Abstract
Members of the substilisin/kexin like proprotein convertase (PCSK) protease family cleave and convert immature pro-proteins into their biologically active forms. By cleaving for example prohormones, cytokines and cell membrane proteins, PCSKs participate in maintaining the homeostasis in a healthy human body. Conversely, erratic enzymatic function is thought to contribute to the pathogenesis of a wide variety of diseases, including obesity and hypercholestrolemia. The first characterized seven PCSK enzymes (PCSK1-2, FURIN, PCSK4-7) process their substrates at a motif made up of paired basic amino acid residues. This feature results in a variable degree of biochemical redundancy in vitro, and consequently, shared substrate molecules between the different PCSK enzymes. This redundancy has confounded our understanding of the specific biological functions of PCSKs. The physiological roles of these enzymes have been best illustrated by the phenotypes of genetically engineered mice and patients that carry mutations in the PCSK genes. Recent developments in genome-wide methodology have generated a large amount of novel information on the genetics of the first seven proprotein convertases. In this review we summarize the reported genetic alterations and their associated phenotypes.
Keywords: Proprotein convertase, Genetics, FURIN, Association.
INTRODUCTION
Many secreted proteins, enzymes and receptors are initially synthesized in cells as inactive precursors that need to be proteolytically processed into biologically active forms. Recently, much research has focused on understanding the biology of an enzyme class that is responsible for this evolutionarily conserved endoproteolysis, namely proprotein convertases (PCSK, proprotein convertase subtilisin/kexin). PCSK enzymes belong to the serine endoprotease superfamily, and they catalyze the hydrolytic cleavage of a wide variety of substrate molecules that range from secreted growth factors (e.g. proVEGF, proTGFβ1) to extracellular pathogens (e.g. viral envelopes and bacterial toxins) [1-3].
The initially identified seven PCSKs (PCSK1 (ENSG00000175426), PCSK2 (ENSG00000125851), FURIN (ENSG00000140564), PCSK4 (ENSG00000115257), PCSK5 (ENSG00000099139), PCSK6 (ENSG00000140479), PCSK7 (ENSG00000160613)) form a group of partly compensatory and structurally conserved subtilisin/kexin-like serine proteases that function primarily in the secretory pathway, endosomes and on the cell surface (Fig. 1) [2, 4]. Before becoming enzymatically capable of cleaving their target molecules PCSK enzymes need to be activated through a series of autoproteolytic events, which are partially dependent on the
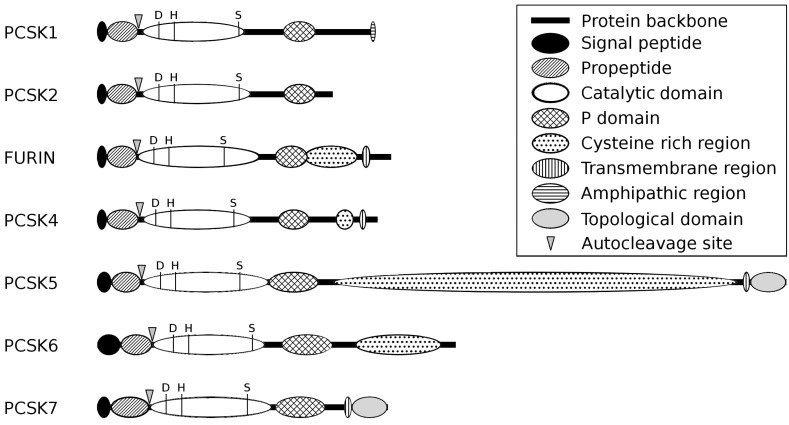
Fig. (1).
Schematic structure of the first seven members of proprotein convertase enzymes. PCSK enzymes possess a highly conserved domain structure, encompassing the N-terminal signal peptide, the inhibitory propeptide (IPR009020), the catalytic peptidase S8/S53 domain (IPR000209) and the P domain (IPR002884). The C-terminus of the PCSK enzymes can contain FURIN-like cysteine rich regions (IPR006212), transmembrane or amphipatic regions, or a topological domain. Triangles represent the autocleavage sites and the three conservative amino acids of the catalytic triad are depicted with the letters D (Aspartate-active site, IPR023827), H (Histidin-active site, IPR022398) and S (Serine-active site, IPR023828). The backbones of the proteins are represented by a black line and the lengths of the individual PCSKs and their domains are shown in proportion to the number of amino acids. (IPR, InterPro database, http://www.ebi.ac.uk/interpro/).
pH and calcium concentration of their environment. The PCSK substrate molecules contain a consensus sequence, which is needed for both substrate recognition and cleavage. The first seven PCSK enzymes act on a stretch of basic amino acids lysine and/or arginine: (K/R)-(X)n-(K/R)↓, with n being 0, 2, 4 or 6 and X any amino acid (Fig. 2). The more recently characterized PCSKs MBTPS1 (ENSG00000 140943) and PCSK9 (ENSG00000169174) do not cleave substrates at basic amino acids. In contrast, MBTPS1 exerts its function on the consensus motif (R/K)-X-(hydrophobic)-X↓, and PCSK9 only has autocatalytic cleavage activity in its prosegment sequence VFAQ152↓. In addition to the target sequences, also some of the flanking amino acids in substrates as well as secondary and tertiary structures serve as important determinants for the proteolytic function of PCSKs [5-8].
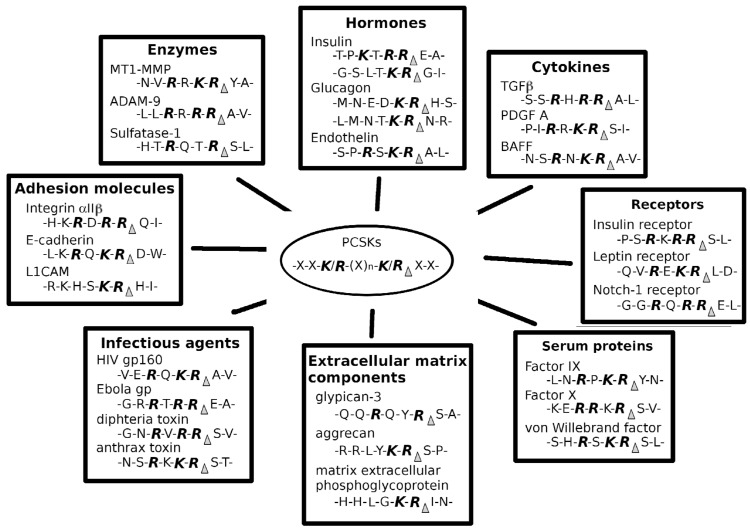
Fig. (2).
Examples of the target molecules of proprotein convertases. The first seven members of the proprotein convertase enzyme family cleave their substrates at the consensus amino acid sequence of (K/R)-(X)n-(K/R), where n is 0, 2, 4 or 6 and X any amino acid. Selected example target molecules and reported cleavage sites are shown.
In vitro experiments have demonstrated that the archetype PCSKs possess closely related, or even redundant biochemical properties and they often share substrate molecules. In contrast, the phenotypes of genetically targeted animals argue for substrate specificity. FURIN [9], PCSK5 [10, 11] and PCSK6 [12] are essential for normal mammalian development, whereas the phenotypes of PCSK1 [13], PCSK2 [14] and PCSK4 [15] deficient mice are more restricted ranging from infertility to defects in the neuro-endocrine system (Table 1). Notably, the biological role of PCSK7 in mammals remained long ill-defined [16, 17], but a recent study showed that PCSK7 deficient mice have an anxiolytic and novelty seeking phenotype that can be partially reversed by a dopamine D2/D4 antagonist [18]. Genetic inactivation has also demonstrated a specific function for the more recently identified PCSK family members MBTPS1 [19] and PCSK9 [20, 21] in cholesterol and lipid metabolism.
Table 1.
Phenotypes of Germ-line PCSK Knock-out Mice
| Germ-line KO | Phenotype | Reference |
|---|---|---|
| PCSK1 | Severe postnatal growth retardation Multiple defects in processing hormone precursors Hyperproinsulinemia, but normal glucagon processing |
[13, 75] |
| PCSK2 | Normal embryonic development, but grow at a slightly reduced rate Reduced adiposity Chronic fasting hypoglycemia, enhanced glucose tolerance Impaired maturation of multiple regulatory peptides or precursor proteins |
[17, 86, 88, 153-160] |
| FURIN | Deficient embryos die between embryonic day 10.5 and 11.5 Failure of ventral closure and axial rotation Cardiovascular defects Absence of chorioallantoic fusion |
[9] |
| PCSK4 | Impaired fertility | [15] |
| PCSK5 | Lethal at birth due to multiple craniofacial and patterning abnormalities | [10, 11] |
| PCSK6 | Complex craniofacial malformations Heterotaxia, combined with pulmonary isomerism |
[12] |
| PCSK7 | Anxiolytic and novelty seeking phenotype | [18, 17] |
Biochemical studies on the first seven members of the PCSK family have offered many important insights into the biological function of these genes. However, due to a significant degree of overlap in biochemical properties and common substrate molecules genetic approaches have been instrumental for fully understanding the biological significance of the conventional PCSKs. The genetics of PCSK enzymes has been increasingly implicated in a multitude of human phenotypes (Table 2). Recent improvements in genome-wide association study (GWAS) arrays and large sample collections have overcome several of the limiting factors of earlier candidate gene approaches that were often tested on small sample sets. We here review the published literature regarding the genetics of the first seven PCSK enzymes in human traits.
Table 2.
Human Traits / Diseases Showing Association with Polymorphisms in Traditional PCSK Genes in Large Genetic Studies
| Gene | Disease / Trait | Reference |
|---|---|---|
| PCSK1 | Fasting glucose-related traits | [52] |
| Body mass index | [71] | |
| Proinsulin levels | [64] | |
| Age at natural menopause | [73] | |
| Proinsulin conversion, glucose homeostasis | [67] | |
| Body mass index and overweight in men | [70] | |
| Obesity | [59, 72] | |
| PCSK2 | Dialysis-related mortality | [99] |
| Age at onset of menarche | [101] | |
| Fibrinogen level | [102] | |
| Total antioxidant level | [96] | |
| Amyotrophic lateral sclerosis | [103] | |
| Maximum common carotid intimal medial thickness | [100] | |
| Chronic kidney disease | [98] | |
| FURIN | Blood pressure | [115] |
| Hypertension | [114] | |
| HPV infection outcome | [111] | |
| PCSK4 | no large genetic association studies reported | |
| PCSK5 | Age at onset of amyotrophic lateral sclerosis | [126] |
| Total ventricular volume | [125] | |
| Height | [130] | |
| Behavioral skills in children that have undergone cardiac surgery | [124] | |
| HDL levels | [128] | |
| PCSK6 | Handedness in dyslexia | [131] |
| Blood pressure | [137] | |
| PCSK7 | Cardiovascular-related traits | [152] |
| Iron homeostasis | [148] |
PCSK1
PCSK1 and PCSK2 are two closely related members of the proprotein convertase enzyme family and they share several functional similarities. Both of them are most actively transcribed in endocrine and neuroendocrine cells. However, recent studies suggest they may also be active in immune cells [22-26]. These proteases localize to the secretory granules, and their activity is regulated by endogenous inhibitors, namely proSAAS for PCSK1 [27] and 7B2 for PCSK2 [28, 29]. The key function of these enzymes is to coordinately process multiple hormone precursors. PCSK1 and PCSK2 targets include proinsulin [30], proopiomelanocortin [31], prorenin [32], proenkephalin [33], prosomatostatin [34], progastrin [35], proglucagon [36] and proghrelin [37].
Because of several important substrates, it is perhaps not surprising that PCSK1 has been linked to many human diseases and endocrinal phenotypes. These include hypogonadism [38-40], adrenal hyperplasias [41], gastrointestinal carcinoids [42], pituitary adenomas [43, 44], hyper- and hypothyroidism [45] and cancers [46-48]. PCSK1 is also abundantly expressed in the hypothalamus [49, 50], the brain center that controls appetite and satiety [51]. Moreover, it is associated with fasting glucose levels [52] and many of its substrates also participate in the regulation of feeding and food processing [53-55].
Along with the cholesterol metabolism regulating convertase PCSK9 [56], PCSK1 is the only proprotein convertase that is known to be mutated in humans [40, 57, 58]. These individuals suffer from a profound endocrinal syndrome characterized by monogenic obesity, hypoadrenalism, a dysregulation of glucose homeostasis and elevated levels of different circulating prohormones.
In addition to monogenic obesity caused by a defictive PCSK1 protein, also polygenic obesity and its connection with polymorphisms in PCSK1 have been investigated. Statistical evidence strongly links three common nonsynonymous SNP variants (rs6232 (N221D) in exon 6, rs6235 (S690T) and rs6234 (Q665E) in exon 14) to common obesity [59]. The N222D substitution is located in the catalytic domain of PCSK1 and reduces its activity, while the S609T and Q665E polymorphisms are located in the C-terminal region of the protein and together they show a high linkage disequilibrium (r2>0.96). This region defines the enzymatic and physical properties of the protein (half-life time, pH optimum, Ca2+ requirement, inhibitor susceptibility [60]), but amino acid changes do not impair the function of the enzyme [58, 61]. A novel, less common SNP variant rs1799904 (R80Q) has been reported to hamper the maturation and activity of PCSK1 [61]. Several other studies have also provided evidence for the association of PCSK1 with obesity-related traits [62-72]. A significant associations between PCSK1 variants and the age of natural menopause has also been reported [73]. However, PCSK1 variants did not significantly associate with the serum level of anti-Mullerian hormone, a marker for ovarian reserve in females [74].
Despite the findings in human studies, the PCSK1 null mouse is not obese [13], although it does show defects in synthesizing mature insulin [75]. The most prominent phenotype of PCSK1 deficient mice is a severe growth defect, which might indicate that PCSK1 can process the growth hormone releasing hormone (GHRH) precursor [76]. However, when researchers aimed at gaining insight into the human PCSK1-related obesity phenotype by mutating the highly conserved codon at position 222 (N222D), an overweight mouse phenotype with impaired proinsulin processing, hyperphagia and increased metabolic efficiency was reported [77]. The activity of the homozygous N222D mutant PCSK1 is about 45 % of acticvity of the wild type enzyme. Although the amount of the enzyme is the same in the tissue extracts from wild type and PCSK1N222D/N222D animals, the ratio of the isoforms is different, suggesting a defect in the autocatalytic cleavage process, which thus may also have importance for human obesity. The level of the hypothalamic alpha-melanocyte-stimulating hormone (αMSH) was also reduced in those animals. This too may contribute to the obesity phenotype [77].
PCSK2
The expression pattern of PCSK2 significantly overlaps with that of PCSK1 and these enzymes often act in concert to process common substrate proteins [78-80]. Knocking out both PCSK1 and PCSK2 in mice is lethal, further supporting the complementary roles for these two enzymes, which cannot be compensated for by other PCSKs [79]. However, the proteolytic functions of PCSK1 and 2 are different, which may result in to distinct products from the same precursor. For example, proglucagon is cleaved into glucagon-like peptide-1 by PCSK1, but PCSK2 processes the same molecule into glucagon [81]. The expression level differences of these convertases are thought to determine which enzyme plays the primary role in the cleavage of a given substrate [82-84]. A unique feature of PCSK2 is that it requires the 7B2 protein for its maturation [85]. The inactive form of PCSK2 binds to 7B2 in the endoplasmatic reticulum and this facilitates the transport of the protein complex to the Golgi apparatus. 7B2 is needed for the activation of proPCSK2, but it also has an inhibitory effect on PCSK2 function [28, 29].
PCSK2 knockout mice appear normal at birth. However, these animals exhibit retarded growth, chronic fasting hypoglycemia and defects in the processing of various neuroendocrine precursor [86-90]. PCSK2 participates together with PCSK1 in insulin maturation [30, 91]. Consequently, PCSK2 knockout mice exhibit increased levels of proinsulin [86].
In humans, different genetic variants of PCSK2 have been shown to associate with type 2 diabetes and related traits in several studies [92-95]. Other human traits that show genetic association with PCSK2 variants include total antioxidant level (SNP rs6044834 [96]), prevalence of myocardial infarction (SNP rs6080699 [97]), chronic kidney disease (SNP rs6080699 [98]), dialysis survival (SNP rs4814615 [99]) maximum common carotid intimal medial thickness (SNP rs4814615 [100]), age of menarche (SNP rs852069 [101]) and fibrinogen levels (rs6044777 [102]). Also, an association between the incidence of amyotrophic lateral sclerosis and SNP rs6080539, which is close to the PCSK2 gene, was reported in US veterans [103]. At present, the molecular mechanisms behind these associations remain to be described.
FURIN
The FURIN gene was discovered more than 25 years ago at the upstream region of FES (feline sarcoma oncogene) [104]. Subsequent biochemical analyses have demonstrated that FURIN is ubiquitously expressed and that it possesses a plethora of target proteins that regulate a wide variety of biological functions ranging from mammalian development to the activation of prohormones and infective agents. FURIN is often considered as a prototypic proprotein convertase, and a significant degree of functional redundancy with other PCSKs, especially with PCSK5 and PCSK7 is evident in in vitro experiments [105]. However, analyses of FURIN germ-line knockout mice have revealed a non-redundant function in mammalian development: FURIN deficient mouse embryos show defective ventral closure and axial rotation and die during the second week of embryonic development [9]. These data support the existence of FURIN specific substrate molecules or alternatively, a lack of expression of other compensatory PCSK enzymes during mammalian embryogenesis.
Because FURIN is essential for mammalian development it is not surprising that FURIN null human subjects have not been described in the literature. In contrast, the expression of FURIN has been reported to be upregulated in many human diseases. Elevated FURIN levels promote for example the metastatic activity of human head and neck cancer, atherosclerosis and the course of pseudomonas infection in cystic fibrosis [106-108]. FURIN expression is enhanced in trans by growth factors such as TGFβ1 [109] and IL12 [110], but a recent study demonstrated that a polymorphism (rs4932178) in the FURIN promoter also directly affects its mRNA levels [111]. This common SNP associates directly with the course of an HBV infection, and may have importance in predicting the disease outcome. Moreover, another study, which investigated genetic alterations in the TGFβ1 pathway genes in colorectal cancer, found a weak, but significant association between the heterozygocity of this SNP and worse disease outcome [112].
FURIN can directly regulate the renin-angiotensin system and factors that maintain the sodium-electrolyte balance [113]. This led a group of Chinese investigators to consider FURIN as a candidate gene in a Kazakh ethnic group with a high prevalence of hypertension [114]. Sequencing of all exons and the promoter region led to a conclusion that rs2071410 moderately associates with the hypertension phenotype. Importantly, the role of FURIN genetics as a risk factor for hypertensive phenotype was recently confirmed by two recent large-scale genetic association studies. First, using a genome-wide association approach to study more than 200 000 subjects of European descent, The International Consortium for Blood Pressure Genome-Wide Association Studies identified a SNP (rs2521501) in the FURIN-FES loci that was associated with an elevation in both the systolic and diastolic blood pressure [115]. Another multi-center study that genotyped ca. 50 000 SNPs in 2100 candidate genes reported two additional polymorphisms in the FURIN gene region, rs2071410 and rs6227, to associate with the diastolic and systolic blood pressure, respectively [116]. Taken together, FURIN clearly emerges as a promising candidate gene for hypertension in three independent studies.
PCSK4
PCSK4 expression is restricted to testicular [117-119] and ovarian cells [120]. Only a few natural substrates for PCSK4 have been identified so far [121] but the lack of functional PCSK4 results in impaired in vivo and in vitro fertility in male mice [15, 119, 122]. To our knowledge, probably due to its restricted expression profile and biological function, there are no GWAS or other types of genetic studies in humans reporting diseases or other conditions or traits associated with PCSK4 polymorphisms.
PCSK5
The PCSK5 gene encodes two distinct splice variants, PCSK5a and PCSK5b. The gene is expressed ubiquitously but the protein isoforms are sorted distinctly in the cell [123]. The shorter PCSK5a is secreted while the longer canonical PCSK5b is a membrane protein localized to the post-Golgi network. Knocking-out PCSK5 in mice is lethal at birth due to multiple craniofacial and patterning abnormalities [10, 11] demonstrating that this gene is essential for mammalian development.
Polymorphisms in the PCSK5 gene region have been associated with various human traits and phenotypes. In addition to sporadic association findings with various apparently non-related traits, the possible neurological role of this gene is emphasized in several genetic association studies. A suggestive genome-wide association (p=1.11e-6) was found for the intronic PCSK5 SNP rs2261722 with attentiveness and other neurobehavioral skills (Child Behavior Checklist for ages 1.5 to 5 years, CBCL/1.5-5) in children who underwent cardiac surgery before six months of age [124]. This converges with the observation that the intergenic PCSK5 SNP rs10512049 is associated with Alzheimer’s disease related total ventricular volume (genome-wide borderline p value of 3.48e-6) [125]. Also, a recent meta-analysis of GWA studies linked PCSK5 to another neurodegenerative disorder, amyotrophic lateral sclerosis (ALS). When several sample collections were studied together, it was found that two PCSK5 SNPs (rs7047865 and rs1258095) associated with the age of ALS onset [126].
Anorectal atresia is a birth defect that causes morbidity and requires surgical operations. A recent study found SNPs in the PCSK5 region that nominally associated with this malformation [127]. This finding may be explained by the observation that PCSK5 coordinately regulates caudal Hox paralogs via growth differentiation factor 11 (GDF11) to control anteroposterior patterning and anorectal development [11]. Using a multi-stage analysis with linkage and association design Iatan et al. (2009) showed that SNPs in the PCSK5 also influence the levels of high-density lipoprotein cholesterol (HDL-C) [128]. The mechanism behind this might be explained by PCSK5 mediated inactivation of endothelial lipase [129], which is a natural modulator of HDL particles.
Interestingly also, a SNP in the PCSK5 region was among the DNA sequence variants most strongly associated with human height [130]. This study also underlined that pin-pointing significant associations with genomic variants in polygenic traits may require an extensive study protocol with enormous sample collections. Using more than 180000 samples and 2.8 million genome-wide SNPs in a highly heritable polygenic trait, the researchers managed to explain up to 20% of the heritable variation of this trait. So although the SNP rs11144688 in the PCSK5 region has a significant effect, it explains only a minor fraction of height variability in the population.
PCSK6
The PCSK6 gene encodes several transcripts which are ubiquitously expressed. In mice PCSK6 regulates the function of TGFβ family cytokines during central nervous system patterning and left-right axis formation [12]. Consequently, approximately one-quarter of PCSK6 deficient mouse embryos die prenatally. In humans the SNP rs11855415 in the PCSK6 region is the marker most significantly associated with direction of handedness in individuals with dyslexia [131]. It reaches the genome-wide significance level with a p-value of 1.99e-8. PCSK6 is known to process for example the NODAL proprotein [12] which has a role in antero-posterior and left-right axes specification. This role of PCSK6 in embryo patterning might explain also the genetic association with cerebral asymmetry and handedness. However, a very recent study showed that rather than dictating the direction of handedness PCSK6 contributes to mechanisms underlying the establishment of normal brain lateralization and thus the degree of handedness [132].
Osteoarthritis (OA) is the most common joint disorder where the loss of bone and cartilage tissue causes joint pain, tenderness, stiffness, locking, and sometimes effusion. Previous functional investigations have implicated that PCSK6 contributes to the pathogenesis of OA by activating aggrecanases leading to aggrecan breakdown [133, 134]. A recent study using a candidate gene approach proved also that the rs900414 SNP in the PCSK6 gene region consistently associated with the severity of pain in OA [135]. A possible functional explanation for this association comes from the location of the associated SNP. It is located in the intron between exons 22 and 23 and could, in theory, be involved in variable splicing of the gene. Exon 23 encodes a hydrophobic cluster that retards PCSK6 secretion and thus a transcript with exon 23 encodes an intracellular PCSK6 isoform, while the variant lacking exon 23 encodes a secreted version of the protein. [136]. Hence, the altered ratios of these enzyme isoforms might contribute to enzyme function through an altered cellular distribution of the protein.
A small study using Chinese individuals linked PCSK6 also to hypertension. A SNP rs1871977 and a consequent 5 marker haplotype associated statistically significantly with high diastolic blood pressure [137]. Also, two previous segregation analyses have found evidence for increased linkage to blood pressure in the genomic region next to the PCSK6 gene [138, 139].
PCSK7
PCSK7 is a ubiquitously expressed gene [140, 141] and the majority of the protein is concentrated to the trans-Golgi network [142] from where it cycles to the plasma membrane [143]. Interestingly, a translocation breakpoint in the 3’ untranslated region of the gene has been observed, and a resulting fusion protein with the IGH gene product is repeatedly detected in patients with lymphomas [144]. Until recently the biological function of PCSK7 in mammals was largely unknown. Scattered observations in the literature first proposed that PCSK7 deficient mice undergo normal development and adults have little phenotypic abnormalities, but Besnard and colleagues demonstrated in 2012 that a PCSK7 deficiency contributes to behavioral patterns in mice [16-18]. However, a recent study using a Xenopus model system demonstrated that the lack of PCSK7 function results in fundamental defects in brain and eye development suggesting a critical role for this enzyme in the embryogenesis of at least lower vertebrates [145].
A series of in vitro experiments has demonstrated that PCSK7 operates often redundantly with other conventional PCSKs. To date, biochemical and genetic approaches have led to the identification of only a few specific substrates. PCSK7 is non-replaceable in rescuing an unstable MHC I in antigen presenting cells [146] and it mediates the proteolysis of proEGF presumably in an indirect manner [147]. Also, polymorphisms in the human PCSK7 have been associated with two phenotypes in recent GWA studies. Oexle et al. (2011) showed a strong (p=1.1e-27) novel association of SNP rs236918 in PCSK7 intron 9 with the level of soluble transferrin receptor (sTfR) in a meta-analysis of five genome-wide association studies [148]. Mechanistically, two distinct functions for PCSK7 in iron metabolism have been suggested. Firstly, the expression of hepcidin, which is the principal iron regulatory hormone, is downregulated by soluble haemojuvelin (sHJV). Release of sHJV from cellular haemojuvelin requires FURIN or another proprotein convertase [149, 150]. Secondly, PCSK7 can function to release sTfR from membrane-bound TfR. This latter mechanism attained some support from a very recent study showing the capacity of PCSK7 to directly shed human TfR1 at an unusual target site KTECER↓ [151]. This plausible physiological mechanism combined with the strong genetic association makes PCSK7 an actual functional candidate for therapeutics that regulate human iron homeostasis.
Another published GWA study linked PCSK7 to cardiovascular events. Middelberg et al. (2011) searched for genes that simultaneously associate with more than one cardiovascular-related biochemical trait [152]. They found that the PCSK7 SNP rs508487 is associated with levels of triglycerides and low-density lipoproteins LDL (borderline multivariate association of p=2.7e-5). However, the authors did not speculate on the functional relevance of the finding and thus it remains to be clarified whether it is true causation or just results from a physical linkage with other genetic factors.
CONCLUSIONS
Genetic association studies have undergone a fundamental change in design and implementation during the last 10-15 years. Large sample collections and hypothesis-free selection of study markers have uncovered numerous important features of several previously unthinkable phenotypes. Also, members of the proprotein convertase gene family have now been convincingly associated with various human traits. For example, the roles of PCSK1 and PCSK2 in common obesity and related traits, FURIN in hypertension, PCSK6 in osteoarthritis and PCSK7 in iron homeostasis have now been well documented by independent study groups. Dissecting these genetic associations further serves three major aims. It can help to reveal the biological grounds for any phenotype, decode the specific functions and substrates for individual PCSKs and finally offer new candidates for drug development [3]. In the future, expanding the investigations also to the regulatory regions of the PCSK genes located even on different chromosomes will shed more light on the phenotypic associations. These studies are continuously facilitated by developments in more powerful and accurate genome-wide technologies, such as large scale sequencing,
ACKNOWLEDGEMENTS
This study was financially supported by the Academy of Finland (projects 128623, 135980 (MP), a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme (MP), Emil Aaltonen Foundation (MP), Sigrid Jusélius Foundation (MP), Tampere Tuberculosis Foundation (MP) and Competitive Research Funding of the Tampere University Hospital (grants 9M080, 9N056 (MP)).
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
ABBREVIATIONS
- GWA
= Genome wide association
- PCSK
= Proprotein convertase subtilisin/kexin
- SNP
= Single nucleotide polymorphis
REFERENCES
- 1.Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artenstein AW, Opal S M. Proprotein convertases in health and disease. N. Engl. J. Med. 2011;365:2507–2518. doi: 10.1056/NEJMra1106700. [DOI] [PubMed] [Google Scholar]
- 3.Seidah N G, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov . 012:;1:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 4.Seidah N G, Sadr M S, Chretien M, Mbikay M. The multifaceted Proprotein Convertases: their unique, redundant, complementary and opposite functions. J. Biol. Chem. 2013 doi: 10.1074/jbc.R113.481549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrich S, Lindberg I, Bode W, Than M E. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J. Mol. Biol. 2005;345:211–227. doi: 10.1016/j.jmb.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 6.Shiryaev S A, Chernov A V, Golubkov V S, Thomsen: e. R. Chudin: e, Chee M S, Kozlov I A, Strongin A Y, Cieplak P. High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome. PLoS One. 2013;8:e54290. doi: 10.1371/journal.pone.0054290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turpeinen H, Kukkurainen S, Pulkkinen K, Kauppila T, Ojala K, Hytonen V P, Pesu M. Identification of proprotein convertase substrates using genome-wide expression correlation analysis. BMC Genomics. 2011;12:618. doi: 10.1186/1471-2164-12-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paolillo L, Simonetti M, Brakch N, D'Auria G, Saviano M, Dettin M, Rholam M, Scatturin A, Di Bello C, Cohen P. l Evidence for the presence of a secondary structure at the dibasic processing site of prohormone: the pro-ocytocin model. EMBO J. 1992;11:2399–2405. doi: 10.1002/j.1460-2075.1992.tb05304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roebroek A J, Umans L, Pauli I G, Robertsone J, van Leuven F, Van de Ven W J, Constam D B. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development: 1998;125:4863–4876. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- 10.Essalmani R, Zaid A, Marcinkiewicz J, Chamberland A, Pasquato A, Seidah N G, Prat A. In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5750–5755. doi: 10.1073/pnas.0709428105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szumska D, Pieles G, Essalmani R, Bilski M, Mesnard D, Kaur K, Franklyn A, El Omari K, Jefferis J, Bentham J, Taylor J M, Schneider J E, Arnold S J, Johnson P, Tymowska-Lalanne Z, Stammers D, Clarke K, Neubauer S, Morris A, Brown S D, Shaw-Smith C, Cama A, Capra V, Ragoussis J, Constam D, Seidah N G, Prat A, Bhattacharya S. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 2008;22:1465–1477. doi: 10.1101/gad.479408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constam D B, Robertson e J. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X, Zhou A, Dey A, Norrbom C, Carroll R, Zhang C, Laurent V, Lindberg I, Ugleholdt R, Holst J J, Steiner D F. Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10293–10298. doi: 10.1073/pnas.162352599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagnon J, Mayne J, Chen A, Raymond A, Woulfe J, Mbikay M, Chretien M. PCSK2-null mice exhibit delayed intestinal motility, reduced refeeding response and altered plasma levels of several regulatory peptides. Life Sci. 2011;88:212–217. doi: 10.1016/j.lfs.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Mbikay M, Tadros H, Ishida N, Lerner C P, De Lamirande: e. Chen, A. El-Alfy, M. Clermont, Y. Seidah, N. G. Chretien, M. Gagnon, C. Simpson: e M. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6842–6846. doi: 10.1073/pnas.94.13.6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Constam D B, Calfon M, Robertsone J. SPC4 SPC6 and the novel protease SPC7 are coexpressed with bone morphogenetic proteins at disti. doi: 10.1083/jcb.134.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villeneuve P, Feliciangeli S, Croissandeau G, Seidah N G, Mbikay M, Kitabgi P, Beaudet A. Altered processing of the neurotensin/neuromedin N precursor in PC2 knock down mice: a biochemical and immunohistochemical study. J. Neurochem. 2002;82:783–793. doi: 10.1046/j.1471-4159.2002.00988.x. [DOI] [PubMed] [Google Scholar]
- 18.Besnard J, Ruda G F, Setola V, Abecassis K, Rodriguiz R M, Huang X P, Norval S, Sassano M F, Shin A I, Web-ster L A, Simeons F R, Stojanovski L, Prat A, Seidah N G, Constam D B, Bickerton G R, Read K D, Wetsel W C, Gilbert I H, Roth B L, Hopkins A L. Automated design of ligands to polypharmacological profiles. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patra D, Xing X, Davies S, Bryan J, Franz C, Hunzikere B, Sandell L J. Site-1 protease is essential for endochondral bone formation in mice. J. Cell Biol. 2007;179:687–700. doi: 10.1083/jcb.200708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashid S, Curtis D E, Garuti R, Anderson N N, Bashmakov Y, Ho Y K, Hammer R E, Moon Y A, Horton J D. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaid A, Roubtsova A, Essalmani R, Marcinkiewicz J, Chamberland A, Hamelin J, Tremblay M, Jacques H, Jin W, Davignon J, Seidah N G, Prat A. Proprotein convertase subtilisin/kexin type 9 (PCSK9): hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology. 2008;48:646–654. doi: 10.1002/hep.22354. [DOI] [PubMed] [Google Scholar]
- 22.LaMendola J, Martin S K, Steiner D F. Expression of PC3, carboxypeptidase E and enkephalin in human monocyte-derived macrophages as a tool for genetic studies. FEBS Lett. 1997;404:19–22. doi: 10.1016/s0014-5793(97)00078-1. [DOI] [PubMed] [Google Scholar]
- 23.Nakashima M, Nie Y, Li Q L, Friedman T C. Up-regulation of splenic prohormone convertases PC1 and PC2 in diabetic rats. Regul. Pept. 2001;102:135–145. doi: 10.1016/s0167-0115(01)00311-1. [DOI] [PubMed] [Google Scholar]
- 24.Mousa S A, Shakibaei M, Sitte N, Schafer M, Stein C. Subcellular pathways of beta-endorphin synthesis, processing, and release from immunocytes in inflammatory pain. Endocrinology. 2004;145:1331–1341. doi: 10.1210/en.2003-1287. [DOI] [PubMed] [Google Scholar]
- 25.Lansac G, Dong W, Dubois C M, Benlarbi N, Afonso C, Fournier I, Salzet M, Day R. Lipopolysaccharide mediated regulation of neuroendocrine associated proprotein convertases and neuropeptide precursor processing in the rat spleen. J. Neuroimmu-nol. 2006;171:57–71. doi: 10.1016/j.jneuroim.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Refaie S, Gagnon S, Gagnon H, Desjardins R, D'Anjou F, D'Orleans-Juste P, Zhu X, Steiner D F, Seidah N G, Lazure C, Salzet M, Day R. Disruption of proprotein convertase 1/3 (PC1/3) expression in mice causes innate immune defects and uncontrolled cytokine secretion. J. Biol. Chem. 2012;287:14703–14717. doi: 10.1074/jbc.M111.323220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fricker L D, McKinzie A A, Sun J, Currane Qian Y, Yan L, Patterson S D, Courchesne P L, Richards B, Levin N, Mzhavia N, Devi L A, Douglass J. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J. Neurosci. 2000;20:639–648. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou A, Webb G, Zhu X, Steiner D F. Proteolytic processing in the secretory pathway. J. Biol. Chem. 1999;274:20745–20748. doi: 10.1074/jbc.274.30.20745. [DOI] [PubMed] [Google Scholar]
- 29.Muller L, Lindberg I. The cell biology of the prohormone convertases PC1 and PC2. Prog. Nucleic Acid Res. Mol. Biol. 1999;63:69–108. doi: 10.1016/s0079-6603(08)60720-5. [DOI] [PubMed] [Google Scholar]
- 30.Smeekens S P, Montag A G, Thomas G, Albiges-Rizo C, Carroll R, Benig M, Phillips L A, Martin S, Ohagi S, Gardner P. Proinsulin processing by the subtilisin-related proprotein convertases furin, PC2 and PC3. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8822–8826. doi: 10.1073/pnas.89.18.8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjannet S, Rondeau N, Day R, Chretien M, Seidah N G. PC1 and PC2 are proprotein convertases capable of cleaving proopiomelanocortin at distinct pairs of basic residues. Proc. Natl. Acad. Sci. U. S. A.: 1991;88:3564–3568. doi: 10.1073/pnas.88.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjannet S, Reudelhuber T, Mercure C, Rondeau N, Chre-tien M, Seidah N G. Proprotein conversion is determined by a multiplicity of factors including convertase processing, substrate specificity, and intracellular environment.Cell type-specific processing of human prorenin by the convertase PC1. J. Biol. Chem. 1992;267:11417–11423. [PubMed] [Google Scholar]
- 33.Breslin M B, Lindberg I, Benjannet S, Mathis J P, Lazure C, Seidah N G. Differential processing of proenkephalin by prohormone convertases 1(3) and 2 and furin. J. Biol. Chem. 1993;268:27084–27093. [PubMed] [Google Scholar]
- 34.Galanopoulou A S, Kent G, Rabbani S N, Seidah N G, Patel Y C. Heterologous processing of prosomatostatin in constitutive and regulated secretory pathways.Putative role of the endoproteases furin PC1 and PC2. J. Biol. Chem. 1993;268:6041–6049. [PubMed] [Google Scholar]
- 35.Rehfeld J F, Zhu X, Norrbom C, Bundgaard J R, Johnsen A H, Nielsen J E, Vikesaa J, Stein J, Dey A, Steiner D F, Friis-Hansen L. Prohormone convertases 1/3 and 2 together orchestrate the site-specific cleavages of progastrin to release gastrin-34 and gastrin-17. Biochem. J. 2008;415:35–43. doi: 10.1042/BJ20080881. [DOI] [PubMed] [Google Scholar]
- 36.Rouille Y, Kantengwa S, Irminger J C, Halban P A. Role of the prohormone convertase PC3 in the processing of proglucagon to glucagon-like peptide 1. J. Biol. Chem. 1997;272:32810–32816. doi: 10.1074/jbc.272.52.32810. [DOI] [PubMed] [Google Scholar]
- 37.Zhu X, Cao Y, Voogd K, Steiner D F. On the processing of proghrelin to ghrelin. J. Biol. Chem. 2006;281:38867–38870. doi: 10.1074/jbc.M607955200. [DOI] [PubMed] [Google Scholar]
- 38.Cadman S M, Kim S H, Hu Y, Gonzalez-Martinez D, Bouloux P M. Molecular pathogenesis of Kallmann's syndrome. Horm. Res. 2007;67:231–242. doi: 10.1159/000098156. [DOI] [PubMed] [Google Scholar]
- 39.Seminara S B, Oliveira L M, Beranova M, Hayes F J, Crowley W FJr. Genetics of hypogonadotropic hypogonadism. J. Endocrinol. Invest. 2000;23:560–565. doi: 10.1007/BF03343776. [DOI] [PubMed] [Google Scholar]
- 40.Jackson R S, Creemers J W, Farooqi I S, Raffin-Sanson M L, Varro A, Dockray G J, Holst J J, Brubaker P L, Corvol P, Polonsky K S, Ostrega D, Becker K L, Bertagna X, Hutton J C, White A, Dattani M T, Hussain K, Middleton S J, Nicole T M, Milla P J, Lindley K J, O'Rahilly S. Small-intestinal dysfunction accompanies the complex en-docrinopathy of human proprotein convertase 1 deficiency. J. Clin. Invest. 2003;112:1550–1560. doi: 10.1172/JCI18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferraz-de-Souza B, Achermann J C. Disorders of adrenal devel-opment. Endocr Dev. 2008;13:19–32. doi: 10.1159/000134753. [DOI] [PubMed] [Google Scholar]
- 42.Tomita T. Immunocytochemical localization of prohormone con-vertase 1/3 and 2 in gastrointestinal carcinoids. Endocr Pathol. 2001;12:137–145. doi: 10.1385/ep:12:2:137. [DOI] [PubMed] [Google Scholar]
- 43.Jin L, Kulige Qian X, Scheithauer B W, Young W F Jr, Davis D H, Seidah N G, Chretien M, Lloyd R V. Distribu-tion and regulation of proconvertases PC1 and PC2 in human pituitary adenomas. Pituitary. 1999;1:187–195. doi: 10.1023/a:1009909232243. [DOI] [PubMed] [Google Scholar]
- 44.Lloyd R V, Jin L, Qian X, Scheithauer B W, Young W F, r; Davis D H. Analysis of the chromogranin A post-translational cleavage product pancreastatin and the prohormone convertases PC2 and PC3 in normal and neoplastic human pituitaries. Am. J. Pathol. 1995;146:1188–1198. [PMC free article] [PubMed] [Google Scholar]
- 45.Shen X, Li Q L, Brent G A, Friedman T C. Thyroid hormone regulation of prohormone convertase 1 (PC1): regional expression in rat brain and in vitro characterization of negative thyroid hormone response elements. J. Mol. Endocrinol. 2004;33:21–33. doi: 10.1677/jme.0.0330021. [DOI] [PubMed] [Google Scholar]
- 46.Lankat-Buttgereit B, Muller S, Schmidt H, Parhofer K G, Gress T M, Goke R. Knockdown of Pdcd4 results in induction of proprotein convertase 1/3 and potent secretion of chromogranin A and secretogranin II in a neuroendocrine cell line. Biol. Cell. 2008;100:703–715. doi: 10.1042/BC20080052. [DOI] [PubMed] [Google Scholar]
- 47.Cheng M, Watson P H, Paterson J A, Seidah N, Chretien M, Shiu R P. Pro-protein convertase gene expression in human breast cancer. Int. J. Cancer: 1997;71:966–971. doi: 10.1002/(sici)1097-0215(19970611)71:6<966::aid-ijc10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Cheng M, Xu N, Iwasiow B, Seidah N, Chretien M, Shiu R P. Elevated expression of proprotein convertases alters breast cancer cell growth in response to estrogen and tamoxifen. J. Mol. Endocrinol. 2001;26:95–105. doi: 10.1677/jme.0.0260095. [DOI] [PubMed] [Google Scholar]
- 49.Schafer M K, Day R, Cullinan W E, Chretien M, Seidah N G, Watson S J. Gene expression of prohormone and propro-tein convertases in the rat CNS: a comparative in situ hybridization analysis. J. Neurosci. 1993;13:1258–1279. doi: 10.1523/JNEUROSCI.13-03-01258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong W, Seidel B, Marcinkiewicz M, Chretien M, Seidah N G, Day R. Cellular localization of the prohormone convertases in the hypothalamic paraventricular and supraoptic nuclei: selective regulation of PC1 in corticotrophin-releasing hormone parvo-cellular neurons mediated by glucocorticoids. J. Neurosci. 1997;17:563–575. doi: 10.1523/JNEUROSCI.17-02-00563.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaoka K, Saharinen P, Pesu M, Holt V E 3rd, Silvennoinen O, O'Shea J J. The Janus kinases (Jaks). Genome Biol. 2004;5:253. doi: 10.1186/gb-2004-5-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manning A K, Hivert M F, Scott R A, Grimsby J L, Bouatia-Naji N, Chen H, Rybin D, Liu C T, Bielak L F, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga J J, Ingelssone Jackson A U, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad M T, Montasser M E, Navarro P, Perry J R, Rasmussen-Torvik L J, Salo P, Sattar N, Shungin D, Strawbridge R J, Tanaka T, van Duijn C M, An P, de Andrade M, Andrews J S, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann J S, Beilby J P, Bellis C, Bergman R N, Blangero J, Boban M, Boehnke M, Boerwinklee Bonnycastle L L, Boomsma D I, Borecki I B, Bottcher Y, Bouchard C, Brunnere Budimir D, Campbell H, Carlson O, Chines P S, Clarke R, Collins F S, Corbaton-Anchuelo A, Couper D, de Faire U, Dedoussis G V, Deloukas P, Dimitriou M, Egan J M, Eiriksdottir G, Erdos M R, Eriksson J G, Eurye Ferrucci L, Ford I, Forouhi N G, Fox C S, Franzosi M G, Franks P W, Frayling T M, Froguel P, Galan P, de Geuse Gigante B, Glazer N L, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris T B, Hayward C, Heath S, Hercberg S, Hicks A A, Hingorani A, Hofman A, Hui J, Hung J, Jarvelin M R, Jhun M A, Johnson P C, Jukema J W, Jula A, Kao W H, Kaprio J, Kardia S L, Keinanen-Kiukaanniemi S, Kivimaki M, Kolcic I, Kovacs P, Kumari M, Kuusisto J, Kyvik K O, Laakso M, Lakka T, Lannfelt L, Lathrop G M, Launer L J, Leander K, Li G, Lind L, Lindstrom J, Lobbens S, Loos R J, Luan J, Lyssenko V, Magi R, Magnusson P K, Marmot M, Meneton P, Mohlke K L, Mooser V, Morken M A, Miljkovic I, Narisu N, O'Connell J, Ong K K, Oostra B A, Palmer L J, Palotie A, Pankow J S, Peden J F, Pedersen N L, Pehlic M, Peltonen L, Penninx B, Pericic M, Perola M, Perusse L, Peyser P A, Polasek O, Pramstaller P P, Province M A, Raikkonen K, Rauramaa R, Rehnberg: e. Rice, K. Rotter, J. I. Rudan, I. Ruokonen, A. Saaristo, T. Sabater-Lleal, M. Salomaa, V. Savage, D. B. Saxena, R. Schwarz, P. Seedorf, U. Sennblad, B. Serrano-Rios, M. Shuldiner, A. R. Sijbrands: e J, Siscovick D S, Smit J H, Small K S, Smith N L, Smith A V, Stancakova A, Stirrups K, Stumvoll M, Sun Y V, Swift A J, Tonjes A, Tuomilehto J, Trompet S, Uitterlinden A G, Uusitupa M, Vikstrom M, Vitart V, Vohl M C, Voight B F, Vollenweider P, Waeber G, Waterworth D M, Watkins H, Wheelere Widene, Wild S H, Willems S M, Willemsen G, Wilson J F, Witteman J C, Wright A F, Yaghootkar H, Zelenika D, Zemunik T, Zgaga L, Wareham N J, McCarthy M I, Barroso I, Watanabe R M, Florez J C, Dupuis J, Meigs J B, Langenberg C. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki K, Jayasena C N, Bloom S R. Obesity and appetite control. Exp. Diabetes Res. 2012;2012:824305. doi: 10.1155/2012/824305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilaweera K N, Barrett P, Mercer J G, Morgan P J. Precursor-protein convertase 1 gene expression in the mouse hypothalamus: differential regulation by ob gene mutation: energy deficit and administration of leptin and coexpression with prepro-orexin. Neuroscience. 2003;119:713–720. doi: 10.1016/s0306-4522(02)00869-2. [DOI] [PubMed] [Google Scholar]
- 55.Farooqi I S, O'Rahilly S. Mutations in ligands and receptors of the leptin-melanocortin pathway that lead to obesity. Nat. Clin. Pract. Endocrinol. Metab. 2008;4:569–577. doi: 10.1038/ncpendmet0966. [DOI] [PubMed] [Google Scholar]
- 56.Abifadel M, Varret M, Rabes J P, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert:e Chambaz J, Chanu B, Lecerf J M, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah N G, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 57.Jackson R S, Creemers J W, Ohagi S, Raffin-Sanson M L, Sanders L, Montague C T, Hutton J C, O'Rahilly S. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat. Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 58.Farooqi I S, Volders K, Stanhope R, Heuschkel R, White A, Lanke Keogh J, O'Rahilly S, Creemers J W. Hyper-phagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J. Clin. Endocrinol. Metab. 2007;92:3369–3373. doi: 10.1210/jc.2007-0687. [DOI] [PubMed] [Google Scholar]
- 59.Benzinou M, Creemers J W, Choquet H, Lobbens S, Dina C, Durande Guerardel A, Boutin P, Jouret B, Heude B, Balkau B, Tichet J, Marre M, Potoczna N, Horber F, Le Stunff C, Czernichow S, Sandbaek A, Lauritzen T, Borch-Johnsen K, Andersen G, Kiess W, Korner A, Kovacs P, Jacobson P, Carlsson L M, Walley A J, Jorgensen T, Hansen T, Pedersen O, Meyre D, Froguel P. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat. Genet. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Y, Lindberg I. Enzymatic properties of carboxyl-terminally truncated prohormone convertase 1 (PC1/SPC3) and evidence for autocatalytic conversion. J. Biol. Chem. 1994;269:18408–18413. [PubMed] [Google Scholar]
- 61.Pickett L A, Yourshaw M, Albornoz V, Chen Z, Solorzano-Vargas R S, Nelson S F, Martin M G, Lindberg I. Func-tional consequences of a novel variant of PCSK1. PLoS One. 2013;8:e55065. doi: 10.1371/journal.pone.0055065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gjesing A P, Vestmar M A, Jorgensen T, Heni M, Holst J J, Witte D R, Hansen T, Pedersen O. The effect of PCSK1 variants on waist, waist-hip ratio and glucose metabolism is modified by sex and glucose tolerance status. PLoS One. 2011;6:e23907. doi: 10.1371/journal.pone.0023907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willer C J, Speliotese K, Loos R J, Li S, Lindgren C M, Heid I M, Berndt S I, Elliott A L, Jackson A U, Lamina C, Lettre G, Lim N, Lyon H N, McCarroll S A, Papadakis K, Qi L, Randall J C, Roccasecca R M, Sanna S, Scheet P, Weedon M N, Wheelere Zhao J H, Jacobs L C, Prokopenko I, Soranzo N, Tanaka T, Timpson N J, Almgren P, Bennett A, Bergman R N, Bingham S A, Bonnycastle L L, Brown M, Burtt N P, Chines P, Coin L, Collins F S, Connell J M, Cooper C, Smith G D, Dennison: e. M. Deodhar, P. Elliott, P. Erdos, M. R. Estrada, K. Evans, D. M. Gianniny, L. Gieger, C. Gillson, C. J. Guiducci, C. Hackett, R. Hadley, D. Hall, A. S. Havulinna, A. S. Hebebrand, J. Hofman, A. Isomaa, B. Jacobs, K. B. Johnson, T. Jousilahti, P. Jovanovic, Z. Khaw, K. T. Kraft, P. Kuokkanen, M. Kuusisto, J. Laitinen, J. Lakattae, G. Luan, J. Luben, R. N. Mangino, M. McArdle, W. L. Meitinger, T. Mulas, A. Munroe, P. B. Narisu, N. Ness, A. R. Northstone, K. O'Rahilly, S. Purmann, C. Rees, M. G. Ridderstrale, M. Ring, S. M. Rivadeneira, F. Ruokonen, A. Sandhu, M. S. Saramies, J. Scott, L. J. Scuteri, A. Silander, K. Sims, M. A. Song, K. Stephens, J. Stevens, S. Stringham, H. M. Tung, Y. C. Valle, T. T. Van Duijn, C. M. Vimaleswaran, K. S. Vollenweider, P. Waeber, G. Wallace, C. Watanabe, R. M. Waterworth, D. M. Watkins, N. Witteman, J. C. Zeggini: e, Zhai G, Zillikens M C, Altshuler D, Caulfield M J, Chanock S J, Farooqi I S, Ferrucci L, Guralnik J M, Hattersley A T, Hu F B, Jarvelin M R, Laakso M, Mooser V, Ong K K, Ouwehand W H, Salomaa V, Samani N J, Spector T D, Tuomi T, Tuomilehto J, Uda M, Uitterlinden A G, Wareham N J, Deloukas P, Frayling T M, Groop L C, Hayes R B, Hunter D J, Mohlke K L, Peltonen L, Schlessinger D, Strachan D P, Wichmann H E, McCarthy M I, Boehnke M, Barroso I, Abecasis G R, Hirschhorn J N. Wellcome Trust Case Control Consortium Genetic Investigation of ANthropometric Traits Consortium Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strawbridge R J, Dupuis J, Prokopenko I, Barker A, Ahlqviste Rybin D, Petrie J R, Travers M E, Bouatia-Naji N, Dimas A S, Nica A, Wheelere Chen H, Voight B F, Taneera J, Kanoni S, Peden J F, Turrini F, Gustafsson S, Zabena C, Almgren P, Barker D J, Barnes D, Dennisone M, Eriksson J G, Eriksson P, Eurye Folkersen L, Fox C S, Frayling T M, Goel A, Gu H F, Horikoshi M, Isomaa B, Jackson A U, Jameson K A, Kajantiee Kerr-Conte J, Kuulasmaa T, Kuusisto J, Loos R J, Luan J, Makrilakis K, Manning A K, Martinez-Larrad M T, Narisu N, Nastase Mannila M, Ohrvik J, Osmond C, Pascoe L, Payne F, Sayer A A, Sennblad B, Silveira A, Stancakova A, Stirrups K, Swift A J, Syvanen A C, Tuomi T, van 't Hooft F M, Walker M, Weedon M N, Xie W, Zethelius B, Ongen H, Malarstig A, Hopewell J C, Saleheen D, Chambers J, Parish S, Danesh J, Kooner J, Ostenson C G, Lind L, Cooper C C, Serrano-Rios M, Ferranninie Forsen T J, Clarke R, Franzosi M G, Seedorf U, Watkins H, Froguel P, Johnson P, Deloukas P, Collins F S, Laakso M, Dermitzakise T, Boehnke M, McCarthy M I, Wareham N J, Groop L, Pattou F, Gloyn A L, Dedoussis G V, Lyssenko V, Meigs J B, Barroso I, Watanabe R M, Ingelssone Langenberg C, Hamsten A, Florez J C. ophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corpeleijne Petersen L, Holst C, Saris W H, Astrup A, Langin D, MacDonald I, Martinez J A, Oppert J M, Polak J, Pedersen O, Froguel P, Arner P, Sorensen T I, Blaake E. Obesity-related polymorphisms and their associations with the ability to regulate fat oxidation in obese Europeans: the NUGENOB study. Obesity (Silver Spring) 2010;18:1369–1377. doi: 10.1038/oby.2009.377. [DOI] [PubMed] [Google Scholar]
- 66.Kilpelainen T O, Bingham S A, Khaw K T, Wareham N J, Loos R J. Association of variants in the PCSK1 gene with obesity in the EPIC-Norfolk study. Hum. Mol. Genet. 2009;18:3496–3501. doi: 10.1093/hmg/ddp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heni M, Haupt A, Schafer S A, Ketterer C, Thamer C, Machicao F, Stefan N, Staiger H, Haring H U, Fritsche A. Association of obesity risk SNPs in PCSK1 with insulin sensitivity and proinsulin conversion. BMC Med. Genet. 2010; 11:86–2350-11-86. doi: 10.1186/1471-2350-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Renstrom F, Payne F, Nordstrom A, Britoe C, Rolandsson O, Hallmans G, Barroso I, Nordstrom P, Franks P. W. GIANT Consortium Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Hum. Mol. Genet. 2009;18:1489–1496. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rouskas K, Kouvatsi A, Paletas K, Papazoglou D, Tsapas A, Lobbens S, Vatin V, Durande Labrune Y, Delplanque J, Meyre D, Froguel P. Common variants in FTO MC4R TMEM18 PRL AIF1 and PCSK1 show evidence of association with adult obesity in the Greek population. Obesity (Silver Spring) 2012;20:389–395. doi: 10.1038/oby.2011.177. [DOI] [PubMed] [Google Scholar]
- 70.Qi Q, Li H, Loos R J, Liu C, Hu F B, Wu H, Yu Z, Lin X. Association of PCSK1 rs6234 with obesity and related traits in a Chinese Han population. PLoS One. 2010;5:e10590. doi: 10.1371/journal.pone.0010590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wen W, Cho Y S, Zheng W, Dorajoo R, Kato N, Qi L, Chen C H, Delahanty R J, Okada Y, Tabara Y, Gu D, Zhu D, Haiman C A, Mo Z, Gao Y T, Saw S M, Go M J, Takeuchi F, Chang L C, Kokubo Y, Liang J, Hao M, Le Marchand L, Zhang Y, Hu Y, Wong T Y, Long J, Han B G, Kubo M, Yamamoto K, Su M H, Miki T, Henderson B E, Song H, Tan A, He J, Ng D P, Cai Q, Tsunoda T, Tsai F J, Iwai N, Chen G K, Shi J, Xu J, Sim X, Xiang Y B, Maeda S, Ong R T, Li C, Nakamura Y, Aung T, Kamatani N, Liu J J, Lu W, Yokota M, Seielstad M, Fann C S, Wu J Y, Lee J Y, Hu F B, Tanaka T, Taie S, Shu X O. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat. Genet. 2012;44:307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang Y C, Chiu Y F, Shih K C, Lin M W, Sheu W H, Donlon T, Curb J D, Jou Y S, Chang T J, Li H Y, Chuang L M. Common PCSK1 haplotypes are associated with obesity in the Chinese population. Obesity (Silver Spring) 2010;18:1404–1409. doi: 10.1038/oby.2009.390. [DOI] [PubMed] [Google Scholar]
- 73.He C, Kraft P, Chasman D I, Buring J E, Chen C, Hankinson S E, Pare G, Chanock S, Ridker P M, Hunter D J. A large-scale candidate gene association study of age at menarche and age at natural menopause. Hum. Genet. 2010;128:515–527. doi: 10.1007/s00439-010-0878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Dorp W, van den Heuvel-Eibrink M M, Stolk L, Pieters R, Uitterlinden A G, Visser J A, Laven J S. Genetic varia-tion may modify ovarian reserve in female childhood cancer survivors. Hum. Reprod. 2013;28:1069–1076. doi: 10.1093/humrep/des472. [DOI] [PubMed] [Google Scholar]
- 75.Zhu X, Orci L, Carroll R, Norrbom C, Ravazzola M, Steiner D F. Severe block in processing of proinsulin to insulin accompanied by elevation of des-64,65 proinsulin intermediates in islets of mice lacking prohormone convertase 1/3. Proc Natl Acad Sci U S A. 2002;99:10299–10304. doi: 10.1073/pnas.162352799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Posner S F, Vaslet C A, Jurofcik M, Lee A, Seidah N G, Nillni eA. Stepwise posttranslational processing of progrowth hormone-releasing hormone (proGHRH) polypeptide by furin and PC1. Endocrine. 2004;23:199–213. doi: 10.1385/ENDO:23:2-3:199. [DOI] [PubMed] [Google Scholar]
- 77.Lloyd D J, Bohan S, Gekakis N. Obesity hyperphagia and increased metabolic efficiency in Pc1 mutant mice. Hum. Mol. Genet. 2006;15:1884–1893. doi: 10.1093/hmg/ddl111. [DOI] [PubMed] [Google Scholar]
- 78.Yang Y, Hua Q X, Liu J, Shimizue H, Choquette M H, Mackin R B, Weiss M A. Solution structure of proinsulin: connecting domain flexibility and prohormone processing. J. Biol. Chem. 2010;285:7847–7851. doi: 10.1074/jbc.C109.084921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seidah N G. What lies ahead for the proprotein convertases?. Ann. N. Y. Acad. Sci. 2011;1220:149–161. doi: 10.1111/j.1749-6632.2010.05883.x. [DOI] [PubMed] [Google Scholar]
- 80.Cyr N E, Stuart R C, Zhu X, Steiner D F, Nillni eA. Biosynthesis of proTRH-derived peptides in prohormone convertase 1 and 2 knockout mice. Peptides. 2012;35:42–48. doi: 10.1016/j.peptides.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 81.Holst J J. Glucagon and glucagon-like peptides 1 and 2. Results Probl. Cell Differ. 2010;50:121–135. doi: 10.1007/400_2009_35. [DOI] [PubMed] [Google Scholar]
- 82.Day R, Schafer M K, Watson S J, Chretien M, Seidah N G. Distribution and regulation of the prohormone convertases PC1 and PC2 in the rat pituitary. Mol. Endocrinol. 1992;6:485–497. doi: 10.1210/mend.6.3.1316544. [DOI] [PubMed] [Google Scholar]
- 83.Rhodes C J, Thorne B A, Lincoln B, Nielsene Hutton J C, Thomas G. Processing of proopiomelanocortin by insulin secretory granule proinsulin processing endopeptidases. J. Biol. Chem. 1993;268:4267–4275. [PMC free article] [PubMed] [Google Scholar]
- 84.Rehfeld J F, Bundgaard J R, Hannibal J, Zhu X, Norrbom C, Steiner D F, Friis-Hansen L. The cell-specific pattern of cholecystokinin peptides in endocrine cells versus neurons is governed by the expression of prohormone convertases 1/3 2 and 5/6. Endocrinology. 2008;149:1600–1608. doi: 10.1210/en.2007-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seidah N G, Hsi K L, De Serres G, Rochemont J, Hamelin J, Antakly T, Cantin M, Chretien M. Isolation and NH2-terminal sequence of a highly conserved human and porcine pituitary protein belonging to a new superfamily.Immunocytochemical localization in pars distalis and pars nervosa of the pituitary and in the supraoptic nucleus of the hypothalamus. Arch. Biochem. Biophys: 1983; 225:525–534. doi: 10.1016/0003-9861(83)90063-2. [DOI] [PubMed] [Google Scholar]
- 86.Furuta M, Yano H, Zhou A, Rouille Y, Holst J J, Carroll R, Ravazzola M, Orci L, Furuta H, Steiner D F. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6646–6651. doi: 10.1073/pnas.94.13.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Furuta M, Carroll R, Martin S, Swift H H, Ravazzola M, Orci L, Steiner D F. Incomplete processing of proinsulin to insulin accompanied by elevation of Des-31,32 proinsulin intermediates in islets of mice lacking active PC2. J. Biol. Chem.: 1998;273:3431–3437. doi: 10.1074/jbc.273.6.3431. [DOI] [PubMed] [Google Scholar]
- 88.Berman Y, Mzhavia N, Polonskaia A, Furuta M, Steiner D F, Pintar J E, Devi L A. Defective prodynorphin processing in mice lacking prohormone convertase PC2. J. Neurochem. 2000;75:1763–1770. doi: 10.1046/j.1471-4159.2000.0751763.x. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X, Pan H, Peng B, Steiner D F, Pintar J E, Fricker L D. Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J. Neurochem. 2010;112:1168–1179. doi: 10.1111/j.1471-4159.2009.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Furuta M, Zhou A, Webb G, Carroll R, Ravazzola M, Orci L, Steiner D F. Severe defect in proglucagon processing in islet A-cells of prohormone convertase 2 null mice. J. Biol. Chem. 2001;276:27197–27202. doi: 10.1074/jbc.M103362200. [DOI] [PubMed] [Google Scholar]
- 91.Bailyese M, Bennett D L, Hutton J C. Proprotein-processing endopeptidases of the insulin secretory granule. Enzyme: 1991;45:301–313. doi: 10.1159/000468903. [DOI] [PubMed] [Google Scholar]
- 92.Yoshida H, Ohagi S, Sanke T, Furuta H, Furuta M, Nanjo K. Association of the prohormone convertase 2 gene (PCSK2) on chromosome 20 with NIDDM in Japanese subjects. Diabetes: 1995;44:389–393. doi: 10.2337/diab.44.4.389. [DOI] [PubMed] [Google Scholar]
- 93.Leak T S, Keene K L, Langefeld C D, Gallagher C J, Mychaleckyj J C, Freedman B I, Bowden D W, Rich S S, Sale M M. Association of the proprotein convertase subtilisin/kexin-type 2 (PCSK2) gene with type 2 diabetes in an African American population. Mol. Genet. Metab. 2007;92:145–150. doi: 10.1016/j.ymgme.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng X, Ren W, Zhang S, Liu J, Li S, Li J, Yang P, He J, Su S, Li P. Association of type 2 diabetes susceptibility genes (TCF7L2, SLC30A8 PCSK1 and PCSK2) and proinsulin conversion in a Chinese population. Mol. Biol. Rep. 2012;39:17–23. doi: 10.1007/s11033-011-0705-6. [DOI] [PubMed] [Google Scholar]
- 95.Jonsson A, Isomaa B, Tuomi T, Eriksson J G, Groop L, Lyssenko V. Effect of a common variant of the PCSK2 gene on reduced insulin secretion. Diabetologia. 2012;55:3245–3251. doi: 10.1007/s00125-012-2728-5. [DOI] [PubMed] [Google Scholar]
- 96.Comuzzie A G, Cole S A, Laston S L, Voruganti V S, Haack K, Gibbs R A, Butte N F. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fujimaki T, Kato K, Yokoi K, Oguri M, Yoshida T, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, Nozawa Y, Kimura G, Yamada Y. Association of genetic variants in SEMA3F, CLEC16A, LAMA3, and PCSK2 with myocardial infarction in Japanese individuals. Atherosclerosis. 2010;210:468–473. doi: 10.1016/j.atherosclerosis.2009.11.050. [DOI] [PubMed] [Google Scholar]
- 98.Yoshida T, Kato K, Yokoi K, Oguri M, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, Nozawa Y, Ya-mada Y. Association of gene polymorphisms with chronic kidney disease in Japanese individuals. Int. J. Mol. Med. 2009;24:539–547. doi: 10.3892/ijmm_00000263. [DOI] [PubMed] [Google Scholar]
- 99.Murea M, Lu L, Ma L, Hicks P J, Divers J, McDonough C W, Langefeld C D, Bowden D W, Freedman B I. Genome-wide association scan for survival on dialysis in African-Americans with type 2 diabetes. Am. J. Nephrol. 2011;33:502–509. doi: 10.1159/000327985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O'Donnell C J, Cupples L A, D'Agostino R B, Fox C S, Hoffmann U, Hwang S J, Ingellson e, Liu C, Murabito J M, Polak J F, Wolf P A, Demissie S. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI's Framingham Heart Study. BMC Med. Genet. 2007;8 ( Suppl 1):S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Elks C E, Perry J R, Sulem P, Chasman D I, Franceschini N, He C, Lunetta K L, Visser J A, Byrnee M, Cous-miner D L, Gudbjartsson D F, Esko T, Feenstra B, Hottenga J J, Koller D L, Kutalik Z, Lin P, Mangino M, Marongiu M, McArdle P F, Smith A V, Stolk L, van Wingerden S H, Zhao J H, Albrechte Corre T, Ingelssone Hayward C, Magnusson P K, Smithe N, Ulivi S, Warrington N M, Zgaga L, Alavere H, Amin N, Aspelund T, Bandinelli S, Barroso I, Berenson G S, Bergmann S, Blackburn H, Boerwinklee Buring J E, Busonero F, Campbell H, Chanock S J, Chen W, Cornelis M C, Couper D, Coviello A D, d'Adamo P, de Faire U, de Geuse J, Deloukas P, Doring A, Smith G D, Easton D F, Eiriksdottir G, Emilsson V, Eriksson J, Ferrucci L, Folsom A R, Foroud T, Garcia M, Gasparini P, Geller F, Gieger C, Gudnason V, Hall P, Hankinson S E, Ferreli L, Heath A C, Hernandez D G, Hofman A, Hu F B, Illig T, Jarvelin M R, Johnson A D, Karasik D, Khaw K T, Kiel D P, Kilpelainen T O, Kolcic I, Kraft P, Launer L J, Laven J S, Li S, Liu J, Levy D, Martin N G, McArdle W L, Melbye M, Mooser V, Murray J C, Murray S S, Nalls M A, Navarro P, Nelis M, Ness A R, Northstone K, Oostra B A, Peacock M, Palmer L J, Palotie A, Pare G, Parker A N, Pedersen N L, Peltonen L, Pennell C E, Pharoah P, Polasek O, Plump A S, Pouta A, Porcue Rafnar T, Rice J P, Ring S M, Rivadeneira F, Rudan I, Sala C, Salomaa V, Sanna S, Schlessinger D, Schork N J, Scuteri A, Segre A V, Shuldiner A R, Soranzo N, Sovio U, Srinivasan S R, Strachan D P, Tammesoo M L, Tikkanene Toniolo D, Tsui K, Tryggvadottir L, Tyrer J, Uda M, van Dam R M, van Meurs J B, Vollenweider P, Waeber G, Wareham N J, Waterworth D M, Weedon M N, Wichmann H E, Willemsen G, Wilson J F, Wright A F, Young L, Zhai G, Zhuang W V, Bierut L J, Boomsma D I, Boyd H A, Crisponi L, Demerathe W, van Duijn C M, Econs M J, Harris T B, Hunter D J, Loos R J, Metspalu A, Montgomery G W, Ridker P M, Spector T D, Streetene A, Stefansson K, Thorsteinsdottir U, Uitterlinden A G, Widene Murabito J M, Ong K K, Murray A. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zemunik T, Boban M, Lauc G, Jankovic S, Rotim K, Vatavuk Z, Bencic G, Dogas Z, Boraska V, Torlak V, Susac J, Zobic I, Rudan D, Pulanic D, Modun D, Mudnic I, Gunjaca G, Budimir D, Hayward C, Vitart V, Wright A F, Campbell H, Rudan I. Genome-wide association study of biochemical traits in Korcula Island. Croatia. Croat. Med. J. 2009;50:23–33. doi: 10.3325/cmj.2009.50.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kwee L C, Liu Y, Haynes C, Gibson J R, Stone A, Schichman S A, Kamel F, Nelson L M, Topol B, Van den Eeden S K, Tanner C M, Cudkowicz M E, Grasso D L, Lawson R, Muralidhar S, Oddonee Z, Schmidt S, Hauser M A. A high-density genome-wide association screen of sporadic ALS in US veterans. PLoS One. 2012;7:e32768. doi: 10.1371/journal.pone.0032768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roebroek A J, Schalken J A, Leunissen J A, Onnekink C, Bloemers H P, Van de Ven W J. Evolutionary conserved close linkage of the c-fes/fps proto-oncogene and genetic sequences encoding a receptor-like protein. EMBO J. 1986;5:2197–2202. doi: 10.1002/j.1460-2075.1986.tb04484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Remacle A G, Shiryaev S A, Ohe S, Cieplak P, Srinivasan A, Wei G, Liddington R C, Ratnikov B I, Parent A, Desjardins R, Day R, Smith J W, Lebl M, Strongin A Y. Substrate cleavage analysis of furin and related proprotein convertases.A comparative study. J. Biol. Chem. 2008;283:20897–20906. doi: 10.1074/jbc.M803762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bassi D E, Mahloogi H, Al-Saleem L, Lopez De Cicco R, Ridge J A, Kl ein-Szanto A J. Elevated furin expression in aggressive human head and neck tumors and tumor cell lines. Mol. Carcinog. 2001;31:224–232. doi: 10.1002/mc.1057. [DOI] [PubMed] [Google Scholar]
- 107.Turpeinen H, Raitoharjue Oksanen A, Oksala N, Levula M, Lyytikainen L P, Jarvinen O, Creemers J W, Kahonen M, Laaksonen R, Pelto-Huikko M, Lehtimaki T, Pesu M. Proprotein convertases in human atherosclerotic plaques: the over-expression of FURIN and its substrate cytokines BAFF and APRIL. Atherosclerosis. 2011;219:799–806. doi: 10.1016/j.atherosclerosis.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 108.Ornatowski W, Poschet J F, Perkette Taylor-Cousar J L, Deretic V. Elevated furin levels in human cystic fibrosis cells result in hypersusceptibility to exotoxin A-induced cytotoxicity. J. Clin. Invest. 2007;117:3489–3497. doi: 10.1172/JCI31499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Blanchette F, Day R, Dong W, Laprise M H, Dubois C M. TGFbeta1 regulates gene expression of its own converting enzyme furin. J. Clin. Invest. 1997;99:1974–1983. doi: 10.1172/JCI119365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pesu M, Muul L, Kanno Y, O'Shea J J. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood. 2006; 108:983–985. doi: 10.1182/blood-2005-09-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lei R X, Shi H, Peng X M, Zhu Y H, Cheng J, Chen G H. Influence of a single nucleotide polymorphism in the P1 pro-moter of the furin gene on transcription activity and hepatitis B virus infection. Hepatology. 2009;50:763–771. doi: 10.1002/hep.23062. [DOI] [PubMed] [Google Scholar]
- 112.Forsti A, Li X, Wagner K, Tavelin B, Enquist K, Palmqvist R, Altieri A, Hallmans G, Hemminki K, Lenner P. Polymorphisms in the transforming growth factor beta 1 pathway in relation to colorectal cancer progression. Genes Chromosomes Cancer. 2010;49:270–281. doi: 10.1002/gcc.20738. [DOI] [PubMed] [Google Scholar]
- 113.Cousin C, Bracquart D, Contrepas A, Corvol P, Muller L, Nguyen G. Soluble form of the (pro)renin receptor generated by intracellular cleavage by furin is secreted in plasma. Hypertension. 2009;53:1077–1082. doi: 10.1161/HYPERTENSIONAHA.108.127258. [DOI] [PubMed] [Google Scholar]
- 114.Li N, Luo W, Juhong Z, Yang J, Wang H, Zhou L, Chang J. Associations between genetic variations in the FURIN gene and hypertension. BMC Med. Genet. 2010;11:124. doi: 10.1186/1471-2350-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ehret G B, Munroe P B, Rice K M, Bochud M, Johnson A D, Chasman D I, Smith A V, Tobin M D, Verwoert G C, Hwang S J, Pihur V, Vollenweider P, O'Reilly P F, Amin N, Bragg-Gresham J L, Teumer A, Glazer N L, Lau-ner L, Zhao J H, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks A A, Jackson A U, Peden J F, Tanaka T, Wild S H, Rudan I, Igl W, Milaneschi Y, Parker A N, Fava C, Chambers J C, Foxe R, Kumari M, Go M J, van der Harst P, Kao W H, Sjogren M, Vinay D G, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup P H, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen K D, Lehtimaki T, Matullo G, Wu Y, Gaunt T R, Onland-Moret N C, Cooper M N, Platou C G, Orge Hardy R, Dahgam S, Palmen J, Vitart V, Braund P S, Kuznetsova T, Uiterwaal C S, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer N D, Aspelund T, Garcia M, Chang Y P, O'Connell J R, Steinle N I, Grobbee D E, Arking D E, Kardia S L, Morrison A C, Hernandez D, Najjar S, McArdle W L, Hadley D, Brown M J, Connell J M, Hingorani A D, Day I N, Lawlor D A, Beilby J P, Lawrence R W, Clarke R, Hopewell J C, Ongen H, Dreisbach A W, Li Y, Young J H, Bis J C, Kahonen M, Viikari J, Adair L S, Lee N R, Chen M H, Olden M, Pattaro C, Bolton J A, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling T M, Islam M, Jafar T H, Erdmann J, Kulkarni S R, Bornstein S R, Grassler J, Groop L, Voight B F, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw K T, Weder A B, Hunt S C, Sun Y V, Bergman R N, Collins F S, Bonnycastle L L, Scott L J, Stringham H M, Peltonen L, Perola M, Vartiainene Brand S M, Staessen J A, Wang T J, Burton P R, Artigas M S, Dong Y, Snieder H, Wang X, Zhu H, Lohman K K, Rudock M E, Heckbert S R, Smith N L, Wiggins K L, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld C D, Rosengren A, Thelle D S, Corsi A M, Singleton A, Forrester T, Hilton G, McKenzie C A, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann H E, Cho Y S, Kim H L, Lee J Y, Scott J, Sehmi J S, Zhang W, Hedblad B, Nilsson P, Smith G D, Wong A, Narisu N, Stancakova A, Raffel L J, Yao J, Kathiresan S, O'Donnell C J, Schwartz S M, Ikram M A, Longstreth W T Jr, Mosley T H, Seshadri S, Shrine N R, Wain L V, Morken M A, Swift A J, Laitinen J, Prokopenko I, Zitting P, Cooper J A, Humphries S E, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker S J, van Gilst W H, Janipalli C S, Mani K R, Yajnik C S, Hofman A, Mattace-Raso F U, Oostra B A, Demirkan A, Isaacs A, Rivadeneira F, Lakattae G, Orru M, Scuteri A, Ala-Korpela M, Kangas A J, Lyytikainen L P, Soininen P, Tukiainen T, Wurtz P, Ong R T, Dorr M, Kroemer H K, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector T D, Zhai G, Meschia J F, Nalls M A, Sharma P, Terzic J, Kumar M V, Denniff M, Zukowska-Szczechowskae Wagenknecht L E, Fowkes F G, Charchar F J, Schwarz P E, Hayward C, Guo X, Rotimi C, Bots M L, Brande Samani N J, Polasek O, Talmud P J, Nyberg F, Kuh D, Laan M, Hveem K, Palmer L J, van der Schouw Y T, Casas J P, Mohlke K L, Vineis P, Raitakari O, Ganesh S K, Wong T Y, Taie S, Cooper R S, Laakso M, Rao D C, Harris T B, Morris R W, Dominiczak A F, Kivimaki M, Marmot M G, Miki T, Saleheen D, Chandak G R, Coresh J, Navis G, Salomaa V, Han B G, Zhu X, Kooner J S, Melander O, Ridker P M, Bandinelli S, Gyllensten U B, Wright A F, Wilson J F, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller P P, Elosua R, Soranzo N, Sijbrandse J, Altshuler D, Loos R J, Shuldiner A R, Gieger C, Meneton P, Uitterlinden A G, Wareham N J, Gudnason V, Rotter J I, Rettig R, Uda M, Strachan D P, Witteman J C, Hartikainen A L, Beckmann J S, Boerwinklee Vasan R S, Boehnke M, Larson M G, Jarvelin M R, Psaty B M, Abecasis G R, Chakravarti A, Elliott P, van Duijn C M, Newton-Cheh C, Levy D, Caulfield M J, Johnson T, Tang H, Knowles J, Hlatky M, Fortmann S, Assimes T L, Quertermous T, Go A, Iribarren C, Absher D, Risch N, Myers R, Sidney S, Ziegler A, Schillert A, Bickel C, Sinning C, Rupprecht H J, Lackner K, Wild P, Schnabel R, Blankenberg S, Zeller T, Munzel T, Perret C, Cambien F, Tiret L, Nicaud V, Proust C, Dehghan A, Hofman A, Uitterlinden A, van Duijn C, Levy D, Whitteman J, Cupples L A, Demissie-Banjaw S, Ramachandran V, Smith A, Gudnason V, Boerwinklee Folsom A, Morrison A, Psaty B M, Chen I Y, Rotter J I, Bis J, Volcik K, Rice K, Taylor K D, Marciante K, Smith N, Glazer N, Heckbert S, Harris T, Lumley T, Kong A, Thorleifsson G, Thorgeirsson G, Holm H, Gulcher J R, Stefansson K, Andersen K, Gretarsdottir S, Thorsteinsdottir U, Preuss M, Schreiber S, Meitinger T, Konig I R, Lieb W, Hengstenberg C, Schunkert H, Erdmann J, Fischer M, Grosshennig A, Medack A, Stark K, Linsel-Nitschke P, Bruse P, Aherrahrou Z, Peters A, Loley C, Willenborg C, Nahrstedt J, Freyer J, Gulde S, Doering A, Meisinger C, Wichmann H E, Klopp N, Illig T, Meinitzer A, Tomaschitz A, Halperine Dobnig H, Scharnagl H, Kleber M, Laaksonen R, Pilz S, Grammer T B, Stojakovic T, Renner W, Marz W, Bohm B O, Winkelmann B R, Winkler K, Hoffmann M M, O'Donnell C J, Voight B F, Altshuler D, Siscovick D S, Musunuru K, Peltonen L, Barbalic M, Melander O, Elosua R, Kathiresan S, Schwartz S M, Salomaa V, Guiducci C, Burtt N, Gabriel S B, Stewart A F, Wells G A, Chen L, Jarinova O, Roberts R, McPherson R, Dandona S, Pichard A D, Rader D J, Devaney J, Lindsay J M, Kent K M, Qu L, Satler L, Burnett M S, Li M, Reilly M P, Wilensky R, Waksman R, Epstein S, Matthai W, Knouff C W, Waterworth D M, Hakonarson H H, Walker M C, Mooser V, Hall A S, Balmforth A J, Wright B J, Nelson C, Thompson J R, Samani N J, Braund P S, Ball S G, Smith N L, Felix J F, Morrison A C, Demissie S, Glazer N L, Loehr L R, Cupples L A, Dehghan A, Lumley T, Rosamond W D, Lieb W, Rivadeneira F, Bis J C, Folsom A R, Benjamine Aulchenko Y S, Haritunians T, Couper D, Murabito J, Wang Y A, Stricker B H, Gottdiener J S, Chang P P, Wang T J, Rice K M, Hofman A, Heckbert S R, Foxe R, O'Donnell C J, Uitterlinden A G, Rotter J I, Willerson J T, Levy D, van Duijn C M, Psaty B M, Witteman J C, Boerwinklee Vasan R S, Kottgen A, Pattaro C, Boger C A, Fuchsberger C, Olden M, Glazer N L, Parsa A, Gao X, Yang Q, Smith A V, O'Connell J R, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang S J, Johnson A D, Dehghan A, Teumer A, Pare G, Atkinsone J, Zeller T, Lohman K, Cornelis M C, Probst-Hensch N M, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer L J, Harris T B, Rampersaude Mitchell B D, Arking D E, Boerwinklee Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer J, Haritunians T, Lumley T, Siscovick D, Psaty B M, Zillikens M C, Oostra B A, Feitosa M, Province M, de Andrade M, Turner S T, Schillert A, Ziegler A, Wild P S, Schnabel R B, Wilde S, Munzel T F, Leak T S, Illig T, Klopp N, Meisinger C, Wichmann H E, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu F B, Johansson A, Igl W, Zaboli G, Wild S H, Wright A F, Campbell H, Ellinghaus D, Schreiber S, Aulchenko Y S, Felix J F, Rivadeneira F, Uitterlinden A G, Hofman A, Imboden M, Nitsch D, Brandstatter A, Kollerits B, Kedenko L, Magi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Volzke H, Kroemer H K, Nauck M, Volker U, Polasek O, Vitart V, Badola S, Parker A N, Ridker P M, Kardia S L, Blankenberg S, Liu Y, Curhan G C, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner A R, Coresh J, Schmidt R, Ferrucci L, Shlipak M G, van Duijn C M, Borecki I, Kramer B K, Rudan I, Gyllensten U, Wilson J F, Witteman J C, Pramstaller P P, Rettig R, Hastie N, Chasman D I, Kao W H, Heid I M, Fox C S, Vasan R S, Glazer N L, Felix J F, Lieb W, Wild P S, Felix S B, Watzinger N, Larson M G, Smith N L, Dehghan A, Grosshennig A, Schillert A, Teumer A, Schmidt R, Kathiresan S, Lumley T, Aulchenko Y S, Konig I R, Zeller T, Homuth G, Struchalin M, Aragam J, Bis J C, Rivadeneira F, Erdmann J, Schnabel R B, Dorr M, Zweiker R, Lind L, Rodeheffer R J, Greiser K H, Levy D, Haritunians T, Deckers J W, Stritzke J, Lackner K J, Volker U, Ingelssone Kullo I, Haerting J, O'Donnell C J, Heckbert S R, Stricker B H, Ziegler A, Reffelmann T, Redfield M M, Werdan K, Mitchell G F, Rice K, Arnett D K, Hofman A, Gottdiener J S, Uitterlinden A G, Meitinger T, Blettner M, Friedrich N, Wang T J, Psaty B M, van Duijn C M, Wichmann H E, Munzel T F, Kroemer H K, Benjamine J, Rotter J I, Witteman J C, Schunkert H, Schmidt H, Volzke H, Blankenberg S, Chambers J C, Zhang W, Lord G M, van der Harst P, Lawlor D A, Sehmi J S, Gale D P, Wass M N, Ahmadi K R, Bakker S J, Beckmann J, Bilo H J, Bochud M, Brown M J, Caulfield M J, Connell J M, Cook H T, Cotlarciuc I, Davey Smith G, de Silva R, Deng G, Devuyst O, Dikkeschei L D, Dimkovic N, Dockrell M, Dominiczak A, Ebrahim S, Eggermann T, Farrall M, Ferrucci L, Floege J, Forouhi N G, Gansevoort R T, Han X, Hedblad B, Homan van der Heide J J, Hepkema B G, Hernandez-Fuentes M, Hypponene Johnson T, de Jong P E, Kleefstra N, Lagou V, Lapsley M, Li Y, Loos R J, Luan J, Luttropp K, Marechal C, Melander O, Munroe P B, Nordfors L, Parsa A, Peltonen L, Penninx B W, Peruchae Pouta A, Prokopenko I, Roderick P J, Ruokonen A, Samani N J, Sanna S, Schalling M, Schlessinger D, Schlieper G, Seelen M A, Shuldiner A R, Sjogren M, Smit J H, Snieder H, Soranzo N, Spector T D, Stenvinkel P, Sternberg M J, Swaminathan R, Tanaka T, Ubink-Veltmaat L J, Uda M, Vollenweider P, Wallace C, Waterworth D, Zerres K, Waeber G, Wareham N J, Maxwell P H, McCarthy M I, Jarvelin M R, Mooser V, Abecasis G R, Lightstone L, Scott J, Navis G, Elliott P, Kooner J S. International Consortium for Blood Pressure Genome-Wide Association Studies;Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ganesh S K, Tragante V, Guo W, Guo Y, Lanktree M B, Smithe N, Johnson T, Castillo B A, Barnard J, Baumert J, Chang Y P, Elbers C C, Farrall M, Fischer M E, Franceschini N, Gaunt T R, Gho J M, Gieger C, Gong Y, Isaacs A, Kleber M E, Mateo Leach I, McDonough C W, Meijs M F, Mellander O, Molony C M, Nolte I M, Pad-manabhan S, Price T S, Rajagopalan R, Shaffer J, Shah S, Shen H, Soranzo N, van der Most P J, Van Iperene P, Van Setten J A, Vonk J M, Zhang L, Beitelshees A L, Ber-enson G S, Bhatt D L, Boer J M, Boerwinklee Burkley B, Burt A, Chakravarti A, Chen W, Cooper-Dehoff R M, Curtis S P, Dreisbach A, Duggan D, Ehret G B, Fabsitz R R, Fornage M, Foxe Furlong C E, Gansevoort R T, Hofker M H, Hovingh G K, Kirkland S A, Kottke-Marchant K, Kutlar A, Lacroix A Z, Langaee T Y, Li Y R, Lin H, Liu K, Maiwald S, Malik R, CARDIOGRAM M, Mu-rugesan G, Newton-Cheh C, O'Connell J R, Onland-Moret N C, Ouwehand W H, Palmas W, Penninx B W, Pepine C J, Pettinger M, Polak J F, Ramachandran V S, Ranchalis J, Redline S, Ridker P M, Rose L M, Scharnag H, Schork N J, Shimbo D, Shuldiner A R, Srinivasan S R, Stolk R P, Taylor H A, Thorand B, Trip M D, van Duijn C M, Verschuren W M, Wijmenga C, Winkelmann B R, Wyatt S, Young J H, Boehm B O, Caulfield M J, Chasman D I, Davidson K W, Doevendans P A, Fitzgerald G A, Gums J G, Hakonarson H, Hillege H L, Illig T, Jarvik G P, John-son J A, Kastelein J J, Koenig W, Marz W, Mitchell B D, Murray S S, Oldehinkel A J, Rader D J, Reilly M P, Reiner A P, Schadte E, Silverstein R L, Snieder H, Stanton A V, Uitterlinden A G, van der Harst P, van der Schouw Y T, Samani N J, Johnson A D, Munroe P B, de Bakker P I, Zhu X, Levy D, Keating B J, Asselbergs F W. LifeLines Cohort Study Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum. Mol. Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seidah N G, Day R, Hamelin J, Gaspar A, Collard M W, Chretien M. Testicular expression of PC4 in the rat: molecular diversity of a novel germ cell-specific Kex2/subtilisin-like proprotein convertase. Mol. Endocrinol. 1992;6:1559–1570. doi: 10.1210/mend.6.10.1448111. [DOI] [PubMed] [Google Scholar]
- 118.Nakayama K, Kim W S, Torii S, Hosaka M, Nakagawa T, Ikemizu J, Baba T, Murakami K. Identification of the fourth member of the mammalian endoprotease family homologous to the yeast Kex2 protease.Its testis-specific expression. J. Biol. Chem. 1992;267:5897–5900. [PubMed] [Google Scholar]
- 119.Gyamera-Acheampong C, Tantibhedhyangkul J, Weerachatyanu-kul W, Tadros H, Xu H, van de Loo J W, Pelletier R M, Tanphaichitr N, Mbikay M. Sperm from mice genetically deficient for the PCSK4 proteinase exhibit accelerated capacitation, precocious acrosome reaction, reduced binding to egg zona pellucida, and impaired fertilizing ability. Biol. Reprod. 2006;74:666–673. doi: 10.1095/biolreprod.105.046821. [DOI] [PubMed] [Google Scholar]
- 120.Tadros H, Chretien M, Mbikay M. The testicular germ-cell protease PC4 is also expressed in macrophage-like cells of the ovary. J. Reprod. Immunol. 2001;49:133–152. doi: 10.1016/s0165-0378(00)00085-1. [DOI] [PubMed] [Google Scholar]
- 121.Gyamera-Acheampong C, Mbikay M. Proprotein convertase subtilisin/kexin type 4 in mammalian fertility: a review. Hum. Reprod. Update. 2009;15:237–247. doi: 10.1093/humupd/dmn060. [DOI] [PubMed] [Google Scholar]
- 122.Tardif S, Guyonnet B, Cormier N, Cornwall G A. Alteration in the processing of the ACRBP/sp32 protein and sperm head/acrosome malformations in proprotein convertase 4 (PCSK4) null mice. Mol. Hum. Reprod. 2012;18:298–307. doi: 10.1093/molehr/gas009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.De Bie I, Marcinkiewicz M, Malide D, Lazure C, Nakayama K, Bendayan M, Seidah N G. The isoforms of proprotein convertase PC5 are sorted to different subcellular compartments. J. Cell Biol. 1996;135:1261–1275. doi: 10.1083/jcb.135.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim D S, Stanaway I B, Rajagopalan R, Bernbaum J C, Solot C B, Burnham N, Zackaie H, Clancy R R, Nicolson S C, Gerdes M, Nickerson D A, Hakonarson H, Gaynor J W, Jarvik G P. Results of genome-wide analyses on neurodevelopmental phenotypes at four-year follow-up following cardiac surgery in infancy. PLoS One. 2012;7:e45936. doi: 10.1371/journal.pone.0045936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Furney S J, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, Hodges A, Powell J, Wahlund L O, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Spenger C, Lathrop M, Shen L, Kim S, Saykin A J, Weiner M W, Lovestone S. Alzheimer's Disease Neuroimaging Initiative AddNeuroMed Consortium Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer's disease. Mol. Psychiatry. 2011;16:1130–1138. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ahmeti K B, Ajroud-Driss S, Al-Chalabi A, Andersen P M, Armstrong J, Birve A, Blauw H M, Brown R H, Bruijn L, Chen W, Chio A, Comeau M C, Cronin S, Diekstra F P, Soraya Gkazi A, Glass J D, Grab J D, Groene J, Haines J L, Hardiman O, Heller S, Huang J, Hung W Y, Jaworski J M, Jones A, Khan H, Landers J E, Langefeld C D, Leigh P N, Marion M C, McLaughlin R L, Meininger V, Melki J, Miller J W, Mora G, Pericak-Vance M A, Rampersaude Robberecht W, Russell L P, Salachas F, Saris C G, Shatunov A, Shaw C E, Siddique N, Siddique T, Smith B N, Sufit R, Topp S, Traynor B J, Vance C, van Damme P, van den Berg L H, van Es M A, van Vught P W, Veldink J H, Yang Y, Zheng J G. Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 19 1p34 1 Neurobiol. Aging. 2013;34:357: e7–357. doi: 10.1016/j.neurobiolaging.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carter T C, Kay D M, Browne M L, Liu A, Romitti P A, Kuehn D, Conley M R, Caggana M, Druschel C M, Brody L C, Mills J L. Anorectal atresia and variants at predicted regulatory sites in candidate genes. Ann. Hum. Genet. 2013;77:31–46. doi: 10.1111/j.1469-1809.2012.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Iatan I, Dastani Z, Do R, Weissglas-Volkov D, Ruel I, Lee J C, Huertas-Vazquez A, Taskinen M R, Prat A, Seidah N G, Pajukanta P, Engert J C, Genest J. Genetic variation at the proprotein convertase subtilisin/kexin type 5 gene modulates high-density lipoprotein cholesterol levels. Circ. Cardiovasc. Genet. 2009;2:467–475. doi: 10.1161/CIRCGENETICS.109.877811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jin W, Fuki I V, Seidah N G, Benjannet S, Glick J M, Rader D J. Proprotein convertases [corrected] are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 2005;280:36551–36559. doi: 10.1074/jbc.M502264200. [DOI] [PubMed] [Google Scholar]
- 130.Lango Allen H, Estrada K, Lettre G, Berndt S I, Weedon M N, Rivadeneira F, Willer C J, Jackson A U, Vedantam S, Raychaudhuri S, Ferreira T, Wood A R, Weyant R J, Segre A V, Speliotese K, Wheelere Soranzo N, Park J H, Yang J, Gudbjartsson D, Heard-Costa N L, Randall J C, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid I M, Luan J, Thorleifsson G, Winkler T W, Goddard M E, Sin Lo K, Palmer C, Workalemahu T, Aulchenko Y S, Johansson A, Zillikens M C, Feitosa M F, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrechte Ernst F, Glazer N L, Hayward C, Hottenga J J, Jacobs K B, Knowles J W, Kutalik Z, Monda K L, Polasek O, Preuss M, Rayner N W, Robertson N R, Steinthorsdottir V, Tyrer J P, Voight B F, Wiklund F, Xu J, Zhao J H, Nyholt D R, Pellikka N, Perola M, Perry J R, Surakka I, Tammesoo M L, Altmaiere L, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman D I, Chen C, Coin L, Cooper M N, Dixon A L, Gibson Q, Grundberge Hao K, Juhani Junttila M, Kaplan L M, Kettunen J, Konig I R, Kwan T, Lawrence R W, Levinson D F, Lorentzon M, McKnight B, Morris A P, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem R M, Salvie Sanna S, Shi J, Sovio U, Thompson J R, Turchin M C, Vandenput L, Verlaan D J, Vitart V, White C C, Ziegler A, Almgren P, Balmforth A J, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson J G, Freimer N B, Geuse J, Glorioso N, Haiqing S, Hartikainen A L, Havulinna A S, Hicks A A, Hui J, Igl W, Illig T, Jula A, Kajantiee Kilpelainen T O, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki M L, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker A N, Peden J F, Petersmann A, Pichler I, Pietilainen K H, Pouta A, Ridderstrale M, Rotter J I, Sambrook J G, Sanders A R, Schmidt C O, Sinisalo J, Smit J H, Stringham H M, Bragi Walters G, Widene Wild S H, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle W L, Nelis M, Peters M J, Ripatti S, van Meurs J B, Aben K K, Ardlie K G, Beckmann J S, Beilby J P, Bergman R N, Bergmann S, Collins F S, Cusi D, den Heijer M, Eiriksdottir G, Gejman P V, Hall A S, Hamsten A, Huikuri H V, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer L J, Lehtimaki T, Melander O, Mosley T H Jr, Musk A W, Nieminen M S, O'Donnell C J, Ohlsson C, Oostra B, Palmer L J, Raitakari O, Ridker P M, Rioux J D, Rissanen A, Rivolta C, Schunkert H, Shuldiner A R, Siscovick D S, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen G J, Viikari J, Heath A C, Martin N G, Montgomery G W, Province M A, Kayser M, Arnold A M, Atwood L D, Boerwinkle e, Chanock S J, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley A T, Hengstenberg C, Hoffman W, Lathrop G M, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright A F, Assimes T L, Barroso I, Hofman A, Mohlke K L, Boomsma D I, Caulfield M J, Cupples L A, Erdmann J, Fox C S, Gudnason V, Gyllensten U, Harris T B, Hayes R B, Jarvelin M R, Mooser V, Munroe P B, Ouwehand W H, Penninx B W, Pramstaller P P, Quertermous T, Rudan I, Samani N J, Spector T D, Volzke H, Watkins H, Wilson J F, Groop L C, Haritunians T, Hu F B, Kaplan R C, Metspalu A, North K E, Schlessinger D, Wareham N J, Hunter D J, O'Connell J R, Strachan D P, Wichmann H E, Borecki I B, van Duijn C M, Schadte E, Thorsteinsdottir U, Peltonen L, Uitterlinden A G, Visscher P M, Chatterjee N, Loos R J, Boehnke M, McCarthy M I, Ingelssone Lindgren C M, Abecasis G R, Stefansson K, Frayling T M, Hirschhorn J N. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scerri T S, Brandler W M, Paracchini S, Morris A P, Ring S M, Richardson A J, Talcott J B, Stein J, Monaco A P. PCSK6 is associated with handedness in individuals with dyslexia. Hum. Mol. Genet. 2011;20:608–614. doi: 10.1093/hmg/ddq475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arning L, Ocklenburg S, Schulz S, Ness V, Gerding W M, Hengstler J G, Falkenstein M, Epplen J T, Gunturkun O, Beste C. VNTR Polymorphism Is Associated with Degree of Handedness but Not Direction of Handedness. PLoS One. 2013;8:e67251. doi: 10.1371/journal.pone.0067251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Malfait A M, Arnere C, Song R H, Alston J T, Markosyan S, Staten N, Yang Z, Griggs D W, Tortorella M D. Proprotein convertase activation of aggrecanases in cartilage in situ. Arch. Biochem. Biophys. 2008;478:43–51. doi: 10.1016/j.abb.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 134.Tortorella M D, Arnere C, Hills R, Gormley J, Fok K, Pegg L, Munie G, Malfait A M. ADAMTS-4 (aggrecanase-1): N-terminal activation mechanisms. Arch. Biochem. Biophys. 2005;444:34–44. doi: 10.1016/j.abb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 135.Malfait A M, Seymour A B, Gao F, Tortorella M D, Le Graverand-Gastineau M P, Wood L S, Doherty M, Doherty S, Zhang W, Arden N K, Vaughn F L, Leaverton P E, Spector T D, Hart D J, Maciewicz R A, Muir K R, Das R, Sorge R E, Sotocinal S G, Schorscher-Petcu A, Valdes A M, Mogil J S. A role for PACE4 in osteoarthritis pain: evidence from human genetic association and null mutant phenotype. Ann. Rheum. Dis. 2012;71:1042–1048. doi: 10.1136/annrheumdis-2011-200300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsuji A, Higashine K, Hine C, Mori K, Tamai Y, Nagamune H, Matsuda Y. Identification of novel cDNAs encoding human kexin-like protease, PACE4 isoforms. Biochem. Biophys. Res. Commun. 1994;200:943–950. doi: 10.1006/bbrc.1994.1541. [DOI] [PubMed] [Google Scholar]
- 137.Li J P, Wang X B, Chen C Z, Xu X, Hong X M, Xu X P, Gao W, Huo Y. The association between paired basic amino acid cleaving enzyme 4 gene haplotype and diastolic blood pressure. Chin. Med. J. (Engl) 2004;117:382–388. [PubMed] [Google Scholar]
- 138.Xu X, Rogus J J, Terwedow H A, Yang J, Wang Z, Chen C, Niu T, Wang B, Xu H, Weiss S, Schork N J, Fang Z. An extreme-sib-pair genome scan for genes regulating blood pressure. Am. J. Hum. Genet. 1999;64:1694–1701. doi: 10.1086/302405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu X, Yang J, Rogus J, Chen C, Schork N, Xu X. Mapping of a blood pressure quantitative trait locus to chromosome 15q in a Chinese population. Hum. Mol. Genet. 1999;8:2551–2555. doi: 10.1093/hmg/8.13.2551. [DOI] [PubMed] [Google Scholar]
- 140.Bruzzaniti A, Goodge K, Jay P, Taviaux S A, Lam M H, Berta P, Martin T J, Moseley J M, Gillespie M. T. PC8 [corrected] a new member of the convertase family. Biochem. J. 1996;314 (Pt 3):727–731. doi: 10.1042/bj3140727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seidah N G, Hamelin J, Mamarbachi M, Dong W, Tardos H, Mbikay M, Chretien M, Day R. cDNA structure tissue distribution and chromosomal localization of rat PC7 a novel mammalian proprotein convertase closest to yeast kexin-like proteinases. Proc. Natl. Acad. Sci. U. S. A.: 1996;93:3388–3393. doi: 10.1073/pnas.93.8.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Salvas A, Benjannet S, Reudelhuber T L, Chretien M, Seidah N G. Evidence for proprotein convertase activity in the endoplasmic reticulum/early Golgi. FEBS Lett. 2005;579:5621–5625. doi: 10.1016/j.febslet.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 143.Declercq J, Meulemans S, Pletse Creemers J W. Internalization of proprotein convertase PC7 from plasma membrane is mediated by a novel motif. J. Biol. Chem. 2012;287:9052–9060. doi: 10.1074/jbc.M111.306407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Meerabux J, Yaspo M L, Roebroek A J, Van de Ven W J, Lister T A, Young B D. A new member of the proprotein convertase gene family (LPC) is located at a chromosome translocation breakpoint in lymphomas. Cancer Res. 1996;56:448–451. [PubMed] [Google Scholar]
- 145.Senturker S, Thomas J T, Mateshaytis J, Moos MJr. A homolog of subtilisin-like proprotein convertase 7 is essential to anterior neural development in Xenopus. PLoS One. 2012;7:e39380. doi: 10.1371/journal.pone.0039380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leonhardt R M, Fiegl D, Rufere Karger A, Bettin B, Knittler M R. Post-endoplasmic reticulum rescue of unstable MHC class I requires proprotein convertase PC7. J. Immunol. 2010;184:2985–2998. doi: 10.4049/jimmunol.0900308. [DOI] [PubMed] [Google Scholar]
- 147.Rousselete Benjannet S, Marcinkiewicze Asselin M C, Lazure C, Seidah N G. Proprotein convertase PC7 enhances the activation of the EGF receptor pathway through processing of the EGF precursor. J. Biol. Chem. 2011;286:9185–9195. doi: 10.1074/jbc.M110.189936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oexle K, Ried J S, Hicks A A, Tanaka T, Hayward C, Bruegel M, Gogele M, Lichtner P, Muller-Myhsok B, Dor-ing A, Illig T, Schwienbacher C, Minelli C, Pichler I, Fiedler G M, Thiery J, Rudan I, Wright A F, Campbell H, Ferrucci L, Bandinelli S, Pramstaller P P, Wichmann H E, Gieger C, Winkelmann J, Meitinger T. Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum. Mol. Genet. 2011;20:1042–1047. doi: 10.1093/hmg/ddq538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lin L, Nemethe Goodnough J B, Thapa D R, Gabayan V, Ganz T. Soluble hemojuvelin is released by proprotein convertase-mediated cleavage at a conserved polybasic RNRR site. Blood Cells Mol. Dis. 2008;40:122–131. doi: 10.1016/j.bcmd.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron ho-meostasis. Blood. 2008;111:924–931. doi: 10.1182/blood-2007-07-100677. [DOI] [PubMed] [Google Scholar]
- 151.Guillemot J, Canuel M, Essalmani R, Prat A, Seidah N G. Implication of the proprotein convertases in iron homeostasis: Proprotein convertase 7 sheds human transferrin receptor 1 and furin activates hepcidin. Hepatology. 2013;57:2514–2524. doi: 10.1002/hep.26297. [DOI] [PubMed] [Google Scholar]
- 152.Middelberg R P, Ferreira M A, Henders A K, Heath A C, Madden P A, Montgomery G W, Martin N G, Whitfield J B. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med. Genet. 2011;12:123–2350-12-123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Anini Y, Mayne J, Gagnon J, Sherbafi J, Chen A, Kaefer N, Chretien M, Mbikay M. Genetic deficiency for proprotein convertase subtilisin/kexin type 2 in mice is associated with decreased adiposity and protection from dietary fat-induced body weight gain. Int. J. Obes. (Lond) 2010;34:1599–1607. doi: 10.1038/ijo.2010.90. [DOI] [PubMed] [Google Scholar]
- 154.Wang J, Xu J, Finnerty J, Furuta M, Steiner D F, Verchere C B. The prohormone convertase enzyme 2 (PC2) is essential for processing pro-islet amyloid polypeptide at the NH2-terminal cleavage site. Diabetes. 2001;50:534–539. doi: 10.2337/diabetes.50.3.534. [DOI] [PubMed] [Google Scholar]
- 155.Miller R, Aaron W, Toneff T, Vishnuvardhan D, Beinfeld M C, Hook V Y. Obliteration of alpha-melanocyte-stimulating hormone derived from POMC in pituitary and brains of PC2-deficient mice. J. Neurochem. 2003;86:556–563. doi: 10.1046/j.1471-4159.2003.01856.x. [DOI] [PubMed] [Google Scholar]
- 156.Miller R, Toneff T, Vishnuvardhan D, Beinfeld M, Hook V Y. Selective roles for the PC2 processing enzyme in the regulation of peptide neurotransmitter levels in brain and peripheral neuroendocrine tissues of PC2 deficient mice. Neuropeptides. 2003;37:140–148. doi: 10.1016/s0143-4179(03)00027-1. [DOI] [PubMed] [Google Scholar]
- 157.Pan H, Che F Y, Peng B, Steiner D F, Pintar J E, Fricker L D. The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J. Neurochem. 2006;98:1763–1777. doi: 10.1111/j.1471-4159.2006.04067.x. [DOI] [PubMed] [Google Scholar]
- 158.Rehfeld J F, Lindberg I, Friis-Hansen L. Increased synthesis but decreased processing of neuronal proCCK in prohormone con-vertase 2 and 7B2 knockout animals. J. Neurochem. 2002;83:1329. doi: 10.1046/j.1471-4159.2002.01244.x. [DOI] [PubMed] [Google Scholar]
- 159.Johanning K, Juliano M A, Juliano L, Lazure C, Lamango N S, Steiner D F, Lindberg I. Specificity of prohormone convertase 2 on proenkephalin and proenkephalin-related substrates. J. Biol. Chem. 1998;273:22672–22680. doi: 10.1074/jbc.273.35.22672. [DOI] [PubMed] [Google Scholar]
- 160.Allen R G, Peng B, Pellegrino M J, Millere D, Grandy D K, Lundblad J R, Washburn C L, Pintar J E. Altered processing of pro-orphanin FQ/nociceptin and pro-opiomelanocortin-derived peptides in the brains of mice expressing defective prohormone convertase 2. J. Neurosci. 2001;21:5864–5870. doi: 10.1523/JNEUROSCI.21-16-05864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]