Abstract
Aging is a critical determinant of regenerative capacity in many organ systems, but it remains unresolved in the lung. This study examines murine lung cell dynamics during age-dependent lung regeneration. Proliferation of lung progenitor cells (EpCAMneg/Sca-1high lung mesenchymal stromal cells - LMSCs, EpCAMpos/Sca-1low epithelial progenitor cells, proSP-Cpos alveolar type II epithelial cells - AECII, and CD31pos - endothelial cells) was tracked to day 3 or 7 after pneumonectomy (PNX) or SHAM surgery in 3, 9, and 17 month mice. In 3 month mice, post-PNX LMSC proliferation peaked early (3 days), with 50%–80% more BrdU-positive cells than the other cell types, which peaked later (4–7 days). In older mice (9 and 17 month), abundance and post-PNX proliferation of LMSCs at day 3 were reduced (40%–80%). In both young and old mice, LMSCs were isolated and compared phenotypically with whole lung non-LMSCs. Donor age had no qualitative effect on the phenotype (LMSC vs. non-LMSC), with increased expression of CD90/Thy1, CD105/Eng, CD106/Vcam, CD146/Mcam, and Pdgfrα, and up-regulation of mRNA encoding Fap, Eln, Col1a1, Col3a1, Aldh1a3, Arhgef25, Dner, Fgfr1, and Midkine. However, compared with LMSCs isolated from young mice, LMSCs from older mice exhibited reduced mRNA expression of retinoic acid (Aldh1a3, Rbp4), Fgf/Wnt (Fgfr1, Sfrp1, Wnt2, and Ctnnb1), and elastogenesis (Col1a1, Eln, Fbn1, and Sdc2) pathway genes. Isolated LMSCs from older mice also demonstrated lower colony-forming units (−67%), growth potential (−60% by day 7), ALDH activity (−49%), and telomerase activity (−47%). Therefore, age is associated with declining proliferative potential and regenerative functions of LMSCs in the lung.
Introduction
Aging is a significant determinant of regenerative capacity in the lung [1–4]. The cellular and molecular mechanisms that contribute to aging effects in the lung are unknown, although as in other organ systems, age may impact the capacity of stem and progenitor cells to replicate, differentiate, or perform paracrine functions which are relevant to regeneration [5–7]. The lung is known to contain multiple resident cell populations with proliferative potential contributing to tissue regeneration and repair after injury [8]. These populations include alveolar type II epithelial (CD45neg, CD31neg, and pro-SPCpos), endothelial cells (CD45neg, CD31pos), mesenchymal stem cells (CD45neg, CD31neg, Sca-1high, and Epcamneg) [9–12], Clara cells (CCSPpos), bronchiolar epithelial progenitor cells (EpPC) (CCSPpos, Sca-1low) [13,14], colony-forming epithelial cells (CD45neg, CD31neg, Sca1low, and Epcampos) [11], and basal cells (CD45neg, CD31neg, Sca-1high, CD49high, ALDHpos; p63pos, Krt5pos, NGFRpos, and Itg alpha 6pos) [15,16]. However, there are no inclusive reports that have compared the relative cell dynamics of these proliferative populations (ie, progenitor cells) during lung regeneration. Moreover, the effect of aging on cellular dynamics is unknown.
As is seen with many mammals during periods of high somatic growth [1,17], young adult mice are capable of extensive lung regeneration after unilateral pneumonectomy [18,19]. During postpneumonectomy lung regeneration, cell proliferation occurs throughout the lung parenchyma in mesenchymal, endothelial, and epithelial compartments, as demonstrated by DNA labeling studies [19–21]. We have previously demonstrated that lung regeneration (compensatory lung regrowth after unilateral pneumonectomy) is age dependent in the mouse, with regenerative capacity declining as early as 9 months of age [4]. Moreover, aging has a profound effect on self-renewal and phenotype of resident lung mesenchymal cells, which is consistent with their role in modulating regenerative capacity in mice [4]. A previous work suggests that resident lung mesenchymal stem cells (LMSCs) can be identified using a combination of surface markers (CD45neg, CD31neg, Sca-1high, and Epcamneg) [9–12] by flow cytometry. These CD45neg, CD31neg, Sca-1high, and Epcamneg LMSCs are a heterogeneous progenitor cell population that demonstrates enrichment for clonogenic lipofibroblastic and nonlipofibroblastic cells, and they are predominantly located in the distal lung parenchyma [9–12]. In this study, we aimed at investigating the effect of age on cellular dynamics and molecular features related to self-renewal in LMSCs, in a model of age-dependent postpneumonectomy lung regeneration [4].
Materials and Methods
Study design
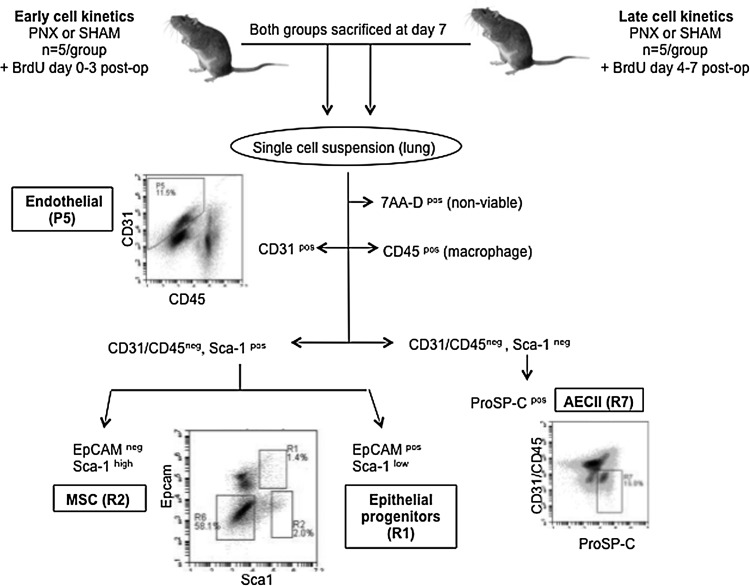
All protocols were approved by the Institutional Animal Care and Use Committee at Tufts University (IACUC protocol # G974-08 and G2011-31). The first objective of this study was to enumerate the pre-PNX abundance and post-PNX proliferation of proliferative cell populations in two discrete time periods (0–3 days; 4–7 days) after PNX. Figure 1 details the experimental design and surface markers that are used to identify each cell type by flow cytometry, including lung mesenchymal stromal cells (LMSCs), EpPC, alveolar epithelial type II cells (AECII), and endothelial cells (EPC). Left unilateral pneumonectomy (PNX) or sham thoracotomy (SHAM) surgeries were performed on 3 month C57BL6 female mice (n=5/group). For the early (day 0–3) group, the mice received BrdU from surgery until the end of day 3 post-PNX. For the late (day 4–7) group, the mice received BrdU from day 4–7. All mice (both groups) were euthanized at day 7 postsurgery for analysis using flow cytometry.
FIG. 1.
Enumeration of post-PNX proliferating cell populations by flow cytometry of 3 month C57BL/6 mice fed BrDU (day 0–3 or day 4–7 post-PNX). Cell enumeration excluded 7AA-D nonviable cells, and included endothelial, (CD45neg, CD31pos), lung mesenchymal stromal cells (MSC - CD45/CD31/EpCamnegSca1high), epithelial progenitor (CD45/CD31neg, EpCampos, Sca1low) and alveolar type II (CD45/CD31/Sca1neg, ProPS-Cpos) cells. Flow cytometry was performed on single-cell lung suspensions using an Accuri C6 and Cflow software v. 1.0.208.2; ≥100K total events per sample.
The second objective of the study was to specifically compare the abundance and post-PNX proliferation of progenitor cell populations in aged versus young mice. Using the same experimental design (Fig. 1), left unilateral pneumonectomy (PNX) or sham thoracotomy (SHAM) surgery was performed on young (3 month), middle-aged (9 month), and old (17 month) C57BL6 female mice (n=5/group). BrdU was administered to all mice (both intraperitoneal injection immediately, after surgery, and ad libitum in the drinking water) from day 0–3 (ie, “early”), and the mice were euthanized at day 7 postsurgery for analysis using flow cytometry.
The third objective of the study was to establish potential mechanisms for the age-dependent alterations in cell kinetics through a better understanding of the potential contributions of age-related changes in phenotype, proliferation capacity, self-renewal gene expression, ALDH activity, and telomerase activity of LMSCs. First, signature mesenchymal surface epitopes were compared in LMSCs isolated from young (3 month), middle-aged (9 month), and old (17 month) mice. Since the surface phenotype of LMSCs isolated from different age donors were very similar, and given past studies [4] indicating that proliferation and myofibroblastic differentiation in lung fibroblasts (LFs) decline as early as 9 months of age in mice, the remaining experiments focused on LMSCs isolated from young (3 month) and middle-aged (12 month) mice. Pro-regenerative and baseline differentiation markers were investigated using a functional gene expression profile established for LMSCs using microarray analysis (Affymetrix microarrays) on LMSCs isolated from 3 month mice, compared with non-LMSCs (depleted whole lung). The gene expression profile generated for LMSCs isolated from young mice was validated using qPCR, then compared with LMSCs isolated from middle-aged (12 month) mice. Finally, age-dependent determinants of cell proliferation were investigated in LMSCs isolated from 3 and 12 month mice using colony-forming unit–fibroblast (CFU-F), growth curves, ALDH activity, and telomerase activity.
Animals and surgical procedures used for LMSC studies
Mice used in this study were either young adult (11 week), middle-aged (9–12 months), or old (17 month) female C57BL/6 (20–35 g depending on age) obtained from Jackson Laboratories. Mice were aged in-house for both the 9 and 17 month groups (obtained as retired breeders at 7 months from Jackson Labs). The 11 week mice were also acclimatized in-house for 2 weeks before surgery. Although the young mice were virgins, all the older mice were retired breeders (purchased at 8 months of age) in this study. Since previous studies have established that an earlier pregnancy does not alter compensatory lung regrowth [22], it was assumed that aging was the major factor influencing cell behavior in this study. For surgery, mice were anesthetized by an intraperitoneal injection of ketamine (50–75 mg/kg) and xylazine (5 mg/kg), and they then received 2 mL of warmed normal saline and 100 mg/kg sodium ampicillin subcutaneously. Orotracheal intubation was performed under direct visualization using a 20-gauge catheter (BD Insyte catheter; Becton, Dickinson and Co) over a flexible stylet. Mice were secured in supine position, and mechanically ventilated (AUT6110; Buxco Electronics) at 200 tidal breaths of 0.3 mL of room air per minute, at a positive end-expiratory pressure of 3 cm H2O during surgery and recovery. After achieving adequate anesthetic depth (absence of response to toe-pinch), the left unilateral pneumonectomy (PNX) surgery was performed as previously described by our laboratory [18]. Sham-operated animals (SHAM) underwent an identical procedure, except that after the thoracotomy, the chest was left open for 5 min to simulate the conditions of the pneumonectomy group without removal of the left lung, then closed as described. For the early (day 0–3) group, BrdU was administered once by an intraperitoneal injection immediately after surgery (200 μL of 4 mg/mL BrdU in PBS), as mice receive subcutaneous fluids at the time of surgery, and, therefore, do not drink much water in the first 12 h after surgery, and then subsequently ad libitum in the drinking water (0.8 g/L BrdU) until the end of day 3 post-PNX, at which time the BrdU was removed from the drinking water. For the late (day 4–7) group, mice received BrDU ad libitum in the drinking water (0.8 g/L) from day 4–7. All mice (both groups) were euthanized at day 7 postsurgery.
Flow cytometry
The mice were anesthetized as described earlier either at 0 h (baseline) or at 7 days after surgery (PNX or SHAM), then euthanized by cervical dislocation. The pulmonary vasculature was perfused with ice-cold phosphate-buffered saline, and the trachea was cannulated. 1.5 mL of digestion media (2 mg/mL collagenase/dispase, Roche, 10269638001; 100 U/mL DNase, d4513-1vl, Sigma; 10% FBS, HyClone, alpha-MEM, Lonza) was infused through the cannula; the lungs were removed en bloc and digested in media at 37°C for 1.5 h. Cells were sequentially filtered through 100 μm and 40 μm filters, and then resuspended in red blood cell lysis solution (5 min, RT). Cells were centrifuged (120 g, 5 min), resuspended in 5% FBS alpha-MEM (4°C), and counted. 1×106 cells were used per sample, and blocked with antiCD16/32 (24G2; BD Bioscience) for 20 min. Primary and secondary antibodies were applied sequentially for 20 min at 4°C; then, the cells were resuspended in ice-cold 0.15 M sodium chloride and 95% ethanol. For BrdU staining, cells were fixed (1% paraformaldehyde, 0.01% tween-20, in PBS) at room temperature for 30 min, treated with DNase at room temperature for 15 min, resuspended in staining buffer with anti-BrdU antibody, incubated at RT for 30 min, washed 3× with staining buffer, and then run immediately. Cells were analyzed on either an FACSCalibur flow cytometer (BD Biosciences) or a BD Accuri C6 flow cytometer (BD Biosciences).
Immunohistochemistry
The mice were anesthetized as described earlier at 3 days after surgery, then euthanized by cervical dislocation. After median sternotomy, the pulmonary vasculature was perfused with ice-cold Hanks balanced salt solution, the trachea was cannulated, and the lungs were removed en bloc. The lungs were fixed via an intratracheal infusion of 10% buffered formalin at 25 cmH20 overnight. The trachea was then ligated, and the lung was embedded in paraffin. Immunofluorescent staining (IF) was performed on 5 μm paraffin sections. Primary antibodies (all used at 1:100) included rabbit anti-CD45 (Abcam; ab10558), rabbit anti-CD31 (Abcam; ab28364), rabbit anti-EPCAM (Abcam; ab71916), goat anti-Sca1 (R&D systems; AF1226), and rat anti-BrdU (Abcam; ab6326). Tissue sections were deparaffinized and hydrated using standard methods, and antigen retrieval was performed using a citrate buffer (pH 6.0) and vegetable steamer heating (prewarmed buffer, 20 min). Tissues were washed thrice in PBS before blocking for 2 h with 10% donkey serum, and then exposed to the primary antibodies (15–18 h at 4°C). Detection of the primary antibodies was achieved using donkey anti-rabbit Alexa Fluor 488 (green), donkey anti-rat Alexa Fluor 488 or 594, and donkey anti-goat Alexa Fluor 488 or 350 at 1:200 (30 min at 37°C). No primary (isotype only) and secondary-only control assays were performed; nonspecific staining was not observed.
Magnetic-activated cell sorting
For all studies except for the initial microarray analysis (see details below), LMSCs (CD45neg, CD31neg, EpCamneg, and Sca1pos) were isolated using magnetic-activated cell sorting (MACS). Cells were grown, isolated, and counted as described earlier. Cells were then blocked with 24G2 for 20 min. Primary antibodies (Sca-1 FITC, CD45-biotin, CD31-biotin, and EpCAM-biotin) were added and incubated for 20 min. After washing, 10 mL per 107 cells of anti-biotin beads were added and incubated at 4°C for 15 min. Cells were washed again, and processed through the cell sorter (autoMACS Pro Separator; Milteny Biotech) using the deplete option. The unlabeled fraction (CD45neg, CD31neg, and EpCAMneg) was then resuspended, stained as before using anti-FITC beads, and processed through the cell sorter using the possel option. The positive fractions (CD45neg, CD31neg, EpCAMneg, and Sca1pos) were resuspended, counted, and either plated at 2×105 cells/150 mm plate or processed for protein or RNA. To check purities after MACS, and initial cell culture, a small portion of each fraction was stained with streptavidin-linked fluorochrome and assessed for the presence or absence of staining in the appropriate fractions using flow cytometry. CD45neg, CD31neg, EpCAMneg, and Sca1pos cells represented more than 92% of the total cell population (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/scd).
Microarray
After tissue preparation as described earlier, cells were isolated by flow cytometry (CD45neg, CD31neg, and Sca1high) and immediately stored in RNAprotect Cell reagent (Qiagen #76526). Sorted cells from 15 animals were pooled for each group (LMSCs, or depleted whole lung) to minimize biological variability [23]. Total RNA was prepared from the isolated cells using the Qiagen RNeasy plus mini kit (#74134) according to the manufacturer's directions. Total RNA concentrations, A260/A280 ratios, and RIN values were determined using an Agilent 2100 Bioanalyzer (Aglient Technologies). All samples had an RIN number >8. All microarray analysis was performed as described in the Affymetrix GeneChip Expression Analysis Technical Manual using Affymetrix Mouse Genome 430 2.0 GeneChips and the One-Cycle cDNA Synthesis and HT IVT Labeling kits (Affymetrix, Inc.).
Microarray analysis
Biologically pooled samples (1 array/condition) were analyzed statistically using the S-Score algorithm previously described [24–27], and they have shown good sensitivity when compared with many other existing analysis methods without sacrificing specificity (including RMA, dChip, and MAS5). This is particularly applicable and appropriate to our individual time point datasets in which we have only one paired array set for each time point and has been described by our laboratory in previous analyses [19]. By definition, S-score is related to P-value by an exponential relation, and a value of three corresponds to a P-value of 0.003 [24,27]. Genes with an S-score ≥3.0 or ≤−3.0 (P≤0.003) and a fold-change ≥1.5 or ≤−1.5 were selected for further analysis.
Quantitative reverse-transcription PCR validation
Total RNA from cells (LMSCs or depleted whole lung) obtained by MACS from individual mice (n=5/group) was isolated as described earlier. The RNA from each individual (no pooled samples) was then subjected to genomic DNA elimination and first-strand cDNA synthesis using a commercial kit (RT2 First Strand Kit; SA Biosciences #330401) to generate the cDNA templates for PCR amplification. Quality control was performed using the SA Biosciences QC qRT-PCR array (SA Biosciences) to test for any inhibition of cDNA synthesis, or the presence of genomic DNA contamination. Gene expression assays were performed using sets of premade mouse primer pairs (SABiosciences) (see Table 1). Quantitative PCR was performed using a ABI 7500 Detection system, and RT2 qPCR SYBR green PCR Master Mix (SA Biosciences), according to the manufacturer's recommended protocol. Each sample was analyzed in triplicate, and relative gene expression was calculated using the comparative Ct method [28] after normalization to the housekeeping gene Gapdh, which did not show differences in expression between any of the samples analyzed.
Table 1.
Comparison of Fold Changes in Gene Expression of LMSCs Versus LMSC-Depleted Whole Lung Cells Isolated from Either Middle-Aged or Young Mice and Analyzed Using Microarray and Quantitative Polymerase Chain Reaction
| |
3 month LMSC vs. depleted whole lung |
12 month LMSC vs. depleted whole lung |
12 month vs. 3 month LMSC |
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | array | Sscore | qPCR | t-test | qPCR | t-test | qPCR | t-test | SABiosciences Cat# for PCR primers |
|
Aldh1a3 |
11.10 |
4.44 |
145.81 |
1.24E-07 |
99.29 |
2.30E-07 |
−2.02 |
4.47E-06 |
PPM31491A |
|
Arhgef25 |
77.00 |
3.38 |
19.52 |
1.38E-07 |
19.21 |
2.05E-08 |
−2.11 |
5.50E-05 |
PPM26321A |
|
Col1a1 |
13.40 |
3.48 |
41.72 |
7.25E-09 |
53.52 |
2.73E-09 |
−7.85 |
2.15E-07 |
PPM03845F |
|
Col3a1 |
4.60 |
3.36 |
37.31 |
7.44E-08 |
43.03 |
1.62E-05 |
−1.39 |
4.27E-02 |
PPM04784B |
|
Ctnnb1 |
n/s |
n/s |
1.54 |
1.38E-02 |
−1.14 |
2.53E-01 |
−14.63 |
1.78E-07 |
PPM03384A |
|
Dner |
7.00 |
3.64 |
35.23 |
8.29E-11 |
18.38 |
1.26E-06 |
−2.82 |
1.35E-06 |
PPM38109A |
|
Eln |
9.30 |
3.95 |
25.42 |
1.22E-08 |
16.59 |
4.70E-08 |
−6.43 |
3.48E-07 |
PPM36834B |
|
Fbn1 |
36.20 |
4.06 |
35.53 |
1.58E-08 |
24.28 |
6.39E-09 |
−3.29 |
2.10E-05 |
PPM36411E |
|
Fgfr1 |
4.10 |
4.25 |
13.49 |
1.91E-07 |
19.33 |
6.99E-07 |
−601.84 |
3.37E-09 |
PPM03039F |
|
Fn1 |
3.30 |
3.03 |
7.94 |
1.27E-08 |
8.28 |
8.98E-10 |
−1.18 |
1.34E-02 |
PPM03786A |
|
Mdk |
12.30 |
3.74 |
11.31 |
2.37E-07 |
25.23 |
5.06E-08 |
2.31 |
9.63E-06 |
PPM03800D |
|
Mmp2 |
11.20 |
4.23 |
32.91 |
7.30E-09 |
46.06 |
8.77E-10 |
−2.42 |
5.88E-06 |
PPM03642C |
|
Mmp14 |
6.50 |
3.68 |
25.96 |
1.69E-07 |
20.73 |
4.71E-09 |
−2.91 |
4.76E-06 |
PPM03617D |
|
Pdgfrα |
9.00 |
4.11 |
11.27 |
9.97E-10 |
15.75 |
1.90E-08 |
3.21 |
1.81E-06 |
PPM03640D |
|
Peg3 |
13.00 |
3.01 |
17.97 |
3.54E-10 |
19.59 |
3.97E-08 |
−2.80 |
1.24E-06 |
PPM39006A |
|
Pi16 |
205.00 |
4.12 |
179.26 |
1.99E-07 |
49.46 |
2.49E-09 |
−52.04 |
4.35E-07 |
PPM27839A |
|
Rbp4 |
9.60 |
4.11 |
16.49 |
2.61E-09 |
17.68 |
2.18E-08 |
−3.26 |
5.55E-06 |
PPM39030B |
|
Sdc2 |
14.40 |
4.04 |
11.90 |
6.72E-09 |
18.07 |
1.96E-07 |
−22.66 |
2.72E-08 |
PPM34226F |
|
Sfrp1 |
15.30 |
3.61 |
22.23 |
7.33E-10 |
15.69 |
4.30E-07 |
−26.40 |
1.16E-07 |
PPM04438C |
|
Sod3 |
20.70 |
4.32 |
15.33 |
7.15E-10 |
12.95 |
2.39E-08 |
−2.15 |
2.21E-05 |
PPM04365C |
|
Twist2 |
15.00 |
4.14 |
165.54 |
9.54E-08 |
53.79 |
4.25E-07 |
−39.79 |
1.19E-08 |
PPM25525F |
|
Vim |
n/s |
n/s |
3.58 |
2.20E-06 |
3.24 |
4.46E-05 |
1.33 |
3.64E-02 |
PPM04780B |
| Wnt2 | 6.00 | 3.53 | 9.30 | 2.09E-08 | 11.41 | 2.94E-05 | −14.69 | 4.37E-06 | PPM03833A |
Note: S-score statistics (see Materials and Methods section) were used to compare the microarray data for LMSCs versus depleted whole lung, while comparative paired t-tests were used to compare qPCR data for LMSCs versus depleted whole lung.
LMSCs, lung mesenchymal stromal cells.
Clonogenicity and growth potential
LMSCs from young (3 month) and middle-aged (12 month) mice were isolated using MACS (n=5/group) as described earlier, and then 2000 cells were plated (using alpha-MEM, 15% FBS, 2 mM L-glutamine, penicillin, streptomycin, and amphotericin) on 100 mm polystyrene plates. For the CFU-Fs, 2000 cells/plate, n=3 plates were used for enumeration of colonies of >50 cells (>5 doublings). For the growth curve analysis, sorted cells were plated at different densities (500, 1000, or 2000 cells/plate, n=3 plates), and were analyzed at either day 1, 3, 5, or 7 using crystal violet staining to assess cell growth.
ALDH activity
LMSCs from young (3 month) and middle-aged (12 month) mice were isolated using MACS (n=5/group) as described earlier, plated (using alpha-MEM, 15% FBS, 2 mM L-glutamine, penicillin, streptomycin, and amphotericin) on 100 mm polystyrene plates, and grown to log phase. An assay of ALDH activity (Aldefluor assay) was performed in triplicate according to the manufacturer's directions (Aldagen). Briefly, cells were washed 2× with PBS, tripsonized (TrypLE; Invitrogen) and then washed from the plate with 10% FBS in alpha-MEM. Cells were counted and then resuspended in ALDEFLUOR Assay Buffer at 1×106 cells/mL. Five micro liter DEAB solution was used to control tubes. Cells were incubated in Aldefluor reagent at 37°C for 45 min, and then resuspended in Assay Buffer. At least 100,000 events were recorded for each sample (in triplicate). Aldefluor activity was reported as a percent of fluorescence above baseline.
Telomerase activity
A quantitative PCR-based assay to evaluate endogenous telomerase activity was performed according to the manufacturer's directions (QTD kit #MT3011; Allied Biotech). Briefly, LMSCs from young (3 month) and middle-aged (12 month) mice were isolated using MACS (n=5/group) as described earlier. Total protein was extracted from 1×106 cells, and quantified using a standard Pierce BCA protein assay kit (#23221; Thermo Fisher Scientific). Protein was also extracted from 1×106 mouse embryonic fibroblast cells (MEFs; ATCC) and differentiated LFs (ATCC) as controls. Equal amounts of protein were then used in a one-step assay in which total protein was added to a mastermix containing oligonucleotides for the telomerase step, as well as DNA polymerase and nucleotides needed to amplify the extension products for analysis. The mix was heated to 37°C for 20 min to assess the ability of the endogenous telomerase to add a varied number of telomeric repeats (TTAGGG) onto the 3′ end of the substrate oligonucleotide. These extension products were then quantified by standard SYBR green-based qPCR in the same tube. The Ct values obtained by qPCR were compared with a standard curve generated from controls provided by the manufacturer, which then provides a distribution of telomerase molecules/reaction for each sample.
Results
Postpneumonectomy lung regeneration is associated with early peak proliferation in LMSCs
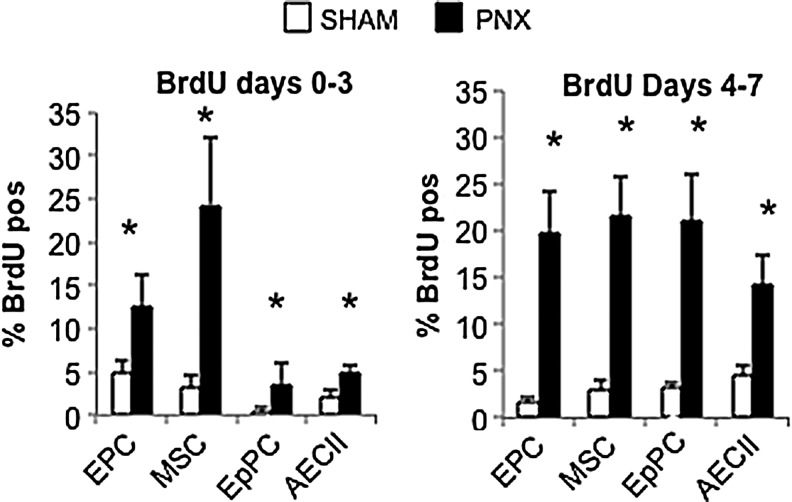
Post-PNX proliferation of distal lung progenitor cell populations, including endothelial (EPC), mesenchymal stromal (LMSC), epithelial (EpPC), and alveolar type II (AECII) cells, was assessed in young adult (3 month) mice, as shown in Fig. 1. As shown in Fig. 2, the post-PNX proliferation of LMSCs reached maximum cell density in the earlier of the two periods (day 0–3). Although all cell types examined showed a significant increase in BrdU incorporation (compared with SHAM) during this period, LMSCs demonstrated approximately 50%–80% more frequent BrdU incorporation than other cell types. In contrast, at the later time point (BrdU incorporation from day 4–7), all cell types examined demonstrated similar levels of post-PNX BrdU uptake. As illustrated in Fig. 3, proliferation of LMSCs early during the regenerative process (day 3 after PNX) occurred predominantly in the alveolar parenchyma. Although Sca1-positive, CD45/CD31/EpCAM negative cells were present in both perivascular and alveolar parenchymal locations, BrdU incorporation in this subset of cells would be found only along alveolar septae (Fig. 3).
FIG. 2.
Enumeration of post-PNX proliferation (BrDUpos) of cells in 3 month mice at two time periods (day 0–3, day 4–7) after PNX or SHAM surgery, including endothelial progenitor cells (EPC - CD45neg, CD31pos), lung mesenchymal stromal cells (MSCs - CD45/CD31/EpCamnegSca1high), epithelial progenitor cells (EpPC - CD45/CD31neg, EpCampos, and Sca1low), and alveolar epithelial type II cells (AECII - CD45/CD31/Sca1neg, ProPS-Cpos). (A) Early (day 0–3) and (B) later (day 4–7) response to PNX. Proliferation of MSCs peaked earliest. For each group n=5, and data are presented as mean and standard deviation (P<0.05* vs. control).
FIG. 3.
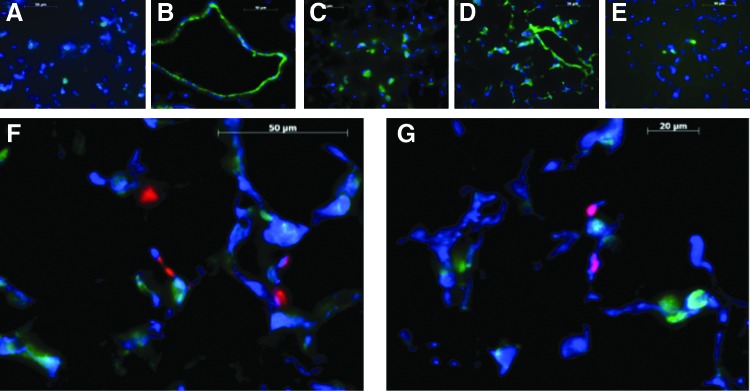
Immunohistochemical locatization of proliferating lung mesenchymal stromal cells (LMSCs) (Sca1-positive, CD45/CD31/EPCAM-negative, BrdU-positive cells) 3 days after pneumonectomy in 3 month mice. (A)–CD45 (green), DAPI (blue); (B)–CD31 (green), DAPI (blue); (C)–EPCAM (green), DAPI (blue); (D)–Sca1 (green), DAPI (blue); (E)–BrdU (green), DAPI (blue); (F), (G)–CD45/CD31/EPCAM (green), Sca1 (blue), BrdU (red). The combined staining illustrates the topographic location of proliferating LMSCs within the lung alveolar parenchyma during postpneumonectomy regeneration. Color images available online at www.liebertpub.com/scd
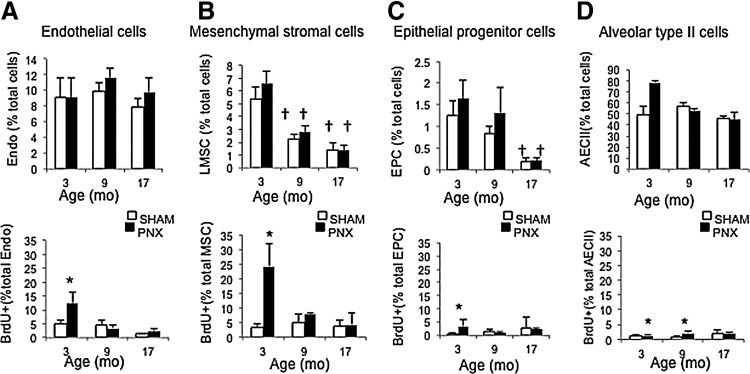
Older mice have lower abundance before PNX and lower post-PNX proliferation of LMSCs
To examine the effect of aging on proliferation of LMSCs, PNX and SHAM surgeries were performed on older (9 and 17 month) mice, and BrdU incorporation was quantified in the early postsurgery period (BrdU given from day 0–3) when LMSC proliferation peaks in younger mice, as shown in Fig. 4. Compared with young adult (3 month) mice, middle-aged (9 month) and old mice (17 month) demonstrated a significant (P<0.05) reduction in both pre-PNX abundance and post-PNX expansion of LMSCs (40%–60%, and 50%–80% reduction, in 9 and 17 month old mice, respectively). Abundance of EpPC cells also decreased with age, and post-PNX proliferation of EpPCs and EPCs was decreased with age. In contrast, post-PNX proliferation of AECII was retained in 9 month mice, although absent in 17 month mice. In summary, both the total abundance and proliferation after PNX of LMSCs (days 0–3) were reduced by middle age (9 months), in accordance with past demonstrations of declining regenerative capacity in similar age groups [4]. Notably, the total abundance of LMSCs and EPCs also decreased with age in the SHAM groups, which may represent age-related differentiation and/or senescence of these populations [4]. The proliferation of AECIIs was maintained in 9 month mice equivalent to 3 month mice, consistent with a previous study using the PNX model [4].
FIG. 4.
Enumeration of total abundance and day 0–3 proliferating (BrDUpos) cells in 3, 9, and 17 month mice after PNX or SHAM surgery. (A) Endothelial cells (Endo - CD45neg, CD31pos), (B) lung mesenchymal stromal cells (LMSCs - CD45/CD31/EpCamnegSca1high), (C) epithelial progenitor cells (EPC - CD45/CD31neg, EpCampos, Sca1low), and (D) alveolar epithelial type II cells (AECII - CD45/CD31/Sca1neg, ProPS-Cpos). Abundance (% total) of LMSCs and EPCs, and proliferation of LMSCs and endothelial cells declined with age. For each group n=5, and data are presented as mean and standard deviation; *P<0.05 versus SHAM, †P<0.05 versus 3 month.
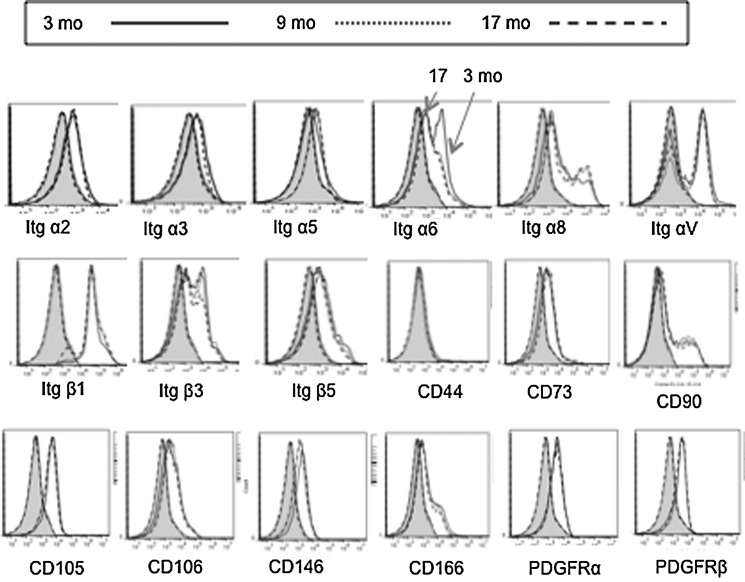
LMSCs isolated from young, middle-aged, and old mice have similar phenotypic profile, but differ in expression of genes encoding self-renewal and differentiation functions
LMSCs isolated from young adult (3 month), middle-aged (9 month), and old (17 month) mice were assessed for surface phenotype, focusing on traditional mesenchymal markers and cell-cell and cell-matrix adhesion molecules, as these may contribute to “outside-in” signaling leading to migration or cell proliferation. As illustrated in Fig. 5, LMSCs (>90%) isolated from both young and old mice expressed Itgα2, Itgα3, Itgα5, Itgα6, Itgα8, ItgαV, Itgβ1, Itgβ3, Itgβ5, CD73, CD90, CD105, CD146, CD166, Pdgfrα, and Pdgfrβ (Fig. 5). Interestingly, the only significant phenotypic difference detected by flow cytometry was Itga6 (laminin receptor), which was expressed with lower frequency in older mice.
FIG. 5.
Molecular phenotype of LMSCs isolated from 3, 9, and 17 month mice (n=5/group) analyzed using flow cytometry. Note that only Itga6 shows significant alteration with age. Flow cytometry was performed on single-cell lung suspensions using an Accuri C6 and Cflow software v. 1.0.208.2; ≥100K total events per sample. The shaded portion of each graph represents fluorescence from the isotype controls.
Mesenchymal stromal cells characterized in other organ systems often express genes encoding functions such as stemness, proliferation, migration, differentiation potential, and cell-matrix interactions [10,29,30]. In order to identify specific functional characteristics of LMSCs isolated in this study that differ from other cells in the lung, a microarray analysis was performed on pooled samples of LMSCs (Sca-1pos CD31neg, CD45neg, and EpCAMneg) isolated from 3 month-old mice (n=15), and compared with all negatively selected cells in the lung. Genes that were highly over-expressed in LMSCs were verified statistically using quantitative PCR, and these genes are listed in Table 1. These genes include the mesenchymal signature genes that encoded Pdgfrα, Fap, Eln, Col1a1, and Col3a1; the pro-regenerative and survival related genes Aldh1a3, Arhgef25, Dner, Fgfr1, Midkine, Peg3, Sod3, and Twist2. Interestingly, two genes that were very highly expressed in LMSCs and therefore may represent unique phenotypic markers include Pi16 and Twist2. Quantitative PCR was then performed on the same populations of cells isolated from middle-aged (12 month) mice, and demonstrated similar over-expression of most of these genes in LMSCs compared with the rest of the lung (Table 1). These data confirm that LMSCs with the same phenotypic profile (compared with the rest of the depleted whole lung) are present in young and middle-aged mice, despite differences in their post-PNX proliferative capacity. However, when gene expression levels were compared directly between LMSCs isolated from 12 versus 3 month mice, there were significant differences in the degree of gene expression. Specifically, all genes involved in retinoic acid signaling (Aldh1a3, Rbp4), genes involved in Fgfr1/Wnt signaling (Fgfr1, Sfrp1, Ctnnb1, and Wnt2), and genes involved in extracellular matrix formation (Col1a1, Eln, Fbn1, and Sdc2) were down-regulated in LMSCs isolated from 12 month mice compared with LMSCs isolated from 3 month mice (Table 1).
LMSCs isolated from middle-aged (12 month) mice exhibit reduced clonogenicity, ALDH activity, and telomerase activity
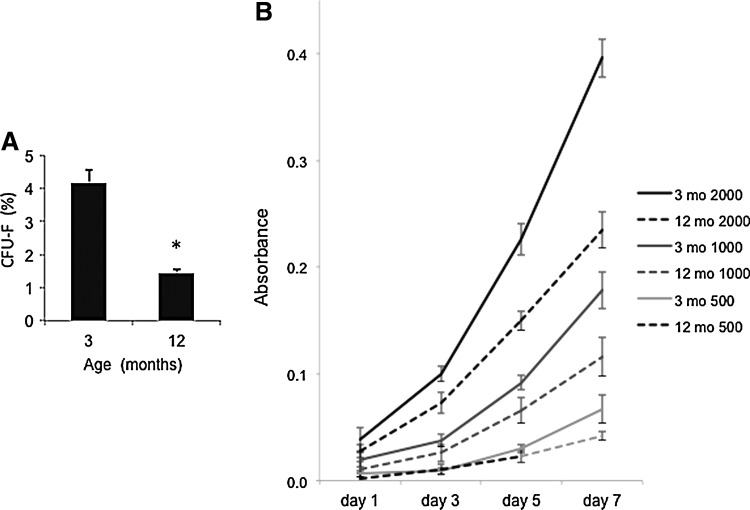
Since both regenerative capacity of the lung [4] and post-PNX proliferation of LMSCs decline at middle age (ie, 9 month), the remainder of the study focused on comparing LMSCs isolated from middle-aged mice (12 month) with those isolated from young adult (3 month) mice. Histologically, LMSCs isolated from adult (3 month) and middle-aged (12 month) mice have similar appearances, as illustrated in Fig. 6. To test the clonogenicity of LMSCs isolated from both 3 month and 12 month mice in vitro, CFU-F were counted in both cell populations before passage. As illustrated in Fig. 7A, LMSCs isolated from 3 month mice exhibited higher CFU-F (4.2%), versus LMSCs isolated from 12 month mice (1.4%, P<0.05). To further examine these differences, growth curves were performed using three different initial cell plating densities (500, 1000, and 2000 cells/plate), and cell proliferation was followed over 7 days. As shown in Fig. 7B, at all three plating densities, and at all time points, cell proliferation was lower in LMSCs isolated from older mice (>60% reduction by day 7). Density dependence of cell proliferation was not observed at either age.
FIG. 6.
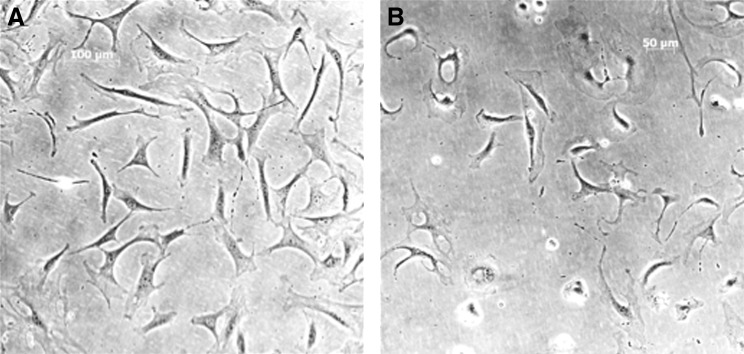
(A) Phase contrast photomicrograph of passage 0 LMSCs isolated from 3 month mice and cultured for 4 days (αMEM). (B) Phase contrast photomicrograph of passage 0 LMSC isolated from 12 month mice and cultured for 4 days (αMEM).
FIG. 7.
(A) Comparison of passage 0 CFU-F using LMSCs (2000 cells/plate) isolated by MACS from 3 and 12 month-old mice. *P<0.05; n=3 experiments/group. For each group, data are presented as mean and standard deviation. (B) Comparison of growth curves from 3 and 12 month passage 0 LMSCs, plated at three different densities (500, 1000, and 2000 cells/plate) and quantified using crystal violet staining at either 1, 3, 5, or 7 days after initial plating. P<0.05 for all time points (except day 1) at all plating densities in both young and old LMSCs. For each group, n=3 experiments/group, and data are presented as mean±standard deviation.
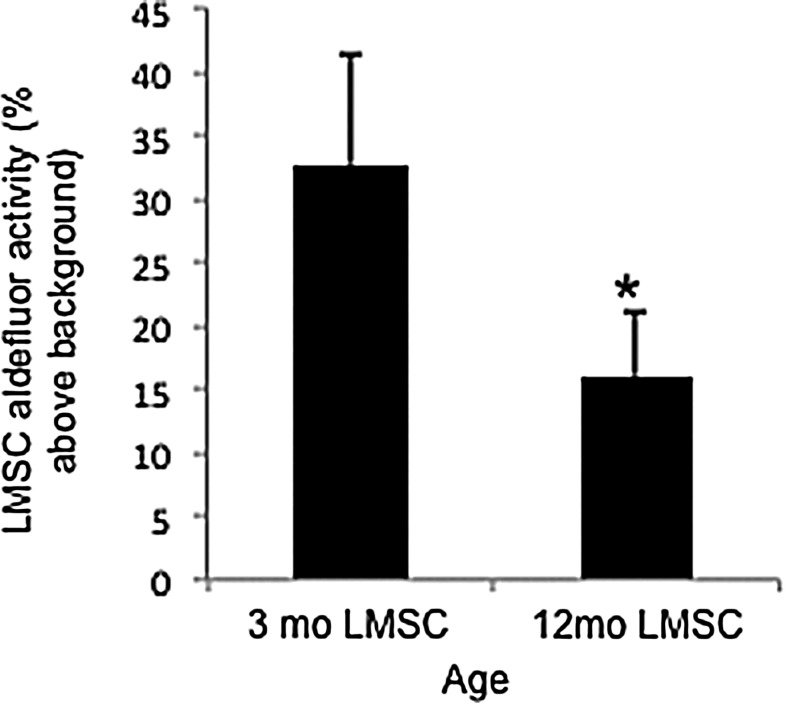
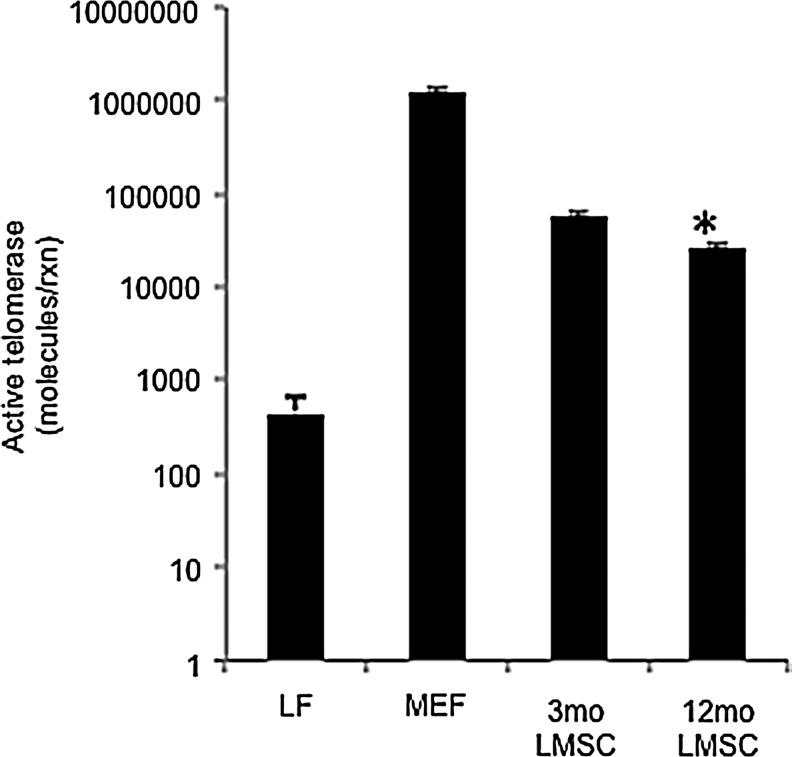
Both ALDH activity and telomerase activity were assessed in freshly isolated and cultured LMSCs obtained from both 3 and 12 month mice. As shown in Fig. 8, LMSCs isolated from 12 month mice exhibited less ALDH activity (−49%) versus 3 month mice. This is consistent with lower mRNA expression of Aldh1a3 in 12 month mice (see Table 1). Figure 9 shows telomerase activity assessed in LMSCs isolated from 3 and 12 month mice, and compared with cell lines with known high (MEFs) and low telomerase activity (differentiated human LFs). LMSCs isolated from 12 month mice exhibited significantly lower telomerase activity (−47%) than 3 month old mice. The two control cell lines flank the LMSCs in terms of telomerase activity as anticipated, with ATCCs human LFs showing extremely low levels and MEFs showing significantly higher telomerase activity. Thus, LMSCs exhibit relatively high telomerase activity that declines with age.
FIG. 8.
Comparison of ALDH activity in LMSCs isolated from either 3 or 12 month mice. Values are expressed as percent above background fluorescence. *P<0.05, n=3 replicates/group. For each group, data are presented as mean and standard deviation.
FIG. 9.
Comparison of telomerase activity in LMSCs isolated from either 3 or 12 month mice. Values are expressed on a log scale as molecules/reaction. P<0.05, n=3 replicates/group. Controls for high telomerase activity (mouse embryonic fibroblasts–MEFs) and low telomerase activity (ATCC differentiated lung fibroblasts - LFs) were included for comparison. For each group, data are presented as mean and standard deviation.
Discussion
In this study, we have demonstrated that lung regeneration after pneumonectomy, also referred to as compensatory lung growth, is associated with a dramatic early (day 0–3) proliferation of Sca-1pos LMSCs; whereas proliferation of other lung parenchymal cell populations, including both endothelial and epithelial cells, peaks later in the post-PNX regenerative process (days 4–7). This represents the first report comparing the dynamics of lung mesenchymal cell proliferation after pneumonectomy with the proliferation of other epithelial and endothelial progenitor cell populations as a function of age, and provides a novel insight into the role of these cell populations during early lung regeneration. These findings are consistent with a previous work demonstrating an early mesenchymal gene signature during post-PNX lung regeneration [20], as well as the pivotal role for LFs in fetal alveolar development and adult regenerative alveolarization [31–34]. A role for proliferation of other cell types after pneumonectomy is also clear from our study, and from previous work [19,35]. However, in this current study, we specifically analyzed the role of proliferating LMSCs during lung regeneration due to their early and marked proliferation in the regenerative process relative to the other cell populations examined.
Phenotypic and functional aspects of LMSCs were examined in this study with a particular reference to their brisk proliferative potential post-PNX in vivo. The phenotype was characterized by immunophenotype and global gene expression (both microarray and focused qPCR). The LMSCs isolated and characterized kinetically in this study possessed a typical mesenchymal stem cell phenotype based on their expression of cell surface markers (Itgα2, Itgα3, Itgα5, Itgα6, Itgα8, ItgαV, Itgβ1, Itgβ3, Itgβ5, CD73, CD90, CD105, CD146, CD166, Pdgfrα, and Pdgfrβ), as well as gene expression profile, including over-expression of genes that encode MSC phenotypic proteins (CD90/Thy1, CD105/Eng, CD106/Vcam, CD146/Mcam, Pdgfrα, Fap, Eln, Col1a1, and Col3a1), self-renewal and survival genes (Aldh1a3, Arhgef25, Dner, Fgfr1, Midkine, Peg3, Sod3, and Twist2). These findings are consistent with previous characterizations of LMSC and LF progenitor cell populations isolated in a variety of ways, including Sca1high Epcamneg cells [11], explant outgrowth MSCs [36], adherent cells isolated from dissociated tissue [29,34], and side population cells [10]. Furthermore, several studies, including this current article, have characterized LMSC populations as having high levels of clonogenicity (CFU-Fs) [4,11,36]. The LMSCs also have the ability to synthesize key provisional and mature matrix proteins, suggesting that early proliferation after PNX may supply cells with synthetic capacity in areas of the lung undergoing intense growth. In addition, we identified very high levels (over 200-fold) of a unique gene (Pi16) that has not been previously attributed to LMSCs. Expression of Pi16 as well as Twist2 may be useful in future studies for lineage tracing LMSCs after PNX, following development of the appropriate transgenic reporter mice. Topographically, we observed that LMSCs are predominantly located in the alveolar parenchyma and perivascular location as previously described [9–12]. However, only those LMSCs along the alveolar septae could be observed to proliferate after PNX, using BrdU incorporation in vivo. Thus, LMSCs defined using immunofluorescence using the same epitope staining strategy employed for magnetic sorting of LMSCs, revealed that LMSCs expand mainly in the alveolar compartment, suggesting that they contribute to the alveolar multiplication and/or elongation that occurs during postpneumonectomy compensatory lung growth in mice. It is possible that the topographical distribution of LMSCs using this staining strategy might differ in aged animals, but this was not addressed in this study. In the future, the development of unique markers such as Pi16 and Twist2, as implied by gene expression, may allow more precise topographical localization of LMSCs within the lung. However, neither of these proteins could be detected using currently available reagents (data not shown).
In this study, we have also demonstrated that LMSCs are less abundant before PNX, and proliferate poorly after PNX in older animals. Specifically, the total abundance of LMSCs dramatically declines as early as 9 months of age, and LMSCs no longer demonstrate the dramatic early (day 3) peak in proliferation after PNX, demonstrating that the age effect on self-renewal in vivo is pronounced at the same time when regenerative capacity slows down [4]. In contrast, total abundance of endothelial and EpPCs as well as alveolar type II cells were unchanged by 9 months, although both endothelial and EpPC populations also failed to proliferate in both 9 and 17 month mice. Interestingly, alveolar type II cells continue to proliferate in response to PNX in 9 month-old mice, suggesting that the proliferation of these cells is not an early age-related (ie, middle age) “rate-limiting feature” of lung regeneration. The dramatic decline in total abundance and post-PNX LMSC proliferation reported here is consistent with our previous work demonstrating that lung regeneration is age dependent and associated with reduced LMSC proliferation in vitro, and increased myofibroblastic differentiation characteristics (eg, alpha smooth muscle actin expression) in vivo and in vitro [4]. An age-dependent decline in the self-renewal capacity and differentiation characteristics of resident progenitor cells has also been identified in other organs [7,37–39], and has been linked to declining regenerative capacity [38]. However, the effects of age on lung progenitor cell biology are not well understood, and are currently limited to the study of epithelial cell populations in the context of idiopathic pulmonary fibrosis [6].
Although LMSCs isolated from older mice exhibit similar surface marker phenotypes as LMSCs isolated from younger mice and thus can be considered the same population of cells, there are significant differences in cell function that may relate to the decline in regenerative responses in aging mice. In particular, there is reduced ALDH activity, reduced telomerase activity, down-regulation of key genes associated with retinoic acid and Fgf/Wnt pathways, as well as ECM (ie, collagen synthesis, elastogenesis). These data suggest that loss of self-renewal in LMSC with age may be associated with both intrinsic mechanisms (senescence) and extrinsic mechanisms related to cell-matrix or cell-matrikine interactions. Therefore, epigenetic mechanisms should be considered in future studies. In this regard, we assumed that LMSCs remained in the same location (niche) in younger versus older mice. However, this assumption should be tested, as translocation of LMSCs with age might introduce microenvironmental influences on their capacity for self-renewal and regeneration. Expression of aldehyde dehydrogenase activity (ALDH) has been associated with stemness in many organs systems, including the lung [40]. Specifically, Aldh1a3 has been specifically implicated as the major contributor to ALDH activity and self-renewal capacity in some cell populations [41]; Aldh1a3 gene expression was significantly up-regulated in LMSCs compared with the rest of the whole lung, and was down-regulated in LMSCs isolated from old mice compared with those isolated from young mice. Interestingly, Aldh1a3 is also recognized as an important member of the retinoic acid signaling pathway [41], which is crucial to appropriate mesenchymal cell proliferation and signaling during developmental alveolarization in the lung, and it has also been implicated in lung regeneration in the adult lung [42–44].
High telomerase activity drives long term self-renewal capacity, for example in tracheal basal cells [15], and can increase the capacity for cell proliferation when ectopically induced in MSCs [45]. The diminished plasticity of aging BM-MSCs is also associated with reduced telomerase levels [46]. Relevant to our data, inhibition of telomerase activity specifically inhibits proliferation of ALDH-positive lung cancer stem cells [40]. Decreased telomerase activity also affects the ability of lungs to undergo compensatory lung regrowth after PNX [47], while increased telomerase activity can reverse tissue degeneration in telomerase-deficient mice [48]. Telomerase activity can also be induced in quiescent alveolar type II cells from adult mice, and it is associated with higher proliferation rates in these cells [47, 49]. Interestingly, a recent study also determined that transcription of the telomerase gene (Tert) is positively regulated by β-catenin [50]. This discovery provides a novel link between Wnt/β-catenin signaling and high telomerase activity in stem cells. A similar mechanism may be involved in the reduced telomerase activity seen here in LMSCs. The reduction in both ALDH activity and telomerase activity in LMSCs isolated from older animals is associated with down-regulated gene expression of Aldh1a3, Wnt2, and Ctnnb1. Furthermore, the dramatic down-regulation of Fgfr1 seen in aging LMSCs is consistent with Wnt pathway regulation by Fgfr1 in mesenchymal progenitor cells during normal lung alveolar development [33], as well as post-PNX alveolarization in Pdgfrα-positive fibroblasts [34]. Interestingly, Fgfr1 signaling is also an important response to RA stimulation during developmental alveolarization [31], and more recently, nuclear accumulation of Fgfr1 in response to RA signaling has been demonstrated as both necessary and sufficient for differentiation of embryonic stem cells [51]. These previous findings support the findings of our current study, and allow us to speculate that that reduced ALDH and telomerase activity may be associated with alterations in aspects of Fgfr, Wnt, and retinoic acid signaling that result in reduced clonogenicity and/or self-renewal of LMSCs in older mice, thus diminishing the ability of these cells to appropriately support early lung regeneration in aging mice.
Supplementary Material
Acknowledgments
Funding - NHLBI 072780-02 (EPI/AMH); HL106625-01 (Daniel J Weiss/AMH).
The authors are grateful to Stephanie LaFollette, Brandon Nguyen, and Trevor Dutton at the College of the Holy Cross for their technical assistance.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Cagle PT, Langston C. and Thurlbeck WM. (1988). The effect of age on postpneumonectomy growth in rabbits. Pediatr Pulmonol 5:92–95 [DOI] [PubMed] [Google Scholar]
- 2.Laros CD. and Westermann CJ. (1987). Dilatation, compensatory growth, or both after pneumonectomy during childhood and adolescence. A thirty-year follow-up study. J Thorac Cardiovasc Surg 93:570–576 [PubMed] [Google Scholar]
- 3.Holmes C. and Thurlbeck WM. (1979). Normal lung growth and response after pneumonectomy in rats at various ages. Am Rev Respir Dis 120:1125–1136 [DOI] [PubMed] [Google Scholar]
- 4.Paxson JA, Gruntman A, Parkin CD, Mazan MR, Davis A, Ingenito EP. and Hoffman AM. (2011). Age-dependent decline in mouse lung regeneration with loss of lung fibroblast clonogenicity and increased myofibroblastic differentiation. PLoS One 6:e23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beers MF. and Morrisey EE. (2011). The three R's of lung health and disease: repair, remodeling, and regeneration. J Clin Invest 121:2065–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chilosi M, Doglioni C, Murer B. and Poletti V. (2010). Epithelial stem cell exhaustion in the pathogenesis of idiopathic pulmonary fibrosis. Sarcoidosis Vasc Diffuse Lung Dis 27:7–18 [PubMed] [Google Scholar]
- 7.Mora AL. and Rojas M. (2008). Aging and lung injury repair: a role for bone marrow derived mesenchymal stem cells. J Cell Biochem 105:641–647 [DOI] [PubMed] [Google Scholar]
- 8.Kubo H. (2011). Molecular basis of lung tissue regeneration. Gen Thorac Cardiovasc Surg 59:231–244 [DOI] [PubMed] [Google Scholar]
- 9.Martin J, Helm K, Ruegg P, Varella-Garcia M, Burnham E. and Majka S. (2008). Adult lung side population cells have mesenchymal stem cell potential. Cytotherapy 10:140–151 [DOI] [PubMed] [Google Scholar]
- 10.Summer R, Fitzsimmons K, Dwyer D, Murphy J. and Fine A. (2007). Isolation of an adult mouse lung mesenchymal progenitor cell population. Am J Respir Cell Mol Biol 37:152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ. and Bertoncello I. (2009). Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells 27:623–633 [DOI] [PubMed] [Google Scholar]
- 12.Hegab AE, Kubo H, Fujino N, Suzuki T, He M, Kato H. and Yamaya M. (2010). Isolation and characterization of murine multipotent lung stem cells. Stem Cells Dev 19:523–536 [DOI] [PubMed] [Google Scholar]
- 13.Teisanu RM, Lagasse E, Whitesides JF. and Stripp BR. (2009). Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells 27:612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teisanu RM, Chen H, Matsumoto K, McQualter JL, Potts E, Foster WM, Bertoncello I. and Stripp BR. (2011). Functional analysis of two distinct bronchiolar progenitors during lung injury and repair. Am J Respir Cell Mol Biol 44:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh M, Helm KM, Smith RW, Giordanengo MS, Li B, Shen H. and Reynolds SD. (2011). A single cell functions as a tissue-specific stem cell and the in vitro niche-forming cell. Am J Respir Cell Mol Biol 45:459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH. and Hogan BL. (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A 106:12771–12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda S, Hsia CC, Wagner E, Ramanathan M, Estrera AS. and Weibel ER. (1999). Compensatory alveolar growth normalizes gas-exchange function in immature dogs after pneumonectomy. J Appl Physiol 86:1301–1310 [DOI] [PubMed] [Google Scholar]
- 18.Nolen-Walston RD, Kim CF, Mazan MR, Ingenito EP, Gruntman AM, Tsai L, Boston R, Woolfenden AE, Jacks T. and Hoffman AM. (2008). Cellular kinetics and modeling of bronchioalveolar stem cell response during lung regeneration. Am J Physiol Lung Cell Mol Physiol 294:L1158–L1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paxson JA, Parkin CD, Iyer LK, Mazan MR, Ingenito EP. and Hoffman AM. (2009). Global gene expression patterns in the post-pneumonectomy lung of adult mice. Respir Res 10:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brody JS, Burki R. and Kaplan N. (1978). Deoxyribonucleic acid synthesis in lung cells during compensatory lung growth after pneumonectomy. Am Rev Respir Dis 117:307–316 [DOI] [PubMed] [Google Scholar]
- 21.Hoffman AM, Shifren A, Mazan MR, Gruntman AM, Lascola KM, Nolen-Walston RD, Kim CF, Tsai L, Pierce RA, Mecham RP. and Ingenito EP. (2010). Matrix modulation of compensatory lung regrowth and progenitor cell proliferation in mice. Am J Physiol Lung Cell Mol Physiol 298:L158–L168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faridy EE, Sanii MR. and Thliveris JA. (1988). Influence of maternal pneumonectomy on fetal lung growth. Respir Physiol 72:195–209 [DOI] [PubMed] [Google Scholar]
- 23.Kendziorski C, Irizarry RA, Chen KS, Haag JD. and Gould MN. (2005). On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci U S A 102:4252–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Wang L, Ravindranathan A. and Miles MF. (2002). A new algorithm for analysis of oligonucleotide arrays: application to expression profiling in mouse brain regions. J Mol Biol 317:225–235 [DOI] [PubMed] [Google Scholar]
- 25.Kerns RT, Zhang L. and Miles MF. (2003). Application of the S-score algorithm for analysis of oligonucleotide microarrays. Methods 31:274–281 [DOI] [PubMed] [Google Scholar]
- 26.Kennedy RE, Kerns RT, Kong X, Archer KJ. and Miles MF. (2006). SScore: an R package for detecting differential gene expression without gene expression summaries. Bioinformatics 22:1272–1274 [DOI] [PubMed] [Google Scholar]
- 27.Kennedy RE, Archer KJ. and Miles MF. (2006). Empirical validation of the S-Score algorithm in the analysis of gene expression data. BMC Bioinformatics 7:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl MW. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricciardi M, Malpeli G, Bifari F, Bassi G, Pacelli L, Nwabo Kamdje AH, Chilosi M. and Krampera M. (2012). Comparison of epithelial differentiation and immune regulatory properties of mesenchymal stromal cells derived from human lung and bone marrow. PLoS One 7:e35639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boxall SA. and Jones E. (2012). Markers for characterization of bone marrow multipotential stromal cells. Stem Cells Int 2012:975871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perl AK. and Gale E. (2009). FGF signaling is required for myofibroblast differentiation during alveolar regeneration. Am J Physiol Lung Cell Mol Physiol 297:L299–L308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White AC, Xu J, Yin Y, Smith C, Schmid G. and Ornitz DM. (2006). FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development 133:1507–1517 [DOI] [PubMed] [Google Scholar]
- 33.Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F. and Ornitz DM. (2008). An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol 319:426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Acciani T, Le Cras T, Lutzko C. and Perl AK. (2012). Dynamic regulation of PDGFRalpha expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol 47:517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding BS, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, Crystal RG, Simons M, Sato TN, et al. (2011). Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell 147:539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman AM, Paxson JA, Mazan MR, Davis AM, Tyagi S, Murthy S. and Ingenito EP. (2011). Lung-derived mesenchymal stromal cell post-transplantation survival, persistence, paracrine expression, and repair of elastase-injured lung. Stem Cells Dev 20:1779–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Votteler M, Kluger PJ, Walles H. and Schenke-Layland K. (2010). Stem cell microenvironments—unveiling the secret of how stem cell fate is defined. Macromol Biosci 10:1302–1315 [DOI] [PubMed] [Google Scholar]
- 38.Silva H. and Conboy IM. (2008). Aging and Stem Cell Renewal. StemBook [Internet]. Harvard Stem Cell Institute, Cambridge, MA, 2008. [PubMed] [Google Scholar]
- 39.Drummond-Barbosa D. (2008). Stem cells, their niches and the systemic environment: an aging network. Genetics 180:1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano D, Bleau AM, Fernandez-Garcia I, Fernandez-Marcelo T, Iniesta P, Ortiz-de-Solorzano C. and Calvo A. (2011). Inhibition of telomerase activity preferentially targets aldehyde dehydrogenase-positive cancer stem-like cells in lung cancer. Mol Cancer 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcato P, Dean CA, Giacomantonio CA. and Lee PW. (2011). Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle 10:1378–1384 [DOI] [PubMed] [Google Scholar]
- 42.Hind M. and Maden M. (2004). Retinoic acid induces alveolar regeneration in the adult mouse lung. Eur Respir J 23:20–27 [DOI] [PubMed] [Google Scholar]
- 43.Shi W, Chen F. and Cardoso WV. (2009). Mechanisms of lung development: contribution to adult lung disease and relevance to chronic obstructive pulmonary disease. Proc Am Thorac Soc 6:558–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boucherat O, Franco-Montoya ML, Thibault C, Incitti R, Chailley-Heu B, Delacourt C. and Bourbon JR. (2007). Gene expression profiling in lung fibroblasts reveals new players in alveolarization. Physiol Genomics 32:128–141 [DOI] [PubMed] [Google Scholar]
- 45.Piper SL, Wang M, Yamamoto A, Malek F, Luu A, Kuo AC. and Kim HT. (2012). Inducible immortality in hTERT-human mesenchymal stem cells. J Orthop Res 30:1879–1885 [DOI] [PubMed] [Google Scholar]
- 46.Asumda FZ. and Chase PB. (2011). Age-related changes in rat bone-marrow mesenchymal stem cell plasticity. BMC Cell Biol 12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson SR, Lee J, Reddy R, Williams GN, Kikuchi A, Freiberg Y, Warburton D. and Driscoll B. (2011). Partial pneumonectomy of telomerase null mice carrying shortened telomeres initiates cell growth arrest resulting in a limited compensatory growth response. Am J Physiol Lung Cell Mol Physiol 300:L898–L909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, et al. (2011). Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469:102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driscoll B, Buckley S, Bui KC, Anderson KD. and Warburton D. (2000). Telomerase in alveolar epithelial development and repair. Am J Physiol Lung Cell Mol Physiol 279:L1191–L1198 [DOI] [PubMed] [Google Scholar]
- 50.Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R. and Kemler R. (2012). Wnt/beta-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336:1549–1554 [DOI] [PubMed] [Google Scholar]
- 51.Lee YW, Terranova C, Birkaya B, Narla S, Kehoe D, Parikh A, Dong S, Ratzka A, Brinkmann H, et al. (2012). A novel nuclear FGF Receptor-1 partnership with retinoid and Nur receptors during developmental gene programming of embryonic stem cells. J Cell Biochem 113:2920–2936 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.