Abstract
Background: Two recent genome-wide association studies (GWASs) identified five single nucleotide polymorphisms (SNPs; rs965513, rs944289, rs966423, rs2439302, and rs116909374) associated with papillary thyroid carcinoma (PTC). Each variant showed highly significant but moderate to low disease risk. Here we assessed the cumulative risk and predictive value of the five SNPs.
Methods: We genotyped two cohorts of individuals, 747 PTC cases and 1047 controls from Ohio and 1795 PTC cases and 2090 controls from Poland. Cumulative genetic risk scores were calculated using unweighted and weighted approaches.
Results: All five SNPs showed significant association with PTC. The average cumulative risk score in cases was significantly higher than in controls (p<2.2×10−16). Each additional risk allele increased the risk of having PTC by 1.51 [95% confidence interval (CI) 1.4, 1.64] in Ohio and by 1.35 [95% CI 1.27, 1.44] in Poland. An analysis was performed weighing risk alleles by effect size and assigning individuals to three weighted risk score groups, low (≤2), medium (2–5), and high (>5). Individuals in the high group were significantly more susceptible to PTC compared with individuals in the low group with an odds ratio of 8.7 [95% CI 5.8, 13.3] in Ohio and 4.24 [95% CI 3.10, 5.84] in Poland. Almost identical results were obtained when follicular variant PTCs and microPTCs were omitted. These five SNPs explained 11% of the familial risk of thyroid cancer in the Ohio cohort and 6% in the Polish cohort.
Conclusion: As the genetic risk score increases, the risk of having PTC increases. However, the predictive power of the cumulative effect of these five variants is only moderately high and clinical use may not be feasible until more variants are detected.
Introduction
In the past several years, technological advances have facilitated large-scale genetic analyzes of heritable genetic variants in search of predisposing genes. In particular, the hypothesis-free genome-wide association studies (GWASs) using anonymous markers, mainly single nucleotide polymorphisms (SNPs), have produced a flood of association data. According to the GWAS database (www.genome.gov/gwastudies), 1414 published investigations involving over 700 diseases and other phenotypes, resulting in some 7500 associations, have been reported. The expected impact of these findings has been anticipated to occur in two major areas: (i) where an underlying biological mechanism leading to the predisposition is clarified, new biological insight is gained; (ii) when the effect size of the association is strong, genotypes can be used in disease prediction by genotyping applied to bedside medicine, counseling, and prevention. Even when the effect size of individual markers is low, it is postulated that the predictive power may be enhanced by combining genotype data from several loci. Our study addressed this point.
The most common form of thyroid cancer, papillary thyroid carcinoma (PTC) accounts for 80%–85% of all thyroid cancers. Several large case–control studies unequivocally suggest that familial occurrence of PTC is common, in fact, one of the highest of all cancers (1–4). Remarkably, in spite of this, only a few predisposing mutations have been convincingly demonstrated (5–9). This suggests that easy-to-find high-penetrance mutations probably do not exist or are rare. In contrast, accumulating evidence from other malignancies (10–12) and multifactorial diseases and traits (13–16) suggests that the genetic predisposition often consists of a multitude of low-penetrance alleles (16,17). The first few years of GWASs appear to have amply confirmed this assumption. However, interestingly, clinical and predictive use of these findings has been slow to occur (18–21).
In this study we asked what predictive powers might already be available as a result of two GWASs in PTC. We studied the predictive value of the five markers detected in two recently published GWASs (22,23). We found that the combined use of the genotypes of these markers shows definite promise as a predictive tool, but that more markers are probably needed before genotyping of markers for PTC can become a routinely applied method in clinical practice.
Materials and Methods
Subjects
All studies were approved by the Institutional Review Boards at The Ohio State University Medical Center (OSUMC), Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, and Medical University of Warsaw, Poland. All subjects gave written informed consent before participation.
Ohio cases (n=747) involved individuals with thyroid cancer enrolled in the Ohio State University Wexner Medical Center's (OSUWMC) endocrine neoplasia repository, a large data and bio-repository of individuals with thyroid neoplasia. Individuals were recruited from a multidisciplinary thyroid tumor clinic at OSUWMC, and all cases were histologically confirmed as PTC (including traditional PTC, follicular variant PTC [FVPTC], and microPTC). Ohio control samples (n=1047) were provided by the OSUWMC's Human Genetics Sample Bank. The Columbus Area Controls Sample Bank is a collection of control samples for use in human genetics research that includes anonymized biological specimens and linked phenotypic data. Recruitment takes place in OSUWMC primary care and internal medicine clinics. All patients representing cases and controls provide written informed consent; complete a questionnaire that includes demographic, medical, and family history information; and donate a blood sample. Relevant clinicopathological data for cases were extracted from the electronic medical record. Polish case patients (n=1795) were recruited from thyroid cancer patients from all over Poland and treated at the Medical University of Warsaw and Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology in Warsaw, Poland. All cases were histologically confirmed as PTC (including traditional PTC and rare variants). Polish control samples (n=2090) were provided by the Department of Medical Genetics, Medical University of Warsaw and consisted of consenting volunteers. The controls chosen for this study in both cohorts reported no thyroid disease. Demographic information for all cases and controls can be found in Table 1.
Table 1.
Summary of Demographic Characteristics of Ohio and Polish Samples
| Cohort | Variable | PTC | Control | p-valuea |
|---|---|---|---|---|
| Ohio |
Total |
747 |
1047 |
|
| |
Female |
77% |
69% |
0.0006 |
| |
Male |
23% |
31% |
|
| |
Age (years) |
39.54±14.5 |
46.89±16.2 |
<2×10−16 |
| Polish |
Total |
1795 |
2090 |
|
| |
Female |
90% |
47% |
<2.2×10−16 |
| |
Male |
10% |
53% |
|
| Age (years) | 49.38±14.5 | 36.3±10.6 | <2.2×10−16 |
The p-values were obtained by applying permutation tests to compare age and chi-square tests to compare sex.
PTC, papillary thyroid carcinoma; SD, standard deviation.
DNA extraction and SNP genotyping
For both cases and controls, germline DNA from blood samples was extracted by a standard phenol–chloroform procedure. To genotype the five selected SNPs, SNaPshot assay (ABI) was used in the Ohio samples as described (24).
The Sequenom genotyping technology (Sequenom) was used for the Polish samples. For each sample, 20 ng of genomic DNA was genotyped using iPLEX Gold system. Primers and probes for the analysis were designed using MassARRAY® Assay Design v3.1. Data were visualized and analyzed in MassARRAY Typer Viewer v4.0.24. All oligonucleotides used in the study were ordered and purchased from Integrated DNA Technologies Inc.
Cumulative genetic risk score
For each SNP, the genotypes were coded as 0, 1, or 2 indicating the number of PTC risk alleles in the genotype. Cumulative genetic risk scores were calculated in two ways, using unweighted and weighted approaches. For the unweighted method, the cumulative genetic risk score (CGRS) of an individual is simply the total count of disease alleles from five SNPs obtained by adding coded genotypes (possible score range of 0–10). Weighted CGRS (wCGRS) denotes the sum of the weighted disease allele counts weighted by logarithm odds ratio, log(OR), of each SNP and scaled by a scaling factor of 5/(w1+w2+w3+w4+w5) to make the range of CGRS and wCGRS comparable (25), where wi=log(OR) for the ith SNP, i=1 to 5. To avoid any bias due to missing data, samples with one or more missing genotypes were not included in the genetic risk score calculations. For the unweighted and weighted genetic score analysis, genotypes from 605 cases and 916 controls from Ohio and 1633 cases and 1663 controls from Poland were available.
Statistical analysis
Statistical analyses were performed using the R software package (www.r-project.org). For each SNP, Hardy–Weinberg equilibrium was tested in the control samples by applying chi-square tests. All SNP variants satisfied Hardy–Weinberg Equilibrium in the control samples from both cohorts, with a p-value ≥0.01. All the demographic characteristics between PTC cases and controls were evaluated by applying a permutation test for continuous data and chi-square test for discrete data.
Effect sizes and the strength of association of each disease allele adjusting for the age and sex differences were obtained by applying multivariate logistic regression analysis, assuming a multiplicative allelic model. Pair-wise SNP interaction analysis was performed under a multiplicative model using logistic regression and likelihood ratio tests.
Distributions of genetic risk scores between PTC cases and controls were compared by applying nonparametric Mann–Whitney test. Moreover, a permutation test was applied to compare average genetic scores between cases and controls. The effect of genetic risk scores on PTC was tested by using multivariate logistic regression analysis adjusting for age and sex. Statistical analysis of genetic scores was performed using both unweighted and weighted cumulative genetic scores separately.
The familial relative risk of PTC attributable to a given SNP is calculated by the formula (26–28)
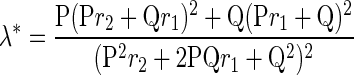 |
where P is the population frequency of the disease allele, Q=1−P, and r1 and r2 are the relative risks (estimated by odds ratios) for heterozygotes and rare homozygotes, relative to common homozygotes. Assuming a multiplicative interaction, the proportion of the familial risk attributable to the SNP is calculated by log(λ*)/log(λ0), where λ0 is the overall familial relative risk (27,28), estimated to be 8.48 for thyroid cancer (1).
To evaluate and compare classifying power of the logistic regression model with unweighted or weighted cumulative genetic risk scores, receiver–operator characteristic (ROC) curves and the area under the curves (AUC) were determined. Since there was a significant difference in age and sex between cases and controls, equal numbers of age- and sex-matched random samples from available cases and controls were selected for the ROC analysis.
Results
Association of each of the five SNPs with PTC risk was confirmed in Ohio and Poland populations
Demographic characteristics of the two cohorts are shown in Table 1 (747 cases/1047 controls from Ohio; 1795 cases/2090 controls from central Poland). Ages are reported as age at diagnosis of PTC in the cases, and as age at blood draw in controls. Controls were significantly older than cases (p<2.2×10−16) in the Ohio cohort and younger than cases (p<2.2×10−16) in the Polish cohort. A statistically significant sex difference, having more females among the cases, also occurred between cases and controls in both cohorts (Ohio, p=0.0006; Poland, p<2.2×10−16). The frequencies of each variant did not show significant difference between males and females (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/thy). Pairwise interactions of risk alleles from five SNPs were evaluated, and no evidence of interactions was found at the p-value=0.05 significance level.
Age- and sex-adjusted odds ratios (OR) and 95% confidence intervals (CIs) were obtained (Table 2) by assuming a multiplicative model. The SNPs, rs965513, rs944289, rs966423, rs2439302, and rs116909374 showed significant association with susceptibility to PTC in the Ohio population with effect sizes [and 95% CI] of 2.09 [1.80, 2.42], 1.25 [1.08, 1.46], 1.30 [1.12, 1.51], 1.46 [1.26, 1.70], and 2.28 [1.57, 3.36], respectively, with p-value <0.005. In the Polish data, all five SNPs showed association at the 0.05 significance level with effect sizes [and 95% CI] of 1.81 [1.59, 2.06], 1.22 [1.09, 1.38], 1.14 [1.01, 1.29], 1.23 [1.09, 1.38], and 1.66 [1.13, 2.44], respectively (Table 2). Of the Ohio samples ∼350 cases and ∼380 controls occurred both in this study and the GWASs (22,23). These cases were not used when allele frequencies and ORs were compared between the studies (Table 2).
Table 2.
Association Analysis Results for Five SNPs in Ohio and Polish Cohorts
| Cohort | SNP ID | Chr. location | Risk allele | Control/ PTC (n) | ORa | 95% CIa | p-valuea | Risk allele freq: PTC | Risk allele freq: control |
|---|---|---|---|---|---|---|---|---|---|
| Ohio |
rs966423 |
2q35 |
C |
548/282b |
1.28 |
[1.04, 1.58] |
2.12×10−2 |
0.496 |
0.429 |
| |
|
|
|
930/639c |
1.30 |
[1.12, 1.51] |
4.49×10−4 |
0.484 |
0.415 |
| |
rs944289 |
14q13 |
T |
653/419b |
1.18 |
[0.99, 1.42] |
7.17×10−2 |
0.617 |
0.575 |
| |
|
|
|
1032/651c |
1.25 |
[1.08, 1.46] |
2.70×10−3 |
0.634 |
0.580 |
| |
rs2439302 |
8p15 |
G |
556/282b |
1.59 |
[1.28, 1.98] |
2.45×10−5 |
0.564 |
0.460 |
| |
|
|
|
938/639c |
1.46 |
[1.26, 1.70] |
5.60×10−7 |
0.555 |
0.467 |
| |
rs116909374 |
14q13 |
T |
559/273b |
2.67 |
[1.61, 4.48] |
1.60×10−4 |
0.066 |
0.026 |
| |
|
|
|
932/622c |
2.28 |
[1.57, 3.36] |
2.11×10−5 |
0.056 |
0.026 |
| |
rs965513 |
9q22 |
A |
583/548b |
2.19 |
[1.84, 2.62] |
<2×10−16 |
0.513 |
0.322 |
| |
|
|
|
941/720c |
2.09 |
[1.80, 2.42] |
<2×10−16 |
0.506 |
0.327 |
| Poland |
rs966423 |
2q35 |
C |
1899/1721 |
1.14 |
[1.01, 1.29] |
2.94×10−2 |
0.461 |
0.425 |
| |
rs944289 |
14q13 |
T |
1913/1699 |
1.22 |
[1.09, 1.38] |
1.06×10−3 |
0.629 |
0.595 |
| |
rs2439302 |
8p15 |
G |
1901/1690 |
1.23 |
[1.09, 1.38] |
9.29×10−4 |
0.541 |
0.485 |
| |
rs116909374 |
14q13 |
T |
2010/1760 |
1.66 |
[1.13, 2.44] |
9.81×10−3 |
0.031 |
0.019 |
| rs965513 | 9q22 | A | 1911/1716 | 1.81 | [1.59, 2.06] | <2×10−16 | 0.461 | 0.352 |
Allelic odds ratios (ORs) with 95% confidence intervals [95% CIs] and p-values, obtained by applying multivariate logistic regression adjusting for age and sex. Derived ORs were used for the weighted risk score analysis.
Ohio cohort used for the validation.
Ohio cohort that contains Ohio samples from the validation cohort (b) and from previous genome-wide association studies (22,23).
SNP, single nucleotide polymorphism.
Cumulative effect of the 5 SNPs
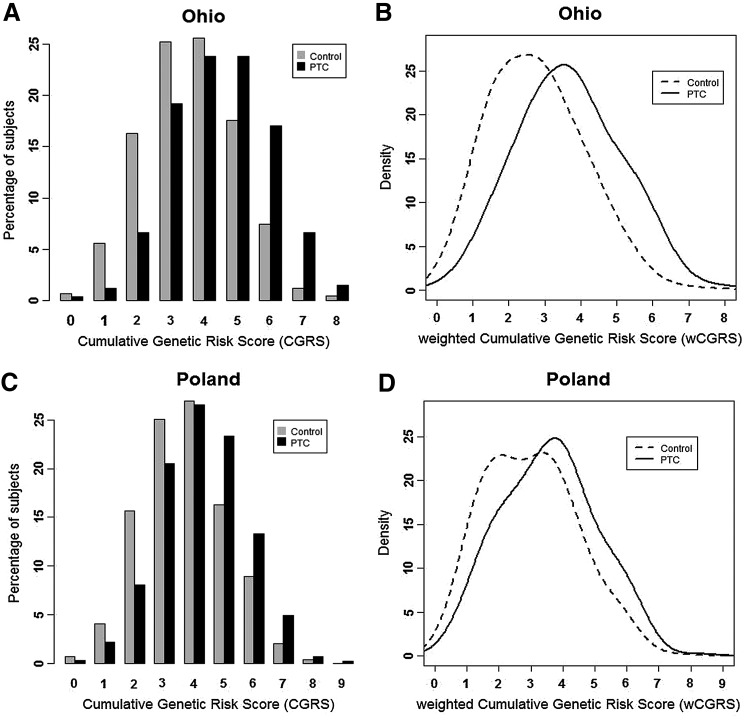
The average (±SD) of cumulative risk scores (CGRS) among OSU cases (4.46±1.47) was significantly higher than controls (3.61±1.42), with a permutation p-value <2.2×10−16 comparing the two groups (Fig. 1A). The average weighted score (wCGRS) of cases (3.68±1.32) in the Ohio cohort was significantly higher than in controls (2.77±1.46), p-value <2.2×10−16. The distribution of wCGRS in cases showed a significant shift towards higher values compared to controls with Mann–Whitney test p-value <2.2×10−16 (Fig. 1B). The Polish data followed a similar pattern, possessing a higher average of CGRS in cases (4.24±1.44) compared to controls (3.70±1.42), p-value <2.2×10−16 (Fig. 1C). The average wCGRS of Polish cases (3.57±1.5) was significantly higher compared to Polish controls (3.0±1.44) p-value <2.2×10−16, and there was a significant shift of wCGRS distribution of the cases to the right with a Mann-Whitney p-value <2.2×10−16 (Fig. 1D).
FIG. 1.
Cumulative risk scores in Ohio and Polish cohorts. (A, C) Distribution of number of risk alleles or cumulative genetic risk scores (CGRSs) between cases and controls. (B, D) Distribution of weighted cumulative genetic risk scores (wCGRSs) between cases and controls. (A, B) Ohio cohort; set of 916 controls and 605 cases without any missing genotypes were used for the analysis. (C, D) Polish cohort; set of 1663 controls and 1633 cases without any missing genotypes were used for the analysis.
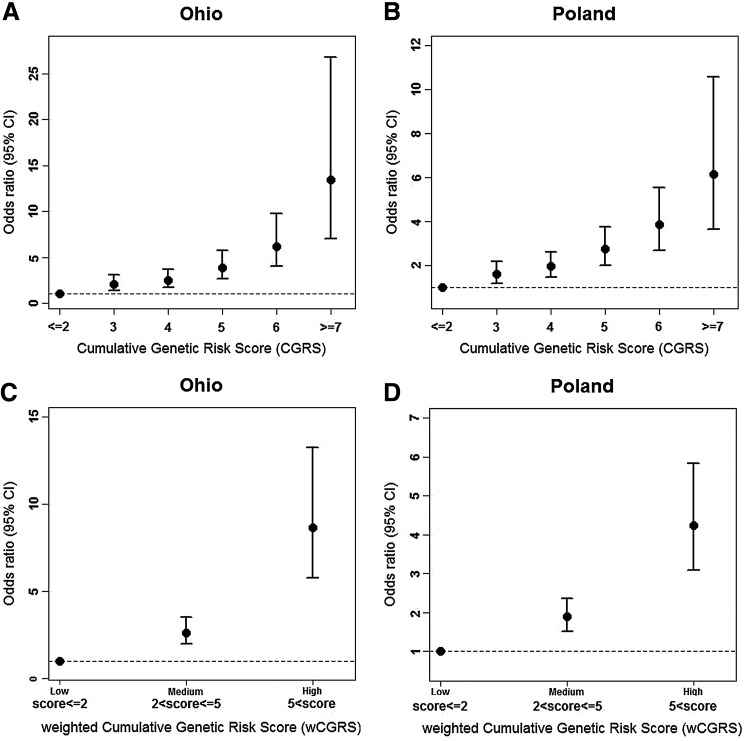
Each additional risk allele increased the odds of the disease by 1.51 [95% CI 1.40, 1.64] in Ohio and 1.35 [95% CI 1.26, 1.44] in the Polish cohort. Both cohorts provided evidence to support that having an additional disease allele increases the odds of having the disease significantly (p-value <2.2×10−16). As the CGRS increased, the odds ratio increased in both cohorts (Table 3). A positive trend in OR compared to the reference group and their 95% CIs is shown in Fig. 2A for Ohio and in Fig. 2B for Poland. As can be seen, in the presence of seven or more risk alleles, the ORs were as high as 13 and 6, respectively.
Table 3.
Association Between the Cumulative Genetic Risk Scores and Papillary Thyroid Carcinoma
| Cohort | CGRS | PTC | Control | Proportion in PTC | Proportion in control | ORa | 95% CIa | p-valuea |
|---|---|---|---|---|---|---|---|---|
| Ohio |
≤2 |
49 |
206 |
0.081 |
0.225 |
Reference |
|
|
| |
3 |
116 |
231 |
0.192 |
0.252 |
2.05 |
[1.40, 3.04] |
3.02×10−4 |
| |
4 |
144 |
235 |
0.238 |
0.257 |
2.49 |
[1.71, 3.66] |
2.56×10−6 |
| |
5 |
144 |
161 |
0.238 |
0.176 |
3.85 |
[2.62, 5.73] |
1.21×10−11 |
| |
6 |
103 |
68 |
0.170 |
0.074 |
6.19 |
[3.40, 9.71] |
8.03×10−16 |
| |
≥7 |
49 |
15 |
0.081 |
0.016 |
13.38 |
[7.02, 26.79] |
2.34×10−14 |
| Polish |
≤2 |
172 |
339 |
0.105 |
0.204 |
Reference |
|
|
| |
3 |
335 |
417 |
0.205 |
0.251 |
1.62 |
[1.20, 2.19] |
1.54×10−3 |
| |
4 |
434 |
449 |
0.266 |
0.270 |
1.96 |
[1.47, 2.62] |
4.90×10−6 |
| |
5 |
381 |
271 |
0.233 |
0.163 |
2.74 |
[2.01, 3.75] |
2.01×10−10 |
| |
6 |
217 |
148 |
0.133 |
0.089 |
3.85 |
[2.68, 5.56] |
4.80×10−13 |
| ≥7 | 94 | 39 | 0.058 | 0.023 | 6.16 | [3.65, 10.57] | 1.91×10−11 |
The OR with 95% confidence interval [95% CI] and p-values, obtained by applying multivariate logistic regression adjusting for age and sex. CGRS≤2 is used as the reference group.
FIG. 2.
Age- and sex-adjusted OR and their 95% CI for the CGRSs (A, B) and wCGRSs (C, D). The groups with CGRS≤2 and wCGRS ≤2 were set as reference groups. (A, C) Ohio cohort. (B, D) Polish cohort.
Individuals were grouped into three categories according to the weighted risk scores, low (wCGRS≤2), medium (2<wCGRS≤5), and high (wCGRS>5). The effect sizes of the medium and high groups were estimated by taking the low group as reference (Figs. 2C, 2D). Individuals in the high group were significantly more susceptible to PTC compared to individuals in the low group, with an odds ratio of 8.7 [95% CI 5.8, 13.3] in the Ohio cohort and 4.24 [95% CI 3.10, 5.84] in the Polish cohort.
Moreover we estimated (see Materials and Methods) that the five SNPS under study explained ∼11% of the familial risk of thyroid cancer in the Ohio cohort and ∼6% of the familial risk in the Polish cohort.
Predictive power of the genetic risk scores
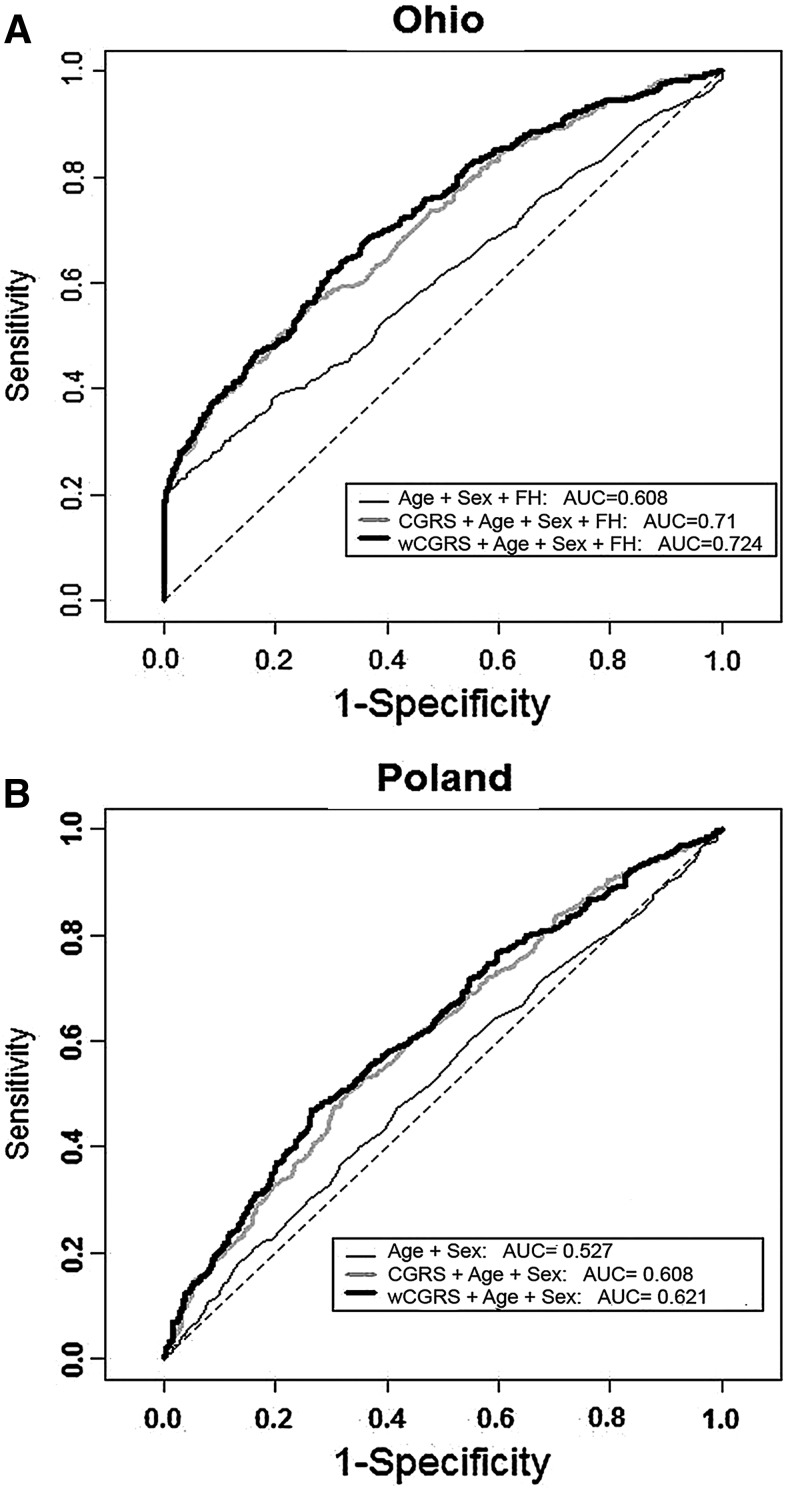
To assess the predictive power of genetic risk score models we applied ROC analysis. To eliminate the effect of the age and sex differences between cases and controls, age- and sex-matched samples were randomly selected from both cohorts. The resulting cohorts comprised a total of 1182 cases and controls from OSU and 1408 cases and controls from Poland. ROC curves and the AUC for the unweighted and weighted genetic scores are provided in Fig. 3A for the OSU samples and Fig. 3B for the Polish samples. The AUC in the models with CGRS and wCGRS are 71% and 72.4% in the OSU cohort and 60.8% and 62.1% in the Polish cohort, both cohorts providing improved models with weighted compared to unweighted scores. Family history information was included in the model for OSU cohort but not available for the Polish cohort.
FIG. 3.
ROC curves. Receiver–operator characteristic (ROC) curves assessing the discriminative power of the unweighted and weighted cumulative genetic risk score models. A random sample of age- and sex-matched cases and controls was used for each analysis. (A) The ohio cohort (cases=591, controls=591). Model was adjusted for the age, sex and family history (FH). (B) The Polish cohort (cases=704, controls=704). Model was adjusted for age and sex.
Inclusion of FVPTC and microPTC had no impact on the cumulative risk assessment
A portion of the cases were diagnosed as FVPTC (n=154 in Ohio and n=172 in Poland). We performed an independent association analysis and calculation of the cumulative risks by excluding these FVPTC cases and obtained very similar results (Supplementary Figs. S1–S3 and Supplementary Tables S2 and S3). A portion of the cases were classified as microPTC (n=118) in the Ohio cohort, but not in the Polish cohort. We performed a similar analysis in the Ohio cohort after exclusion of the microPTC cases. The results were essentially the same (Supplementary Figs. S4–S6 and Supplementary Tables S4 and S5). Thus, we propose that inclusion of FVPTC or microPTC cases did not have an undue influence on the results.
Discussion
It is becoming increasingly clear that the genetic predisposition to common diseases is multifactorial, often resulting from multiple low-penetrance variants. The bulk of information comes from GWASs. The effects of SNPs and other variants implicated in these studies are typically not yet biologically understood, but their predictive values are backed by highly significant statistics. This is the case in our study too. Of the five markers under study, one (rs944289) is well characterized in that it interferes with the transcription of a novel long intergenic noncoding RNA (lincRNAs) gene that appears to act as a tumor suppressor (7). Of the remaining four markers, two (rs966423 and rs2439302) are located in introns of coding genes, but mechanistic data are not yet available. The same is true of the remaining two SNPs, which are both intergenic (22,23). The object of this study was twofold; first, to validate the predictive value of five markers, and second, to assess the additive nature of the markers in prediction. Both aims were in fact amply fulfilled. It should be noted that all results pertain only to individuals of Caucasian, mostly Central European, descent.
PTC differs from many other cancers in several respects. Case–control studies suggest very strong familiality; according to some studies, familiality ratios are among the highest if not the highest of all cancers (1,3,4,29,30). However case–control studies ignore the fact that the environment is shared by most family members. The true heritability of PTC, as defined by twin studies, for example, has not been decisively determined (31). Unknown environmental factors could contribute significantly to the high familiality. The fact that the incidence of PTC is rising (32,33), certainly suggests an important role for changes in detection or environmental factors.
As stated in the Introduction, only a few genes or candidate genes have so far been detected in PTC even though several researchers have done extensive linkage analyses and analyzed functionally plausible candidate genes (34–39). All of these facts are compatible with the genetic predisposition to PTC being multifactorial and mostly of low penetrance. Our study supports this contention in several ways. Importantly, our data confirm the published GWAS findings in that all five SNPs studied displayed odds ratios similar to those reported previously (22,23). The two cohorts we studied were concordant, even though the odds ratios were overall lower in the Polish cohort than in the Ohio cohort. This might be due to many factors, perhaps most likely genuine biological differences between the populations, unknown environmental factors, or subtle differences in the diagnostic criteria. However, the difference could theoretically be related to the age of the studied cohorts; for example, in Ohio the cases were younger (mean 39.5 years) than the controls (46.9 years). Therefore the odds ratios were calculated after proper adjustment for age.
A comparison of our results with similar data from other cancers shows a general concordance in that most published studies of other cancers have documented an additive effect of risk markers. For instance, in a study of seven low penetrance breast cancer variants, ORs in women with the highest numbers of risk alleles rose to 8.69 and the measure of discriminative ability or AUC rose from 0.665 to 0.693 when the genetic risk score information was added to the model with conventional risk factors (10). In a breast cancer study, the AUC rose from 0.63 to 0.667 when genotype data from five markers were added to conventional risk data (11). In comparison, in our study the weighted AUCs were as high as 0.724 (Ohio) and 0.621 (Poland) based on the five SNPs alone. These data suggest that PTC low-penetrance risk alleles play a significant role. This is also evident from data on colorectal cancer, a disease with several well-known high-penetrance predisposing genes. In a study of conventional risk factors and 10 low-penetrance risk loci, the AUC for both combined was 0.59 and for the low-penetrance markers alone, 0.57 (12). The practicality of identifying a subgroup of individuals with defined absolute risk was considered by the authors who nevertheless stated that genotype data in addition to conventional risk data are not currently good enough for individualized risk prediction.
The ultimate goal of our efforts is to provide predictions that are significant enough to be used in counseling or even to inform the clinical handling of PTC patients and their relatives, as well as other individuals being evaluated for thyroid cancer risk. We surmise that this point has not been reached with the available markers. At the same time, the observed rise in overall risk with each additional risk allele makes us view the future with some optimism. For instance, by simply counting the number of risk alleles at five loci we showed that in people with seven or more risk alleles, the OR was as high as 13 in the Ohio cohort, a value that is already remarkably high. We reason that more markers will be detected. Indeed we show here that the five SNPs under study accounted for just 11% of the familial risk of thyroid cancer in Ohio, emphasizing that much remains to be discovered. We tentatively predict that this will indeed allow us to handle PTC risk assessment and intervention in a more accurate and informative way than presently.
Supplementary Material
Acknowledgments
The authors thank the OSU Comprehensive Cancer Center (OSUCCC) Microarray Shared Resource for SNP genotyping with SNaPshot assay and the University of Chicago Comprehensive Cancer Center DNA Sequencing & Genotyping Facility for Sequenom genotyping. This work was supported by National Cancer Institute Grants P30CA16058, P01CA124570, Polish National Science Center Grant NN401584838, and Foundation for Polish Science FOCUS Grant. W.G. and K.J. are supported by the TEAM Programme, co-financed by the Foundation for Polish Science and the European Union European Regional Development Fund.
Author Disclosure Statement
M.D.R. has previously been on a clinical advisory board for Veracyte. All other authors declare no potential conflicts of interest.
References
- 1.Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH.1994Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst 86:1600–1608 [DOI] [PubMed] [Google Scholar]
- 2.Dong C, Hemminki K.2001Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer 92:144–150 [PubMed] [Google Scholar]
- 3.Risch N.2001The genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomarkers Prev 10:733–741 [PubMed] [Google Scholar]
- 4.Albright F, Teerlink C, Werner T, Cannon-Albright L.2012Significant evidence for a heritable contribution to cancer predisposition: a review of cancer familiality by site. BMC Cancer 12:138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Chapelle A, Jazdzewski K.2011MicroRNAs in thyroid cancer. J Clin Endocrinol Metab 96:3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonora E, Tallini G, Romeo G.2010Genetic predisposition to familial nonmedullary thyroid cancer: an update of molecular findings and state-of-the-art studies. J Oncol 2010:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A.2012The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA 109:8646–8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A.2008Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105:7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A.2009Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA 106:1502–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sueta A, Ito H, Kawase T, Hirose K, Hosono S, Yatabe Y, Tajima K, Tanaka H, Iwata H, Iwase H, Matsuo K.2012A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Res Treat 132:711–721 [DOI] [PubMed] [Google Scholar]
- 11.Dai J, Hu Z, Jiang Y, Shen H, Dong J, Ma H, Shen H.2012Breast cancer risk assessment with five independent genetic variants and two risk factors in Chinese women. Breast Cancer Res 14:17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi L-Y, Domingo E, Smillie C, Henrion M, Frampton M, Martin L, Grimes G, Gorman M, Semple C, Ma YP, Barclay E, Prendergast J, Cazier J-B, Olver B, Penegar S, Lubbe S, Chander I, Carvajal-Carmona LG, Ballereau S, Lloyd A, Vijayakrishnan J, Zgaga L, Rudan I, Theodoratou E, Starr JM, Deary I, Kirac I, Kovacevic D, Aaltonen LA, Renkonen-Sinisalo L, Mecklin J-P, Matsuda K, Nakamura Y, Okada Y, Gallinger S, Duggan DJ, Conti D, Newcomb P, Hopper J, Jenkins MA, Schumacher F, Casey G, Easton D, Shah M, Pharoah P, Lindblom A, Liu T, Smith CG, West H, Cheadle JP, Midgley R, Kerr DJ, Campbell H, Tomlinson IP, Houlston RS.2012Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet 44:770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Haan HG, Bezemer ID, Doggen CJM, Le Cessie S, Reitsma PH, Arellano AR, Tong CH, Devlin JJ, Bare LA, Rosendaal FR, Vossen CY.2012Multiple SNP testing improves risk prediction of first venous thrombosis. Blood 120:656–663 [DOI] [PubMed] [Google Scholar]
- 14.Go MJ, Hwang JY, Kim DJ, Lee HJ, Jang HB, Park KH, Song J, Lee JY.2012Effect of genetic predisposition on blood lipid traits using cumulative risk assessment in the Korean population. Genomics Inform 10:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lango H, Palmer CNA, Morris AD, Zeggini E, Hattersley AT, McCarthy MI, Frayling TM, Weedon MN.2008Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes 57:3129–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, Manning AK, Florez JC, Wilson PWF, D'Agostino RB, Cupples LA.2008Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med 359:2208–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Zhao JH, Luan Ja, Luben RN, Rodwell SA, Khaw K-T, Ong KK, Wareham NJ, Loos RJ.2010Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr 91:184–190 [DOI] [PubMed] [Google Scholar]
- 18.Witte JS.2010Genome-wide association studies and beyond In: Fielding JE, Brownson RC, Green LW. (eds) Annual Review of Public Health. Vol. 31 Annual Reviews, Palo Alto, CA, pp. 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam RK, Zhang WW, Trachtenberg J, Seth A, Klotz LH, Stanimirovic A, Punnen S, Venkateswaran V, Toi A, Loblaw DA, Sugar L, Siminovitch KA, Narod SA.2009Utility of incorporating genetic variants for the early detection of prostate cancer. Clin Cancer Res 15:1787–1793 [DOI] [PubMed] [Google Scholar]
- 20.Park J-H, Gail MH, Greene MH, Chatterjee N.2012Potential usefulness of single nucleotide polymorphisms to identify persons at high cancer risk: an evaluation of seven common cancers. J Clin Oncol 30:2157–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Walsh K, DeWan A, Hoh J, Wang Z.2011Disease risk prediction with rare and common variants. BMC Proc 5(Suppl 9):61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K.2009Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet 41:460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Masson G, He H, Jonasdottir A, Sigurdsson A, Stacey SN, Johannsdottir H, Th Helgadottir H, Li W, Nagy R, Ringel MD, Kloos RT, de Visser MCH, Plantinga TS, den Heijer M, Aguillo E, Panadero A, Prats E, Garcia-Castano A, De Juan A, Rivera F, Walters GB, Bjarnason H, Tryggvadottir L, Eyjolfsson GI, Bjornsdottir US, Holm H, Olafsson I, Kristjansson K, Kristvinsson H T M.agnusson O, Thorleifsson G, Gulcher JR, Kong A, Kiemeney LALM, Jonsson T, Hjartarson H, Mayordomo JI, Netea-Maier RT, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Rafnar T, Stefansson K.2012Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat Genet 44:319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He H, Olesnanik K, Nagy R, Liyanarachchi S, Prasad ML, Stratakis CA, Kloos RT, de la Chapelle A.2005Allelic variation in gene expression in thyroid tissue. Thyroid 15:660–667 [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Song K, Lim N, Yuan X, Johnson T, Abderrahmani A, Vollenweider P, Stirnadel H, Sundseth S, Lai E, Burns D, Middleton L, Roses A, Matthews P, Waeber G, Cardon L, Waterworth D, Mooser V.2009Risk prediction of prevalent diabetes in a Swiss population using a weighted genetic score—the CoLaus Study. Diabetologia 52:600–608 [DOI] [PubMed] [Google Scholar]
- 26.Houlston RS, Ford D.1996Genetics of coeliac disease. QJM 89:737–743 [DOI] [PubMed] [Google Scholar]
- 27.Broderick P, Chubb D, Johnson DC, Weinhold N, Forsti A, Lloyd A, Olver B, Ma YP, Dobbins SE, Walker BA, Davies FE, Gregory WA, Child JA, Ross FM, Jackson GH, Neben K, Jauch A, Hoffmann P, Muhleisen TW, Nothen MM, Moebus S, Tomlinson IP, Goldschmidt H, Hemminki K, Morgan GJ, Houlston RS.2012Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Nat Genet 44:58–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, Lissowska J, Brinton L, Peplonska B, Southey MC, Hopper JL, McCredie MR, Giles GG, Fletcher O, Johnson N, dos Santos Silva I, Gibson L, Bojesen SE, Nordestgaard BG, Axelsson CK, Torres D, Hamann U, Justenhoven C, Brauch H, Chang-Claude J, Kropp S, Risch A, Wang-Gohrke S, Schurmann P, Bogdanova N, Dork T, Fagerholm R, Aaltonen K, Blomqvist C, Nevanlinna H, Seal S, Renwick A, Stratton MR, Rahman N, Sangrajrang S, Hughes D, Odefrey F, Brennan P, Spurdle AB, Chenevix-Trench G, Beesley J, Mannermaa A, Hartikainen J, Kataja V, Kosma VM, Couch FJ, Olson JE, Goode EL, Broeks A, Schmidt MK, Hogervorst FB, Van't Veer LJ, Kang D, Yoo KY, Noh DY, Ahn SH, Wedren S, Hall P, Low YL, Liu J, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Sigurdson AJ, Stredrick DL, Alexander BH, Struewing JP, Pharoah PD, Easton DF.2007A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet 39:352–358 [DOI] [PubMed] [Google Scholar]
- 29.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thorsteinsdottir U, Stefansson K.2004Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS Med 1:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czene K, Lichtenstein P, Hemminki K.2002Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-cancer database. Int J Cancer 99:260–266 [DOI] [PubMed] [Google Scholar]
- 31.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K.2000Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85 [DOI] [PubMed] [Google Scholar]
- 32.Albores-Saavedra J, Henson DE, Glazer E, Schwartz AM.2007Changing patterns in the incidence and survival of thyroid cancer with follicular phenotype—papillary, follicular, and anaplastic: a morphological and epidemiological study. Endocr Pathol 18:1–7 [DOI] [PubMed] [Google Scholar]
- 33.Yu GP, Li JC, Branovan D, McCormick S, Schantz SP.2010Thyroid cancer incidence and survival in the national cancer institute surveillance, epidemiology, and end results race/ethnicity groups. Thyroid 20:465–473 [DOI] [PubMed] [Google Scholar]
- 34.Malchoff CD, Sarfarazi M, Tendler B, Forouhar F, Whalen G, Joshi V, Arnold A, Malchoff DM.2000Papillary thyroid carcinoma associated with papillary renal neoplasia: genetic linkage analysis of a distinct heritable tumor syndrome. J Clin Endocrinol Metab 85:1758–1764 [DOI] [PubMed] [Google Scholar]
- 35.McKay JD, Lesueur F, Jonard L, Pastore A, Williamson J, Hoffman L, Burgess J, Duffield A, Papotti M, Stark M, Sobol H, Maes B, Murat A, Kaariainen H, Bertholon-Gregoire M, Zini M, Rossing MA, Toubert ME, Bonichon F, Cavarec M, Bernard AM, Boneu A, Leprat F, Haas O, Lasset C, Schlumberger M, Canzian F, Goldgar DE, Romeo G.2001Localization of a susceptibility gene for familial nonmedullary thyroid carcinoma to chromosome 2q21. Am J Hum Genet 69:440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavaco BM, Batista PF, Sobrinho LG, Leite V.2008Mapping a new familial thyroid epithelial neoplasia susceptibility locus to chromosome 8p23.1-p22 by high-density single-nucleotide polymorphism genome-wide linkage analysis. J Clin Endocrinol Metab 93:4426–4430 [DOI] [PubMed] [Google Scholar]
- 37.He H, Nagy R, Liyanarachchi S, Jiao H, Li W, Suster S, Kere J, de la Chapelle A.2009A susceptibility locus for papillary thyroid carcinoma on chromosome 8q24. Cancer Res 69:625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh I, Filetti S, Vriens MR, Guerrero MA, Tumino S, Wong M, Shen WT, Kebebew E, Duh Q-Y, Clark OH.2009Distinct loci on chromosome 1q21 and 6q22 predispose to familial nonmedullary thyroid cancer: A SNP array-based linkage analysis of 38 families. Surgery 146:1073–1080 [DOI] [PubMed] [Google Scholar]
- 39.Canzian F, Amati P, Harach HR, Kraimps JL, Lesueur F, Barbier J, Levillain P, Romeo G, Bonneau D.1998A gene predisposing to familial thyroid tumors with cell oxyphilia maps to chromosome 19p13.2. Am J Hum Genet 63:1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.