Abstract
Cancer stem cells (CSCs) are a subset of tumor cells that has the ability to self-renew and to generate the diverse cells that comprise the tumor mass. The cell-surface glycoprotein CD44 is one of the most common surface markers used to identify CSCs. Aptamers are synthetic oligonucleotides selected from pools of random sequences that can bind to a wide range of targets with high affinity and specificity. In this study, the systematic evolution of ligands by exponential enrichment (SELEX) technology was used to isolate RNA aptamers using human recombinant full-length CD44 protein and 2′-F-pyrimidine modified RNA library with a complexity of around 1014 different molecules. Following 11 iterative rounds of SELEX, the selected aptamers were cloned and sequenced. Three different sequences were identified. The binding specificities for one of these RNA aptamers was assessed using representative breast cancer cell lines expressing CD44; namely, MDA-MB-231, MCF7, and T47D. The selected RNA aptamer (Apt1) was found to interact specifically with such cancer cells when analyzed by flow cytometry and fluorescent microscopy, with different intensities of fluorescence reflecting the level of CD44 expression on the surface of these cells. It can be concluded that the selected aptamers can be used to target CD44 positive cells, including cancer stem cells, for detection, sorting, and enrichment and for drug delivery purposes.
Introduction
Cancer stem cells (CSCs) are a subset of tumor cells that has the ability to self-renew and to generate the diverse cells that comprise the tumor (Visvader and Lindeman, 2008). Cancer stem cells (CSCs) were first observed in hematological malignancies (Hurt and Farrar, 2008) but have now been identified in solid tumors of breast, prostate, brain, colon, and pancreas (Tang et al., 2007). Cancer stem cells are thought to be resistant to conventional chemotherapy, which makes them potential targets for cancer research and drug development (Tang et al., 2007; Deonarain et al., 2009).
Several cluster of differentiation (CD) markers have been identified specifically on cancer stem cells (WILLIAMS, 2012). The most commonly used surface markers to identify CSCs include CD44, EPCAM, and CD133 (Jaggupilli and Elkord, 2012). CD44 is a cell-surface glycoprotein expressed on lymphocytes, monocytes, and granulocytes, which have been identified as a stem cell marker in some solid tumors, including breast and head and neck cancers (ORIAN-ROUSSEAU, 2010).
CD44 is the receptor for hyaluronan (HA), which is a major component of the extracellular matrix (Wang and Bourguignon, 2011). It is a multistructural and multifunctional cell-surface molecule involved in cell proliferation, cell differentiation, cell migration, angiogenesis, and presentation of cytokines, chemokines, and growth factors to the corresponding receptors (Naor et al., 2002). Hyalronan binding of CD44 seems to prevent apoptosis of tumor cells, rather than promote their migration or invasiveness (Afify et al., 2009).
The CD44, which is also called metastasis-associated protein, is a reliable indicator of tumor load and disease activity (Liu and Jiang, 2006). It also plays an important role in the invasion of a variety of tumor cells, including breast, prostate, and mesotheliomas, and has been positively correlated with the number of circulating prostate cancer cells in the bloodstream (Marhaba et al., 2008; Baumann and Krause, 2010).
Aptamers are an interesting class of high affinity ligands (Jayasena, 2009). They are short, single-stranded (ss) DNA or RNA oligonucleotides, typically isolated from combinatorial libraries by a process of in vitro evolution, termed SELEX (systematic evolution of ligands by exponential enrichment) (Soontornworajit and Wang, 2010). The SELEX procedure is a selection process that allows the isolation of aptamers with unique binding properties from a large library of oligonucleotides through iterative cycles of interaction with the target molecule, separation of bound from unbound aptamer species, elution of bound aptamers, and polymerase chain reaction (PCR) amplification of the binding aptamers for further selection rounds (Stoltenburg et al., 2007).
The present study describes the development of RNA nuclease-resistant aptamer capable of specifically binding to CD44, not only as a purified protein but also by binding on representative breast cancer cells lines.
Materials and Methods
Pool design and library synthesis
An ssDNA library (5′-GGGATGGATCCAAGCTTACTGG (45N) GGG AAGCT TCG ATAGG AATTCGG-3′) was synthesized with a 45-nucleotide random region, with the following forward primer (APT-FT7 5′-GCTAATACGA CTCACTAT AGGGATGG ATC C AAGCTTACTGG-3′ and reverse primer APT-R 5′-CCGAATTCC TATCGAAGCTTCCC-3′). The forward primer APT-FT7 contains a T7 promoter sequence (underlined) for in vitro transcription. The PCR mix contained 10 micrograms of ssDNA library, 5× Gotaq green buffer (Promega), 200 μM of each dNTPs mix, 1 μM of each primer, 2.5 mM MgCl2, and 2.5 U of Taq DNA polymerase. The PCR program starts with 5 minutes at 95°C. The cycling begins with a short denaturation step for 15 seconds at 95°C; the primers are annealed for 20 seconds at 55°C followed by an extension time of 20 seconds at 72°C. These cycles were repeated six times, followed by a final elongation step of 5 minutes at 72°C. The PCR products were purified using 6% PAGE using crush and soak elution method. Following purification, 10 μg of double-stranded DNA were in vitro transcribed into RNA library using DuraScribe T7 Transcription Kit (Epicentre Technologies) according to the manufacturer's instructions and incubated overnight at 37°C, then treated with 10 units of DNase 1 at 37°C for 30 minutes. RNA transcripts were analyzed by 4% agarose gel and purified using 6% PAGE using crush and soak method.
In vitro selection of CD44 proteins using 2′-F-modified RNA library
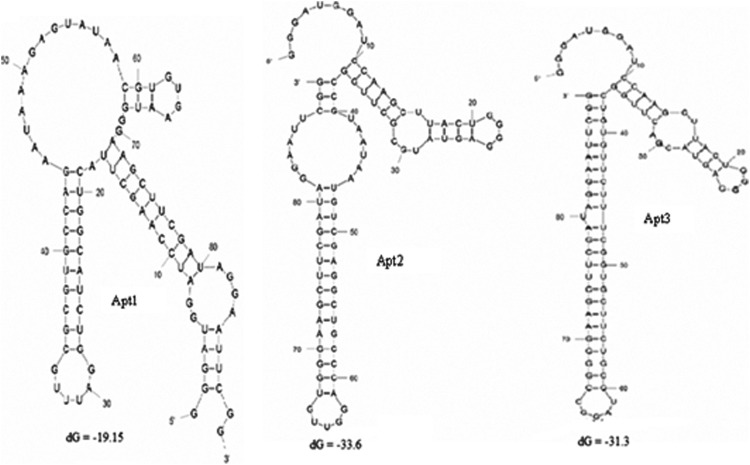
The 2′-F-modified random RNA library was heated at 70°C for 5 minutes in the binding buffer (4.2 mM Na2HPO4, 2 mM K2HPO4, 140 mM NaCl, 10 mM KCl, 2.5 mM MgCl2), then rapidly cooled on ice for 5 minutes, after which it was equilibrated at room temperature for about 10 minutes. The RNA library was then incubated with the glutathione S-transferase (GST)-tagged CD44 recombinant proteins (Abnova Corporation) for 1 hour at 37°C, followed by the separation of nonbounded sequences using GST magnetic beads as negative selection. The selected sequences were eluted from the target, reverse transcribed by SuperScript 3 reverse transcriptase (Invitrogen), amplified by RT-PCR, and then transcribed as described previously to produce RNA transcripts. After 11 iterative rounds, the resultant aptamers were cloned using the pGEM-T Easy vector (Promega). Around 25 of the isolated clones were subsequently sequenced by Macrogen Inc. Sequences analysis and alignments were carried out using the software ClustalW from the website. The most stable secondary structures for the aptamer were predicted using the Mfold program (Fig. 1).
FIG. 1.
The secondary structures of three selected aptamers predicted by the Mfold program.
Synthesis of modified and fluorescein-labeled RNA aptamers
One of the resulted RNA aptamer sequences (Apt1) was synthesized via chemical synthesis using cyanoethylphosphoroamitide chemistry (Midland) on a 50-nanomole scale. The first three nucleotides on each end of the aptamer were synthesized with 2′-O-methyl modification (indicated by lower case letters) to enhance its resistance to nucleases, fluorescein isothiocyanate (FITC)-labeled at the 5′ end and purified by HPLC. To rule out nonspecific cell binding, a control ssDNA random library was synthesized and labeled with FITC as a negative control.
Cellular assays
The human breast adenocarcinoma cell lines MDA-MB-231 and MCF7 and the human ductal breast epithelial tumor cell line T47D were obtained from ATCC (American Type Culture Collection) and cultured in Dulbecco's modified Eagle medium (DMEM): nutrient mixture F-12 (Lonza) supplemented with 10% heat inactivated fetal bovine serum (FBS; Invitrogen), 100 IU/mL penicillin-streptomycin (Invitrogen), and 5mM l-Glutamine (Lonza). The cells were maintained at 37°C in a 5% CO2 atmosphere. Cells were passaged when grown to ∼80% confluency with 1× trypsin (0.05%, Lonza), then cultured in 12-well culture plates containing glass coverslips at a density of 60,000 cells per well, and were grown under standard culture conditions. After 24 hours of incubation, the wells were washed by decanting the culture media and rinsing three times with 1 mL cold 1× phosphate-buffered saline (PBS) per well. Cells were then fixed using 1 mL 4% paraformaldehyde for 10 minutes at room temperature. After fixation, the cells were rinsed two times in 1× PBS, after which nonspecific binding was blocked using 3% bovine serum albumin in PBS for 30 minutes. Cells were then incubated with folded Apt1CD44 aptamer probe (200 nM) in 300 μL of binding buffer (DPBS containing 5 mM MgCl2, 0.1 mg/mLtRNA, and 0.1 mg/mL salmon sperm DNA) in the dark at room temperature for 30 minutes. An ssDNA library labeled with FITC was used as a negative control. After incubation, the cells were washed twice with binding buffer to remove unbound sequences and to reduce the fluorescent background after which coverslips were transferred to slides and dipped in mounting media containing 6-diamidino-2-phenylindole dihydrochloride (DAPI) solution (Cytocell) for nuclear counterstaining. The images were then acquired by flourescent microscopy (Carl Zeiss) with filters for FITC (excitation BP450–490, emission BP515–565) and DAPI (excitation D360/40, emission D460/50). The objective used for imaging was a 60× oil-immersion objective.
Cells of the selected cell lines were cultured until they reached 80% confluency. They were detached with 1× trypsin, centrifuged, and resuspended in 1 mL permeabilizing solution for 10 minutes, then washed twice with binding buffer (DPBS containing 5 mM MgCl2, 0.1 mg/mL tRNA, and 0.1 mg/mL salmon sperm DNA) (Shigdar et al., 2011). The cells were then resuspended in 5 mL assay buffer (DPBS supplemented with 5 mM MgCl2, 0.1 mg/mLtRNA, 0.1 mg/mL salmon sperm DNA, 0.2% sodium azide, and 5% FBS) and incubated for 30 minutes at 4°C, after which cells were washed twice, and 105 cells of each cell line were incubated with 200 nM of FITC-labeled aptamer in a total volume of 200 μL. The binding was carried out in the dark at room temperature for 30 minutes. The same amounts of cells were incubated with phycoerythrin (PE)-labeled anti-CD44 antibody separately as a positive control, and 200nM ssDNA FITC-labeled library was used as a negative control. Finally, the cells were washed three times and resuspended in 250 μL of assay buffer with 2.5 mM MgCl2 extra concentration and analyzed using the FACSCalibur flow cytometer (Becton Dickinson). Typically 10,000 events were counted per sample, and the data was analyzed using CellQuest version 3.3 (BD Biosciences) and WinMDI version 2.8 software.
Binding affinity
The binding affinity constant (Kd) of Apt1 was determined by incubating FITC-labeled Apt1 (350 nM) with CD44 protein (from 45 to 450 nM) for 1 hour at 37°C in 100 μL binding buffer. After incubation, The Apt1-CD44 complexes were immobilized on GST magnetic beads (Promega) for 20 minutes at room temperature and then washed three times with 100 μL of binding buffer to remove unbound Apt1. The Apt1-CD44 complexes were then eluted from the beads using 100 μL elution buffer and incubated for 30 minutes. The fraction of bound Apt1 was quantified by the detection of fluorescence (excitation 494 nm, emission 517 nm) in the elution buffer using a FP-750 spectrofluorimeter (JASCO). The same quantity of FITC-labeled Apt1 (350 nM) was incubated with the GST beads without CD44 protein and used as a negative control to determine the nonspecific binding background. The binding data were analyzed to get a saturating curve by performing a regression analysis fitting model for one binding site.
Cytotoxicity assays
Cellular toxicity of RNA aptamer was assessed using the CellTiter 96-Cell Proliferation Assay (Promega) according to the manufacturer's instructions. Cells of selected cell lines, with a density of 1×104 cells per well, were cultured in 96-well microtiter plates in 200 μL DMEM-F12 complete growth medium overnight for attachment, and during the next day, various RNA aptamers concentrations (50, 100, 250, 500, 1000, 2500 ng) were added in duplicates for 72 hours at the same culture conditions. After that, 100 μL of the media was removed from each well, and 15 μL of (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution was added, mixed by agitating the plate, and incubated for 4 hours at 37°C in the dark. Four hours later, 100 μL of stop solution was added for each well to stop the reaction. Plates were read using an enzyme-linked immunosorbent assay reader, and absorbance was measured at 570 nm (background wavelength is 630 nm). The viability of cells was calculated based on a comparison of untreated cells and those treated with MTT under the same conditions. Cells were incubated for 72 hours prior to counting the number of viable cells.
RNA aptamer stability assay
Cell culture media with 10% FBS was used to detect RNA stability by incubation of 12 μg of each of the O-methyl-modified aptamers with 0.5 mL of cell culture media at 37°C. Samples of 50 μL were taken out and collected after 0, 0.5, 1, 1.5,2, 3, 4, 6, and 24 hours. RNA was reverse transcribed as described above, and 2 μL complementary DNA were amplified using GoTaq® qPCR Master (Promega), and 25 pmole of forward and reverse primers. The conditions for the PCR were 95°C for 10 minutes as initial denaturation, 95°C for 20 seconds, 55°C for 20 seconds, and 72°C for 30 seconds, for 40 cycles.
Results
SELEX results, folding prediction, and binding affinity
Following 11 iterative SELEX rounds and successful cloning and sequencing of the selected aptamers, the sequence homology was determined. Based on the sequencing analysis, three sequences were represented more than once in the 25 clones that were analyzed. Full-length sequences and frequencies of the three selected aptamers are shown in Table 1. One of the sequences (Apt1) was synthesized and used in the cellular assays. Apt1 was synthesized and labeled with FITC, and the first three nucleotides on each end of the aptamer sequence were modified with 2′O-methyl group for better nuclease resistance.
Table 1.
Full Sequences of the Three Selected Aptamers
| Name | Frequency of sequence (%) | No. of clones | Sequence |
|---|---|---|---|
| Apt1 |
76 |
19 |
GGGAUGGAUCCAAGCUUACUGGCAUCUGGAUUUGCGCGUGCCAGAAUAAAGAGUAUAACGUGUGAAUGGGAAGCUUCGAUAGGAAUUCGG |
| Apt2 |
12 |
3 |
GGGAUGGAUCCAAGCUUACUGGGGAGUAUGCGCUUGGCCGUAAUAAUGUCGAGGCUGCCCAGGUUGUGGGAAGCUUCGAUAGGAAUUCGG |
| Apt3 | 12 | 3 | GGGAUGGAUCCAAGCUUACUGGGGAGUACGACUUGGCUGUGUUCUUUCGGUGCUUCUGCGUAGGCCGGGGAAGCUUCGAUAGGAAUUCGG |
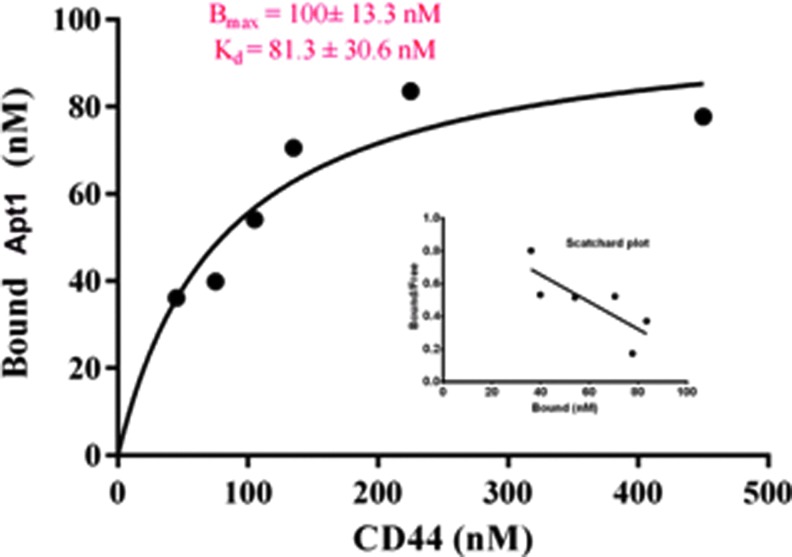
Secondary structures of the selected aptamers were predicted using the Mfold software (Fig. 1). Each sequence showed different free energy values (dG) that reflect the stability of folding structures. The results showed that all selected sequences can form hairpin loop structures with the random region forming the loop and the primer regions making up the stem regions. We also tested the binding affinity for the dominant aptamer Apt1 to CD44 protein using GST magnetic beads separation technique. The results clearly demonstrate the ability of Apt1 to bind CD44 with high affinity (Kd=81.3±30.6 nM) as shown in Fig. 2.
FIG. 2.
Fitting curve to measure the affinity binding of fluorescein isothiocyanate (FITC)-labeled Apt1 to cell surface glycoprotein (CD44). The data were fit with regression analysis for one binding site using GraphPad Prism 6 software. The figure insert shows the scatchared plot. Color images available online at www.liebertpub.com/nat
Cellular assays
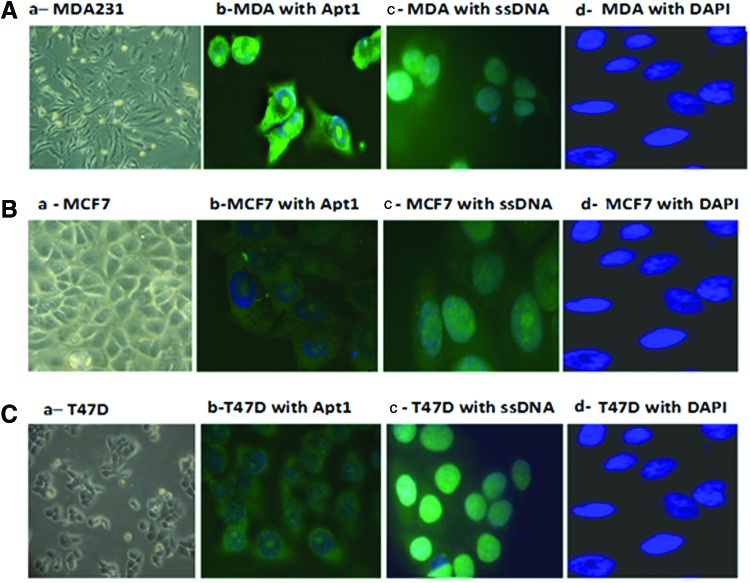
The binding specificity of the selected aptamers was evaluated using the Apt1 FITC-labeled RNA aptamer and CD44+ human cancer cell lines. The main cell line was the breast adenocarcinoma-derived cell line MDA-MB-231, which expresses a large amount of CD44 molecules on the cell surface (Ricardo et al., 2011). The MCF7 cell line and the ductal breast epithelial tumor-derived cell line T47D, which have lower levels of CD44 expression, were also used (Ricardo et al., 2011). The fluorescence intensities of all cell lines were compared using the fluorescent microscope (Fig. 3). The MDA-MB-231 cells clearly showed higher FITC fluorescence intensities as compared to MCF7 and T47D. It is worth mentioning here that the synthesized aptamer had no effect on cell growth and proliferation as assessed by the MTT assay.
FIG. 3.
Detection of CD44 by fluorescence microscopy using FITC-Apt1. Figures A–C represent results for MDA-MB-231, MCF7, and T47D cell lines, respectively. Frames a–d for each cell line represent the following: (a) light microscopy (20×) view of cultured cells before aptamer binding, (b) the binding of Apt1 to the cultured cells, (c) the binding of FITC labeled non-specific single-stranded (ss) DNA library, and (d) the DAPI nuclear DNA counterstain. These 60× fluorescent photographs (b–d), clearly show the higher expression of CD44 by MDA-MB-231 cells as compared with MCF7 and T47D cells. Color images available online at www.liebertpub.com/nat
Flow cytometry analysis
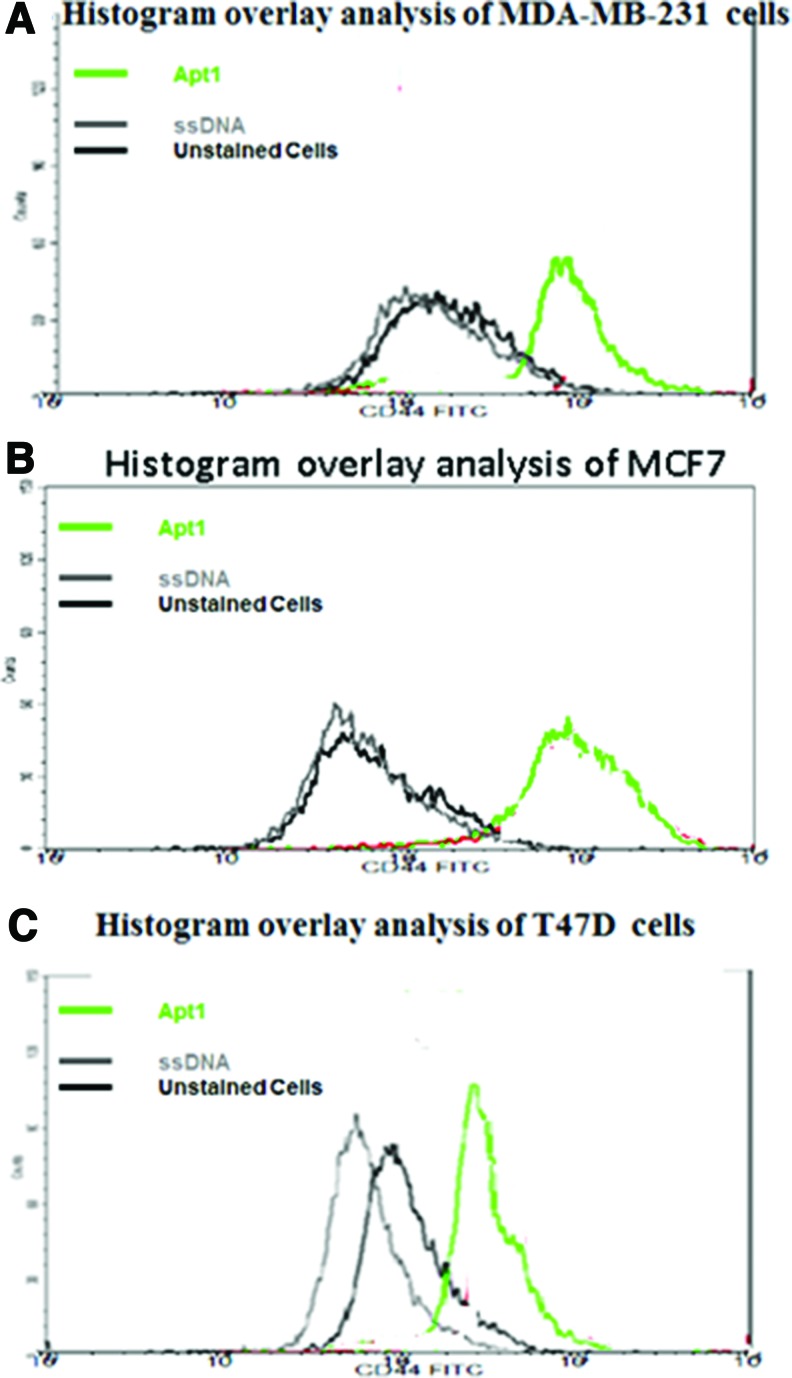
Flow cytometry was used to confirm that the interaction observed between Apt1 and human cancer cells and to rule out nonspecific binding of RNA aptamers to cultured cells (Fig. 4). The binding specificity of Apt1 was evidenced by comparing the measured fluorescence intensities on the histograms shown in Fig. 4. Histogram analysis of the reacted sequences was specific with an observed higher affinity to Apt1 binding. The randomized FITC-labeled ssDNA probe, used as a negative control had little to no binding capacity to the used cancer cells. Unstained cells showed background fluorescence due to autofluorescence.
FIG. 4.
FACscan histogram analysis of the binding of Apt1 to breast cancer cell lines. (A) Histogram analysis for the binding of selected aptamer to MDA-MB231 cells, (B) MCF7 cells, and (C) T47D cells. Green curve represents the binding of Apt1 FITC-labeled aptamers to the cells. Unselected ssDNA library was used as a negative control (gray), and unstained cells were used as a fluorescence background (black). Color images available online at www.liebertpub.com/nat
Aptamer modification and stability
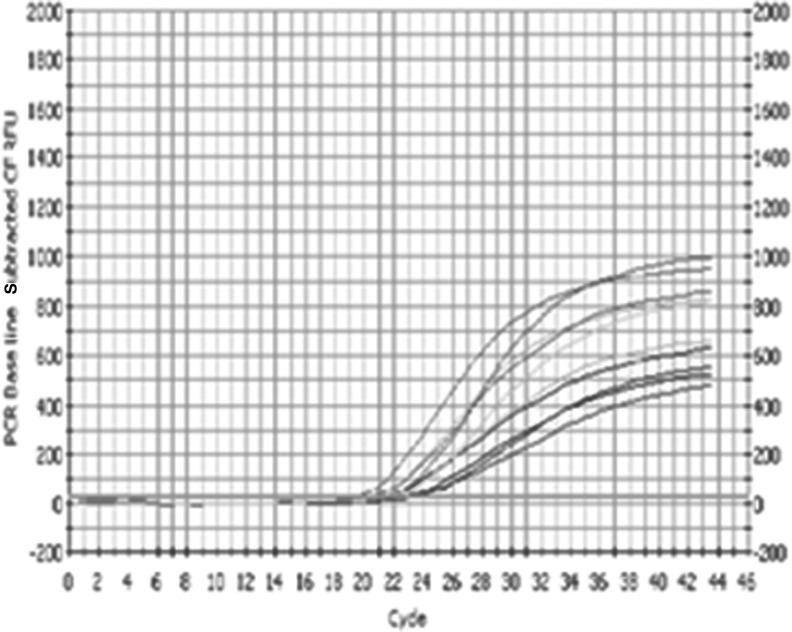
For effective potential use in therapeutic or diagnostic applications, aptamers must resist the degradation by exo- and endonucleases. To achieve adequate in vivo stability of aptamers, 2′O-methyl modifications have been incorporated. To test the stability of 2′O-methyl-modified Apt1, stability assays was carried out by incubating the modified aptamers in growth media containing 10% Fetal Bovine Serum (FBS) for 36 hours. Real time PCR results confirmed that the modified RNA aptamers was very stable when incubated at 37°C for more than 24 hours, as there was a little difference between cycle thresholds for the amplified aptamers at different time points (Fig. 5).
FIG. 5.
RNA stability determination for Apt1 by real time polymerase chain reaction (PCR) at different times (0–24 hours). The starting quantity of DNA template is proportional to the threshold cycles. Real time PCR results also confirmed that the Apt1 RNA aptamer was very stable, as there was little difference between cycle thresholds at different time points.
Discussion
In this study, the SELEX methodology was used to select modified RNA aptamers capable of targeting the CD44 receptor protein. To the best of our knowledge, the three aptamers described here are the first RNA aptamers developed against the cancer stem cell marker CD44. These aptamers were selected with the eventual future goal of using them for targeted delivery of therapeutic drugs to cancer cells expressing CD44.
Fluorescent microscopy showed that Apt1 CD44 aptamer probes did bind specifically to CD44 expressing cells, which was demonstrated by the staining of the cell membranes of the representative cancer cell lines. The MDA-MB-231 cell line demonstrated the highest fluorescence when bound to Apt1, whereas MCF7 and T47D showed lower fluorescence intensity. Such results reflect the previously reported differential levels of CD44 expression on the surface of these cell lines (Ricardo et al., 2011). It should be emphasized here that not all aptamer sequences developed by SELEX against purified form of their target proteins would automatically bind to their targets in their cellular context.
It was noticed that the Apt1 CD44 aptamer probes stained the cell membranes and, more diffusely, the cytoplasm of these cancer cell lines. This staining pattern may be due to the better penetration and more efficient reaction of aptamer probes within the fixed cells. Successful cellular internalization has been reported for a number of aptamers, including anti-PSMA, which targets PSMA in prostate cancer cells, and Sgc8, which targets PTK7 in acute leukemia cells (Lupold et al., 2002; Xiao et al., 2008; Zhou and Rossi, 2009). Twenty-five-base-long, synthetic single-stranded DNA aptamers were derived to bind to known internalized tumor markers such as CD33, CEA, MUC1, and Tn antigens (Orava et al., 2010) and were imported through these surface portals into cancer cells. Zhang et al. reported the development of a cancer-cell-specific DNA aptamer probe, KMF2-1a, using the cell-SELEX method that was successfully internalized to the endosome of target breast cancer cells (Zhang et al., 2012). These results further support the finding that aptamers can serve as promising agents for cell-type-specific intracellular delivery with both diagnostic and therapeutic implications.
The selected aptamers were labeled with FITC and modified with 2′-O-methyl group to inhibit their degradation by nucleases. It has been established that aptamers can be directly modified at either the 5′ or 3′ end using several fluorescent dyes without interfering with the aptamer folding and its ability to bind its target (López-Colón et al., 2011).
Fluorescein-labeled aptamers have been used for imaging studies of cultured cells by fluorescence microscopy, as well as by flow cytometry for a variety of cell types, including leukemia, lung, liver, ovarian, and colorectal cancer cells, as well as virus-infected cells (López-Colón et al., 2011). Somasunderam et al. (2010) reported the selection of ssDNA thioaptamers that can bind hyaluronic acid binding domain of the CD44 receptor. The results of this study showed that the selected thioaptamers did bind the CD44 protein with a higher affinity (180–295 nM) compared with hyaluronic acid (Kd>61 μM). Moreover, the specificity of the selected aptamers confirmed by its ability to bind positive human ovarian cancer cell lines (SKOV3, IGROV, and A2780) while failing to bind the CD44 negative NIH3T3 cell line.
In our study, flow cytometry analysis of FITC-labeled CD44 RNA aptamers with breast cancer cells showed that there is an evident binding between Apt1 and cancer cells, in terms for specificity as shown by histogram analysis. This also confirms the specific binding of CD44 aptamers to CD44-positive cancer cells. These results demonstrate the potential application of the selected aptamers to distinguish between normal and tumor tissue sections.
Aptamers, especially RNA-based ones, are susceptible to degradation by environmental nucleases, largely limiting their clinical value. In this study, the selected aptamers were modified with 2′-O-methyl group to improve the nuclease resistance of the RNA-based aptamers, which significantly increased the stability of these aptamers making them less susceptible to nuclease degradation. These features improve the capabilities of aptamers when used for diagnostic and therapeutic applications.
On the basis of the findings of this study, it is concluded that the isolated RNA aptamers bind specifically to CD44 molecules on the representative cell lines expressing this marker. With further characterization, the developed CD44 aptamers might be exploited in therapeutic applications as vehicles for targeted delivery of cytotoxic drugs against cancer stem cells, as well as in diagnostic assays aimed at detecting such cells in cancer patients. Such further characterization would include first establishing the binding affinities for selected aptamers, and truncating the aptamers to reach the smallest functional sequence.
Acknowledgments
The authors wish to thank all staff members of the molecular biology research lab and the immunology laboratory at Jordan University Hospital, especially Dr. Malek Sallam for his help in flow cytometry and immunofluorescence. The authors also wish to thank all staff members of the genetics laboratory at the National Center for Diabetes, Endocrinology, and Genetics, especially Wesam Mohammad.
Disclosure Statement
No competing financial interests exist.
References
- AFIFY A., PURNELL P., and NGUYEN L. (2009). Role of CD44s and CD44v6 on human breast cancer cell adhesion, migration, and invasion. Exp. Mol. Pathol. 86,95–100 [DOI] [PubMed] [Google Scholar]
- BAUMANN M., and KRAUSE M., (2010). CD44: a cancer stem cell related biomarker with predictive potential for radiotherapy. Clin. Cancer Res. 16,5091–5093 [DOI] [PubMed] [Google Scholar]
- DEONARAIN M.P., KOUSPAROU C.A., and EPENETOS A.A. (2009). Antibodies targeting cancer stem cells: a new paradigm in immunotherapy? MAbs. 1,12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HURT E.M, and FARRAR W.L. (2008). Cancer stem cells: the seeds of metastasis? Mol Interv. 8,140–142 [DOI] [PubMed] [Google Scholar]
- JAGGUPILLI A., and ELKORD E. (2012). Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin. Dev. Immunol. 10.1155/2012/708036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAYASENA S.D. (1999). Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45,1628–1650 [PubMed] [Google Scholar]
- LIU J., and JIANG G. (2006). CD44 and hematologic malignancies. Cell Mol. Immunol. 3,359–365 [PubMed] [Google Scholar]
- LÓPEZ-COLÓN D., JIMÉNEZ E., YOU M., GULBAKAN B., and TAN W., (2011). Aptamers: turning the spotlight on cells. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 3,328–340 [DOI] [PubMed] [Google Scholar]
- LUPOLD S.E., HICKE B.J., LIN Y., and COFFEY D.S. (2002). Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 62,4029–4033 [PubMed] [Google Scholar]
- MARHABA R., KLINGBEIL P., NUEBEL T., NAZARENKO I., BUECHLER M.W., and ZOELLER M. (2008). CD44 and EpCAM: cancer-initiating cell markers. Curr. Mol. Med. 8,784–804 [DOI] [PubMed] [Google Scholar]
- NAOR D., NEDVETZKI S., GOLAN I., MELNIK L., and FAITELSON Y. (2002). CD44 in cancer. Crit. Rev. Clin. Lab. Sci. 39,527–579 [DOI] [PubMed] [Google Scholar]
- ORAVA E.W., CICMIL N., and GARIEPY J. (2010). Delivering cargoes into cancer cells using DNA aptamers targeting internalized surface portals. Biochim. Biophys. Acta 1798,2190–2200 [DOI] [PubMed] [Google Scholar]
- ORIAN-ROUSSEAU V. (2010). CD44, a therapeutic target for metastasising tumours. Eur. J. Cancer 46,1271–1277 [DOI] [PubMed] [Google Scholar]
- RICARDO S., VIEIRA A.F., GERHARD R., LEITÃO D., PINTO R., CAMESELLE-TEIJEIRO J.F., MILANEZI F., SCHMITT F., and PAREDES J., (2011). Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 64,937–946 [DOI] [PubMed] [Google Scholar]
- SHIGDAR S., LIN J., YU Y., PASTUOVIC M., WEI M., and DUAN W. (2011). RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 102,991–998 [DOI] [PubMed] [Google Scholar]
- SOMASUNDERAM A., THIVIYANATHAN V., TANAKA T., LI X., NEERATHILINGAM M., LOKESH G.L., MANN A., PENG Y., FERRARI M., KLOSTERGAARD J., and GORENSTEIN D.G. (2010). Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of human CD44. Biochemistry 26,9106–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOONTORNWORAJIT B., and WANG Y. (2011). Nucleic acid aptamers for clinical diagnosis: cell detection and molecular imaging. Anal. Bioanal. Chem. 399,1591–1599 [DOI] [PubMed] [Google Scholar]
- STOLTENBURG R., REINEMANN C., and STREHLITZ B. (2007). SELEX: a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 24,381–403 [DOI] [PubMed] [Google Scholar]
- TANG C., ANG B.T., and PERVAIZ S., (2007). Cancer stem cell: target for anti-cancer therapy. FASEB J. 21,3777–3785 [DOI] [PubMed] [Google Scholar]
- VISVADER J.E., and LINDEMAN G.J. (2008). Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 8,755–768 [DOI] [PubMed] [Google Scholar]
- WANG S.J., and BOURGUIGNON L.Y. (2011). Role of hyaluronan-mediated CD44 signaling in head and neck squamous cell carcinoma progression and chemoresistance. Am. J. Pathol. 178,956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS J.L. (2012). Cancer stem cells. Clin. Lab. Sci. 25,50–57 [PubMed] [Google Scholar]
- XIAO Z., SHANGGUAN D., CAO Z., FANG X., and Tan W. (2008). Cell-specific internalization study of an aptamer from whole cell selection. Chem. Eur. J. 14,1769–1775 [DOI] [PubMed] [Google Scholar]
- ZHANG K., SEFAH K., TANG L., ZHAO Z., ZHU G., YE M., SUN W., GOODISON S., and TAN W. (2012). A novel aptamer developed for breast cancer cell internalization. ChemMedChem 7,79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU J., and ROSSI J.J. (2009). The therapeutic potential of cell internalizing aptamers. Curr. Top. Med. Chem. 9,1144–1157 [DOI] [PubMed] [Google Scholar]