Abstract
Genome-wide association studies (GWAS) identify genetic variants that distinguish a control population from a population with a specific trait. Two challenges in GWAS are: (1) identification of the causal variant within a longer haplotype that is associated with the trait; (2) identification of causal variants for polygenic traits that are caused by variants in multiple genes within a pathway. We review recent methods that use information in protein–protein and protein–DNA interaction networks to address these two challenges.
Introduction
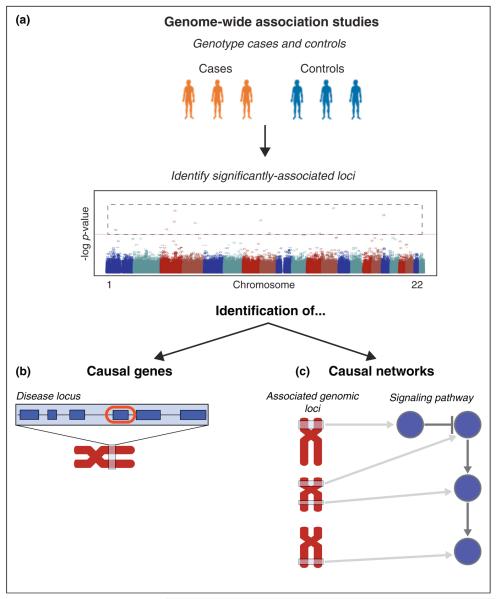
Genome-wide association studies (GWAS) aim to identify genetic variants that distinguish a population of individuals, or cases, that have a particular phenotype/trait (typically a disease) from control individuals [1]. In its simplest form, analysis of a GWAS is a logistic regression where for each genotyped single-nucleotide polymorphism (SNP) the number of copies of the non-reference allele is regressed onto disease status for all individuals. The resulting P-value for each SNP is then corrected for multiple tests, and SNPs with alleles significantly enriched in controls are identified (Figure 1a).
Figure 1.
Two applications of network-based analyses of GWAS. (a) GWAS analysis computes the association between a SNP and case/control, reporting a P-value for each SNP. (b) Casual gene identification is the problem of identifying a single causal gene (circled in red) for the phenotype from a larger locus of candidate genes that is significantly associated with the phenotype. (c) Causal network identification is the problem of finding a group of interacting genes (e.g. a signaling pathway or protein complex) containing SNPs that distinguish cases and controls.
There are two major challenges in using GWAS to identify the genomic underpinnings of complex phenotypes (Figure 1). First, GWAS-identified SNPs are generally not located in the gene(s) underlying the phenotype of interest, but rather, are in linkage disequilibrium with causal genes or SNPs. Thus, one challenge is to identify causal genes within a GWAS-implicated locus (Figure 1b). One solution to this challenge is to use interaction networks to rank genes within a haplotype according to interactions with other genes known to be associated to the phenotype of interest or to similar phenotypes.
A second challenge is that GWAS-detected variants do not explain most of the genetic effects found in affected individuals – even for diseases known to have a strong genetic component, such as obesity and diabetes. This has been termed the “missing heritability problem”) [2–5]. An underexplored cause of missing heritability is genetic heterogeneity: the concept that different collections of causal variants are present in different patients. Genetic heterogeneity manifests itself on two levels. First, affected individuals may harbor distinct causal variants within a given causal gene. Second, causal variants may be distributed across different genes within a pathway (signaling, regulatory, metabolic) or protein complex [6]. This review focuses on the second type of genetic heterogeneity.
Genetic heterogeneity resulting from pathways and protein complexes complicates GWAS because for any specific causal gene, only a subset of the cases will contain a variant in that gene, while other cases will have causal variants in other genes in the pathway. This reduces the power of tests of association between single genes and the phenotype. Unraveling such genetic heterogeneity requires testing the association between the phenotype of interest and different combinations of genes containing putative causal variants. The goal is to identify sets of genes with the property that each affected individual contains a causal variant in at least one gene in the set. It is also possible to consider the case where an affected individual contains multiple causal variants in different genes in the set, but we will not consider this case here. The naive approach of exhaustively testing all combinations of variants is not computationally or statistically feasible. For example, one cannot exhaustively test all 1020 combinations of 5 genes and retain statistical power without data from an astronomical number of individuals.
In this review, we describe recent work using interaction networks to address these two challenges in GWAS, focusing on three specific applications:
Causal gene identification. It has been observed that different causal genes for the same or similar phenotypes often interact, either directly or via common interaction partners. Network approaches use this observation to select putative causal gene(s) from haplotypes by finding genes that are close or related in a network to other known causal genes.
Causal gene identification for expression phenotypes. pt?>Gene expression is a phenotype of particular interest because it is readily measured from micro-arrays or RNA-Seq. Because gene expression is a molecular phenotype, network approaches are attractive as they may provide a mechanistic explanation for a causal variant.
Causal network identification. GWAS of genetically heterogeneous or polygenic diseases require testing groups of genes that are known to participate in the same biological process. Standard gene set enrichment or ranking statistics have been used to test known pathways in GWAS [6]. Interaction networks provide an alternative source of information that can be used profitably to identify combinations of causal variants without limiting analysis to known pathways.
In this review, we focus on the use of interaction networks in GWAS, and more specifically in common variant association studies (CVAS). However, we also briefly summarize some of the approaches used for the analogous causal network identification problem in cancer genome sequencing studies [7,8].
Network approaches
Interaction networks
Large-scale interaction networks incorporate the results of both molecular and high-throughput experiments to describe different biochemical relationships between genes and the proteins they encode. These networks take the form of a graph G = (V, E). The vertices V represent genes and their corresponding protein products. The edges E join pairs of vertices whose corresponding proteins exhibit a specific biochemical interaction (e.g. physical association, phosphorylation, etc.). In some cases, the edges may have a direction corresponding to the directionality of the biological interaction. Commonly used protein–protein interaction (PPI) networks include HPRD [9], BioGRID [10], STRING [11], iRefIndex [12], and Reactome [13], most of which combine literature-curated interactions and interactions derived from high-throughput experiments [14–18]. More recently, Multinet [19•] also integrates protein–DNA interactions from ENCODE.
Causal gene identification
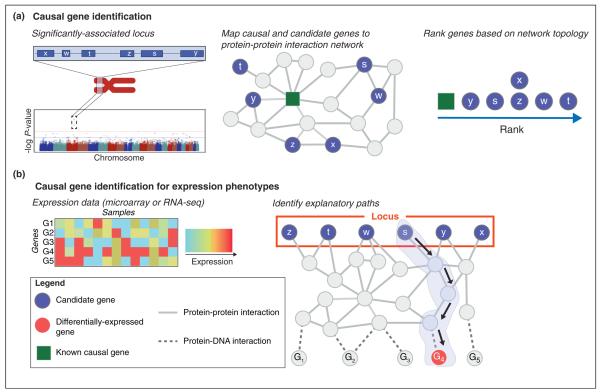
The most common use of interaction networks in GWAS analysis is to identify the causal gene inside a haplotype block (Figure 2 and Table 1a). While GWAS identify haplotype blocks associated with a particular disease or phenotype, they typically do not have the resolution to identify the causal gene within the associated block. A network approach to causal gene identification is motivated by the observation that the protein products of causal genes often directly interact with, or share many interacting partners with, the protein products of other causal genes for the disease. Thus, given prior knowledge of causal genes for a phenotype, one may identify new causal genes by finding the gene in the haplotype block that is closest on the network to the known causal genes.
Figure 2.
Schematic of methods for causal gene identification. (a) Candidate causal genes in a locus (or haplotype block) identified as significantly associated with a phenotype by a GWA study are mapped (blue circles) to a protein–protein interaction network. Each candidate gene is ranked in relation to a set of known causal genes (green squares; for simplicity, only one causal gene is shown) using a network distance measure. Different network distance functions that incorporate different features of network topology have been proposed including connectivity (e.g. direct interactions), network flow, random walks, and topological similarity (e.g. diffusion “profiles”). (b) Methods for identifying causal genes for expression phenotypes identify a causal gene from a locus of candidate genes (blue circles) that explain a differentially-expressed gene (red circle). Network methods find explanatory path(s) from the causal gene to the differentially expressed gene through an integrated network of protein–protein and protein–DNA interactions that provide a mechanistic explanation for the change in expression. In this example, candidate gene s is identified as upstream of differentially-expressed gene G4 with explanatory path (blue) from s to G4 terminates in a protein–DNA interaction.
Table 1.
Network analysis methods for GWAS
| Algorithmic approach | Reference | Interactome | Genetic/phenotypic data |
|---|---|---|---|
| a. Causal gene identification | |||
| Direct neighbors | Oti et al. [20] | HPRD + high-throughput experiments | Causal genes |
| CIPHER [21] | HPRD + OPHID + BIND + MINT | Causal genes + phenome | |
| Lee et al. [55] | HumanNet | GWAS SNPs | |
| Network flow & random walks | GeneWanderer [23] | HPRD, BIND, BioGrid, IntAct, DIP, STRING | Causal genes |
| PRINCE [24] | HPRD + high-throughput experiments (weighted) |
Causal genesa + phenotype similarity scores |
|
| MAXIF [29] | HPRD | Causal genes + phenome | |
| Zhu et al. [25] | HPRD | Causal genes + phenome | |
| Topological similarity | AlignPI [27] | HPRD | Causal genes + phenome |
| VAVIEN [26] | NCBI Entrez Gene (weighted) | Causal genes + phenotype similarity scores |
|
| b. Causal gene identification for expression phenotypes |
|||
| Topological properties | Kreimer and Pe’er [35] |
HPRD | eSNPs |
| Network flow | Tu et al. [30] | PPI: yeast | eQTLs |
| PDI: yeast | |||
| ResponseNet [32] | PPI: yeast | eQTLs | |
| PDI: yeast (weighted) | |||
| ResponseNet2.0 [33] | PPI: BioGRID + DIP + MINT + IntAct | eQTLs | |
| PDI: TRANSFAC (weighted) | |||
| Conductance | eQED [31•] | Yeast (weighted) | eQTLs |
| Kim et al. [34] | PPI: MINT + IntAct + Reactome + HPRD + others PDI: TRED |
eQTLs | |
| c. Causal network identification | |||
| Seed and extend | PINBPA [36,42•] | iRefIndex filtered for high-confidence interactions |
GWAS SNPs |
| dmGWAS [41] | MINT + IntAct + DIP + BioGRID + HPRD + MIPS | GWAS SNPs | |
| NETBAG [39] | BIND + BioGRID + DIP + HPRD + InNetDB + IntAct + BiGG + MINT + MIPS |
De novo CNVs | |
| NETBAG + [40•] | BIND + BioGRID + DIP + HPRD + InNetDB + IntAct + BiGG + MINT + MIPS |
De novo CNVs + SNVs + GWAS-implicated loci |
|
| Exhaustive search of 2-step networks |
NIMMI [44] | BioGRID | GWAS SNPs |
GeneCards is the source of causal gene information for PRINCE. For all other methods, OMIM is the source of causal gene information.
Early methods used a simple definition of network distance, examining only nearest neighbors on a protein interaction network [20,21]. However, most biological interaction networks have a heavy-tailed degree distribution [22], meaning that most pairs of proteins are connected via short paths. This property makes nearest neighbors or shortest paths less desirable distance measures. The first method to utilize a more sophisticated measure of network distance that considers the overall topology of the network, GeneWanderer [23], ranks candidate genes based on the probability that a random walk starting from a known disease gene will finish at each candidate gene. Similar approaches measure network distance using information flow and network propagation [23–25].a Two other methods select candidate causal genes based on their topological similarity to known causal genes [26,27] rather than their network distance.
Several of these methods also improve upon early approaches by incorporating phenotype similarity scores between diseases based on the overlap of their OMIM medical subject heading descriptions (described in [28]). Some methods incorporate phenotype similarity scores only for disease pairs including the disease for which causal genes are sought [24,26], while others integrate a “phenome” network in which phenotypes are nodes and weighted edges between all phenotype pairs represent their similarity [21,25,29]. Incorporating this information enables causal gene identification for diseases for which there are no previously known causal genes.
Causal gene identification for expression phenotypes
An important subproblem of causal gene identification arises when the phenotype of interest is gene expression; loci associated to a gene expression phenotype are sometimes referred to as expression quantitative trait loci (eQTL) or expression SNPs (eSNPs). Network approaches have been used to provide mechanistic explanations for observed correlations between a locus containing one or more source genes and a target gene that is differentially expressed between cases and controls (Table 1b). These methods find high-scoring paths in a combined protein–protein and protein–DNA interaction (PDI) network between one of the source genes and the target gene (Figure 2b). To explain the change in expression, the final edge in these paths is a protein–DNA interaction between a transcription factor that regulates the target gene. Three of the first such methods [30,31•,32] analyzed eQTLs in yeast. The eQED algorithm of [31•] used an electrical resistance model to find high-weight explanatory paths that connect SNPs to differentially expressed genes through known signaling and regulatory interactions. In comparison, ResponseNet [32] and ResponseNet2.0 [33] formulate the problem as a minimum-cost network flow, which is mathematically related to electrical resistance. Kim et al. [34] further extended these ideas, applying them to human cancer data and adding additional steps to identify causal genes from multiple explanatory paths. More recently, Kriemer [35] analyzed eSNPs identified in human whole-genome and RNA-Seq data, and found that source and target genes are generally closer on the PPI network. However, in contrast to the work above, they did not use protein–DNA interactions to find explanatory paths for these associations.
Causal network identification
A third use of interaction networks in GWAS analysis is to identify causal networks, or sets of interacting genes containing causal variants. This approach complements popular pathway-based tests that restrict attention to groups of variants in known pathways or gene sets using enrichment statistics [6,36,37•]. Network approaches address three limitations of gene set analysis. First, gene sets do not model the topology and type of interactions between genes, and instead treat all genes in the set as equivalent. Second, gene set methods perform a separate statistical test on each gene set and do not consider the interconnection of pathways in larger signaling and regulatory networks. Third, by restricting attention to known pathways, gene set methods are unable to discover novel groups of interacting genes that are associated to the phenotype.
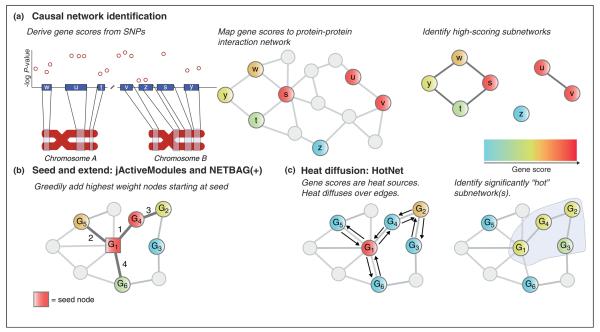
Several algorithms have been introduced to find causal networks in protein–protein interaction networks (Figure 3a and Table 1c) [36,38,39,40•,41,42•]. Authors [36,42•] use the jActiveModules plug-in [43] in Cytoscape to analyze multiple sclerosis GWAS data on the iRefIndex protein–protein interaction network [17]. jActiveModules provides a general approach to find high-scoring subnetworks in a vertex-weighted network (Figure 3b). dmGWAS is a similar approach [41]. The NETBAG [39] and NETBAG + algorithms [40•] – used to identify subnetworks affected by rare and de novo variants in autism and schizophrenia, respectively – are also related but analyze an edge-weighted interaction network. All of these methods use a greedy heuristic (“seed and extend”) to find high-scoring subnetworks by iteratively adding to a subnetwork those genes that increase the subnetwork’s score (Figure 3b). These approaches compute the statistical significance of the resulting subnetworks by comparing to an empirical distribution of subnetwork scores.
Figure 3.
Schematic of methods for causal network identification and examples of two algorithms. (a) Proteins in the protein–protein interaction network are scored using the association P-values within or near their corresponding gene. In this example, nodes are colored using a blue-to-red gradient where blue represents low scores and red represents high scores. Proteins without scores (i.e. those that were not tested in the GWA study or had no significant associations) are colored gray but remain in the network for analysis due to their effect on the network’s topology. High-scoring subnetworks are then reported, taking into account both the protein scores and the network topology. (b) jActiveModules, NETBAG, and NETBAG + all use a greedy heuristic (seed and extend) to identify causal networks by iteratively adding to a subnetwork genes that increase the subnetwork score. jActiveModules uses a vertex-weighted graph where each vertex has an associated Z-score, and the score of a subnetwork with k nodes is the normalized sum of Z-scores. In the original application of jActiveModules, the Z-score of a gene indicated its differential expression in microarray experiments. For the application to GWAS, [36,42•] transform gene-level P-values (from VEGAS [46•]) of association into Z-scores. NETBAG algorithms [39,40•] analyze a weighted graph with edge weights determined by naïve Bayes integration of protein interaction and protein complex databases, protein sequence alignment, and co-evolution. In the vertex-weighted graph shown, G1 is the seed gene, and genes G4, G5, G2, and G6 are added to the subnetwork in that order (as indicated with labels on the edges) G3 is not added because it has a low score. (c) HotNet uses heat diffusion in order to identify causal networks. Heat is assigned to each gene in proportion to its score and diffuses over the edges of the network. The heat diffusion process takes into account the topology of the network so that genes with high-degree pass proportionally less heat to their neighbors than genes with low degree. In the example shown, G4 and G3 are initially cold (indicated by light blue), while G1 and G2 are “hot” (indicated by red and orange, respectively). After heat diffuses along the edges, G1, G2, and G4 have the same heat, while G3 is colder than G4 because it is not directly connected to G1. The remaining nodes G5 and G6 are initially cold and remain cold because they are only connected to the high-degree G1. A hot subnetwork of genes G1, G2, G3, and G4 is identified.
An additional approach is the Network Interface Miner for Multigenic Interactions (NIMMI) [44]. NIMMI employs a modified version of the PageRank algorithm for webpage ranking [45] to compute a weight for each gene that represents its network centrality. These weights are combined with gene-wise P-values from VEGAS [46•], and an exhaustive search is performed of all subnetworks consisting of paths of length 2 from a starting node.
In comparison to the number of methods for causal gene identification, there remain relatively few methods for causal network identification. However, an analogous problem occurs in cancer genome sequencing studies, where the challenge is to identify signaling/regulatory/metabolic networks harboring more somatic aberrations than expected by chance [7,8]. One algorithm introduced for this task, NetBox [47], decomposes a network into modules of mutated genes that are either directly connected or connected through single linker genes. Another algorithm, HotNet [48], uses a heat diffusion model to identify significantly mutated subnetworks as “hotspots” on the network (Figure 3c). Heat is assigned to each node in proportion to its mutation frequency, and this heat then diffuses over the edges of the graph, either for a fixed time [49] or until equilibrium [48]. Hot subnetworks are found by removing cold edges and the statistical significance of the number and size of the resulting hot subnetworks is computed. Thus, HotNet simultaneously considers both the score assigned to each gene and the global topology of the network, in contrast to most of the methods above that use these two features sequentially. Despite the generality of these two algorithms, neither has yet been used to analyze GWAS data. We discuss prospects for adapting these methods for GWAS analysis in the next section.
Challenges and future prospects
A number of challenges remain in network analysis of GWAS. First, network methods are limited by the coverage and quality of protein–protein and protein–DNA interaction networks. High-quality experimental interaction data are laborious to obtain. Consequently, existing network databases have many missing interactions, and these reduce the sensitivity of network analyses. High-throughput interaction data, combined with additional experimental validation, will be crucial to increase sensitivity. Conversely, interaction databases also contain false positive interactions. Some of these are a result of incorrect predictions, errors in data curation, or experimental noise. Others result from the fact that most interaction networks are a superposition of interactions measured in different cell types and conditions, only a subset of which may be active in the tissue of the disease. Authors of [50,51] demonstrated that tissue-specific protein–protein interaction networks can improve disease-gene prioritization results.
Second, the dramatic decline in the cost of DNA sequencing is enabling whole-exome and whole-genome sequencing of cases and controls. Sequencing allows the analysis of de novo variants and rare variants in both coding and non-coding regions. A promising exampleof this type of analysis is demonstrated by Gulsuner et al. [52••], who identified causal subnetworks of interaction networks that contain significant numbers of de novo variants in schizophrenia patients. However, the challenge of extending causal network and causal gene identification approaches to rare variants requires additional methodological advances. For example, since causal rare variants may be randomly associated with different common haplotypes in sampled individuals, most rare variant association study (RVAS) analyses require sensible methods to pool variants across a gene or locus [53]. These approaches help address the problem of genetic heterogeneity resulting from different causal variants within a specific causal gene, but leave open the issue of rare causal variants across genes in a pathway/complex. A combination of pooled rare variants within a locus and network approaches across a locus is a promising direction.
In addition to a role for network approaches in CVAS, RVAS and de novo variant studies, network analyses have proven useful in the analysis of somatic mutations in cancer genomes. Cancer genome sequencing studies face an analogous problem of genetic heterogeneity where causal somatic mutations, or driver mutations, are distributed across multiple genes in a pathway [7,8]. As noted above, several network methods have been introduced for this problem [47–49]. While some of these methods may prove useful for germline variants, there are notable differences in the analyses of somatic vs. germline variants. First, somatic mutations, as well as de novo germline mutations, arise independently in each individual, and thus can be analyzed without considering ancestry and population structure. In contrast, analyses of common and/or rare variants require additional techniques to control for spurious associations with ancestry. Second, analysis of somatic mutations in tumors face issues such as intratumor heterogeneity that do not have parallels in germline studies. Despite these differences, both types of analyses can benefit from greater exchange of methodology.
Looking outside genes, network analysis of non-coding SNPs requires additional information about regulatory interactions, non-coding RNAs, among others. The ENCODE project [49] is an important first step in the generation of such information, but more data are needed. Network analysis will play an increasingly important role in prioritizing candidate causal variants for further experimental validation. Ultimately, the combination of computational and experimental approaches will yield mechanistic insights into the process by which a genetic variant, or a combination of variants, affect a complex phenotype.
Acknowledgements
BJR is supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, an Alfred P. Sloan Research Fellowship, a grant from the National Human Genome Research Institute (R01HG005690), an NSF CAREER Award (CCF-1053753) and NSF grant IIS-1016648. SR is supported by an Alfred P. Sloan Research Fellowship and by the Pew Charitable Trusts as a Pew Scholar in the Biomedical Sciences. MDML is supported by NSF Graduate Research Fellowship DGE 0228243. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Ref. [56] performed benchmarking confirming that methods taking into account global network topology outperform connectivity methods in causal gene identification.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JPA, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. [Internet] Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. [Internet] Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. [Internet] Proc Natl Acad Sci U S A. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. [Internet] Nat Rev Genet. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClellan J, King M-C. Genetic heterogeneity in human disease. [Internet] Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 6.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. [Internet] Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Science. Vol. 339. New York, N.Y.: 2013. Cancer genome landscapes. [Internet] pp. 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garraway LA, Lander ES. Lessons from the cancer genome. [Internet] Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database-2009 update. [Internet] Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. [Internet] Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein–protein interaction networks, with increased coverage and integration. [Internet] Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. [Internet] BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. [Internet] Nucleic Acids Research. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing RM, et al. Large-scale mapping of human protein– protein interactions by mass spectrometry. Molecular Systems Biology. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchins JRa, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rual J-F, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 17.Stelzl U, et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 18.Yu H, et al. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19 •.Khurana E, Fu Y, Chen J, Gerstein M. Interpretation of genomic variants using a unified biological network approach. [Internet] PLOS Comput Biol. 2013;9:e1002886. doi: 10.1371/journal.pcbi.1002886. Presents Multinet, the first protein–protein interaction (PPI) network to incorporate ENCODE data.
- 20.Oti M, Snel B, Huynen Ma, Brunner HG. Predicting disease genes using protein–protein interactions. [Internet] J Med Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Jiang R, Zhang MQ, Li S. Network-based global inference of human disease genes. [Internet] Mol Syst Biol. 2008;4:189. doi: 10.1038/msb.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner A. The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes [Internet] Mol Biol Evol. 2001;18:1283–1292. doi: 10.1093/oxfordjournals.molbev.a003913. [DOI] [PubMed] [Google Scholar]
- 23.Köhler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. [Internet] Am J Human Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. Associating genes and protein complexes with disease via network propagation. [Internet] PLOS Comput Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu J, Qin Y, Liu T, Wang J, Zheng X. Prioritization of candidate disease genes by topological similarity between disease and protein diffusion profiles. [Internet] BMC Bioinformatics. 2013;14(Suppl. 5):S5. doi: 10.1186/1471-2105-14-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erten S, Bebek G, Koyutürk M. Vavien: an algorithm for prioritizing candidate disease genes based on topological similarity of proteins in interaction networks. [Internet] J Comput Biol. 2011;18:1561–1574. doi: 10.1089/cmb.2011.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Liu Q, Jiang R. Align human interactome with phenome to identify causative genes and networks underlying disease families. [Internet] Bioinformatics (Oxford, England) 2009;25:98–104. doi: 10.1093/bioinformatics/btn593. [DOI] [PubMed] [Google Scholar]
- 28.Van Driel Ma, Bruggeman J, Vriend G, Brunner HG, Leunissen JaM. A text-mining analysis of the human phenome. [Internet] Eur J Human Genet (EJHG) 2006;14:535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Jiang T, Jiang R. Uncover disease genes by maximizing information flow in the phenome-interactome network. [Internet] Bioinformatics (Oxford, England) 2011;27:i167–i176. doi: 10.1093/bioinformatics/btr213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tu Z, Wang L, Arbeitman MN, Chen T, Sun F. An integrative approach for causal gene identification and gene regulatory pathway inference. [Internet] Bioinformatics (Oxford, England) 2006;22:e489–e496. doi: 10.1093/bioinformatics/btl234. [DOI] [PubMed] [Google Scholar]
- 31 •.Suthram S, Beyer A, Karp RM, Eldar Y, Ideker T. eQED: an efficient method for interpreting eQTL associations using protein networks. [Internet] Mol Syst Biol. 2008;4:162. doi: 10.1038/msb.2008.4. Rephrases the random walk formulation of Tu et al. as an electric circuit, decreasing computational time and improving prediction accuracy. Demonstrates utility of predicting explanatory pathways for eQTLs.
- 32.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. [Internet] Nat Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basha O, Tirman S, Eluk A, Yeger-Lotem E. ResponseNet2.0: revealing signaling and regulatory pathways connecting your proteins and genes—now with human data. [Internet] Nucleic Acids Res. 2013;41:W198–W203. doi: 10.1093/nar/gkt532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y-A, Wuchty S, Przytycka TM. Identifying causal genes and dysregulated pathways in complex diseases. [Internet] PLOS Comput Biol. 2011;7:e1001095. doi: 10.1371/journal.pcbi.1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreimer A, Pe’er I. Variants in exons and in transcription factors affect gene expression in trans. [Internet] Genome Biol. 2013;14:R71. doi: 10.1186/gb-2013-14-7-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BMJ, Kappos L, Polman CH, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. [Internet] Hum Mol Genet. 2009;18:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37 •.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. [Internet] PLOS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. Provides a comprehensive review of pathway analysis approaches culminating in network-based techniques.
- 38.Ideker T, Dutkowski J, Hood L. Boosting signal-to-noise in complex biology: prior knowledge is power. [Internet] Cell. 2011;144:860–863. doi: 10.1016/j.cell.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. [Internet] Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40 •.Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. [Internet] Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. Demonstrates an algorithm to find high-scoring causal networks in a weighted interaction network incorporating multiple sources of information and extending earlier approach (Gilman et al. 2011). Applies new NETBAG + algorithm to identify networks of variants associated with schizophrenia.
- 41.Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein–protein interaction networks. [Internet] Bioinformatics (Oxford, England) 2011;27:95–102. doi: 10.1093/bioinformatics/btq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42 •.Consortium IMSG Network-based multiple sclerosis pathway analysis with GWAS Data from 15,000 Cases and 30,000 Controls. [Internet] Am J Hum Genet. 2013;92:854–865. doi: 10.1016/j.ajhg.2013.04.019. Describes a large-scale application of a network-based approach to the causal network identification problem.
- 43.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks [Internet] Bioinformatics. 2002;18:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 44.Akula N, Baranova A, Seto D, Solka J, Nalls MA, Singleton A, Ferrucci L, Tanaka T, Bandinelli S, Cho YS, et al. A network-based approach to prioritize results from genome-wide association studies. [Internet] PLOS ONE. 2011;6:e24220. doi: 10.1371/journal.pone.0024220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brin S, Page L. The anatomy of a large-scale hypertextual Web search engine [Internet] Comput Netw ISDN Syst. 1998;30:107–117. [Google Scholar]
- 46 •.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, et al. A versatile gene-based test for genome-wide association studies. [Internet] Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. Introduces VEGAS, a commonly-used technique for assigning gene-based P-values by pooling SNP P-values from GWAS.
- 47.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLOS ONE. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 49.Vandin F, Clay P, Upfal E, Raphael B. Discovery of mutated subnetworks associated with clinical data in cancer [Internet] Pac Symp Biocomput. 2012;17:55–66. [PubMed] [Google Scholar]
- 50.Jiang B, Wang J, Wang Y, Xiao J. Gene prioritization for Type 2 diabetes in tissue-specific protein interaction networks. The Third International Symposium on Optimization and Systems Biology (OSB ’09) 2009:319–328. [Google Scholar]
- 51.Magger O, Waldman YY, Ruppin E, Sharan R. Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks. [Internet] PLOS Comput Biol. 2012;8:e1002690. doi: 10.1371/journal.pcbi.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52 ••.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. Analysis of de novo variants in schizophrenia cases that uses both physical interaction and gene co-expression networks to identify causal networks.
- 53.Li B, Leal S. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. [Internet] Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis Ca, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. [Internet] Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. [Internet] Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Navlakha S, Kingsford C. The power of protein interaction networks for associating genes with diseases. [Internet] Bioinformatics (Oxford, England) 2010;26:1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]