Abstract
Methods to detect DNA and RNA (collectively xNA) are easily plagued by noise, false positives, and false negatives, especially with increasing levels of multiplexing in increasing complex assay mixtures. We describe assay architectures that mitigate these problems, here by converting standard xNA analyte sequences into sequences that incorporate non-standard nucleotides (Z and P). Z and P are extra DNA building blocks that form tight non-standard base pairs without cross-binding to natural oligonucleotides containing G, A, C, and T (GACT). The resulting improvements are assessed in an assay that inverts the standard Luminex xTAG® architecture, placing a biotin on a primer (rather than on a triphosphate). This primer is extended on the target to create a standard GACT extension product that is captured by a CTGA oligonucleotide attached to a Luminex bead. Using conversion, a polymerase incorporates dZTP opposite template dG in the absence of dCTP. This creates a Z-containing extension product that is captured by a bead-bound oligonucleotide containing P, which binds selectively to Z. The assay with conversion produces higher signals than the assay without conversion, possibly because the Z:P pair is stronger than the C:G pair. These architectures improve the ability of the Luminex instruments to detect xNA analytes, producing higher signals without the possibility of competition from any natural oligonucleotides, even in complex biological samples.
Keywords: Detection of Nucleic Acids, Expanded Genetic Alphabet, Conversion Strategy, ZiP-TAG Technology, Luminex xTAG Technology
Assays that target DNA and RNA (collectively xNA) are the gold standards for diagnosing infectious disease.1,2 This is so, in part, because such assays directly detect the causative agent for the disease, rather than some derived molecules. Further, the xNA molecules from an infectious agent, often scarce in a biological sample, can be amplified using PCR, reverse transcriptase PCR (RT-PCR), and other in vitro nucleic acid amplification tools. 3 Finally, detection can be coupled with amplification (as in real-time PCR) and can take advantage of the specificity of sequence hybridization in high throughput assays (as with microarrays).4,5
However, many of the features that make xNA molecules good analytes also create problems. For instance, the amplified xNA molecules from one sample easily become the source of contamination for another sample. Further, in high throughput assays, cross-hybridization between mismatched species can generate false positive signals.6,7 Also, hybridization between probes and analytes is easily interrupted by background nucleic acids to generate false negative signals, especially in complex mixtures. To mitigate these problems, much effort has been applied to select the target, design primers and probes, optimize amplification conditions, and refine detection procedures.8
In principle, expanded genetic systems with additional nucleotide “letters” might also mitigate these problems.9 A six-letter genetic alphabet system (G, A, C, T, X, and Y) offers higher information density than standard genetic system, with 6 n different sequences of length n rather than 4n, the number with just four nucleotides. Therefore, hybridization of xNA strands built from a six-letter alphabet (GACTXY) should be cleaner than with a 4-letter alphabet (GACT).
This abstract notion can today be implemented, as many additional nucleotide "letters" that form additional nucleobase pairs have been delivered by synthetic biologists over the past two decades.10 In many cases, these new base pairs form "orthogonally", meaning that the additional nucleotides do not pair with standard nucleotides. Further, in many cases, DNA polymerases are now available that synthesize duplex DNA containing non-standard base pairs via template-directed polymerization.
This orthogonality has already had considerable impact on diagnostics. For example, 2'-deoxyisoguanosine (isoG) and 5'-methyl-2'-isodeoxycytidine (isoC) form an unnatural base pair that is orthogonal to the standard G:C and A:T pairs, allowing it lower background noise in FDA approved bDNA assays to measure viral load in HIV and hepatitis patient blood.11,12,13 Further, this unnatural base pair supports real-time and multiplexed PCR-based assays in MultiCode® Technology (Luminex), where specific base pairing allows the incorporation of quenchers at specific sites in a DNA duplex.14,15,16
Unfortunately, isoC and isoG have significant disadvantages. Due to a minor tautomer that allows isoG to pair with T, the isoG:isoC base pair is replaced by the standard A:T base pair during PCR. 17 Further, the isoC nucleoside suffers decomposition in acid, making oligonucleotides containing it challenging to prepare, especially to meet FDA specifications.
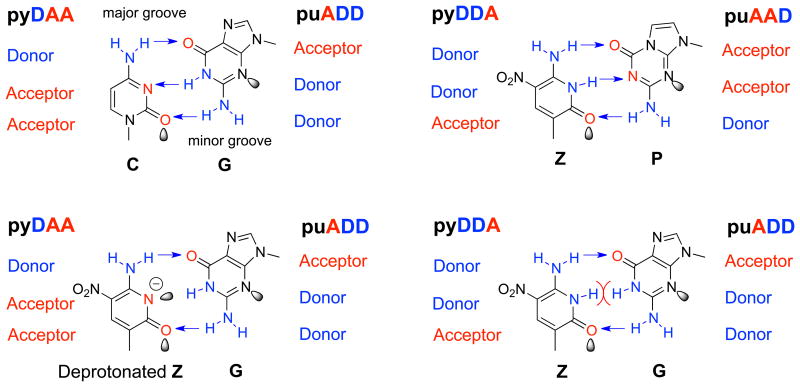
Accordingly, much of our recent work has focused on two other non-standard nucleotides, 2-amino-8-(1′-β-D-2′-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)one (Figure 1, trivially called P), and 6-amino-5-nitro-3-(1- β-D-2′-deoxyribofuranosyl)-2(1H)-pyridone (Figure 1, trivially called Z). These can form the orthogonal non-standard Z:P base pair.18 Further, a range of molecular biological tools has been developed to use them, including polymerases,19 restriction endonucleases,20 and sequencing technology.21
Figure 1.
(Top) dC:dG and dZ:dP pair with Watson-Crick geometry via three hydrogen bonds (the arrows). Deprotonated dZ forms a pair with dG (bottom, right), supporting conversion. Normal dZ cannot form a pair with dG (bottom, left). Purine analogs are indicated by "pu". Pyrimidine analogs are indicated by "py", with hydrogen-bonding acceptor (A) and donor (D) groups.
Of course, neither Z nor P is found in standard xNA analytes. Therefore, to use the Z:P pair in standard xNA diagnostics, architectural innovations must be developed to build them into amplification, capture, and detection schemes.
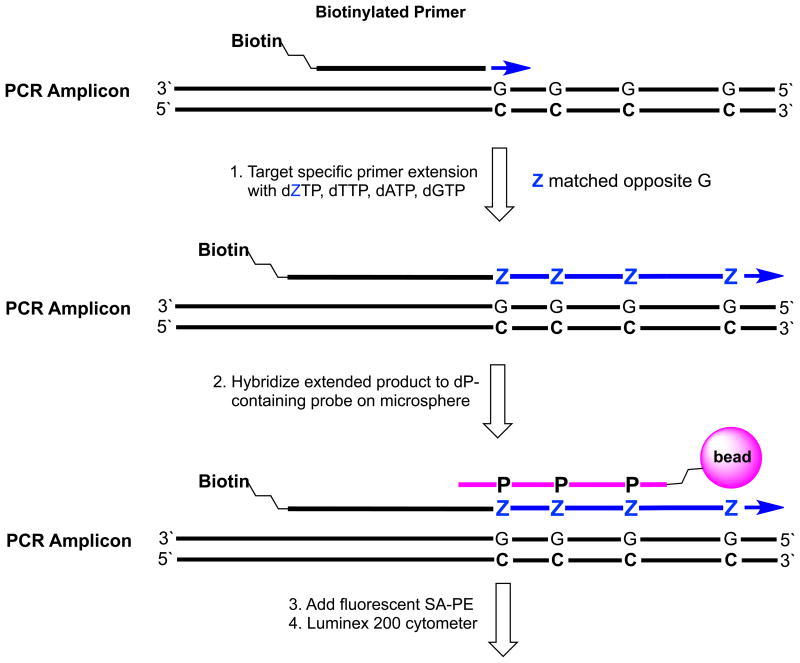
Here, we describe a way to include Z:P pairs in xNA-targeted analyses. This ZiP-TAG™ conversion architecture (Figure 2) exploits the Luminex instrument, and was inspired by the Luminex xTAG system.22,23 However, in the ZiP-TAG assays, the xTAG architecture (Figure 3B) is inverted, and special conditions are exploited to allow polymerases to incorporate dZTP opposite G in a template, rather than dCTP. This “conversion” creates a Z-containing primer extension product that can be orthogonally captured by a bead-bound oligonucleotide containing P. First, we show that the inverted architecture performs, in some ways, better than the standard Luminex xTAG architectures, even without conversion. Then, we show that by adding conversion, consistently stronger signals are seen in multiplexed environments using probe-tag combinations that cannot suffer interference from any standard DNA.
Figure 2.
The ZiP-TAG™ conversion architecture. A target-specific biotinylated primer is extended with dGTP, dATP, dZTP, and dTTP to create a GAZT extension sequence (step 1). The biotinylated product is captured by a color-coded Luminex microsphere via hybridization of bead-bound CTPA probe with its complementary GAZT extension (step 2). The captured product is labeled with fluorescent streptavidine-phycoerythrin (SAPE) and analyzed by Luminex cytometer (steps 3 and 4).
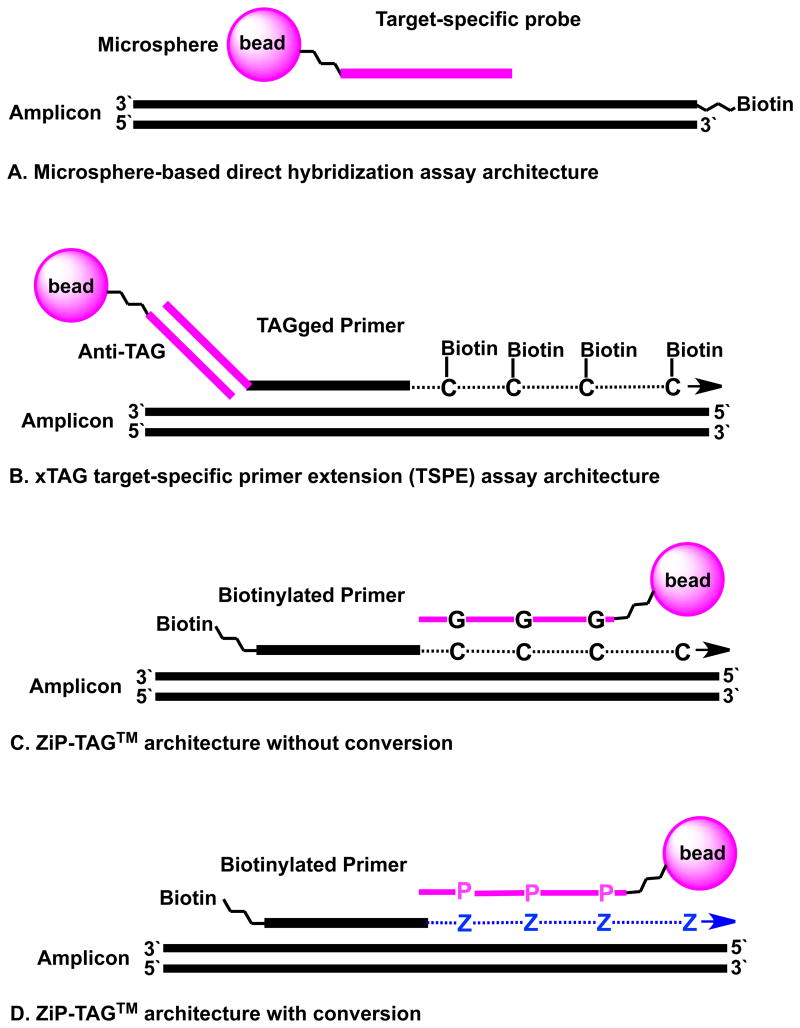
Figure 3. Four assay architectures exploit Luminex microspheres carrying oliogonucleotide probes.
(A) Microsphere-bound probe complements a region of the biotin-labeled amplicon. In a symmetric PCR reaction, the antisense strand of the amplicon competes with the probe for binding to the “sense” biotinylated strand of the amplicon. Asymmetric PCR lowers this competition.
(B) xTAG TSPE extends a TAGged primer binding a segment within amplicon. Anti-TAG on the microsphere complementary to the xTAG captures biotinylated products.
(C) ZiP-TAG™ without conversion. Biotinylated primers are extended with standard triphosphates to give GACT segments complementary to microsphere-bound probes built from standard CTGA nucleotides. This differs from the outcome of architecture (A) in the placement of the biotin, and thereby supports more diverse details, including the ability to use common primers in a multiplexed PCR amplification and the exploitation of two levels of specificity (target-specific primer extension and capture the extended segment).
(D) ZiP-TAG™ with conversion. Biotinylated primers are extended with dZ incorporated opposite template dG to create a GAZT segment complementary to a microsphere-bound probe built from CTPA nucleotides.
Materials and methods
Oligonucleotides
All standard oligonucleotides, 5'-amino (NH2)-modified, and 5′-biotinylated standard oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). All dZ and dP containing oligonucleotides (Supplementary Tables S1 and S3) were synthesized in-house on an ABI 394 DNA Synthesizer using protected dZ and dP phosphoramidites (www.firebirdbio.com, Cat. # DZPhosphor-101, Cat. # DPPhosphor-102). Microsphere-bound anti-TAG sequences (probes) were amino (NH2)-modified and purified by rp-HPLC. TAG (target) sequences used to validate hybridization were biotinylated and purified by rp-HPLC. Primers without modifications were purified by standard desalting. Chimeric target-specific primer extension (TSPE) primers, which had a 24mer TAG sequence 5′ to the target-specific sequence (Supplementary Table S5), were from IDT and purified by PAGE.
Reagents
Bst, Taq, VentR® (exo-), Deep VentR™(exo-)™, 9°N™ (modified), Therminator ™ and Therminator ™ II DNA polymerases were from New England Biolabs. Jumpstart ™ Taq DNA polymerase was from Sigma-Aldrich (Louis, MO). Platinum® GenoTYPE Tsp DNA polymerase and Biotin-14-dCTP were from Invitrogen Corporation. ExoSAP-IT® was from Affymetrix, Inc. (Cleveland, Ohio). All buffers were provided by the supplier of the corresponding enzyme. Individual deoxyribonucleoside triphosphates (excluding dZTP) were from Promega (Madison, WI). dZTP (Cat. # DZTP-101) was from Firebird Biomolecular Sciences (Gainesville, FL). MicroPlex™ Microspheres and Microplex®-xTAG® Microspheres were from Luminex Corporation (Austin, TX). The EDC was from Pierce. MES, 100 g/L sodium dodecyl sulfate, NaCl, Tris, Triton X-100, Tween 20, and Tris-EDTA buffer were from EM Science. The streptavidin-conjugated phycoerythrin (2 mg/μL) was from Molecular Probes, Inc.
Monoplexed and Five-Fold Multiplexed PCR Amplification of Five Target Genes
Monoplexed and five-fold multiplexed PCRs (50 μL) targeted human genomic DNA (Male, 200 ng, Promega), dNTPs (200 μM each), JumpStart Taq DNA polymerase (2.5 units, Sigma), and 1x reaction buffer. Monoplexed reactions contained one set of forward and reverse primers (0.4 μM of each, see Supplementary Table S2); five-fold multiplexed PCR contained five sets of forward and reverse primers (0.4 μM of each). PCR: Initial denaturation was done at 95 °C for 1 min, followed by 31 cycles of (95 °C for 20 s, 60 °C for 30 s, 72 °C for 30 s), and final 72 °C for 10 min. Upon completion, samples (8 μL) from each PCR were mixed with 6x agarose loading dye (2 μL, Promega) and resolved by 3% agarose gel electrophoresis (Figure S6).
PCR products (15 μL) were mixed with ExoSAP-IT (6 μL, USB), the mixture was incubated at 37 °C for 30 min, followed by incubating at 80 °C for 20 min to inactivate enzymes. Treated PCR products (6 μL) were added directly to TSPE reaction.
Target Specific Primer Extension (TSPE)
5′-Biotinylated primer extended with four standard dNTPs (non-conversion) or dZTP + dATP + dTTP + dGTP (ZiP-TAG conversion)
Monoplexed and five-fold multiplexed TSPE were done with ExoSAP-treated PCR products (6 μL, final ca. 25 nM), 5'-biotinylated target-specifically internal primers (final 50 nM for each, Supplementary Tables S4 and S5), Therminator DNA polymerase (1 unit, NEB), and ThermoPol Buffer (20 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM KCl, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8 at 25 °C). Without conversion, four standard dNTPs (final 0.2 mM each) were added. For conversion, nucleoside triphosphates (dATP, dTTP, dGTP, and dZTP, final 0.2 mM of each) were added. TSPE reactions (40 μL final) were at 94 °C for 1 min, followed by 10 cycles (94 °C for 20 s, 58 °C for 30 s, 72 °C for 60 s), and last 72 °C for 1 min. Reactions were then held at 4 °C and quenched with dH2O (160 μL containing 1 μL of 0.5 M EDTA). The final concentration of each 5′-biotinylated primer was ∼10 fmoles/μL.
Luminex xTAGged primer extended with dATP, dTTP, dGTP, and Biotin-14-dCTP (xTAG)
Monoplexed and five-fold multiplexed TSPE were executed with ExoSAP-treated PCR products (6 μL, final ca. 25 nM). xTAGged primers (each 50 nM final, Supplementary Table S3), Platinum Tsp DNA Polymerase (3 units, Invitrogen), and Platinum Tsp Buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl, 1.5 mM MgCl2). Nucleoside triphosphates (dATP, dTTP, dGTP, Biotin-14-dCTP, each 10 μM final) were added. The mixtures (40 μL final) were incubated at 94 °C for 1 min, and then subjected to 40 cycles of PCR (94 °C for 20 s, 58 °C for 30 s, 72 °C for 60 s); and last 72 °C (1 min). Mixtures were held at 4 °C and quenched with 160 μL of dH2O (containing 1 μL 500 mM EDTA). The final concentration of each TAGged primer was ca. 10 fmoles/μL.
Array Sorting
5'-Amino-modified GACT target-specific probes and 5'-amino-modified GACTP target-specific probes were coupled to carboxylated microspheres as described in the “microsphere coupling” section. Microspheres carrying universal anti-TAG probes (Microplex®-xTAG®) were purchased from Luminex. GACT probes, GACTP probes, and anti-TAG probe sequences are listed in Supplementary Table S3, S4, and S5. Each hybridization reaction used ∼5000 microspheres of each microsphere type (Bead 1, Bead 3, Bead 9, Bead 11, and Bead 19). The microspheres were recovered by centrifugation (≥ 8000 × g 2 minutes), the supernatant was removed, and the microspheres were re-suspended in 2x Tm Hybridization Buffer (0.4 M NaCl, 0.2 M Tris, 0.16% Triton X-100, pH 8.0, final 100 microsphere per μL). Aliquots (50 μL) of the microsphere suspension were added to each hybridization reaction with dH2O (50 μL); the background signal was measured. Aliquots (20 μL) obtained by 5-fold serial dilutions (200 fmoles, 40 fmoles, 8 fmoles, or 1.6 fmoles of each analyte) were added to the sample tubes and sorted by the Luminex instrument. The volume was adjusted to 100 μL with dH2O. Heating (95°C, 2 min) and cooling (to 37°C at 0.1°C/s) was followed by hybridization (37°C, 10 min). Then at room temperature, aliquots (50 μL) of 1X Tm Hybridization Buffer containing streptavidin-R-phycoerythrin (6 μg/mL) were added (final volume 150 μL). Each sample was incubated at 37 °C for (10 min). Samples (40 μL) were analyzed at 37 °C on the Luminex 200™ System in triplicate. The instrument was set to read a minimum of 100 events per microsphere with the gate setting established before samples were run. Some results are shown in Figure 4 and 5, as well as in the Supplementary Figures.
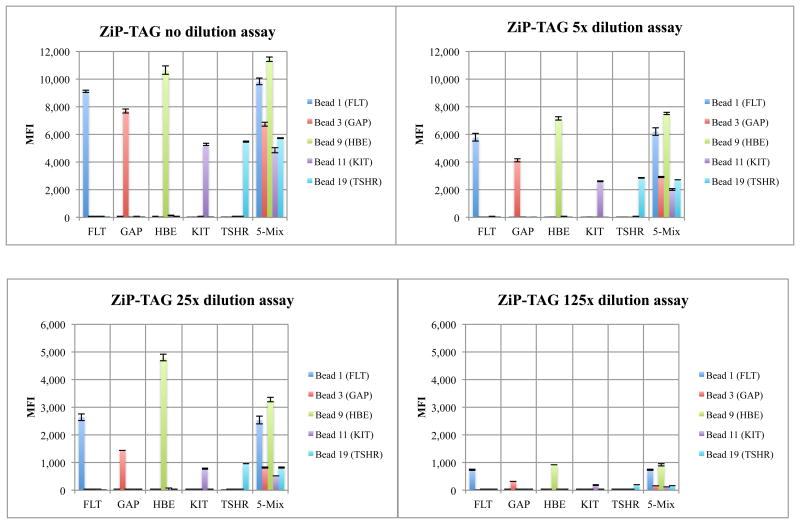
Figure 4.
Luminex detection using the ZiP-TAG architecture with conversion gives a linear dose-response for different amount of analyte (200 fmoles, 40 fmoles, 8 fmoles, and 1.6 fmoles). In each cluster, the 5 bars represent mean fluorescent intensities (MFIs) for five microsphere colors. Clusters 1-5 contain analyte derived by monoplexed PCR. Cluster 6 shows results from five-fold multiplexed PCR.
Figure 5.
Luminex detection using ZiP-TAG architectures without conversion gives a linear dose-response for different amount of analyte (200 fmoles, 40 fmoles, 8 fmoles, and 1.6 fmoles). In each cluster, the 5 bars represent mean fluorescent intensities (MFIs) for the five microsphere colors. Clusters 1-5 contain analyte derived by monoplexed PCR. Cluster 6 shows results from five-fold multiplexed PCR.
Results
Enzymes to Incorporate dZTP Opposite Template dG in the Absence of dCTP
To implement the ZiP-TAG™ conversion architecture (Figure 2), we sought polymerases that might efficiently mismatch dZTP opposite template dG during primer extension (Supplementary Figure S1). In these extension reactions, dZTP was replaced by dCTP, which was absent.
Of the polymerases tested, Therminator most effectively incorporated dZTP opposite isolated template dGs in the absence of dCTP. Therefore, Therminator was examined in detail to determine whether it could support conversion with wide sequence diversity. This was done by challenging Therminator to incorporate multiple dZTPs opposite multiple consecutive template dGs. All primers were extended to give full-length products with negligible pausing, even with four consecutive dGs (data not shown). Further, the efficiency of incorporation of dZTP opposite template dG by Therminator depends on pH. The higher the pH of the buffer (ThermoPol, varying from pH 8.0 to 9.0), the more facile the incorporation of dZTP opposite dG.
Hybridization of GACT Duplexes Compared with Hybridization of GACTZP Duplexes
To demonstrate hybridization, biotinylated GACT and GACTZ DNA analytes were synthesized with sequences complementary to corresponding capture probes (Supplementary Table S1). Formation of GACTZP DNA duplexes at pH 8 (consistent with standard Luminex procedure) was shown on the microspheres; cross-hybridization between standard GACT and GACTZP DNA was also measured. Microspheres carrying standard GACT capture probes were hybridized with either their perfectly complementary GACT analytes or GACTZ analytes. Strong signals indicated formation of perfectly matched hybrids (Lanes 1 and 5, Figure S2A). Weaker signals indicated weaker hybridization between probe-target pairs with five and four dG:dZ mismatches (Lanes 2 and 6, Figure S2A).
We then evaluated microspheres carrying non-standard GACTP capture probes. Negligible hybridization was observed between probe-analyte pairs containing five and four dP:dC mismatches (Lanes 3 and 7, Figure S2A). However, strong hybridization was seen between probe-target pairs with five and four dP:dZ matches (Lanes 4 and 8, Figure S2A).
To further explore the specificity of GACTZP hybridization, biotinylated GACTZ analytes were hybridized separately with a mixture of CTGAP capture probes, each attached to a unique microsphere population. Individual GACTZ analyte recognized only their specific microsphere-immobilized CTGAP complements (Supplementary Figure S2B, Lanes 10, 11, and 12) with specificities similar to those seen with standard GACT DNA (Figure S2B, Lanes 1, 2, and 3).
Validation of the Conversion Strategy in the ZiP-TAG Architecture
To assess the value of conversion in a ZiP-TAG Luminex assay, three targets were designed to simulate an amplicon (TAG10-Temp-Std, TAG14-Temp-Std, and TAG19-Temp-Std, Supplementary Figure S3). Each carried a 5'-anti-TAG sequence (24 nucleotide long) and a 3'-primer-binding segment. To allow direct comparison with standard Luminex architectures, the 5'-anti-TAG sequences were identical to the Luminex “universal” xTAGs.
These targets then served as templates for the extension of a target-specific biotinylated primer using Therminator and various triphosphate mixtures. In the negative control, dCTP was omitted, and no full-length products were produced (Figure S3A, Lane 1). In the presence of dCTP, biotinylated primers were extended to give full-length products containing only standard nucleotides (GACT) (Figure S3A, Lane 2). In parallel, extension-with-conversion was achieved by replacing dCTP by dZTP. Here, full-length products were produced by incorporation of dZ opposite template dGs at approximately the same rate as seen with extension without conversion (Figure S3A, Lane 3).
In all cases, after completion of primer extension, reactions were quenched with EDTA. Biotinylated extended products were diluted (ca. 10 nM) and hybridized to the standard or dP-containing anti-TAG sequences attached to Luminex microspheres (Type A or Type B mixture, Supplementary Figure S3).
Without conversion, the specific signal of each individual biotinylated analyte ranged from 930 to 1450 “median fluorescence intensity” (MFI) units; non-specific signal ranged from 50 to 90 MFI (Supplementary Figure S3B, Lane 1, 2, and 3, top). The signals obtained from multiplex analysis of a mixture of all three analytes (Figure S3B, Lane 4, top) were similar to those obtained in singleplexed analysis.
With conversion, however, the signals were stronger and signal/noise ratios were higher. MFIs for the correctly captured analytes ranged from 3000 to 4165 MFI, while the non-specific signal dropped to 19-67 MFI, the noise (Supplementary Figure S3B, Lanes 1, 2, and 3, bottom). The signal intensities for the mixture of three analytes (Figure S3B, Lanes 4, bottom) were also similar to those seen in singleplex assays.
Three hypotheses account for the stronger signals obtained with the GACTZ products, formed by conversion, compared with GACT products obtained without conversion. First, the dP:dZ pair is generally stronger than the dG:dC pair (18). Further, the CGTA template does not compete with the CPTA probes to lower binding to the microspheres. Finally, the dP:dZ pair is “orthogonal” to standard pairs. This orthogonality should lower noise, as the biotinylated analytes with GAZT extensions hybridize to their complementary CTPA probes free of competition. These three features synergistically contribute to the strong signals of the ZiP-TAG architecture with conversion. In contrast, biotinylated analyte without conversion can bind to either GACT capture probe or re-anneal to its template.
Comparing the ZiP-TAG Architecture to the Luminex xTAG Architecture
We then developed an assay to capture five target genes amplified from human genomic DNA: FMS-like tyrosine kinase receptor-3 (FLT3, important in colon cancer), heparin-binding EGF-like growth factor (HBEGF, important in ovarian cancer), v-kit Hardy-Zuckerman (KIT, important in blood cell cancer), thyroid stimulating hormone (TSH, important in thyroid cancer), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, a housekeeping gene). Primers (Supplementary Table S2) targeting these genes produce 70-150 nucleotide amplicons in regions where single nucleotide polymorphisms are seen.
These were tested in monoplexed and five-fold multiplexed PCRs with human genomic DNA. All PCRs gave the expected electrophoretic bands (Supplementary Figure S4). As in a standard Luminex xTAG TSPE assay, PCR products were treated with ExoSAP-IT to remove excess triphosphates and primers. The treated products (∼ 1 pmole) were served as templates for three assays: ZiP-TAG with conversion, ZiP-TAG without conversion, and Luminex xTAG (Figure 3).
For the ZiP-TAG conversion, biotinylated primers (Supplementary Table S3) were extended (10 cycles, Therminator) using triphosphate mixtures (dGTP, dATP, dTTP, and dZTP) lacking dCTP. The biotinylated products were serially diluted, hybridized with five sets of Luminex microspheres, incubated with SAPE, and analyzed at 37°C (Figure 4).
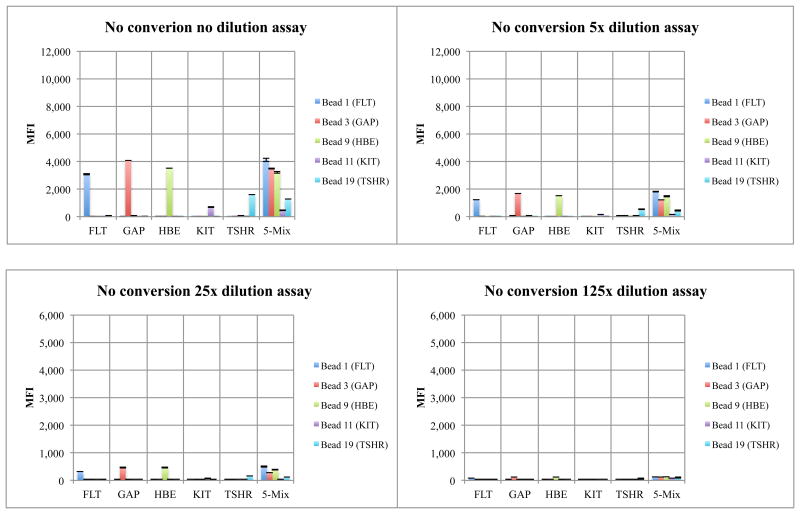
For the architecture without conversion, four standard nucleoside triphosphates were used to extend the same biotinylated primers to give biotinylated products with a GACT extension, complementary to CTGA probes attached to Luminex microspheres (Supplementary Table S4). MFI signals being approximately a third that seen with conversion. In both cases, the desired dose-response relation was observed (Figure 4 and 5). A negative control omitted nucleoside triphosphates. The absence of a signal (data not shown) showed that the ExoSAP treatment removed excess dNTPs that might have been left over from the PCR.
These results were compared with the results from a standard xTAG architecture, where unbiotinylated primers carrying a GACT "universal" oligonucleotide TAG (Supplementary Table S5) were extended on PCR products with biotin-14-dCTP, again either singly or in a mixture. Here, no changes were made in the standard procedure; the Luminex-recommended Platinum Tsp DNA Polymerase with 40 cycles (instead of 10 cycles) was used on same amplicons. Each sample was diluted, hybridized, and then analyzed at 37°C as before. The results are shown in Supplementary Figure S5.
With the standard xTAG architecture, an additional separation step was required to get the desired dose-response relation. When the mixture was analyzed without a wash to remove the excess biotin-14-dCTP, a non-linear dose-response curve was seen (Supplementary Figure S5A). This was attributed to a competition between incorporated and non-incorporated biotinylated ytosine for the reporter SAPE. Consistent with this attribution, a linear dose-response curve was obtained upon washing (Supplementary Figure S5B). The signal, however, was less uniform, but in some cases ∼2 fold higher than with the conversion assay (Figure 4). This is presumably due to the incorporation of a non-uniform number of biotin-14-dCTP units for each target.
Discussion
We introduce here two new ZiP-TAG™ architectures for xNA detection using Luminex instruments. One does not involve conversion (Figure 3C); the other does (Figure 3D). Both use bead-bound probes to hybridize to a sequence newly formed by the extension of a biotinylated primer on a sequence internal to the amplicon. The second, however, lowers cross-reactivity in complex xNA mixtures using a polymerase-based "conversion" strategy.
These results can be set within a larger context of the "liquid arrays" of the Luminex system, which has been used for over a decade to support multiplexed detection of xNA analysts.24 Generally, a bead of a particular “color” is used to detect a particular xNA analyte, and carries an oligonucleotide probe that is complementary to the analyte's sequence. An architecture is constructed so that when the analyte is present, a biotin moiety becomes associated with the bead. Then, the attached biotin captures a fluorescent reporter (generally streptavidin-phycoerythrin) on the microsphere via a streptavidin-biotin interaction. The Luminex instrument then detects the fluorescence of the phycoerythrin associated with the color of the bead.
Different architectures differ in how the biotin becomes associated with the Luminex bead. In the “direct hybridization” assay (Figure 3A), biotin is introduced on one PCR primer. The microsphere-bound probe is designed to be complementary to the biotinylated PCR product. This architecture is summarized as {Phycoerythrin-Streptavidin} {Biotin-PCR primer-Amplicon} {Hybridization probe-Microsphere}, where hyphens indicate covalent bonds, and the species within brackets {} indicate non-covalent complexes with other bracketed species.
Direct hybridization has a simple workflow, but it has many disadvantages, including a nonlinear dose-response behavior. 25 Here, the signal rises (rather than falls) upon dilution, presumably because of binding between the sense and anti-sense PCR products competes with binding to the bead-bound probe (Supplementary Figure S6). Dilution of the sample has been recommended as one "fix" for the problem. Alternatively, asymmetric PCR or other ways of separation of the sense and antisense strands have been applied, adding a step to the workflow.26,27
As an alternative, "allele specific" or "target specific" primer extension (ASPE or TSPE), introduces biotin by the extension of a TAGged primer using biotinylated dCTP. The bead-bound probe sequence (anti-TAG) is specific for the TAG sequence of the target, but is designed to bind to nothing else in the assay by being depleted in dG.28 The result is a complex: {Phycoerythrin-Streptavidin} {Biotinylated-dC-primer-TAG} {anti-TAG-Microsphere}. The universal TAG and anti-TAG sequences allowed the re-use of coupled beads for many different assays, and allow for uniform hybridization conditions. However, off-target primer-extension from the biotinylated primer, and possibly primer dimer formation, can lead to false positive signals and background. Further, excess biotinylated dCTP must be removed to have reasonable signals, adding a step to the workflow.
In the new architectures described here, biotinylated triphosphate and the oligonucleotide TAG are omitted. Instead, the {SAPE} {biotin-primer-extension} complex becomes attached to the microsphere by hybridization of the extension to a complementary oligonucleotide attached to the microsphere. This architecture allows for either extension with standard triphosphates (Figure 3C, without conversion) or non-standard triphosphates (replacing dCTP by dZTP, Figure 3D, with conversion). In addition to avoiding an expensive biotinylated triphosphate in large excess, this extension creates a new sequence that is not present in the primer pool. This means that off-target extension does not generate noise.
This architecture was further improved by adding conversion. When the extension of the biotinylated primer is done with dZTP and without dCTP, the biotinylated species is appended to a GAZT tag that cannot complement any natural oligonucleotide in a mixture, no matter how complex. This further suppresses noise and also provides high signals.
Thus, the ZiP-TAG architectures incorporate several specificity principles:
The microsphere-bound CTPA probes cannot hybridize to anything but GAZT extensions.
No TAG sequences need to be chosen to avoid hybridization with adventitious DNA.
No off-target priming leads to the creation of a TAGged primer that was extended with biotinylated dCTP, a mis-priming that generates noise.
The signals from different targets are uniform, since exactly one biotin is bound to the microsphere regardless of the target sequence.
Which of these are most valuable depends on the circumstances. For example, in an individual patient infected with only one respiratory virus, only one amplicon will end up in large amounts to be detected, notwithstanding the fact that the assay has multiplexing capability. In this case, the orthogonality of the GACTZP system is incompletely exploited. However, for more complex assay mixtures, including those containing multiple amplicons, all of the specificity elements should prove valuable. For example, patients with immune suppression (e.g., those suffering from AIDS) are susceptible to multiple infectious diseases, implying that multiple amplicons might be generated from a sample from a single individual.29 Likewise, genetic profiling of tumors will involve multiple amplicons, also using all of these specificity elements.
These studies have not, of course, tested the full potential for Luminex multiplexed detection, which can (in principle) be increased to 100 targets (or more). By exploiting other reagent innovations (in particular, self-avoiding molecular recognition systems, or SAMRS) and other architectural innovations (such as nested PCR), the upstream PCR amplification can also be highly multiplexed.30,31 The combination of multiplexed amplification and detection has been done for 12x detection of insect-borne RNA viruses, 15x detection of respiratory disease panel, and 8x detection of HIV targets, these work will be reported separately.
Supplementary Material
Acknowledgments
This work was supported in part by DARPA under its ADEPT program (HR0011-11-2-0018), and the NIAID under its program to analyze chronic and recent HIV infections (R01AI098616).
Footnotes
Supplementary information: Screening enzymes to incorporate dZTP opposite template dG, hybridization of GACTZP duplexes, and validation of the conversion strategy in the ZiP-TAG architecture. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes: The authors declare no competing financial interest.
References
- 1.Mahony JB, Petrich A, Smieja M. Crit Rev Clin Lab Sci. 2011;48:217. doi: 10.3109/10408363.2011.640976. [DOI] [PubMed] [Google Scholar]
- 2.Yang S, Rothman RE. Lancet Infect Dis. 2004;4:337. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milbury CA, Li J, Makrigiorgos GM. Clin Chem. 2009;55:632. doi: 10.1373/clinchem.2008.113035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benes V, Castoldi M. Methods. 2010;50:244. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Miller MB, Tang YW. Clin Microbiol Rev. 2009;22:611. doi: 10.1128/CMR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarder P, Nehorai A, Davis PH, Stanley SL. IEEE Trans Nanobiosci. 2008;7:142. doi: 10.1109/TNB.2008.2000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson AW, Wolf LK, Georgiadis RM. J Am Chem Soc. 2002;124:14601. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 8.Emmadi R, Boonyaratanakornkit JB, Selvarangan R, Shyamala V, Zimmer BL, Williams L, et al. The Journal of molecular diagnostics: JMD. 2011;13(6):583. doi: 10.1016/j.jmoldx.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benner SA. Acc Chem Res. 2004;37:784. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 10.Kimoto M, Cox RS, Hirao I. Expert Rev Mol Diagn. 2011;11:321. doi: 10.1586/erm.11.5. [DOI] [PubMed] [Google Scholar]
- 11.Collins ML, Irvine B, Tyner D, et al. Nucleic Acids Res. 1997;25:2979. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trimoulet P, Halfon P, Pohier E, et al. J Clin Microbiol. 2002;40:2031. doi: 10.1128/JCM.40.6.2031-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbeik T, Surtihadi J, Destree M, Gorlin J, Holodniy M, Jortani SA, et al. J Clin Microbiol. 2004;42:563. doi: 10.1128/JCM.42.2.563-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherrill CB, Marshall DJ, Moser MJ, Larsen CA, Daude SL, Prudent JR. J Am Chem Soc. 2004;126:4550. doi: 10.1021/ja0315558. [DOI] [PubMed] [Google Scholar]
- 15.Nolte FS, Marshall DJ, Rasberry C, Schievelbein S, Banks GG, Storch GA, et al. J Clin Microbiol. 2007;45:2779. doi: 10.1128/JCM.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balada-Llasat JM, LaRue H, Kelly C, Rigali L, Pancholi P. J Clin Virol. 2011;50:42. doi: 10.1016/j.jcv.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. Nucleic Acids Res. 2004;32:1937. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang ZY, Hutter D, Sheng PP, Sismour AM, Benner SA. Nucleic Acids Res. 2006;34:6095. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang ZY, Sismour AM, Sheng PP, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Yang ZY, Yan M, Alvarado B, Wang G, Benner SA. Nucleic Acids Res. 2011;39:3949. doi: 10.1093/nar/gkq1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang ZY, Chen F, Alvarado JB, Benner SA. J Am Chem Soc. 2011;133:15105. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merante F, Yaghoubian S, Janeczko R. J Clin Virol. 2007;40:S31. doi: 10.1016/S1386-6532(07)70007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krunic N, Yager TD, Himsworth D, Merante F, Yaghoubian S, Janeczko R. J Clin Virol. 2007;40:S39. doi: 10.1016/S1386-6532(07)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunbar SA. Clinica Chimica Acta. 2006;363:71. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong B, Stewart M, Mazumder A. Cytometry. 2000;40:102. [PubMed] [Google Scholar]
- 26.Deregt D, Gilbert SA, Dudas S, Pasick J, Baxi S, Burton KM, Baxi MK. J Virol Methods. 2006;136:17. doi: 10.1016/j.jviromet.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 27.Sriprakash KS, Lundh N, Mooonhuh M, Radding CM. J Biol Chem. 1975;250:5438. [PubMed] [Google Scholar]
- 28.Bortolin S, Black M, Modi H, Boszko I, Kobler D, Fieldhouse D, et al. Clin Chem. 2004;50:2028. doi: 10.1373/clinchem.2004.035071. [DOI] [PubMed] [Google Scholar]
- 29.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Proc Natl Acad Sci USA. 2008;105:7552. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshika S, Chen F, Leal NA, Benner SA. Angew Chem Int Edit. 2010;49:5554. doi: 10.1002/anie.201001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang ZY, Chen F, Chamberlin SG, Benner SA. Angew Chem Int Edit. 2010;49:177. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.