.5465-5470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Oldstone MB. Genetic mapping of lymphocytic choriomeningitis virus pathogenicity: virulence in guinea pigs is associated with the L RNA segment. J. Virol. 1985;55:704–9 . doi: 10.1128/jvi.55.3.704-709.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tortorici M A, Ghiringhelli P D, Lozano M E, Albariño C G, Romanowski V. Zinc-binding properties of Junín virus nucleocapsid protein. J.Virol. 2001;82:121–8. doi: 10.1099/0022-1317-82-1-121. [DOI] [PubMed] [Google Scholar]
- 33.Levingston Macleod JM, D’Antuono A, Loureiro ME, Casabona JC, Gomez G a, Lopez N. Identification of two functional domains within the arenavirus nucleoprotein. J.Virol. 2011;85:2012–23. doi: 10.1128/JVI.01875-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capul A. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains. . J.Virol . 2007; 81:9451–60. doi: 10.1128/JVI.00499-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyer WR, Pöpplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. 2003;77:2866–72. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12701–5. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowden T . Unusual molecular architecture of the machupo virus attachment glycoprotein. J Virol . 2009; 83:8259–65. doi: 10.1128/JVI.00761-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonhomme CJ, Capul AA, Lauron EJ, Bederka LH, Knopp KA, Buchmeier MJ. Glycosylation modulates arenavirus glycoprotein expression and function. Virology. 2011;409:223–33. doi: 10.1016/j.virol.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albariño CG, Bird BH, Chakrabarti AK, Dodd KA, White D M, Bergeron E, Shrivastava-Ranjan P, Nichol S T. Reverse genetics generation of chimeric infectious Junin/Lassa virus is dependent on interaction of homologous glycoprotein stable signal peptide and G2 cytoplasmic domains. J Virol. 2011;85:112–22. doi: 10.1128/JVI.01837-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson S M, Clegg J C. Sequence analysis of the S RNA of the African arenavirus Mopeia: an unusual secondary structure feature in the intergenic region. Virology: 1991;180:543–52. doi: 10.1016/0042-6822(91)90068-m. [DOI] [PubMed] [Google Scholar]
- 41.Franze-Fernández M T, Iapalucci S, López N, Rossi C. Subgenomic RNAs of Tacaribe virus. In The Arenaviridae: 1993:113–132. [Google Scholar]
- 42.Polyak SJ, Zheng S, Harnish DG. 5' termini of Pichinde arenavirus S RNAs and mRNAs contain nontemplated nucleotides. J. Virol. 1995;69:3211–5. doi: 10.1128/jvi.69.5.3211-3215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcin D, Kolakofsky D. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J Virol. 1992;66:1370–6. doi: 10.1128/jvi.66.3.1370-1376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flanagan ML, Oldenburg J, Reignier T, Holt N, Hamilton G A, Martin VK, Cannon PM. New world clade B arenaviruses can use transferrin receptor 1 (TfR1) dependent and independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J Virol. 2008;82:938–48 . doi: 10.1128/JVI.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goñi SE, Stephan BI, Iserte JA, Contigiani M S, Lozano M E, Tenorio A. Viral diversity of Junín virus field strains. Virus Res. 2011;160:150–8 . doi: 10.1016/j.virusres.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Abraham J, Kwong JA, Albariño CG, Lu JG, Radoshitzky S R, Salazar-Bravo J, Farzan M, Spiropoulou CF, Choe H. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic new world clade B arenaviruses. PLoS pathogens. 2009;5:e1000358. doi: 10.1371/journal.ppat.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Marriott KA, Harnish DG, Aronson J F. Reassortant analysis of guinea pig virulence of pichinde virus variants. Virology. 2001;290:30–8. doi: 10.1006/viro.2001.1127. [DOI] [PubMed] [Google Scholar]
- 48.Rimmelzwaan GF, Berkhoff EGM, Nieuwkoop NJ, Smith D J, Fouchier RM, Osterhaus a DE. Full restoration of viral fitness by multiple compensatory co-mutations in the nucleoprotein of influenza A virus cytotoxic T-lymphocyte escape mutants. J.Virol. 2005;86:1801–5. doi: 10.1099/vir.0.80867-0. [DOI] [PubMed] [Google Scholar]
- 49.Parks CL, Lerch RA, Walpita P, Wang HP, Sidhu MS, Udem SA. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 2001;75:910–20. doi: 10.1128/JVI.75.2.910-920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fehling SK, Lennartz F, Strecker T. Multifunctional nature of the arenavirus RING finger protein Z. Viruses. 2012;4:2973–3011. doi: 10.3390/v4112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Igonet S, Vaney MC, Vonhrein C, Bricogne G, Stura EA, Hengartner H, Eschli B, Rey F. a X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19967–72. doi: 10.1073/pnas.1108910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morin B, Coutard B, Lelke M, Ferron F, Kerber R, Jamal S, Frangeul A, Baronti C, Charrel R, de Lamballerie X, Vonrhein C, Lescar J, Bricogne G, Günther S, Canard B. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathogens. 2010;6:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Li L, Liu X, Dong S, Wang W, Huo T, Guo Y, Rao Z, Yang C. Crystal structure of Junin virus nucleoprotein. J. Virol. 2013;94(Pt 10 ):2175–83. doi: 10.1099/vir.0.055053-0. [DOI] [PubMed] [Google Scholar]