Abstract
Background & objectives:
Enterococci have emerged as important nosocomial pathogens and emergence of resistance to many of the antimicrobials used for Gram-positive organisms has made the management of infections due to Enterococcus species difficult. Resistance to glycopeptide antibiotics, especially vancomycin is of special concern. This study was undertaken to perform a phenotypic and genotypic characterization of vancomycin resistant Enterococcus (VRE) isolates obtained from clinical samples in a tertiary care hospital in southern India.
Methods:
Susceptibility testing was performed for Enterococcus isolates collected over a period of one year (November 2008-October 2009). Minimum inhibitory concentrations (MIC) of vancomycin and teicoplanin were determined for the isolates by the agar dilution method. Genotypic characterization of VRE isolates was done by performing multiplex polymerase chain reaction (PCR) for detecting the various vancomycin resistance genes.
Results:
Of the 367 isolates of Enterococcus species isolated, 32 were found to be resistant to vancomycin after MIC testing. VanA was the commonest phenotype of vancomycin resistance and the commonest genotype was vanA. Among the other important findings of the study was the presence of heterogeneity in isolates of VRE with the vanA gene cluster with regards to resistance to teicoplanin and the coexistence of vanA and vanC1 gene clusters in an isolate of E. gallinarum which conferred high level glycopeptide resistance to the isolate.
Interpretation & conclusions:
Enterococcus species have emerged as important nosocomial pathogens in our patients with a capacity to cause a variety of infections. The vancomycin resistance among Enterococcus isolates was 8.7 per cent in our study which was high compared to other Indian studies. VanA was the commonest phenotype of glycopeptide resistance and vanA was the commonest vancomycin resistance gene. The study also demonstrates phenotypic as well as genotypic heterogeneity among isolates of VRE from clinical specimens.
Keywords: Enterococcus, linezolid, MIC, multiplex PCR, resistance, vancomycin
Enterococci have emerged as important nosocomial pathogens in the last few decades and the major reason for this is the trend of increasing antimicrobial resistance seen in these organisms1. Enterococci have been implicated in clinical conditions like bacteraemias, urinary tract infections, peritonitis, surgical site infections, etc., especially in the hospital settings worldwide. In western countries enterococci have been found to be the second most common cause of nosocomial urinary tract infections and the third most common cause of nosocomial bacteraemia1. In the Indian scenario, enterococci are emerging nosocomial pathogens isolated from a variety of clinical conditions like urinary tract infections and bacteraemias2.
The propensity of Enterococcus species to easily acquire resistance genes and the presence of some unique mechanisms conferring resistance to antibiotics like aminoglycosides and glycopeptides have severely limited the choices available for treating serious infections due to these organisms1. The emergence of multi-drug resistant enterococci has lead to a scenario which is almost as bad as the preantibiotic era since many of these multi-drug resistant (MDR) strains have developed resistance to practically all available antibiotics.1 Vancomycin resistance among Enterococcus isolates is a major problem in most of the western world, especially in the United States where according to the National Nosocomial Infections Surveillance(NNIS) data, more than 28 per cent of all nosocomial enterococcal strains are vancomycin resistant3.
In the Indian context, aminoglycoside resistance in enterococci has been dealt with in a few studies4,5. There is especially a dearth of information on the genetic basis of vancomycin resistance among isolates of enterococci from India. The present study was aimed at detecting the antimicrobial resistance pattern among Enterococcus isolates obtained from clinical specimens in a tertiary care centre in south India with a special emphasis on vancomycin resistance in enterococci and its genetic basis.
Material & Methods
This study was carried out in the department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, India, from November 2008 to October 2009. All isolates of Enterococcus species obtained from sterile body fluids like blood, CSF, pleural fluid, peritoneal fluid, etc. over the study period were included. Isolates of Enterococcus from urine samples, wound swabs and pus samples were also included in the study. The study protocol was approved by the institutional ethics committee.
Enterococcus isolates were identified to the species level following Facklam and Collin's phenotypic characterization scheme for Enterococci6. All isolates of Enterococcus were tested for their antimicrobial susceptibility patterns using the standard guidelines issued by the Clinical Laboratories Standards Institute (CLSI)7. For studying the antimicrobial susceptibility pattern in enterococcal isolates, three methods were used Kirby-Bauer disk diffusion technique, screening agar method for aminoglycosides and vancomycin and minimum inhibitory concentration (MIC) testing by the agar dilution method7. Kirby Bauer disk diffusion method was used for determining the susceptibility of the isolates to the commonly used antibiotics against Enterococcus spp. The antibiotics tested were ampicillin (10 μg), ciprofloxacin (5 μg), gentamicin high content (120 μg), tetracycline (30 μg), vancomycin (30 μg), teicoplanin (30 μg), linezolid (30 μg), and chloramphenicol (30 μg). For urine isolates, susceptibility testing for nitrofura ntoin (300 μg) was also done.
Isolates were screened for high level gentamicin (HLG) resistance, high level streptomycin (HLS) resistance and vancomycin resistance. The concentrations of the screening agars for HLG resistance and HLS resistance were 500 and 2000 μg/ml, respectively, while the concentration of vancomycin in vancomycin screening agar was 6 μg/ml. The quality control strains used for vancomycin screening were Staphylococcus aureus ATCC 25923, Enterococcus faecalis ATCC 29212 and E. faecalis ATCC 51299 (LGC Promochem India Pvt, Bangalore, India).
Minimum inhibitory concentration testing (Agar dilution method): Minimum inhibitory concentrations of the glycopeptide antibiotics vancomycin and teicoplanin against the Enterococcus isolates were determined by agar dilution method according to the CLSI guidelines7. The antibiotic powders used in the procedure were from Hi-media laboratories, Mumbai, India. E. faecalis ATCC 29212 was included as a quality control strain.
Genotypic characterization of vancomycin resistance genes: Multiplex PCR was carried out to detect the presence of genes encoding for vancomycin resistance. Of the many genotypes of vancomycin resistance described in enterococci, attempt was made to identify the commonest ones, i.e., vanA, vanB and vanC genotypes (vanC1 gene or vanC2/C3 gene). The assay chosen was based on specific amplification of internal fragments of genes encoding D-alanine-d-alanine ligases and related proteins responsible for glycopeptide resistance.
The PCR conditions and the primers used for the genotypic characterization of vancomycin resistant strains were as previously described8. The following pairs of primers were used.
vanA A1 5’-GGGAAAACGACAATTGC-3’
A2 5’-GTACAATGCGGCCGTTA-3’
vanB B1 5’-ATGGGAAGCCGATAGTC-3’
B2 5’-GATTTCGTTCCTCGACC-3’
vanC-1 C1 5’-GGTATCAAGGAAACCTC-3’
C2 5’-CTTCCGCCATCATAGCT-3’
vanC2/C3 D1 5’-CTCCTACGATTCTCTTG-3’
D2 5’-CGAGCAAGACCTTTAAG-3’
rrs (16SrRNA)G1 5’-GGATTAGATACCCTGGTAGTCC-3’
G2 5’-TCGTTGCGGGACTTAACCCAAC-3’
PCR amplicons were custom sequenced to confirm the identity of the vancomycin resistance gene clusters responsible for glycopeptide resistance (Macrogen Inc, Seoul, South Korea).
Statistical analysis: GraphPad InStat3 software (GraphPadInc SanDiego, USA) was used for statistical analysis. Fisher's exact test was used for comparing variables like ICU (intensive care unit) stay, mortality, etc.
Results
A total of 367 isolates of Enterococcus species were isolated over the one year period from various clinical specimens. The maximum number of isolates were from urine specimens (59.1%), followed by isolates from exudates (37%). Only 14 isolates (3.8%) were from blood samples. Two hundred and ninety one (79.3%) of the 367 Enterococcus isolates were obtained from inpatients and the rest from outpatients.
Antimicrobial resistance among Enterococcus isolates: Of the 367 isolates of Enterococcus, 170 (46.33%) were resistant to ampicillin. 136 (37%) of all Enterococcus isolates were found to show high-level gentamicin resistance by disk diffusion method. Two isolates which were identified as sensitive to HLG by disk diffusion testing were eventually identified as resistant by the screening agar with 500 μg/ml of gentamicin. Only 17 per cent (63 isolates) of the Enterococcus isolates were found to be resistant to streptomycin by the screening agar method with 2000 μg/ml of streptomycin. The degree of resistance to ciprofloxacin was quite high at 273 (74.38%) of all isolates. Resistance to tetracycline was also high at 262 (71.38%). Nitrofurantoin was tested only for the urine isolates and 69 (29%) of all urine isolates showed in vitro resistance to it. Glycopeptide resistance, i.e., resistance to vancomycin and teicoplanin among the enterococcal isolates by disk diffusion was 9.26 and 7.6 per cent, respectively. Thirty two isolates (8.7%) were found to be resistant to vancomycin by the vancomycin screening agar method compared to 9.26 per cent (34 isolates) by the disk diffusion method. Therefore, the concordance between the two methods was more than 94 per cent. None of the Enterococcus isolates were resistant to linezolid.
Thirty one isolates showed MIC of vancomycin ≥128 μg/ml and were resistant to vancomycin by agar dilution method. Only one isolate was intermediate to vancomycin with a MIC of 8 μg/ml. A majority of the enterococcal isolates (326 isolates) showed MIC of vancomycin less than 2 μg/ml. Therefore, 32 isolates were identified as vancomycin resistant enterococci (VRE). Among the 28 isolates of Enterococcus resistant to teicoplanin by MIC testing, eight had MIC >128 μg/ml whereas for 14 isolates teicoplanin MIC was 128 μg/ml. For five isolates, the teicoplanin MIC was 64 μg/ml, whereas only a single isolate had teicoplanin MIC of 32μg/ml. All VRE isolates confirmed by MIC testing had been correctly identified by the vancomycin screening agar.
Of the 32 VRE isolates, seven were from pus samples and wound swabs followed by urine samples (6 isolates). Only four isolates were from blood. Three VRE isolates were obtained from CSF samples from cases of meningitis. Other specimens from which VRE were isolated included peritoneal fluid, synovial fluid, tissue biopsies, etc.
All the VRE isolates were recovered from inpatients. VRE isolates were more likely to be obtained from inpatients rather than outpatients as compared to vancomycin sensitive Enterococcus (VSE) isolates (P<0.001).
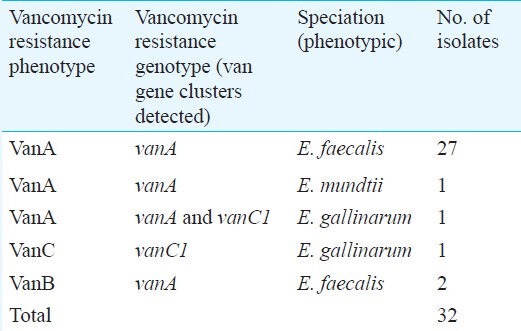
Phenotypic characterization of vancomycin resistant enterococci (VRE): Among the Enterococcus isolates showing resistance to vancomycin and teicoplanin, 29 were identified as E. faecalis. One was identified as E. mundtii, two as E. gallinarum.
Of the 32 isolates of Enterococcus found to be resistant to vancomycin by MIC testing, 29 were found to be resistant to teicoplanin also. These 29 isolates (90.6% of VRE), therefore, showed the VanA type of phenotype (resistance to both vancomycin and teicoplanin). On speciation, 27 of these isolates were identified as E. faecalis and one each as E.gallinarum and E. mundtii. Two isolates of E. faecalis showed the VanB type of phenotype (resistant to vancomycin, sensitive to teicoplanin). A single isolate showed a low degree of vancomycin resistance (MIC=8μg/ml) while being sensitive to teicoplanin. This isolate was speciated as E. gallinarum (Table).
Table.
Phenotypic and genotypic characterization of VRE isolates
All isolates of VRE were resistant to ciprofloxacin and tetracycline. Seven VRE isolates were sensitive to high level gentamicin by disk diffusion. Of these, six were found to have MIC<500μg/ml on further testing and were, therefore, sensitive to high level gentamicin. Only one VRE isolate was resistant to streptomycin with MIC >2000 μg/ml. Three isolates of VRE were sensitive to ampicillin. Among the 13 urinary VRE isolates, only two were sensitive to nitrofurantoin. Chloramphenicol was tested against all VRE isolates and 12 (37.5%) were sensitive. All VRE isolates were sensitive to linezolid.
Genetic basis of vancomycin resistance by polymerase chain reaction: Of the 32 isolates of VRE subjected to multiplex PCR for detecting vancomycin resistance genes, 31 (96.87%) were found to possess the vanA gene (Fig. 1). Of these, two showed VanB phenotype and the remaining showed high level resistance to both vancomycin and teicoplanin (VanA phenotype). One isolate showing VanA phenotype was found to possess both vanC1 and vanA genes by multiplex PCR. (Fig. 2). One isolate was found to be positive for the vanC1 gene associated with intrinsic vancomycin resistance. The VRE isolates found positive for vanC1 gene were later tested with primer sets specific for E.gallinarum and gave positive results.
Fig. 1.
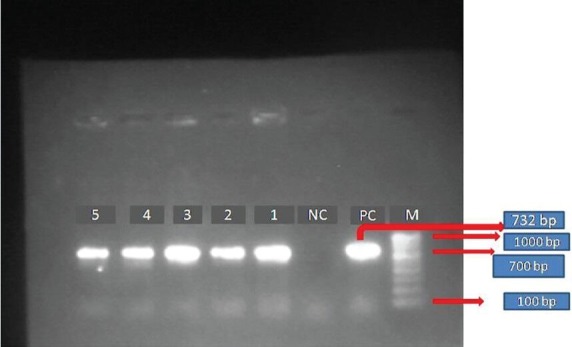
Gel electrophoresis of amplified products by PCR for vancomycin resistance genes. Agarose gel electrophoresis of the amplified products by PCR for Vancomycin resistance genes. M-100 base pair DNA ladder (Bangalore Genei, India). PC, positive control for vanA gene; NC, negative control; Lanes 1-5 positive for vanA gene (732 bp).
Fig. 2.

Gel electrophoresis of amplified products by multiplex PCR for detection of vancomycin resistance genes. Agarose gel electrophoresis of amplified products by PCR for vancomycin resistance genes. M-100 bp DNA ladder; Lane 9-Positive for vanA (732bp), vanC1 (822bp) and 16SrRNA internal control (320 bp)-multiplex PCR; Lane 8-Positive only for 16SrRNA internal control (no van resistance genes) by multiplex PCR; Lane 6-Positive for vanC1 (822 bp) by PCR targeting only vanC1 gene; Lanes 2 & 4-Positive for E.gallinarum (173 bp) by using species-specific primers.
PCR results were confirmed by sequencing and a majority were found to have 99-100 per cent identity with the partial plasmid encoding for vancomycin resistance protein A of E. faecalis (Gen Bank Accession no.ABN05630.1). The vanC1 amplicons had 100 per cent identity with gene encoding for vancomycin resistance protein VanC (Gen Bank Accession no. NZ_GG670287.1).
Discussion
In the last two decades, the emergence of VRE and their increasing prevalence worldwide has made it difficult to treat serious enterococcal infections. VRE was first reported by Uttley et al in 1989 from Great Britain9 and after that has been reported from many countries of the world. It is especially a big problem in the western world. Studies from the United States in the past decade reported vancomycin resistance in as many as 17 per cent of all Enterococcus strains and in up to 28 per cent of all nosocomial Enterococcus species strains3,10.
Although the prevalence of VRE infections in India is much lower than in the western world, it has been increasing in the past one decade. Mathur et al11 from New Delhi were the first to report VRE from India in 1999. Another study from north India reported vancomycin resistance in only 1 per cent of the Enterococcus species strains2, followed by a study from Chandigarh in which 5.5 per cent of 144 Enterococcus isolates from urine specimens were identified as VRE12. However, all isolates of VRE in their study showed a low-level vancomycin resistance ranging from 8-32 μg/ml. In a study done on enterococcal isolates from blood in New Delhi, only four isolates of E. faecium showed low degree resistance to vancomycin13. In another study from north India, 2 per cent of all enterococcal isolates were vancomycin resistant14.
In our study, vancomycin resistance was found among 8.7 per cent Enterococcus isolates. This was high compared to many other Indian studies. A majority of the VRE isolates (96.8%) were found to have a high-level vancomycin resistance. This was consistent with the low-level intrinsic vancomycin resistance seen in motile Enterococcus species like E. casseliflavus and E. gallinarum. Later, this particular isolate was identified as E. gallinarum, both by phenotypic and genotypic characterization. All the VRE isolates in our study were from patients who had been admitted to the wards or intensive care units.
The commonest phenotype seen among VRE strains is the VanA phenotype in which high level inducible resistance to both vancomycin and teicoplanin is seen (MICs ≥64 μg/ml)15. VanA phenotype was seen in 87.5 per cent of all VRE isolates in our study. Two VRE isolates showed VanB phenotype (resistant to vancomycin and sensitive to teicoplanin). VanB phenotype is the second most common phenotype of vancomycin resistance reported worldwide15. VanA and VanB phenotypes are due to acquisition of new genetic elements and have been mostly reported in E. faecalis and E. faecium isolates whereas VanC phenotype is constitutive low level vancomycin resistance seen in motile species of Enterococcus like E. gallinarum and E.casseliflavus15. There are many resistance genes associated with these glycopeptide resistant phenotypes of Enterococcus species and these are denoted as vanA, vanB, vanC1, vanC2/C3, vanD, etc. Other gene clusters responsible for vancomycin resistance which have been reported recently include vanG, vanL, vanM and vanN16.
vanA genotype strains showing susceptibility to teicoplanin have been reported in parts of East Asia like China, Japan and South Korea17. Reports of such isolates also exist from other parts of the world like Brazil18. The term VanB phenotype-vanA genotype VRE has been used for such strains by some authors19. Park et al from Seoul, South Korea20, reported an outbreak at a tertiary care hospital where six VanB phenotype vanA genotype E. faecium isolates with heterogenous expression of teicoplanin resistance were isolated. Such isolates have not yet been reported from India. The mechanism responsible for such heteroresistance to teicoplanin in Enterococcus isolates carrying the vanA gene is not yet clear. Some authors have opined that such heterogeneity is due to the presence of mutations, either in the vanA gene cluster or in the vanS regulatory element21,22. In a study from South Korea, the presence of an insertion sequence IS1216V in the coding region of the vanS gene has been suggested as a possible mechanism for this heterogeneity20.
Both the vanA and vanC1 genes were detected in a isolate of E. gallinarum. The occurrence of vanA and vanC1 genes in a single Enterococcus isolate was reported for the first time by Dutka-Malen et al23 from the faeces of a patient under oral therapy with vancomycin. VanC1 gene is specific for motile enterococci like E. gallinarum which show low-level intrinsic resistance to vancomycin. The in vivo acquisition of plasmids carrying the vanA gene cluster confers high-level resistance to vancomycin. This finding is important because motile species of Enterococcus like E.gallinarum or E. casseliflavus can be found in the environment and their detection in a patient is not an indication for strict isolation precautions for the patient in spite of their being intrinsically vancomycin resistant. This is because, intrinsic vancomycin resistance is not transferable. However, the evidence that these organisms can take up other resistance genes which not only make them highly resistant to vancomycin, but also make them capable of spreading this resistance to other Enterococcus strains, makes the control of these organisms important. There have been a few reports of E. gallinarum harboring both the vanA and vanC1 genes24. VRE strains possessing vanB gene in addition to the vanC1 gene have also been reported25. The possession of both vanA and vanC1 gene clusters will alter the resistance phenotype of an VRE isolate. In our study, the E. gallinarum isolate which possessed both the vanA and vanC1 genes showed a VanA phenotype with high level resistance to both vancomycin and teicoplanin. Detection of motile enterococci with additional resistance genes implies that phenotypic identification of enterococci to species level and determination of glycopeptide MICs do not necessarily predict the genotype.
Among the various antimicrobials available and evaluated for treatment of serious infections with vancomycin resistant enterococci are quinupristin dalfopristin, linezolid, daptomycin, chloramphenicol, etc. In our study, we have evaluated the in vitro activity of linezolid and chloramphenicol against VRE isolates.
Linezolid was the first oxazolidinone to be available for clinical use in 2000. It has activity against both E. faecium and E. faecalis26. Another advantage of this drug is that it can be administered both intravenously and orally. All the VRE isolates in our study were found to be sensitive to linezolid. Though linezolid resistance in Enterococcus species has not yet been reported from India, but from other parts of the world27. Outbreaks due to linezolid resistant enterococci, though rare, have been reported recently28. However, cases of linezolid-resistant vancomycin-resistant E. faecium infection without any prior exposure to linezolid have been reported29.
Chloramphenicol has proved to be useful in some instances for treating VRE infections30. In our study, 37.5 per cent of the VRE isolates were found to be susceptible to chloramphenicol. However, due to concerns regarding its ototoxicity and nephrotoxicity, streptomycin is not commonly used for enterococcal infections.
In conclusion, the vancomycin resistance rate among the Enterococcus isolates was 8.7 per cent in our study which was high compared to other reports from India. The commonest phenotype of glycopeptide resistance seen in our study was the VanA phenotype (resistance to both vancomycin and teicoplanin). Other phenotypes seen were VanB (resistant to vancomycin, sensitive to teicoplanin) and VanC (low level intrinsic resistance to vancomycin). The presence of both vanA and vanC1 genes in an E. gallinarum isolate indicated that phenotypic and genotypic characterization of glycopeptide resistance might not always correspond. The detection of vanA gene cluster in two isolates of E. faecalis showed VanB phenotype of glycopeptide resistance. Glycopeptide resistance among our isolates was high, probably reflecting the increased use of vancomycin in our hospital over the past few years. This fact highlights the importance of strict enforcement of antibiotic policies coupled with greater adherence to infection control measures to prevent emergence and spread of antibiotic resistant bacteria.
References
- 1.Arias CA, Murray BE. Antibiotic resistant bugs in the 21st Century - A clinical super-challenge. N Engl J Med. 2009;360:439–43. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 2.Mathur P, Kapil A, Chandra R, Sharma P, Das B. Antimicrobial resistance in Enterococcus faecalis at a tertiary care centre of northern India. Indian J Med Res. 2003;118:25–8. [PubMed] [Google Scholar]
- 3.NNIS. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32:470–85. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal J, Kalyan R, Singh M. High level aminoglycoside resistance and beta-lactamase production in Enterococci at a tertiary care hospital in India. Jpn J Infect Dis. 2009;62:158–9. [PubMed] [Google Scholar]
- 5.Mendiratta DK, Kaur H, Deotale V, Thamke DC, Narang R, Narang P. Status of high level aminoglycoside resistant Enterococcus faecalis and Enterococcus faecium in a rural hospital of Central India. Indian J Med Microbiol. 2008;26:369–71. doi: 10.4103/0255-0857.43582. [DOI] [PubMed] [Google Scholar]
- 6.Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–4. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.18th ed. Wayne, USA: CLSI; 2008. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 8.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–7. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uttley AHC, George RC, Naidoo J, Woodford N, Johnson AP, Calms CH. High-level vancomycin-resistant enterococci causing hospital infections. Epidemiol Infect. 1989;103:173–81. doi: 10.1017/s0950268800030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Low DE, Keller N, Barth A, Jones RN. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32(Suppl 2):S133–45. doi: 10.1086/320185. [DOI] [PubMed] [Google Scholar]
- 11.Mathur P, Chaudhary R, Dhawan B, Sharma N, Kumar L. Vancomycin-resistant Enterococcus bacteraemia in a lymphoma patient. Indian J Med Microbiol. 1999;17:194–5. [Google Scholar]
- 12.Taneja N, Rani P, Emmanuel R, Sharma M. Significance of vancomycin resistant enterococci from urinary specimens at a tertiary care centre in northern India. Indian J Med Res. 2004;119:72–4. [PubMed] [Google Scholar]
- 13.Kapoor L, Randhawa VS, Deb M. Antimicrobial resistance of enterococcal blood isolates at a pediatric care hospital in India. Jpn J Infect Dis. 2005;58:101–3. [PubMed] [Google Scholar]
- 14.Kaur N, Chaudhary U, Aggarwal R, Bala K. Emergence of VRE and their antimicrobial sensitivity pattern in a tertiary care teaching hospital. J Med Biol Sci. 2009;8:26–32. [Google Scholar]
- 15.Centinkaya Y, Yalk P, Mayhall CG. Vancomycin - resistant Enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S. d-Ala-d-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 2011;55:4606–12. doi: 10.1128/AAC.00714-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu L, Cao B, Liu Y, Guo P, Song S, Li R. A new Tn1546 type of VanB phenotype-vanA genotype vancomycin-resistant Enterococcus faecium isolates in mainland China. Diagn Microbiol Infect Dis. 2009;63:70–5. doi: 10.1016/j.diagmicrobio.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Zanella RC, Castro Lima MJ, Tegani LS, Hitomi A, de Cunto Brandileone MC, Palazzo IC. Emergence of VanB phenotype-vanA genotype in vancomycin-resistant Enterococci in Brazilian hospital. Braz J Microbiol. 2006;37:117–8. [Google Scholar]
- 19.Qu T, Zhang J, Zhou Z, Wei Z, Yu Y, Chen Y. Heteroresistance to teicoplanin in Enterococcus faecium harboring the vanA gene. J Clin Microbiol. 2009;47:4194–6. doi: 10.1128/JCM.01802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park IJ, Lee WG, Shin JH, Lee KW, Woo GJ. VanB phenotype-vanA genotype Enterococcus faecium with heterogeneous expression of teicoplanin resistance. J Clin Microbiol. 2008;46:3091–3. doi: 10.1128/JCM.00712-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WG, Huh JY, Cho SR, Lim YA. Reduction in glycopeptide resistance in vancomycin-resistant enterococci as a result of vanA cluster rearrangments. Antimicrob Agents Chemother. 2004;48:1379–81. doi: 10.1128/AAC.48.4.1379-1381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eom JS, Hwang IS, Hwang BY, Lee JG, Lee YJ, Cheong HJ, et al. Emergence of vanA genotype vancomycin-resistant enterococci with low or moderate levels of teicoplanin resistance in Korea. J Clin Microbiol. 2004;42:1785–6. doi: 10.1128/JCM.42.4.1785-1786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutka-Malen S, Blaimont B, Wauters G, Courvalin P. Emergence of high-level resistance to glycopeptides in Enterococcus gallinarum and Enterococcus casseliflavus. Antimicrob Agents Chemother. 1994;38:1675–7. doi: 10.1128/aac.38.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camargo ILBC, Barth AL, Pilger K, Seligman BGS, Machado ARL, Darini ALC. Enterococcus gallinarum carrying the vanA gene cluster: first report in Brazil. Braz J Med Biol Res. 2004;37:1669–71. doi: 10.1590/s0100-879x2004001100012. [DOI] [PubMed] [Google Scholar]
- 25.Schooneveldt JM. Detection of a vanB determinant in Enterococcus gallinarum in Australia. J Clin Microbiol. 2000;38:3902. doi: 10.1128/jcm.38.10.3902-3902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zirakzadeh A, Patel R. Vancomycin-resistant Enterococci: Colonization, infection, detection, and treatment. Mayo Clin Proc. 2006;81:529–36. doi: 10.4065/81.4.529. [DOI] [PubMed] [Google Scholar]
- 27.Pai MP, Rodvold KA, Schreckenberger PC, Gonzales RD, Petrollati JM, Quinn JP. Risk factors associated with the development of infection with linezolid- and vancomycin-resistant Enterococcus faecium. Clin Infect Dis. 2002;35:1269–72. doi: 10.1086/344177. [DOI] [PubMed] [Google Scholar]
- 28.Ntokou E, Stathopoulos C, Kristo I, Dimitroulia E, Labrou M, Vasdeki A, et al. Intensive care unit dissemination of multiple clones of linezolid-resistant Enterococcus faecalis and Enterococcus faecium. J Antimicrob Chemother. 2012;67:1819–23. doi: 10.1093/jac/dks146. [DOI] [PubMed] [Google Scholar]
- 29.Rahim S, Pillai SK, Gold HS, Venkataraman L, Inglima K, Press RA. Linezolid-resistant, vancomycin-resistant Enterococcus faecium infection in patients without prior exposure to linezolid. Clin Infect Dis. 2003;36:e146–8. doi: 10.1086/374929. [DOI] [PubMed] [Google Scholar]
- 30.Murray BE. Vancomycin resistant enterococcal infections. N Engl J Med. 2000;342:710–21. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]


