Rare EGFR mutations were collected from 10117 non-small cell lung cancer samples analyzed by ERMETIC-IFCT French network. Among 1047 EGFR-mutated samples, 102 (10%) harboured rare mutations (41 in exon 18, 49 in exon 20, 12 with others). Association with smoking differed between exon 18 and 20, and control rate to EGFR-TKI was 47%, strongly varied between patients requiring individual assessment.
Keywords: epidermal growth factor receptor mutations, exon 18 mutations, exon 20 mutations, non-small-cell lung cancer, tyrosine-kinase inhibitors
Abstract
Background
There is scarce data available about epidermal growth factor receptor (EGFR) mutations other than common exon 19 deletions and exon 21 (L858R) mutations.
Patients and methods
EGFR exon 18 and/or exon 20 mutations were collected from 10 117 non-small-cell lung cancer (NSCLC) samples analysed at 15 French National Cancer Institute (INCa)-platforms of the ERMETIC-IFCT network.
Results
Between 2008 and 2011, 1047 (10%) samples were EGFR-mutated, 102 (10%) with rare mutations: 41 (4%) in exon 18, 49 (5%) in exon 20, and 12 (1%) with other EGFR mutations. Exon 20 mutations were related to never-smoker status, when compared with exon 18 mutations (P < 0.001). Median overall survival (OS) of metastatic disease was 21 months [95% confidence interval (CI) 12–24], worse in smokers than in non-smoker patients with exon 20 mutations (12 versus 21 months; hazard ratio [HR] for death 0.27, 95% CI 0.08–0.87, P = 0.03). Under EGFR-tyrosine kinase inhibitors (TKIs), median OS was 14 months (95% CI 6–21); disease control rate was better for complex mutations (6 of 7, 86%) than for single mutations (16 of 40, 40%) (P = 0.03).
Conclusions
Rare EGFR-mutated NSCLCs are heterogeneous, with resistance of distal exon 20 insertions and better sensitivity of exon 18 or complex mutations to EGFR-TKIs, probably requiring individual assessment.
introduction
In 2004, identification of somatic mutations in the epidermal growth factor receptor (EGFR) provided a clinically relevant driver oncogene of non-small-cell lung cancer (NSCLC) [1–3]. About 10%–15% of all NSCLC in Western Europeans and 30% in East Asians are EGFR mutated. The most common (85%–90%) EGFR mutations are in-frame deletions around the LeuArgGluAla motifs (LREA residues 746–750) of exon 19 (45%–50%) and the Leu858Arg (L858R) substitution in exon 21 (40%–45%) [4]. They result in the preferential binding of EGFR tyrosine kinase inhibitors (TKIs), i.e., gefitinib, erlotinib, or afatinib, leading to >60% overall response rates (ORR), median progression-free survival (PFS) >9 months, and overall survival (OS) of >20 months, better than with chemotherapy [5–10]. Epidemiological data, correlations with clinical characteristics, EGFR-TKI response, and prognostic value remain unclear in NSCLC cases with uncommon EGFR mutations [11–19].
Therefore, the ERMETIC-IFCT network decided to report its results of EGFR exon 18 and 20 mutations based on 10 117 analyses carried out between 2008 and 2011 [20].
methods
centres and molecular analysis
Centres and technics are detailed in supplementary data, available at Annals of Oncology online [21, 22] (supplementary Table S1, available at Annals of Oncology online). Rare EGFR mutations were defined as mutations at exon 18 and/or 20; complex mutations were defined as mutations at more than one exon: double mutation at exon 18 and 20 or one mutation at exon 18 or 20 with one mutation in another exon (19 or 21) and were compared with COSMIC (Catalog of Somatic Mutations in Cancer) [23], somaticmutations-EGFR.org [4] and PubMed.
clinical data
French National Cancer Institute [24] required that clinical data be collected for testing, such as demographic information, clinical staging [25], and lung cancer histology (WHO classification) [26]. Never smokers were defined as <100 cigarettes in life time. French National Cancer Institute (ClinicalTrials.gov, number NCT01700582) established requirements for clinical information on patient follow-up under treatment, including response to treatment (RECIST criteria) [27] and survival.
statistical analysis
Categorical variables were compared using chi-square tests, or Fisher's exact tests when necessary. Significance was determined at P < 0.05. OS was calculated from the date of lung cancer diagnosis to death from any cause or was censored at the last follow-up date. The median follow-up was 26 months (1–110 months). Progression-free survival (PFS) was defined as the time from the date of EGFR-TKI treatment initiation to the date of disease progression or death and was censored at the date of last tumour assessment (when carried out). Survival curves were estimated using Kaplan–Meier method for OS. A Cox model was applied to estimate hazard ratios (HRs) and 95% confidence interval (CI). Analyses were undertaken using SAS version 9.1.3 (SAS Institute, Cary, NC).
results
EGFR mutation frequency
On 10 117 NSCLC samples, n = 9070 (90%) were EGFR wild-type and n = 1047 (10%) were EGFR-mutated. EGFR exon 18 and 20 mutations were observed in 102 (10%) EGFR-mutated samples, 1% of the 10 117 NSCLC samples analysed, without relationship between the frequencies of the whole and rare EGFR mutations detected by each platform (supplementary Figure S1, available at Annals of Oncology online). Rare EGFR mutations included in exon 18 n = 41 (4% of all EGFR-mutated cases), exon 20 n = 49 (5%), and complex mutations n = 12 (1%) (supplementary Figure S2, available at Annals of Oncology online). Histological characteristics are provided in the supplementary data, available at Annals of Oncology online.
molecular epidemiology of rare EGFR mutations
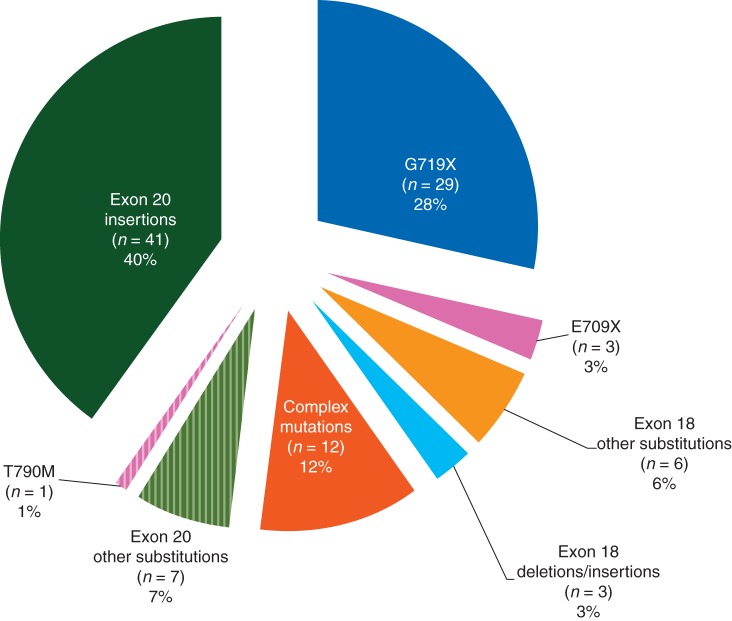
Rare EGFR mutations are described in Figure 1 and supplementary data (supplementary Tables S2 and S3, available at Annals of Oncology online).
Figure 1.
Distribution of the 102 rare EGFR mutations.
clinical data of patients with rare EGFR mutations
All patients were Caucasian, none had received EGFR-TKI before DNA sequencing (Table 1 and supplementary Table S3, available at Annals of Oncology online). Surprisingly, the majority of patients (n = 46, 62%) were smokers, with former/current smokers (n = 27/19), in contrast to never smokers (n = 26). In univariate analysis, early stage was a good prognostic marker (HR for death 0.203, 95% CI 0.075–0.553; P = 0.002) (data not shown).
Table 1.
Clinical characteristics and survival of patients with rare EGFR mutations (n = 74); best response and survival to reversible EGFR-TKIs (n = 50)
| Patients, n (%) | Patients with exon 18 EGFR mutations, n (%) | Patients with exon 20 EGFR mutations, n (%) | Patients with complex exon 18 and exon 20 EGFR mutations, n (%) | |
|---|---|---|---|---|
| 74 | 28 (38) | 38 (51) | 8 (14) | |
| Age (years), median (range) | 62 (24–85) | 62.7 (49–84) | 60.9 (34–85) | 63.5 (24–71) |
| Gender | ||||
| Female | 42 (57) | 14 (50) | 23 (61) | 5 (62) |
| Male | 32 (43) | 14 (50) | 15 (39) | 3 (38) |
| Smoking status* | ||||
| Never smoker | 26 (35) | 4 (15) | 21 (57) | 1 (12) |
| Former smoker | 27 (36) | 14 (52) | 10 (27) | 3 (38) |
| Current smoker | 19 (26) | 9 (33) | 6 (16) | 4 (50) |
| MD | 2 | 1 | 1 | |
| Histological type | ||||
| Adenocarcinoma | 64 (88) | 24 (89) | 33 (87) | 7 (88) |
| Squamous cell carcinoma | 5 (7) | 2 (7) | 2 (5) | 1 (12) |
| Large cell carcinoma | 4 (5) | 1 (6) | 3 (8) | 0 (0) |
| MD | 1 | 1 | ||
| Stage | ||||
| I–IIIA | 28 (38) | 14 (50) | 13 (34) | 1 (12) |
| IIIB–IV | 46 (62) | 14 (50) | 25 (66) | 7 (88) |
| OS (months), median (95% CI) | 24 (18–43) | 41 (9–92) | 21 (14-not estimated) | 23 (11–50) |
| OS, stage I–IIIB | 92 (36-not estimated), n = 28 | 92 (9–92), n = 10 | not reached, n = 16 | 12.5 (2.5–22), n = 2 |
| OS, stage IV | 21 (12–24), n = 41 | 27 (6–43), n = 13 | 14 (10–21), n = 22 | 24 (23–50), n = 6 |
| EGFR-TKI usage, erlotinib/gefitinib | 50 | 18 | 25 | 7 |
| First-line | 11 (22) | 1 (6) | 9 (36) | 1 (14) |
| Second-line | 33 (66) | 13 (72) | 15 (60) | 5 (72) |
| Third-line | 4 (8) | 2 (11) | 1 (4) | 1 (14) |
| Upper | 2 (4) | 2 (11) | 0 (0) | 0 (0) |
| Best response to EGFR-TKI | ||||
| PR | 7 (15) | 1 (7) | 2 (8) | 4 (57)** |
| SD | 15 (32) | 4 (27) | 9 (36) | 2 (29)*** |
| PD | 25 (53) | 10 (66) | 14 (56) | 1 (14) |
| MD | 3 | 3 | 0 | 0 |
| Progression-free survival (months), median (95% CI) | 4 (2-not estimated) | 3 (1-not estimated), | 2 (1-not estimated) | not reached, |
| <3 months | 22 (48) | 6 (43) | 16 (64) | 0 (0) |
| ≥3, <6 months | 13 (28) | 5 (36) | 4 (16) | 4 (57) |
| ≥6 months | 11 (24) | 3 (21) | 5 (20) | 3 (43) |
| Unknown | 4 | 4 | 0 | 0 |
| Overall survival from EGFR-TKI (months), median (95% CI) | 14 (6–21) | 22 (1–44) | 9·5 (4–15) | 14 (5–23) |
*P = 0.003; when stratifying tobacco status between never smokers and former/current smokers, P < 0.001.
**P = 0.004 for objective response; ***P = 0.03 for disease control.
EGFR: epidermal growth factor receptor; CI, confidence interval; MD, missing data; TKIs, tyrosine-kinase inhibitors; PR, partial response; SD, stabilised disease; PD progression disease.
Mostly men (n = 14, 50%) showed single exon 18 mutations and mostly women (n = 23, 61%) showed single exon 20 mutations. Never smokers were significantly fewer among patients with exon 18 mutations than with exon 20 mutations (P < 0.001). Survival could be different depending on smoking habits with median OS in metastatic patients with exon 20 mutations worse in smokers than in non-smokers (12 versus 21 months, HR for death 0.27, 95% CI 0.08–0.87; P = 0.03).
EGFR-TKI response in patients with rare EGFR mutations
Fifty patients had been known to be treated with EGFR-TKI (erlotinib n = 40, gefitinib n = 9, gefinitib following by erlotinib n = 1) (Table 1). The best response was assessable in patients (n = 47), with partial responses (n = 7, 15%), stable disease (n = 15, 32%), and progressive disease (n = 25, 53%), with a disease control rate (DCR) at 47%. Primary resistance to EGFR-TKI is suggested in patients with PFS under 3 months (n = 22, 48%).
ORR and DCR under EGFR-TKI appeared not different between exon 18 (n = 1, 7% and n = 5, 34%) and exon 20 (n = 2, 8% and n = 11, 44%), but higher for complex mutations (n = 4, 57% and n = 6, 86%) compared with single exon 18 or exon 20 mutations (P = 0.004 and P = 0.03, respectively). Median OS from EGFR-TKI was better for patients with exon 18 (22 months, 95% CI 1–44) than for patients with exon 20 mutations (9.5 months, 95% CI 4–15) (supplementary Figure S3, available at Annals of Oncology online).
discussion
This report refers to the first multicentre exhaustive study on rare EGFR mutations that was conducted on a national basis [15]. This kind of collaborative effort involving numerous cancer centres provides a more accurate estimate of the frequency of rare and uncommon EGFR mutations, thereby avoiding the biases inherent in the compilation of cases involving just one mutation type. In addition, we analysed complex EGFR mutations, such as compound mutations or co-mutations. Furthermore, we assessed the response rates to EGFR-TKI in order to avoid overestimating the clinical effects of EGFR inhibitors in a particular genotype.
EGFR mutations were observed in 10% of NSCLC samples, in line with the mutation percentage habitually found in Caucasian populations [28], as in France in 2011 (INCa: http://www.e-cancer.fr). Of these EGFR mutations, 102 were classified as rare and represented 1% of all NSCLC tested samples or 10% of the EGFR-mutated cases. For these rare mutations, most clinicians are reluctant to prescribe EGFR-TKIs [29]. Furthermore, we described 22 EGFR mutations that had not been reported before. Of these, 10 were harboured by tumour patients undergoing EGFR-TKI therapy, with seven patients presenting progressive disease, one stable disease, and the remaining two showing partial response.
The rare EGFR mutations that were the most frequently seen were exon 20 insertions (4%), in line with previous publications reporting their presence in 1%–17% of all EGFR mutations, depending on the selected populations [12–17, 30, 31]. These observations should encourage centres to search for these EGFR exon 20 insertions, as they are easy to detect using fragment length analysis. Although EGFR exon 20 insertions are more frequent than EGFR exon 19 insertions, they are found with the same predominance of never smokers NSCLC as other insertions, such as HER2 [19, 32]. In our study, OS was shown to be significantly poorer in current/former smokers with advanced disease exhibiting EGFR exon 20 mutations compared with never smokers. These data suggest that NSCLC carcinogenesis differs between smokers and non-smokers, with potential impact on therapeutic strategies [33–36]. As opposed to EGFR exon 19 insertions, EGFR exon 20 mutations have been associated with de novo resistance to EGFR-TKI. However, in line with our study findings, a few cases showing improved clinical response to EGFR-TKI have been reported [12–14, 17, 30, 31]. Molecular description showed that EGFR exon 20 insertions varied in length (from 3 to 12 bp) and position within the exon, in contrast to EGFR exon 19 or HER2 exon 20 insertions [14, 30, 31, 37]. As previously reported, we observed a heterogeneity of insertions, with 18 different variants and only 6 occurring more than once [14, 30, 31]. The most frequent mutation in our study was A767V769dupASV (12% of exon 20 insertions) rather than V769D770insASV, as previously reported [30, 31]. Yet, as is often the case, the preferential location of our exon 20 insertions followed the C-helix (A767 to C775) as distal insertions, which play a critical role in catalytic activity, as well as drug affinity resulting in resistance to EGFR-TKIs [14, 31, 37]. Among our 27 patients with EGFR distal exon 20 insertions, 19 were treated with EGFR-TKI; of these, 12 presented with progressive disease, 6 with stable disease, and only 1 exhibited partial response: ORR (n = 1; 5%) and DCR (n = 7; 36%). These data confirm patient resistance to EGFR-TKIs in this mutation type. More proximal insertions that affect E762 to Y764 could be associated with disease control under EGFR-TKI [14, 17, 30], which is backed up by our study findings showing one patient with partial response and one exhibiting stable disease in the two presenting with proximal exon 20 insertion. These results suggest that the specific sequence of the insertion variant should be detailed for having more information about EGFR-TKI sensitivity. Lastly, rare EGFR exon 20 point mutations were associated with EGFR-TKIs resistance, with the exception of one of our cases involving R776H as previously published [4].
In our study, former/current smokers more frequently exhibited EGFR exon 18 mutations, as known to other TP53 or KRAS point mutations [33]. Of all the rare mutations found, EGFR exon 18 G719X mutations were the second most frequently reported in our study, in line with previous reports, with slightly less sensitivity than the common EGFR mutations [13, 17]. In our study, 4 of 11 (36%) patients with G719S or G719A mutations undergoing EGFR-TKI therapy presented disease control. E709X mutations were shown to be less sensitive than G719X mutants in vitro experiments [38]. Nevertheless, one of our patients with a G709A + G719S mutation showed prolonged disease control, and a partial response to neratinib has recently been reported in case of such mutation (supplementary reference S9, available at Annals of Oncology online).
The EGFR rare mutations exhibiting the highest sensitivity appeared to be complex mutations, which represented 12% of mutation cases in our study. This can be compared with published literature indicating a frequency of compound mutations ranging from 4% to 14%, according to chosen detection method of EGFR mutations or the definition of these types of complex mutations (13, supplementary reference S15, available at Annals of Oncology online, 18). G719X complex mutations are the most frequently reported, in line with our study findings (58% of our complex mutations), followed by L858R complex mutations, as in our study (51% of our complex mutations), S768I (25% of our complex mutations), with EGFR exon 19 deletion detected much less frequently (only one case found in our cohort) [13, 18]. Overall, EGFR-TKI efficacy in patients with complex mutations was similar to or slightly poorer than that in patients with single common EGFR mutations, as seen for G719S + L861Q and G719S + S768I in our cohort [39]. It should be noted that, in cases with the S768I mutation alone, sensitivity appears to be maintained [17]. The complex mutation G719A + S768I that we identified in one patient exhibiting partial response to erlotinib has previously been reported by others, notably in two cases showing partial response [18] and in two other cases showing no response at all (12, supplementary reference S27, available at Annals of Oncology online). The complex mutations involving L858R appeared to affect the response to EGFR-TKIs in differing ways according to the other associated mutations [18]. In our cohort, one patient with L858R + S768I presented disease control, whereas another patient with L858R + S768D770dupSVD presented progressive disease. Unexpectedly, we observed disease control in one patient with L858R + T790M.
These types of mutations are heterogeneous and require novel targeted therapeutic approaches, which are possible if the mutations are searched for, and if the variant is documented. For rare EGFR mutations, two different approaches could be advised: one possibility would be to compile databases with a maximum of data concerning these types of cases (16; Biomarker France Study), and another involving in vitro experiments or computer biology approaches for testing EGFR-TKIs [31].
fundings
This work was supported by the French National Cancer Institute (INCa) and Ligue Nationale Contre le Cancer (LNCC).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
L. HAJOUJI (CH, Colmar), D. HOLTEA (CH, Mulhouse), J. MAZIERES (CHU, Toulouse), A-C. METIVIER (APHP Bichat, Paris), R. SCHOTT (CLCC, Strasbourg), C. DANIEL (Institut Curie, Paris), C. FOA (St-Joseph Hospital, Marseille), M. PIBAROT (St-Joseph Hospital, Marseille), D. PLANCHARD (IGR, Villejuif), E. PLUQUET (CH - Tourcoing), J-C SORIA (IGR, Villejuif), C. CLAROT (CH, Abbeville), E. DANSIN (CLCC, Lille), D. DEBIEUVRE (CH, Mulhouse), S. FRIARD (Foch Hospital, Suresnes), L. GREILLIER (APHM Nord, Marseille), J-J. LAFITTE (CHU, Lille), A. MADROSZYK (CLCC, Marseille), G. MASSARD (NHC, Strasbourg), J. MEDIONI (APHP HEGP, Paris), B. MENNECIER (NHC, Strasbourg), L. MOREAU (CH, Colmar), F. PAGNIER (CH, Douai), J-L. PUJOL (CHU, Montpellier), X. QUANTIN (CHU Montpellier), J-M ZIPPER (CH, Mulhouse).
references
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, et al. EGFR Mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, et al. EGF Receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray S, Dahabreh IJ, Linardou H, et al. Somatic mutations of the tyrosine kinase domain of epidermal growth factor receptor and tyrosine kinase inhibitor response to TKIs in non-small cell lung cancer: an analytical database. J Thorac Oncol. 2008;3:832–839. doi: 10.1097/JTO.0b013e31818071f3. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 6.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 8.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 9.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for european patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 10.Yang JC, Shih JY, Su WC, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-lung 2): a phase 2 trial. Lancet Oncol. 2012;13:539–548. doi: 10.1016/S1470-2045(12)70086-4. [DOI] [PubMed] [Google Scholar]
- 11.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 12.Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 13.Wu JY, Yu CJ, Chang YC, et al. Effectiveness of tyrosine kinase inhibitors on ‘uncommon’ epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17:3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 14.Yasuda H, Kobayashi S, Costa DB. EGFR Exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13:e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 15.Oxnard GR, Jänne PA. Power in numbers: meta-analysis to identify inhibitor-sensitive tumor genotypes. Clin Cancer Res. 2013;19:1634–1636. doi: 10.1158/1078-0432.CCR-13-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh P, Chen H, Andrews J, et al. DNA-Mutation Inventory to refine and enhance cancer treatment (DIRECT): a catalog of clinically relevant cancer mutations to enable genome-directed anticancer therapy. Clin Cancer Res. 2013;19:1894–1901. doi: 10.1158/1078-0432.CCR-12-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massarelli E, Johnson FM, Erickson HS, et al. Uncommon epidermal growth factor receptor mutations in non-small cell lung cancer and their mechanisms of EGFR tyrosine kinase inhibitors sensitivity and resistance. Lung Cancer. 2013;80:235–241. doi: 10.1016/j.lungcan.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi S, Canepa HM, Bailey AS, et al. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2013;8:45–51. doi: 10.1097/JTO.0b013e3182781e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He M, Capelletti M, Nafa K, et al. EGFR Exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res. 2012;18:1790–1797. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowak F, Soria JC, Calvo F. Tumour molecular profiling for deciding therapy-the French initiative. Nat Rev Clin Oncol. 2012;9:479–486. doi: 10.1038/nrclinonc.2012.42. [DOI] [PubMed] [Google Scholar]
- 21.Beau-Faller M, Degeorges A, Rolland E, et al. Cross-validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non-small cell lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR-TKIs in non-small cell lung carcinoma [ERMETIC] project–part 1) J Thorac Oncol. 2011;6:1006–1015. doi: 10.1097/JTO.0b013e318211dcee. [DOI] [PubMed] [Google Scholar]
- 22.Cadranel J, Mauguen A, Faller M, et al. Impact of systematic EGFR and KRAS mutation evaluation on progression-free survival and overall survival in patients with advanced Non-small-cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project-part 2) J Thorac Oncol. 2012;7:1490–1502. doi: 10.1097/JTO.0b013e318265b2b5. [DOI] [PubMed] [Google Scholar]
- 23.Forbes SA, Tang G, Bindal N, et al. COSMIC (the catalogue of somatic mutations in cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Institut National du Cancer: Bonnes pratiques pour la recherche à visée théranostique de mutations somatiques dans les tumeurs solides. 2010 Août http://www.e-cancer.fr/expertises-publications-de-l-inca/rapports-et-expertises/soins. (8 January 2010, date last accessed) [Google Scholar]
- 25.Vallières E, Shepherd FA, Crowley J, et al. The IASLC lung cancer staging project: proposals regarding the relevance of TNM in the pathologic staging of small cell lung cancer in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4:1049–1059. doi: 10.1097/JTO.0b013e3181b27799. [DOI] [PubMed] [Google Scholar]
- 26.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European organization for research and treatment of cancer, National cancer institute of the United States, National cancer institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 28.Cadranel J, Zalcman G, Sequist L. Genetic profiling and epidermal growth factor receptor-directed therapy in nonsmall cell lung cancer. Eur Respir J. 2011;37:183–193. doi: 10.1183/09031936.00179409. [DOI] [PubMed] [Google Scholar]
- 29.Yatabe Y, Pao W, Jett JR. Encouragement to submit data of clinical response to EGFR-TKIs in patients with uncommon EGFR mutations. J Thorac Oncol. 2012;7:775–776. doi: 10.1097/JTO.0b013e318251980b. [DOI] [PubMed] [Google Scholar]
- 30.Oxnard GR, Lo PC, Nishino M, et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thor Oncol. 2013;8:179–184. doi: 10.1097/JTO.0b013e3182779d18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arcila ME, Nafa K, Chaft JE, et al. EGFR Exon 20 insertion mutations in lung adenocarcinomas : prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arcila ME, Chaft JE, Nafa K, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18:4910–4918. doi: 10.1158/1078-0432.CCR-12-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mounawar M, Mukeria A, Le Calvez F, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non-small cell lung cancers in relation to smoking history. Cancer Res. 2007;67:5667–5672. doi: 10.1158/0008-5472.CAN-06-4229. [DOI] [PubMed] [Google Scholar]
- 34.Lee YJ, Shim HS, Kang YA, et al. Dose effect of cigarette smoking on frequency and spectrum of epidermal growth factor receptor gene mutations in Korean patients with non-small cell lung cancer. J Cancer Res Clin Oncol. 2010;136:937–944. doi: 10.1007/s00432-010-0853-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol. 2011;29:2066–2070. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couraud S, Zalcman G, Milleron B, et al. Lung cancer in never smokers–a review. Eur J Cancer. 2012;48:1299–1311. doi: 10.1016/j.ejca.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Costa DB, Yasuda H, Sng NY, et al. Sensitivity to EGFR inhibitors based on location of EGFR exon 20 insertions mutations within the tyrosine kinase domain of EGFR. J Clin Oncol. 2012;30 (15_Suppl): 7523. [Google Scholar]
- 38.Kancha RK, Peschle C, Duyster J. The epidermal growth factor receptor-L861Q mutation increases kinase activity without leading to enhanced sensitivity toward epidermal growth factor receptor kinase inhibitors. J Thorac Oncol. 2011;6:387–392. doi: 10.1097/JTO.0b013e3182021f3e. [DOI] [PubMed] [Google Scholar]
- 39.Chen YR, Fu YN, Lin CH, et al. Distinctive activation patterns in constitutively active and gefitinib-sensitive EGFR mutants. Oncogene. 2006;25:1205–1215. doi: 10.1038/sj.onc.1209159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.