Abstract
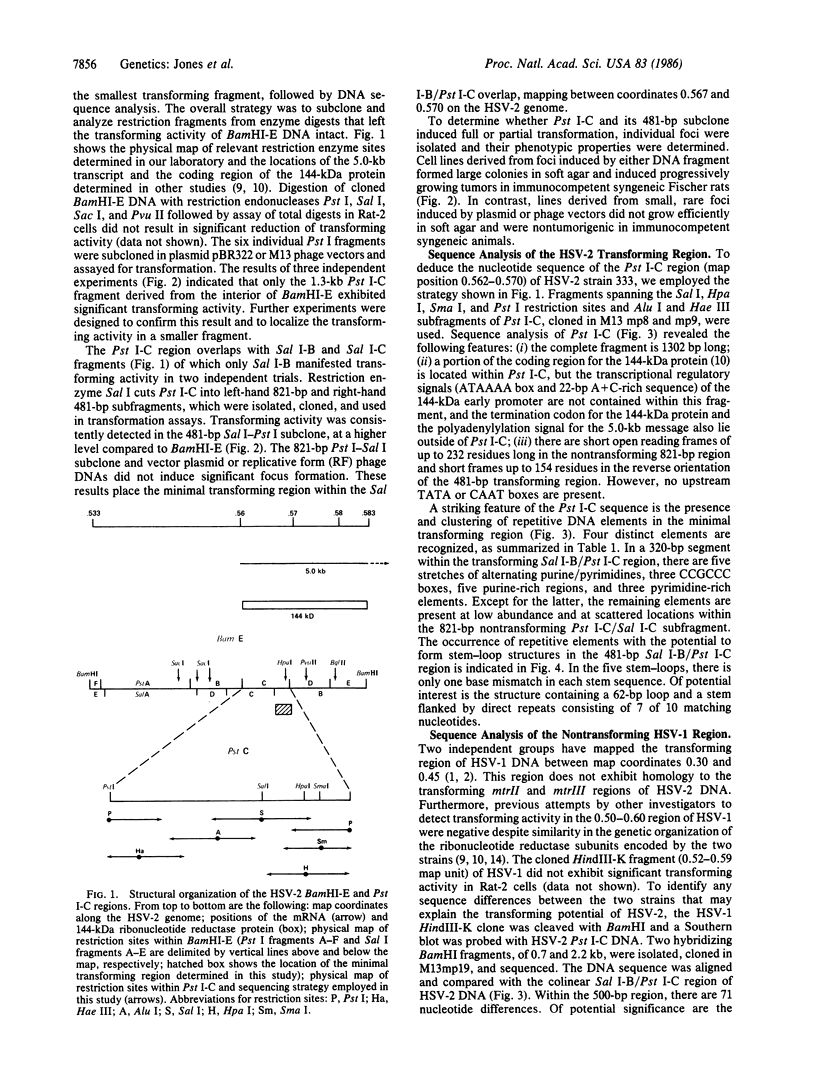
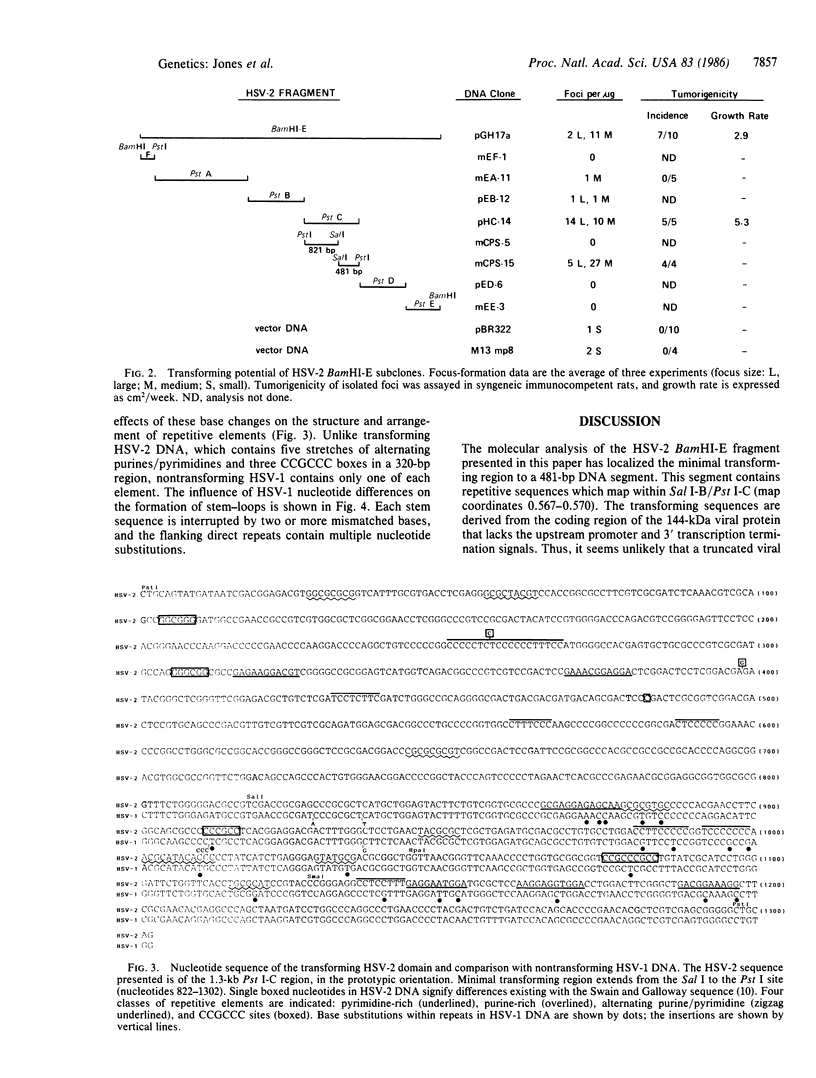
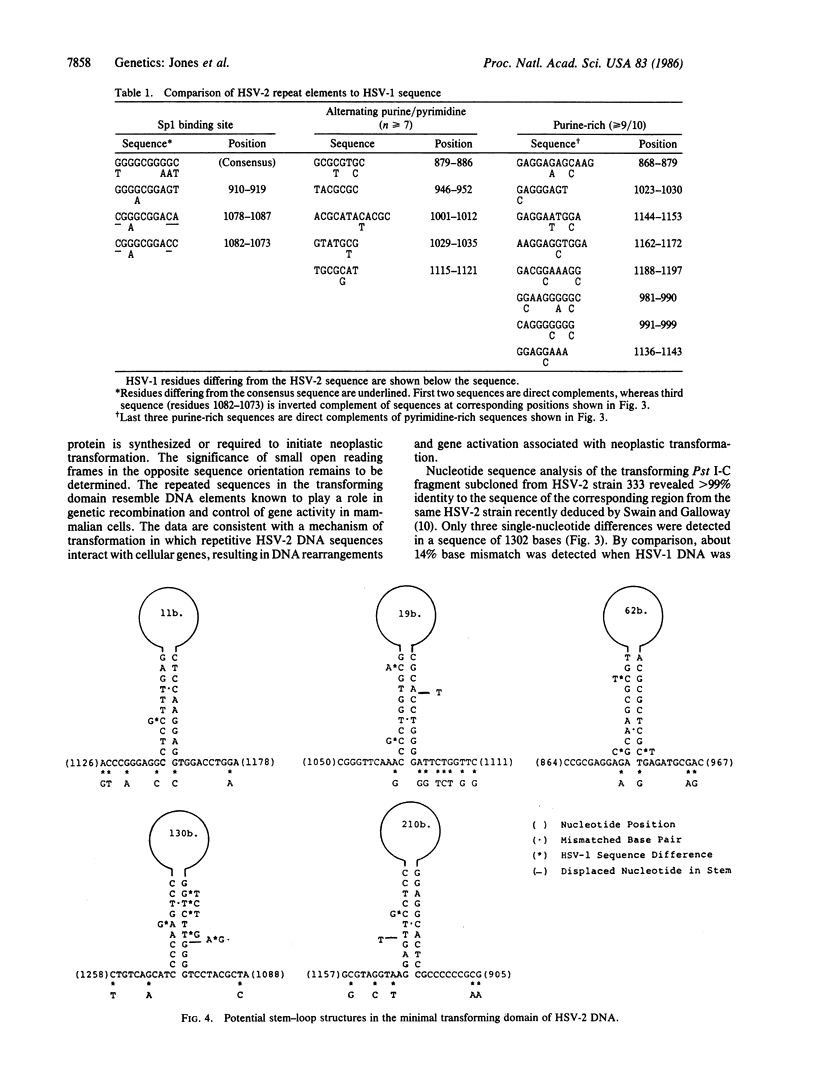
The 7.5-kilobase BamHI E fragment (BamHI-E) of herpes simplex virus type 2 (HSV-2) DNA (map position 0.533-0.583) encodes the 144-kDa subunit of ribonucleotide reductase and induces the neoplastic transformation of immortalized cell lines. To define the minimal transforming region of BamHI-E, a series of subclones were constructed that spanned the entire fragment. These subclones were assayed for focus formation in Rat-2 cells. Removal of the promoter region from the viral 144-kDa-protein gene left the transforming activity of DNA clones intact. A 481-bp Pst I-Sal I subclone of BamHI-E was capable of inducing focus formation and tumorigenic conversion. The nucleotide sequence of this fragment and the colinear nontransforming region of HSV-1 DNA was determined and compared. Striking differences were detected in the structure and organization of repeated sequence elements. Specifically, transforming HSV-2 DNA contains multiple regions of alternating purines and pyrimidines, G + C-rich sequences that are potential binding sites for transcription factor Sp1, and insertion-like sequence elements that are interrupted by base substitutions in nontransforming HSV-1 DNA. These results define a distinct transforming domain in HSV-2 DNA composed of repetitive elements implicated in gene rearrangement and activation.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson K. P., Frink R. J., Devi G. B., Gaylord B. H., Costa R. H., Wagner E. K. Detailed characterization of the mRNA mapping in the HindIII fragment K region of the herpes simplex virus type 1 genome. J Virol. 1981 Mar;37(3):1011–1027. doi: 10.1128/jvi.37.3.1011-1027.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M., Weber F., Jahn G., Dorsch-Häsler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985 Jun;41(2):521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Camacho A., Spear G. Transformation of hamster embryo fibroblasts by a specific fragment of the herpes simplex virus genome. Cell. 1978 Nov;15(3):993–1002. doi: 10.1016/0092-8674(78)90283-0. [DOI] [PubMed] [Google Scholar]
- Cameron I. R., Park M., Dutia B. M., Orr A., Macnab J. C. Herpes simplex virus sequences involved in the initiation of oncogenic morphological transformation of rat cells are not required for maintenance of the transformed state. J Gen Virol. 1985 Mar;66(Pt 3):517–527. doi: 10.1099/0022-1317-66-3-517. [DOI] [PubMed] [Google Scholar]
- Galloway D. A., Nelson J. A., McDougall J. K. Small fragments of herpesvirus DNA with transforming activity contain insertion sequence-like structures. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4736–4740. doi: 10.1073/pnas.81.15.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Iwasaka T., Smith C. C., Aurelian L., Lewis G. K., Ts'o P. O. Multistep transformation by defined fragments of herpes simplex virus type 2 DNA: oncogenic region and its gene product. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8493–8497. doi: 10.1073/pnas.82.24.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar D., Bacchetti S. Is ribonucleotide reductase the transforming function of herpes simplex virus 2? Nature. 1983 Mar 3;302(5903):76–79. doi: 10.1038/302076a0. [DOI] [PubMed] [Google Scholar]
- Ishii S., Kadonaga J. T., Tjian R., Brady J. N., Merlino G. T., Pastan I. Binding of the Sp1 transcription factor by the human Harvey ras1 proto-oncogene promoter. Science. 1986 Jun 13;232(4756):1410–1413. doi: 10.1126/science.3012774. [DOI] [PubMed] [Google Scholar]
- Jariwalla R. J., Aurelian L., Ts'o P. O. Immortalization and neoplastic transformation of normal diploid cells by defined cloned DNA fragments of herpes simplex virus type 2. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5902–5906. doi: 10.1073/pnas.80.19.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jariwalla R. J., Aurelian L., Ts'o P. O. Tumorigenic transformation induced by a specific fragment of DNA from herpes simplex virus type 2. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2279–2283. doi: 10.1073/pnas.77.4.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jariwalla R. J., Tanczos B., Jones C., Ortiz J., Salimi-Lopez S. DNA amplification and neoplastic transformation mediated by a herpes simplex DNA fragment containing cell-related sequences. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1738–1742. doi: 10.1073/pnas.83.6.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. M., Jelinek W. R. Replication of a plasmid bearing a human Alu-family repeat in monkey COS-7 cells. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4660–4664. doi: 10.1073/pnas.83.13.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986 Feb 28;44(4):545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Leavitt J., Gunning P., Kedes L., Jariwalla R. Smooth muscle alpha-action is a transformation-sensitive marker for mouse NIH 3T3 and Rat-2 cells. 1985 Aug 29-Sep 4Nature. 316(6031):840–842. doi: 10.1038/316840a0. [DOI] [PubMed] [Google Scholar]
- Macnab J. C., Orr A., La Thangue N. B. Cellular proteins expressed in herpes simplex virus transformed cells also accumulate on herpes simplex virus infection. EMBO J. 1985 Dec 1;4(12):3223–3228. doi: 10.1002/j.1460-2075.1985.tb04069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Clements J. B. DNA sequence homology between two co-linear loci on the HSV genome which have different transforming abilities. EMBO J. 1983;2(11):1953–1961. doi: 10.1002/j.1460-2075.1983.tb01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol J., Behe M., Felsenfeld G. Effect of the B--Z transition in poly(dG-m5dC) . poly(dG-m5dC) on nucleosome formation. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1771–1775. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes G. R., LaFemina R., Hayward S. D., Hayward G. S. Morphological transformation by DNA fragments of human herpesviruses: evidence for two distinct transforming regions in herpes simplex virus types 1 and 2 and lack of correlation with biochemical transfer of the thymidine kinase gene. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):629–641. doi: 10.1101/sqb.1980.044.01.066. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier P. H., Cortese R. A fast and simple method for sequencing DNA cloned in the single-stranded bacteriophage M13. J Mol Biol. 1979 Mar 25;129(1):169–172. doi: 10.1016/0022-2836(79)90068-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Swain M. A., Galloway D. A. Herpes simplex virus specifies two subunits of ribonucleotide reductase encoded by 3'-coterminal transcripts. J Virol. 1986 Mar;57(3):802–808. doi: 10.1128/jvi.57.3.802-808.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]


