Abstract
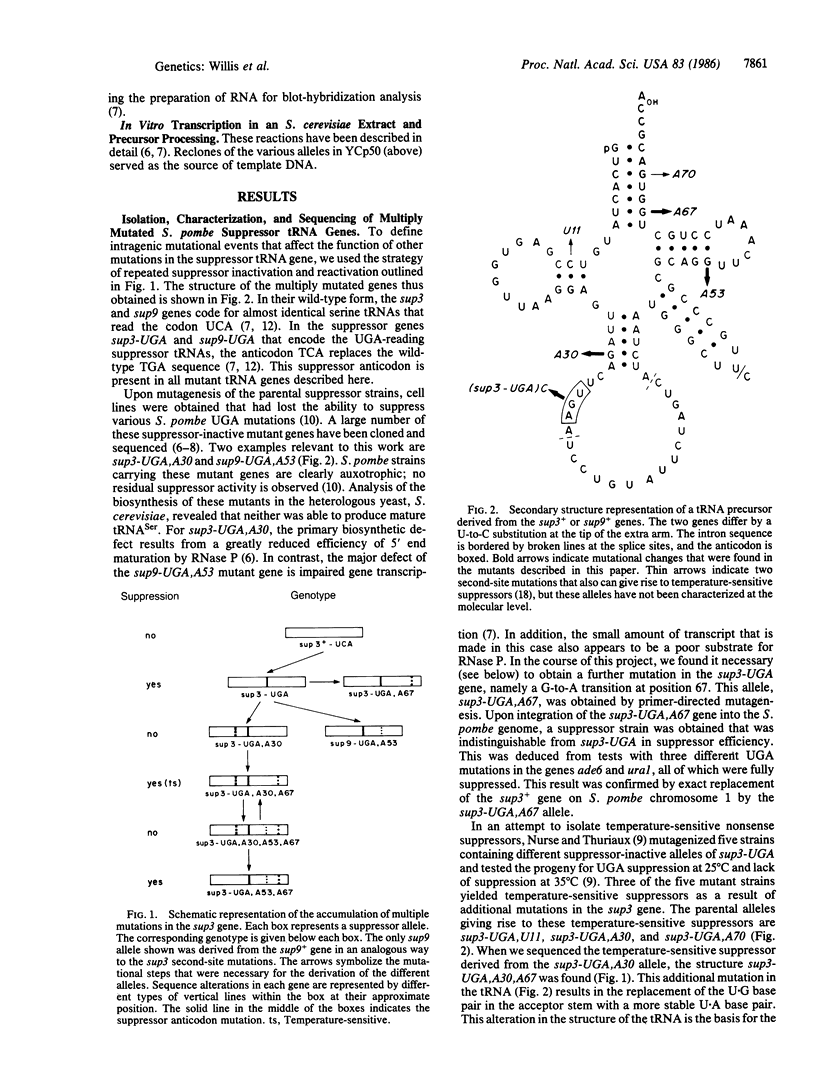
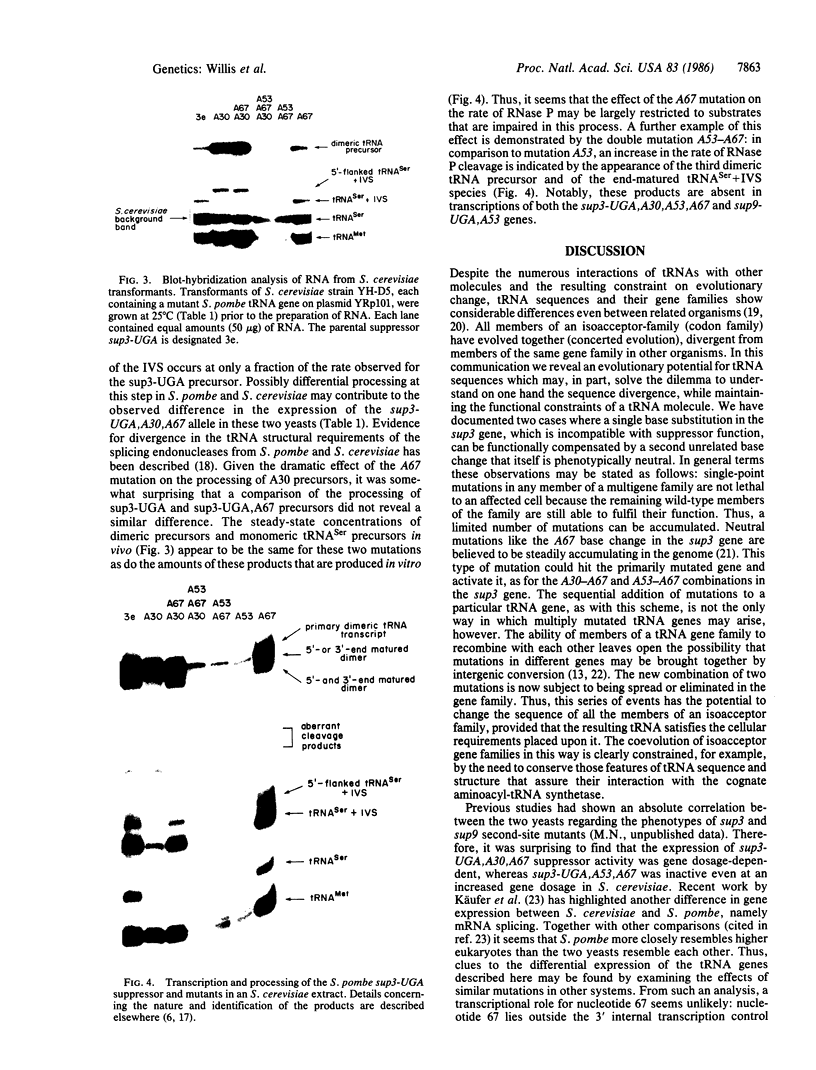
Successive rounds of mutagenesis of a Schizosaccharomyces pombe strain bearing the UGA-reading sup3 tRNASer suppressor have been carried out for two cycles of inactivation and reactivation of the suppressor. The suppressor phenotype at each stage was found to involve different combinations of three mutations, A30, A53, and A67, in the sup3-UGA gene. Single mutations A30 and A53 inactivate the suppressor as does the presence of all three mutations. A67 by itself is phenotypically neutral, but in combination with either A30 or A53 suppressor function is restored. The frequency with which these and other complementation events occur in S. pombe demonstrates a significant potential for nucleotide sequence evolution in tRNA. Differential expression of the S. pombe genes in Saccharomyces cerevisiae suggests that the two yeasts have diverged at the transcriptional and RNA processing level. Processing of the mutant tRNA precursors in S. cerevisiae reveals a hierarchy of structural domains within the tRNA that vary in their importance for RNase P cleavage.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amstutz H., Munz P., Heyer W. D., Leupoid U., Kohli J. Concerted evolution of tRNA genes: intergenic conversion among three unlinked serine tRNA genes in S. pombe. Cell. 1985 Apr;40(4):879–886. doi: 10.1016/0092-8674(85)90347-2. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. Processing of tRNA in prokaryotes and eukaryotes. CRC Crit Rev Biochem. 1984;17(1):45–71. doi: 10.3109/10409238409110269. [DOI] [PubMed] [Google Scholar]
- Heyer W. D., Munz P., Amstutz H., Aebi R., Gysler C., Schuchert P., Szankasi P., Leupold U., Kohli J., Gamulin V. Inactivation of nonsense suppressor transfer RNA genes in Schizosaccharomyces pombe. Intergenic conversion and hot spots of mutation. J Mol Biol. 1986 Apr 5;188(3):343–353. doi: 10.1016/0022-2836(86)90159-2. [DOI] [PubMed] [Google Scholar]
- Hottinger H., Pearson D., Yamao F., Gamulin V., Cooley L., Cooper T., Söll D. Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Mol Gen Genet. 1982;188(2):219–224. doi: 10.1007/BF00332678. [DOI] [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Käufer N. F., Simanis V., Nurse P. Fission yeast Schizosaccharomyces pombe correctly excises a mammalian RNA transcript intervening sequence. Nature. 1985 Nov 7;318(6041):78–80. doi: 10.1038/318078a0. [DOI] [PubMed] [Google Scholar]
- Munz P., Amstutz H., Kohli J., Leupold U. Recombination between dispersed serine tRNA genes in Schizosaccharomyces pombe. Nature. 1982 Nov 18;300(5889):225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schaack J., Söll D. Transcription of a Drosophila tRNAArg gene in yeast extract: 5'-flanking sequence dependence for transcription in a heterologous system. Nucleic Acids Res. 1985 Apr 25;13(8):2803–2814. doi: 10.1093/nar/13.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Smith J. D. Transcription and processing of transfer RNA precursors. Prog Nucleic Acid Res Mol Biol. 1976;16:25–73. doi: 10.1016/s0079-6603(08)60755-2. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Moll J., Meissner F., Hartmann T. Compilation of tRNA sequences. Nucleic Acids Res. 1985;13 (Suppl):r1–49. doi: 10.1093/nar/13.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Vorderwülbecke T., Hartmann T. Compilation of sequences of tRNA genes. Nucleic Acids Res. 1985;13 (Suppl):r51–104. doi: 10.1093/nar/13.suppl.r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. S., Söll D., Sharp S. Point mutations in the 5' ICR and anticodon region of a Drosophila tRNAArg gene decrease in vitro transcription. Nucleic Acids Res. 1985 Jan 25;13(2):435–447. doi: 10.1093/nar/13.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner-Smith M., Hottinger H., Willis I., Koch T. L., Arentzen R., Söll D. The sup8 tRNALeu gene of Schizosaccharomyces pombe has an unusual intervening sequence and reduced pairing in the anticodon stem. Mol Gen Genet. 1984;197(3):447–452. doi: 10.1007/BF00329941. [DOI] [PubMed] [Google Scholar]
- Tobian J. A., Drinkard L., Zasloff M. tRNA nuclear transport: defining the critical regions of human tRNAimet by point mutagenesis. Cell. 1985 Dec;43(2 Pt 1):415–422. doi: 10.1016/0092-8674(85)90171-0. [DOI] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell. 1984 Jan;36(1):179–187. doi: 10.1016/0092-8674(84)90087-4. [DOI] [PubMed] [Google Scholar]
- Willis I., Frendewey D., Nichols M., Hottinger-Werlen A., Schaack J., Söll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986 May 5;261(13):5878–5885. [PubMed] [Google Scholar]
- Willis I., Hottinger H., Pearson D., Chisholm V., Leupold U., Söll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984 Jul;3(7):1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]