Abstract
Disease susceptibility for type 1 diabetes is strongly associated with the inheritance of specific HLA alleles. However, conventional allele frequency analysis can miss HLA associations because many alleles are rare. In addition, disparate alleles that have similar peptide-binding sites, or shared epitopes, can be missed. To identify the HLA shared epitopes associated with diabetes, we analyzed high-resolution genotyping for class I and class II loci. The HLA epitopes most strongly associated with susceptibility for disease were DQB1 A57, DQA1 V76, DRB1 H13, and DRB1 K71, whereas DPB1 YD9,57, HLA-B C67, and HLA-C YY9,116 were more weakly associated. The HLA epitopes strongly associated with resistance were DQB1 D57, DQA1 Y80, DRB1 R13, and DRB1 A71. A dominant resistance phenotype was observed for individuals bearing a protective HLA epitope, even in the presence of a susceptibility epitope. In addition, an earlier age of disease onset correlated with significantly greater numbers of susceptibility epitopes and fewer resistance epitopes (P < 0.0001). The prevalence of both DQ and DR susceptibility epitopes was higher in patients than in control subjects and was not exclusively a result of linkage disequilibrium, suggesting that multiple HLA epitopes may work together to increase the risk of developing diabetes.
Introduction
Type 1 diabetes is a multifactorial disease with a strong genetic component. More than 40 genetic linkages have been associated with this disease, but linkage to HLA class II is the most dominant (1). A number of studies have shown that HLA-DRB1*03:01, *04:01, *04:02, and *04:05 are most strongly linked to diabetes, whereas DRB1*04:03 is resistant. In addition, DQB1*02:01 and *03:02 are strongly linked to diabetes, whereas DQB1*06:02 is resistant (2–8). The fact that DQB1*03:02 is associated with diabetes when linked to DRB1*04:05 but resistant when linked to DRB1*04:03 suggests that both loci contribute to disease susceptibility (9). However, the strong linkage disequilibrium between many DRB1 and DQB1 alleles makes it difficult to sort out the relative influence of each locus. In addition, analysis of HLA susceptibility is confounded because disparate HLA alleles may share peptide-binding motifs, often referred to as shared epitopes. For example, alanine in position 57 of the DQ β-chain (DQB1 A57), which is present in DQB1*02:01, *02:02, *03:02, *03:04, and *03:05, has a stronger association with diabetes than any of the individual alleles. In contrast, an aspartic acid residue in the same position is strongly associated with resistance (10). Conventional frequency analysis of the HLA alleles associated with diabetes is limited because it does not account for these HLA shared epitopes. Thus, a systematic analysis of all possible HLA class I and class II epitopes should provide a better understanding of genetic susceptibility for type 1 diabetes.
HLA epitopes can be single amino acids or groups of contiguous or noncontiguous amino acids. They are mostly found in the highly polymorphic antigen-binding groove of HLA class I and class II molecules. Class I molecules (HLA-A, -B, and -C) present endogenous peptides to CD8+ T cells. The peptide-binding groove has six distinct pockets comprising various combinations of polymorphic amino acids. Class II molecules (HLA-DR, -DQ, and -DP) are heterodimers that present exogenous peptides to CD4+ T cells and have four major pockets comprising polymorphic β-chain amino acids in conjunction with α-chain residues (11). The HLA-DR polymorphism is found in the β-chain because the α-chain is effectively conserved in the peptide-binding region. In contrast, both the α- and β-chains of HLA-DQ and -DP are polymorphic (12). The repertoire of peptides that can bind in the peptide-binding groove for each HLA allele is determined by the amino acids that line the pockets. These polymorphic amino acid residues can potentially combine to form millions of different HLA epitopes.
We have developed the HLA Epitope Analysis Program (HEAP) to analyze the millions of shared HLA epitopes comprising one to four noncontiguous amino acids between patient and control populations. This program was used previously to demonstrate that the HLA-DRB1 epitope LA67,74 is most strongly associated with susceptibility for rheumatoid arthritis (13). In the present study, we used HEAP to identify all the class I and class II HLA epitopes associated with diabetes susceptibility or resistance. We also analyzed the effect of multiple susceptibility HLA epitopes on the risk of disease.
Research Design and Methods
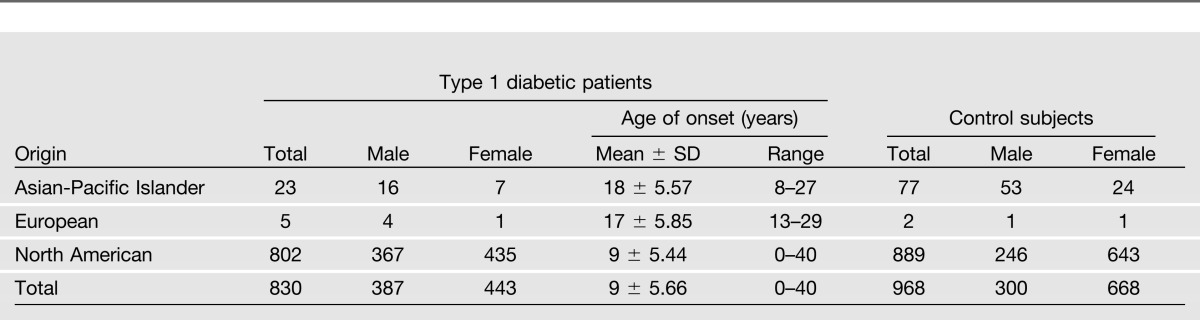
This study used a dataset provided by the Type 1 Diabetes Genetics Consortium (T1DGC), made available by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Central Repository (www.niddkrepository.org). The T1DGC is a large, worldwide, collaborative study with the aim of identifying genes that determine an individual’s risk of type 1 diabetes. The T1DGC had previously enrolled case (type 1 diabetic patients) and control subjects (no history of type 1 diabetes) from populations with a low prevalence of the disease (see www.T1DGC.org for details). High-resolution HLA genotyping was performed at eight classical major histocompatibility complex loci by four genotyping centers using standardized typing protocols, reagents, and quality control procedures (14). In the present study, we analyzed 830 type 1 diabetic patients and 968 control subjects. The characteristics of both groups are summarized in Table 1.
Table 1.
Descriptive characteristics of the study subjects
HLA data from the type 1 diabetes component of the International Histocompatibility Working Group (IHWG) (www.ncbi.nlm.nih.gov/projects/gv/mhc/ihwg.cgi) were also analyzed to validate the findings from the T1DGC. IHWG is a worldwide association of investigators who collaborated to collect HLA genotype, microsatellite, clinical, and demographic data for type 1 diabetic patients and healthy individuals. Of the 2,957 subjects provided, we censored those who did not have allele-level HLA typing. For race-matched control subjects, we used high-resolution HLA typing from 1,644 consecutive subjects typed at ClinImmune Laboratories (Aurora, CO).
HLA Epitope and Allele Frequency Analysis
The HEAP was used to identify susceptibility and resistance HLA epitopes by comparing the frequency of each amino acid and all combinations of up to four amino acids of the HLA molecule between patients and control subjects. Only polymorphic amino acids at positions 2–192 (class I) and positions 8–93 (class II) were analyzed. Amino acids outside these ranges were not considered because they are unlikely to influence interactions with either the peptide or the T-cell receptor (TCR). The number of patients or control subjects carrying at least one copy of an epitope was compared with the number of individuals not carrying the epitope. The distribution of these epitopes was calculated with 2 × 2 contingency tables and analyzed with either Pearson χ2 test or Fisher exact test, as appropriate. The resulting P value for each epitope was corrected for multiple comparisons by the false discovery rate method described by Benjamini and Yekutieli (15). We analyzed 2,332,039 HLA-A epitopes, 3,954,218 HLA-B epitopes, 346,175 HLA-C epitopes, 360,801 DRB1 epitopes, 67,818 DQA1 epitopes, 186,413 DQB1 epitopes, 1,075 DPA1 epitopes, and 37,782 DPB1 epitopes. A conventional allele frequency analysis was also performed for comparison.
Criteria for Susceptibility and Resistance HLA Epitopes
HLA epitopes with a corrected P value ≤0.001 and an odds ratio (OR) ≥1.5 (susceptibility) or ≤0.5 (resistance) were defined as statistically significant. In addition, the positions of the susceptibility and resistance epitopes were mapped on existing crystal structures for HLA-B (16), -C (17), -DR (18), -DQ (19), and -DP (20) to determine whether they could potentially influence peptide binding and/or interact with the TCR.
Results
HLA Epitopes Associated With Susceptibility
We used a dataset of 830 type 1 diabetic patients and 968 control subjects from whom high-resolution HLA genotyping at all eight classical HLA loci (HLA-A, -B, -C for class I and HLA-DRB1, -DQA1, -DQB1, -DPA1, and -DPB1 for class II) had previously been obtained (14). This dataset was analyzed with HEAP to identify HLA epitopes associated with type 1 diabetes. By focusing the analysis on the polymorphic amino acid residues, we identified 7,286,321 epitopes comprising between one and four amino acids in the patient and control groups. The susceptibility epitopes with the lowest P value and highest OR for each locus are summarized in Table 2. A conventional allele frequency analysis was also performed for comparison (Supplementary Tables 1 and 2).
Table 2.
Shared HLA epitopes associated with susceptibility to type 1 diabetes
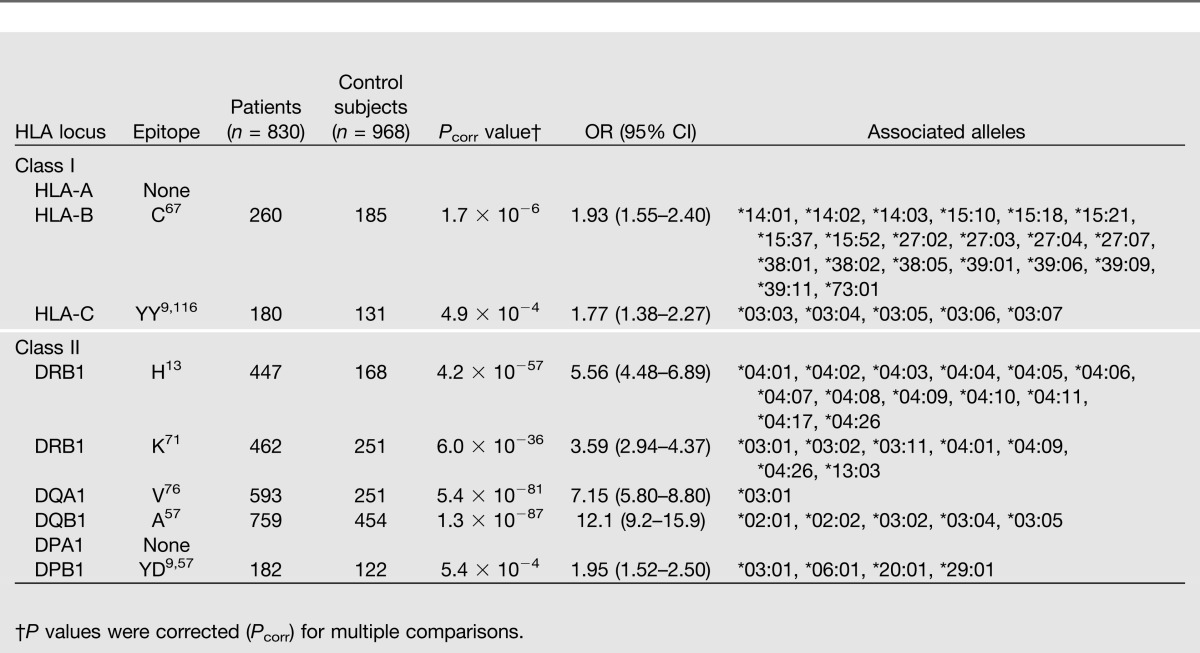
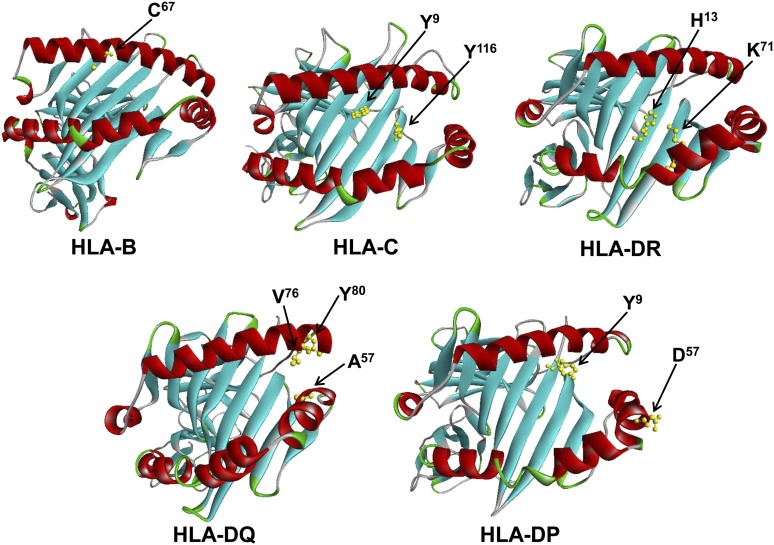
The previously reported HLA-DQB1 epitope A57 showed the strongest association with type 1 diabetes (P = 1.3 × 10−87, OR 12.1) and is found in five alleles (Table 2). Three of the alleles, DQB1*02:01, *02:02, and *03:02, were more common in patients than in control subjects and were also identified in the allele frequency analysis (Supplementary Table 1). However, the other two alleles that share A57, DQB1*03:04 and *03:05, were not statistically significant because of low frequency in the population. HLA-DQB1 A57 is located at the end of the peptide-binding groove and points inward where it may interact with peptides (Fig. 1). Four DQA1 single amino acid epitopes (S26, Q47, R56, and V76) were most strongly associated with diabetes and had the same P value (5.4 × 10−81) and OR (7.15). These epitopes are only found in DQA1*03:01, which was also identified in the allele frequency analysis. On the basis of the crystal structure of HLA-DQ, S26, Q47, and R56 were unlikely to influence peptide-binding or to interact with the TCR. However, DQA1 V76 points down toward the peptide-binding region and could possibly interact with peptides that bind (Fig. 1).
Figure 1.
HLA epitope positions on crystal structures of HLA molecules. The susceptibility epitopes for each HLA molecule are shown in yellow. In addition, the resistance epitope HLA-DQA1 Y80 is highlighted. The resistance epitopes for HLA-DRB1 and -DQB1 are found in the same positions as the susceptibility epitopes.
The DRB1 locus contained the next most significant HLA epitopes associated with susceptibility for diabetes. DRB1 H13 had the strongest association (P = 4.2 × 10−57, OR 5.56) and is positioned on the floor of the peptide-binding groove, pointing upward (Fig. 1). In addition, DRB1 K71 demonstrated a significant association with type 1 diabetes (P = 6.0 × 10−36, 3.59). This epitope was chosen because the most significant resistance DRB1 epitope A71 is found in the same position. DRB1 K71 points into the peptide-binding region of HLA-DR where it may influence the peptide repertoire (Fig. 1). Two of the alleles that share these DR epitopes, DRB1*04:01 and DRB1*03:01, were also identified by the allele frequency analysis; the others were too rare in the dataset (Supplementary Table 1). No increase in statistical significance was observed for DQB1, DQA1, or DRB1 when analyzing combinations of two or more amino acids.
Weaker associations with susceptibility for diabetes were found in the DPB1 and HLA-B and -C loci (Table 2). Analysis of the DPB1 locus identified a two-amino acid epitope, YD9,57 (P = 5.4 × 10−4, OR 1.95). This HLA epitope is found in DPB1*03:01, which was also identified in the allele frequency analysis (Supplementary Table 1). DPB1 Y9 is found in the peptide-binding groove, whereas DPB1 D57 is facing upward near the end of the groove where it may affect TCR recognition (Fig. 1). HLA-B C67 (P = 1.7 × 10−6, 1.93) is positioned near the middle of the peptide groove, pointing down into the peptide-binding region. Twenty HLA-B alleles share this epitope, but the allele frequency analysis could only identify one allele, B*15:10 (Supplementary Table 2). Extending the epitope analysis to include combinations of two or more amino acids did not increase the statistical significance. HLA-C YY9,116 (P = 4.9 × 10−4, 1.77) is found in five alleles, including C*03:04, which was identified in the allele frequency analysis (Supplementary Table 2). Both HLA-C Y9 and HLA-C Y116 are found in the peptide-binding groove, pointing upward (Fig. 1). The HEAP analysis did not find any combinations of amino acids that were significantly associated with diabetes in the HLA-DPA1 or HLA-A loci.
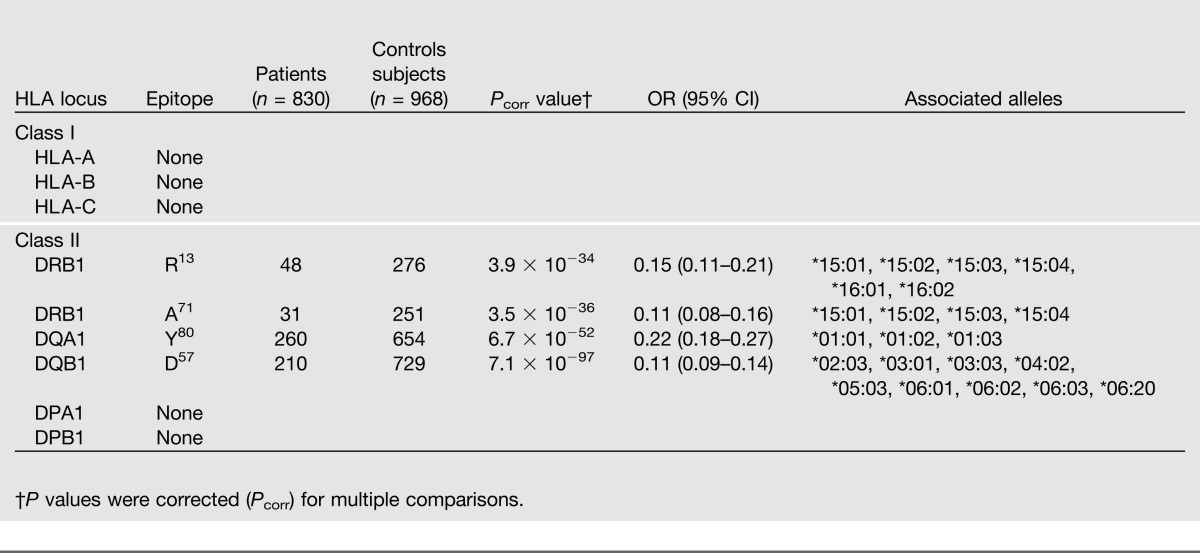
HLA Epitopes Associated With Resistance
The most strongly resistance-associated epitope was DQB1 D57 (P = 7.1 × 10−97, OR 0.11), and it is in the same position as the susceptibility epitope DQB1 A57. Similarly, the HLA-DRB1 resistance epitopes R13 (P = 3.9 × 10−34, 0.15) and A71 (P = 3.5 × 10−36, 0.11) are found at the same positions as the DRB1 susceptibility epitopes. The associated DQ and DR alleles that share these resistance HLA epitopes include disparate alleles that were not identified by allele frequency analysis (Supplementary Table 1). Unlike DRB1 and DQB1, the resistance epitope DQA1 Y80 (P = 6.7 × 10−52, 0.22) was not found in the same position as the susceptibility epitope (Table 3). This protective epitope points into the peptide-binding groove and potentially restricts the peptide repertoire that can bind (Fig. 1). DQA1*01:01, *01:02, and *01:03 share this epitope, and these alleles were also identified in the allele frequency analysis (Supplementary Table 1). Extending the epitope analysis to include combinations of two or more amino acids did not increase the statistical significance. The HEAP analysis did not find any resistance epitopes in the class I or the HLA-DP loci.
Table 3.
Shared HLA epitopes associated with resistance to type 1 diabetes
HLA Susceptibility and Resistance to Type 1 Diabetes Have a Strong Gene-Dose Effect
We counted the number of type 1 diabetic patients and control subjects who had inherited homozygous or heterozygous copies of the HLA-B, -C, -DRB1, -DQA1, -DQB1, and -DPB1 epitopes and calculated the OR for each combination (Table 4). Two copies of DQB1 A57 had the strongest association with type 1 diabetes (P = 5.8 × 10−96, OR 11.3). In contrast, homozygous inheritance of DQB1 D57 had the greatest negative association with disease (P = 4.7 × 10−50, 0.08). Resistance appeared to be dominant because individuals who had inherited one copy of DQB1 D57 had a lower probability (OR ≤ 0.5) of diabetes (Table 4). Even when the susceptibility epitope DQB1 A57 was inherited with the resistance-associated epitope, the probability of disease was still low (P = 3.8 × 10−5, 0.62). A similar gene–dose effect was observed with DRB1. Homozygous inheritance of DRB1 K71 was strongly associated with type 1 diabetes (P = 3.6 × 10−14, 4.88), whereas two copies of DRB1 A71 were protective (P = 2.8 × 10−6, 0.05). The dominant effect of the resistance-associated epitope DRB1 A71, even in the presence of DRB1 K71, was apparent (P = 0.0008, 0.32).
Table 4.
Homozygous vs. heterozygous inheritance of shared HLA epitopes
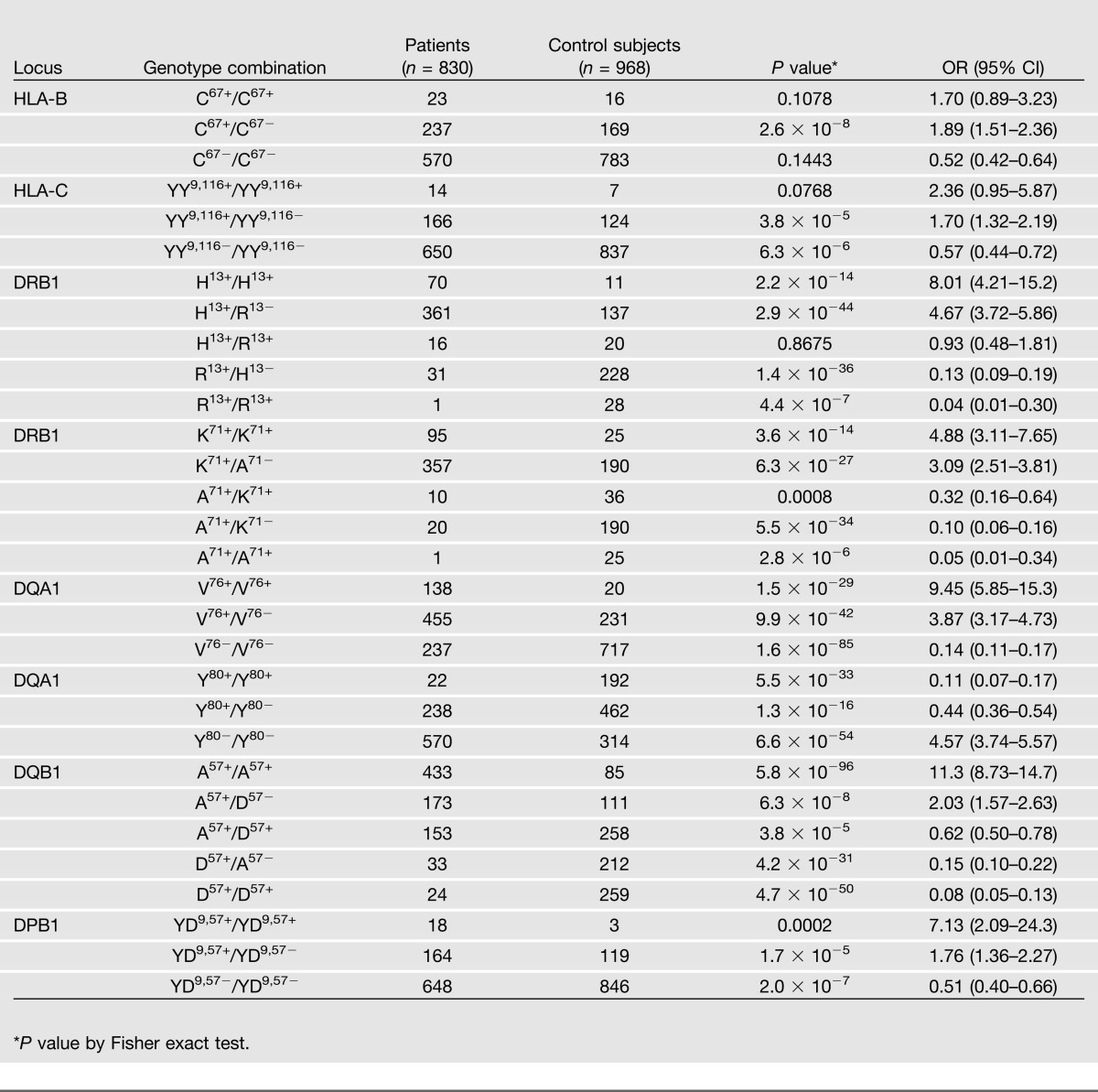
Not every resistance-associated HLA epitope had a dominant protective effect. Individuals who were homozygous for DRB1 H13 (P = 2.2 × 10−14, OR 8.01) had a greater risk for type 1 diabetes, whereas those homozygous for DRB1 R13 (P = 4.4 × 10−7, 0.04) were protected (Table 4). However, heterozygous individuals (H13+/R13+) had an OR close to 1.0, suggesting that the two epitopes may neutralize each other with respect to disease susceptibility or resistance. Two copies of the susceptibility-associated HLA-B C67, HLA-C YY9,116, DQA1 V76, or DPB1 YD9,57 epitopes had a higher OR for disease. However, there was still an increased risk (OR ≥ 1.5) when only one copy of the susceptibility epitope was inherited, suggesting that susceptibility was dominant for these loci.
Validation of HLA Epitopes With an International Dataset
The T1DGC dataset comprises study subjects who were primarily of North American descent (Table 1). To validate the findings, we analyzed data from an international cohort of type 1 diabetic patients with significantly greater ethnic diversity (see www.ncbi.nlm.nih.gov/projects/gv/mhc/ihwg.cgi for study details). This larger dataset only contained high-resolution HLA-typing for the class II loci DRB1 and DQB1. The HEAP analysis identified the same HLA epitopes with the strongest associations with susceptibility: DQB1 A57 (P = 8.1 × 10−192, OR 10.9), DRB1 H13 (P = 2.1 × 10−166, 7.42), and DRB1 K71 (P = 4.9 × 10−173, 9.01). Similarly, the same HLA epitopes with the strongest association with resistance were found: DQB1 D57 (P = 1.9 × 10−194, 0.13), DRB1 R13 (P = 3.1 × 10−99, 0.09), and DRB1 A71 (P = 4.6 × 10−101, 0.06) (Supplementary Table 3). In addition, the dominance of the resistance-associated HLA epitopes was evident (Supplementary Table 4). This was true even for the DRB1 epitope H13, which in the analysis of the T1DGC dataset appeared neutral (Table 4).
Multiple Susceptibility Epitopes Correlate With Earlier Age of Disease Onset
We counted the number of alleles for each patient or control subject that contained an HLA epitope associated with susceptibility or resistance (see Tables 2 and 3 for associated alleles) and correlated this total with age of disease onset. If both the DRB1 H13 and K71 susceptibility epitopes were found in the same allele, they were counted only once. The maximum number of class I susceptibility epitopes per person was 4 (2 each for HLA-B and HLA-C) and for class II, 8 per person (2 each for DRB1, DQA1, DQB1, and DPB1) for a total of 12 possible. A similar computation was performed for the resistance HLA epitopes. Because only DRB1, DQA1, and DQB1 had significantly associated resistance epitopes, the maximum number was six for any individual (Table 3). With this system, the number of epitopes for each subject in the study was totaled. Some patients and control subjects had up to nine susceptibility HLA epitopes, whereas others had none. The number of resistant HLA epitopes varied from zero to six. Overall, 96.5% (801 of 830) of the type 1 diabetic patients and 71.8% (695 of 968) of the control subjects had at least one susceptibility HLA epitope. In contrast, 48.2% (400 of 830) of the patients and 91.6% (887 of 968) of the control subjects had at least one resistance HLA epitope.
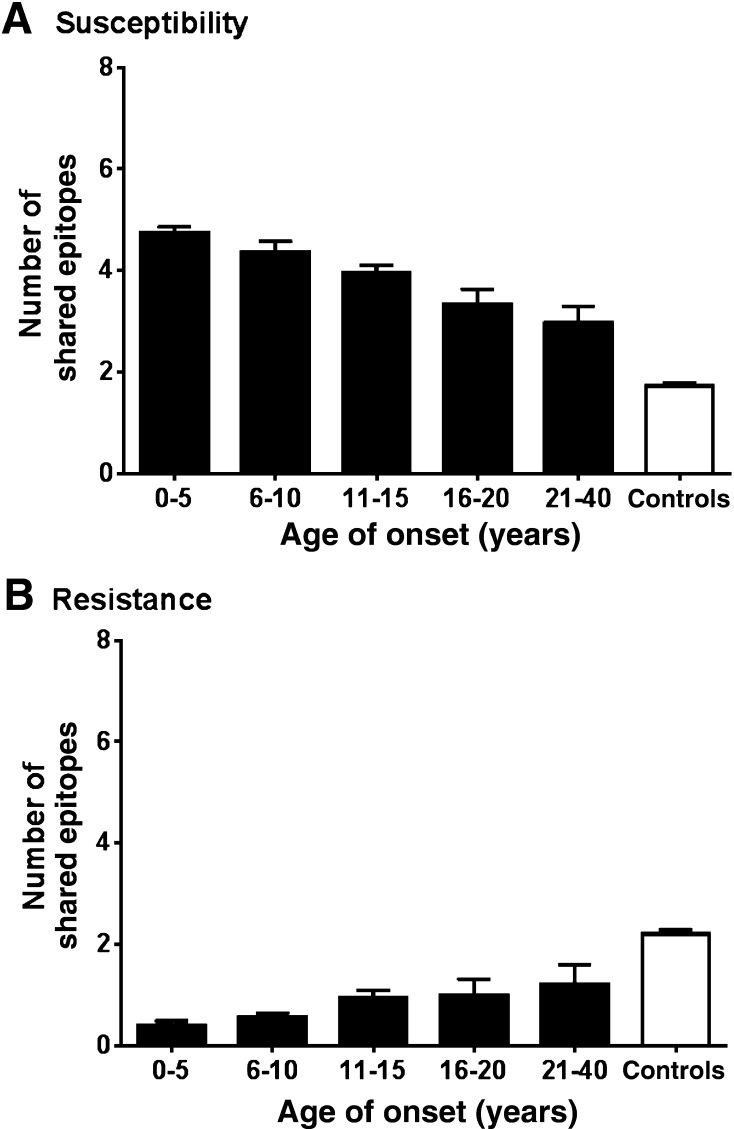
The average number of susceptibility and resistance HLA epitopes correlated with the age of disease onset. Patients in the earliest diagnosis group, between 0 and 5 years, had the highest average number of susceptibility HLA epitopes (Fig. 2A) and the lowest average number of resistance epitopes (Fig. 2B). The average number of predisposing epitopes was decreased and the number of resistance epitopes increased in patients with diabetes in whom the disease developed at an older age. In the control subjects, the average number of susceptibility epitopes was lower and the average number of resistance epitopes higher than in any of the patient groups.
Figure 2.
Susceptibility and resistance HLA epitopes have an additive effect. A: The number of susceptibility epitopes was calculated for each type 1 diabetic patient and control subject. The maximum number for any individual was 12 (two alleles each for HLA-B, -C, -DRB1, -DQA1, -DQB1, and -DPB1). The data were correlated with the age of disease onset. For each age range, the average number of epitopes and 95% CI are shown. No ages were available for the control subjects. B: The number of resistance epitopes was calculated and correlated with the age of disease onset as in A. The maximum number for each individual was six (two alleles each for HLA-DRB1, -DQA1, and -DQB1). A one-way ANOVA revealed significant differences between the groups [F(5.2, 1,792) = 192.9 (P < 0.0001) and F(5,1972) = 159.2 (P < 0.0001) for A and B, respectively].
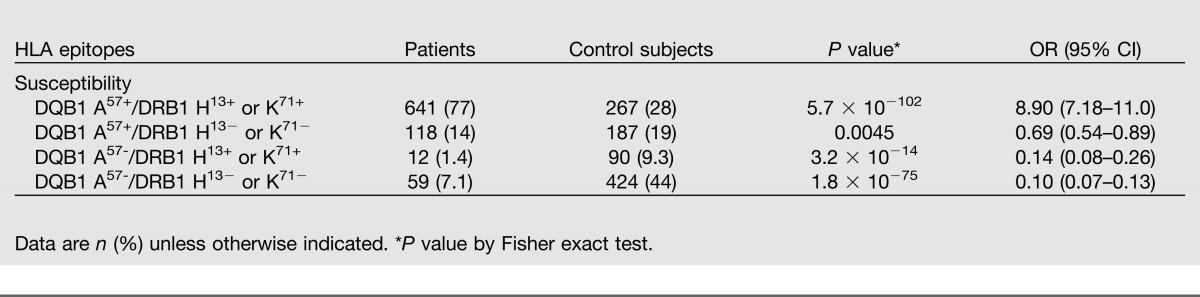
Linkage Disequilibrium Between DQ and DR Epitopes in Patients and Control Subjects
The prevalence of having both the DQB1 A57 and the DRB1 H13 or DRB1 K71 epitopes (DQB1 A57-DRB1 H13/K71) was compared between patients and control subjects (Table 5). We found a 77% concordance in diabetic patients compared with 28% in the control subjects (P = 5.7 × 10−102, OR 8.9). The percentage of those who only had DQB1 A57 was similar between patients and control subjects (14 vs. 19%), and the proportion of those who had DRB1 H13/K71 was lower in patients than in control subjects (1.4 vs. 9.3%). To verify this finding, we analyzed the larger IWHG international dataset and found a 91% concordance in patients with diabetes for DQB1 A57-DRB1 H13/K71 compared with 39% of the nondiabetic individuals (P = 4.2 × 10−252, 15.6). The percentage of patients versus nondiabetic individuals who only had DQB1 A57 was 2.7 vs. 14.2%, and that for only the DRB1 H13/K71 epitope was 3.2 vs. 9.6% (data not shown).
Table 5.
Linkage disequilibrium of DQ/DR in type 1 diabetic patients and control subjects
Discussion
Conventional allele frequency analysis can overlook alleles with relatively rare frequencies. In addition, traditional analysis of HLA population genetics does not account for shared HLA epitopes that allow disparate alleles to present the same antigen. Our analysis found HLA epitopes associated with susceptibility and resistance to disease. In addition, rare alleles were identified because HLA epitopes were first determined at the amino acid level and then the associated alleles identified.
A previous group that used the same dataset from the T1DGC identified a number of susceptible, neutral, and protective HLA-DR-DQ haplotypes associated with type 1 diabetes (9). Instead of analyzing the disease associations by individual loci, the authors looked at specific combinations of DRB1, DQA1, and DQB1 alleles. They established that the most susceptible haplotypes were DRB1*03:01-DQA1*05:01-DQB1*02:01, DRB1*04:05-DQA1*03:01-DQB1*03:02, DRB1*04:01-DQA1*03:01-DQB1*03:02, and DRB1*04:02-DQA1*03:01-DQB1*03:02. When they compared the amino acid differences between the closely related DRB1*04:01-DQ and DRB1*04:04-DQ haplotypes to determine why DRB1*04:01 was more predisposing, they found that these differ at position 71, identifying the same DRB1 K71 epitope found in the present study. In addition, the risk conferred by certain DR alleles, in combination with DQB1*03:02, was demonstrated.
In the same study, the authors reported that the most protective DR-DQ haplotypes were DRB1*15:01-DQA1*01:02-DQB1*06:02, DRB1*14:01-DQA1*01:01-DQB1*05:03, and DRB1*07:01-DQA1*02:01-DQB1*03:03 (9). The resistance DRB1 R13 and DRB1 A71 epitopes are found in DRB1*15:01. The present HEAP analysis also identified DRB1 H60 (P = 3.3 × 10−7, OR 0.15), which is found in DRB1*14:01 and DRB1 I67 (P = 9.2 × 10−25, 0.35), which is expressed by DRB1*07:01 (data not shown). However, these epitopes were not included in Table 2 because we listed those with the lowest P values and highest ORs. The DQB1*05:03, *06:02, and *03:03 alleles share the highly protective DQB1 D57 epitope that we identified. Therefore, our analysis found susceptibility and resistance HLA epitopes shared by the same alleles previously associated with disease. However, the HEAP analysis also identified disparate alleles that share these HLA epitopes, which can be missed in the conventional allele frequency analysis.
Although DR and DQ may play the strongest role in susceptibility for type 1 diabetes, HLA class I has also been shown to contribute (21). A previous study reported that the most significantly associated alleles were HLA-B*57:01 and B*39:06. In addition, the B*39:06 allele seemed to modulate risk on all DRB1-DQA1-DQB1 haplotypes on which it resided, suggesting a class I effect that was independent of class II (21). We identified the susceptibility epitope HLA-B C67, which is found in B*39:06 (Table 2) and appears to be dominant, even when the epitope is heterozygous (P = 2.6 × 10−8, OR 1.89). We also identified HLA-C YY9,116, which is present in five alleles: C*03:03, *03:04, *03:05, *03:06, and *03:07. Inheritance of this susceptibility epitope also appears dominant (P = 3.8 × 10−5, 1.70). These HLA-C alleles have not been previously reported as associated with type 1 diabetes.
DPB1*03:01 and *02:02 have been associated with susceptibility to type 1 diabetes, whereas DPB1*04:02 has been associated with resistance (22–24). In addition, these associations have been found across different ethnicities (25). We identified DPB1 YD9,57 (P = 5.4 × 10−4, OR 1.95), which is expressed in DPB1*03:01. Inheritance of this epitope had a dominant effect on disease susceptibility (Table 4). In addition, the present analysis found DPB1 EL35,85 with a very high OR (P = 0.002, 8.10) (data not shown). The HLA alleles that share this epitope include DPB1*02:02 and two rare alleles, DPB1*100:01 and *34:01. Therefore, HEAP identified the same susceptibility DPB1 alleles previously reported (24) but extended the finding to include disparate alleles that share the epitope. No resistance epitope associated with any DPB1 allele, including DPB1*04:02, was identified in the present analysis.
The linkage between HLA-DR and HLA-DQ has been associated with the possible presence of a DQA1*05:01-DQB1*03:02 transdimer. Previous studies suggested that the presence of the transdimer is associated with the highest risk of developing type 1 diabetes (9,26–29). Because the DR-DQ region is separated by <100 kb, it is difficult to analyze the two loci independently (30). Consequently, the high concordance we found between the HLA-DQ and HLA-DR susceptibility epitopes in patients could be a result of an HLA-DR linkage with DQA1*05:01. In the T1DGC dataset, 56.7% of the patients who have DRB1 H13 or K71 also have DQA1*05:01. However, only 18.8% of the patients with diabetes in the study have both DQB1*03:02 and DQA1*05:01 alleles and could potentially express the transdimer. These data suggest that the presence of DRB1 and DQB1 epitopes correlates more strongly with diabetes than does the possible presence of the DQA1*05:01-DQB1*03:02 transdimer. How the HLA-DQ and HLA-DR epitopes act in concordance with each other to increase susceptibility for diabetes requires further investigation.
In conclusion, HLA epitopes play an important role in the predisposition for type 1 diabetes. Because the main function of HLA molecules is to present peptides to T cells, HLA epitopes are presumably involved in peptide-binding and/or TCR interactions. In a study investigating DQB1, alleles with the resistance-associated D57 had a fourfold greater affinity for a GAD peptide than the susceptibility-associated A57 (10). Others have shown that this polymorphism can either enhance or abrogate peptide binding, depending on the peptide involved (31–34). Experiments testing peptide binding to susceptibility and resistance HLA epitopes are necessary to better understand how these HLA epitopes influence disease susceptibility.
Article Information
Funding. This research used resources provided by the T1DGC, a collaborative clinical study sponsored by the NIDDK, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, National Institute of Child Health and Human Development, and JDRF International and supported by U01-DK-062418. The U.K. case series collection was also funded by the JDRF and Wellcome Trust and the National Institute for Health Research, Cambridge Biomedical Centre, Cambridge Institute for Medical Research, U.K., which is in receipt of a Wellcome Trust Strategic Award (079895). The data from the T1DGC study were supplied by the NIDDK Central Repositories.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. C.L.R. analyzed the data and wrote the manuscript. K.M.A. analyzed data and edited the manuscript. L.J.S. and R.P.S. wrote the computer program. M.T.A. contributed to data analysis and interpretation and discussion and reviewed the manuscript. B.M.F. reviewed and edited the manuscript. B.M.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the American Society for Histocompatibility and Immunogenetics 39th Annual Meeting, Chicago, Illinois, 17–21 November 2013.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-1153/-/DC1.
This manuscript was not prepared in collaboration with investigators of the T1DGC study and does not necessarily reflect the opinions or views of the T1DGC study, the NIDDK Central Repositories, or the study sponsors.
References
- 1.Barrett JC, Clayton DG, Concannon P, et al. Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ettinger RA, Papadopoulos GK, Moustakas AK, Nepom GT, Kwok WW. Allelic variation in key peptide-binding pockets discriminates between closely related diabetes-protective and diabetes-susceptible HLA-DQB1*06 alleles. J Immunol 2006;176:1988–1998 [DOI] [PubMed] [Google Scholar]
- 3.Bugawan TL, Klitz W, Alejandrino M, et al. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens 2002;59:452–469 [DOI] [PubMed] [Google Scholar]
- 4.Cucca F, Muntoni F, Lampis R, et al. Combinations of specific DRB1, DQA1, DQB1 haplotypes are associated with insulin-dependent diabetes mellitus in Sardinia. Hum Immunol 1993;37:85–94 [DOI] [PubMed] [Google Scholar]
- 5.Erlich HA, Zeidler A, Chang J, et al. HLA class II alleles and susceptibility and resistance to insulin dependent diabetes mellitus in Mexican-American families. Nat Genet 1993;3:358–364 [DOI] [PubMed] [Google Scholar]
- 6.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet 1996;59:1134–1148 [PMC free article] [PubMed] [Google Scholar]
- 7.Sheehy MJ, Scharf SJ, Rowe JR, et al. A diabetes-susceptible HLA haplotype is best defined by a combination of HLA-DR and -DQ alleles. J Clin Invest 1989;83:830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parry CS, Brooks BR. A new model defines the minimal set of polymorphism in HLA-DQ and -DR that determines susceptibility and resistance to autoimmune diabetes. Biol Direct 2008;3:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erlich H, Valdes AM, Noble J, et al. Type 1 Diabetes Genetics Consortium HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato AK, Sturniolo T, Sinigaglia F, Stern LJ. Substitution of aspartic acid at beta57 with alanine alters MHC class II peptide binding activity but not protein stability: HLA-DQ (alpha1*0201, beta1*0302) and (alpha1*0201, beta1*0303). Hum Immunol 1999;60:1227–1236 [DOI] [PubMed] [Google Scholar]
- 11.Castelli FA, Buhot C, Sanson A, et al. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol 2002;169:6928–6934 [DOI] [PubMed] [Google Scholar]
- 12.Robinson J, Waller MJ, Fail SC, et al. The IMGT/HLA database. Nucleic Acids Res 2009;37(Database issue):D1013–D1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed BM, Schuyler RP, Aubrey MT. Association of the HLA-DRB1 epitope LA(67, 74) with rheumatoid arthritis and citrullinated vimentin binding. Arthritis Rheum 2011;63:3733–3739 [DOI] [PubMed] [Google Scholar]
- 14.Mychaleckyj JC, Noble JA, Moonsamy PV, et al. T1DGC HLA genotyping in the international Type 1 Diabetes Genetics Consortium. Clin Trials 2010;7(Suppl.):S75–S87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat 2001;29:1165–1188 [Google Scholar]
- 16.Kumar P, Vahedi-Faridi A, Saenger W, et al. Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens. J Biol Chem 2009;284:29784–29797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 2000;405:537–543 [DOI] [PubMed] [Google Scholar]
- 18.Dessen A, Lawrence CM, Cupo S, Zaller DM, Wiley DC. X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity 1997;7:473–481 [DOI] [PubMed] [Google Scholar]
- 19.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol 2001;2:501–507 [DOI] [PubMed] [Google Scholar]
- 20.Dai S, Murphy GA, Crawford F, et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc Natl Acad Sci U S A 2010;107:7425–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noble JA, Valdes AM, Varney MD, et al. Type 1 Diabetes Genetics Consortium HLA class I and genetic susceptibility to type 1 diabetes: results from the Type 1 Diabetes Genetics Consortium. Diabetes 2010;59:2972–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baschal EE, Aly TA, Babu SR, et al. HLA-DPB1*0402 protects against type 1A diabetes autoimmunity in the highest risk DR3-DQB1*0201/DR4-DQB1*0302 DAISY population. Diabetes 2007;56:2405–2409 [DOI] [PubMed] [Google Scholar]
- 23.Noble JA, Valdes AM, Thomson G, Erlich HA. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes 2000;49:121–125 [DOI] [PubMed] [Google Scholar]
- 24.Varney MD, Valdes AM, Carlson JA, et al. Type 1 Diabetes Genetics Consortium HLA DPA1, DPB1 alleles and haplotypes contribute to the risk associated with type 1 diabetes: analysis of the type 1 diabetes genetics consortium families. Diabetes 2010;59:2055–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz TD, Valdes AM, Santiago A, et al. DPB1 alleles are associated with type 1 diabetes susceptibility in multiple ethnic groups. Diabetes 2004;53:2158–2163 [DOI] [PubMed] [Google Scholar]
- 26.Koeleman BP, Lie BA, Undlien DE, et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun 2004;5:381–388 [DOI] [PubMed] [Google Scholar]
- 27.Moustakas AK, Papadopoulos GK. Molecular properties of HLA-DQ alleles conferring susceptibility to or protection from insulin-dependent diabetes mellitus: keys to the fate of islet beta-cells. Am J Med Genet 2002;115:37–47 [DOI] [PubMed] [Google Scholar]
- 28.Pociot F, McDermott MF. Genetics of type 1 diabetes mellitus. Genes Immun 2002;3:235–249 [DOI] [PubMed] [Google Scholar]
- 29.van Lummel M, van Veelen PA, Zaldumbide A, et al. Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire. J Biol Chem 2012;287:9514–9524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kockum I, Sanjeevi CB, Eastman S, Landin-Olsson M, Dahlquist G, Lernmark A. Complex interaction between HLA DR and DQ in conferring risk for childhood type 1 diabetes. Eur J Immunogenet 1999;26:361–372 [DOI] [PubMed] [Google Scholar]
- 31.Kwok WW, Domeier ME, Johnson ML, Nepom GT, Koelle DM. HLA-DQB1 codon 57 is critical for peptide binding and recognition. J Exp Med 1996;183:1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwok WW, Domeier ML, Raymond FC, Byers P, Nepom GT. Allele-specific motifs characterize HLA-DQ interactions with a diabetes-associated peptide derived from glutamic acid decarboxylase. J Immunol 1996;156:2171–2177 [PubMed] [Google Scholar]
- 33.Kwok WW, Nepom GT, Raymond FC. HLA-DQ polymorphisms are highly selective for peptide binding interactions. J Immunol 1995;155:2468–2476 [PubMed] [Google Scholar]
- 34.Nepom BS, Nepom GT, Coleman M, Kwok WW. Critical contribution of beta chain residue 57 in peptide binding ability of both HLA-DR and -DQ molecules. Proc Natl Acad Sci USA 1996;93:7202–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]