Abstract
Type I interferons (IFNs) exert their effects through the induction of hundreds of IFN-stimulated genes (ISGs), many of which function by inhibiting viral replication and modulating immune responses. ISG15, a di-ubiquitin-like protein, is one of the most abundantly induced ISGs and is critical for control of certain viral and bacterial infections. Like ubiquitin, ISG15 is covalently conjugated to target proteins. In addition, free unconjugated ISG15 is present both intra- and extracellularly. Although much remains to be learned about conjugated ISG15, even less is known about the 2 free forms of ISG15. This article focuses on the role that ISG15 plays during the host response to pathogen challenge, in particular on the recent observations describing the immunomodulatory properties of free ISG15 and its potential implication in disease pathogenesis.
Introduction
Infected cells carry out signaling cascades that serve to eliminate the invading microbes and simultaneously alert neighboring cells to the threat of potential infection. This response is initiated by detection of pathogen-associated molecular patterns by host pattern-recognition receptors (PRRs), including Toll-like receptors (TLRs), RIG-I-like receptors (RLRs), and nucleic acid sensors, such as DAI, IFI16, DHX9, and DHX36 (Kim and others 2010; Unterholzner and others 2010; Tang and others 2012). Engagement of PRRs triggers signaling cascades that lead to the induction of several genes that restrict pathogen replication and modulate the ensuing immune response. Central among this response is the type I interferon (IFN) system and the hundreds of effector proteins it induces. In humans and mice, the type I IFN family is composed of 16 related members: 12 IFN-α subtypes, IFN-β, IFN-ɛ, IFN-κ, and IFN-ω, all of which bind to the type I IFN receptor (IFNAR) (Gonzalez-Navajas and others 2012). Binding of type I IFN to IFNAR results in activation of the receptor-associated tyrosine kinases Jak1 and Tyk2, which leads to STAT1- and STAT2-dependent transcription of hundreds of IFN-stimulated genes (ISGs) (Gonzalez-Navajas and others 2012). ISGs encode proteins that utilize diverse mechanisms to limit the spread of infection. These mechanisms include restriction of various stages of microbial replication, as well as immunomodulatory functions. In recent years, there has been an explosion of research focused on understanding the function of individual ISGs and how these genes contribute to the host antiviral response. Included among these ISGs is the ubiquitin-like protein, ISG15.
ISG15: A Ubiquitin-Like ISG
ISG15 is one of the most robustly induced transcripts after type I IFN stimulation (Der and others 1998). It is also upregulated by TLR stimulation, viral infection, and TNF-α stimulation (Haas and others 1987; Malakhova and others 2002; Chairatvit and others 2012). ISG15 was identified in IFN-treated lysates as a 15 kDa protein with both sequence homology and immuno-crossreactivity to ubiquitin (Farrell and others 1979; Blomstrom and others 1986; Haas and others 1987). The crystal structure of ISG15 verified its similarity to ubiquitin by revealing that it is composed of 2 ubiquitin-like (Ubl) domains connected by a short linker (Narasimhan and others 2005). Each Ubl domain assumes a β-grasp fold that can be superimposed onto that of the ubiquitin structure.
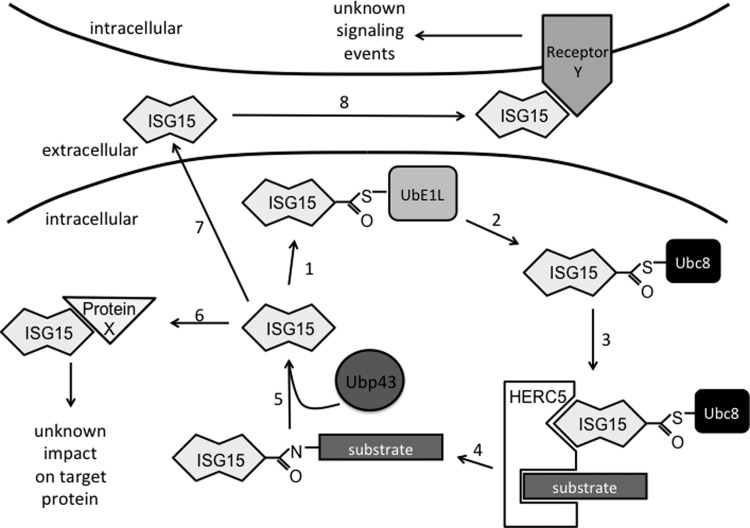
ISG15 is synthesized as a 17 kDa precursor that is proteolytically processed into the mature 15 kDa form (Knight and others 1988; Potter and others 1999). Like ubiquitin, mature ISG15 contains a C-terminal LRLRGG motif that is necessary for mediating its covalent formation of isopeptide bonds to lysine residues on target proteins (Loeb and Haas 1992; Lenschow and others 2005). ISG15 conjugation, or ISGylation, occurs through an enzymatic cascade that is similar to, yet distinct from that of ubiquitin conjugation (Fig. 1). The activating E1 enzyme, UbE1L, forms an ATP-dependent thioester bond with ISG15 (Yuan and Krug 2001). After activation, ISG15 is transferred to the active-site cysteine of the conjugating E2 enzyme, Ubc8 (Kim and others 2004; Zhao and others 2004). The E3 ligases HERC5 (human), HERC6 (mouse), HHARI, and Efp transfer ISG15 to lysine residues on specific substrates (Dastur and others 2006; Nakasato and others 2006; Takeuchi and others 2006a; Wong and others 2006; Zou and others 2007; Versteeg and others 2010; Ketscher and others 2012; Oudshoorn and others 2012). Unlike the ubiquitin conjugation pathway, in which hundreds of E3s ubiquitinate individual substrates, HERC5 in humans and HERC6 in mice appear to function as the dominant E3s for the ISG15 system because specific knockdown of their expression with siRNA abrogates nearly all ISG15 conjugates induced by IFN (Dastur and others 2006; Wong and others 2006; Ketscher and others 2012; Oudshoorn and others 2012). HERC5 is associated with polyribosomes and appears to cotranslationally modify proteins with ISG15 (Durfee and others 2010). Colocalization with the translational machinery may explain why such a broad range of target proteins are conjugated by a single E3 ligase. In contrast to ubiquitin, there currently is no evidence that the ISG15 E3s can add ISG15 “chains” onto target proteins. Rather, a single ISG15 molecule appears to be conjugated to a lysine residue, although multiple lysines may be modified within the same protein. Finally, conjugation to ISG15 is not a permanent post-translational modification. Rather, it is reversible due to specific removal of ISG15 from conjugated proteins by the isopeptidase Ubp43 (Malakhova and others 2002; Kim and Zhang 2005). A screen for additional deubiquitinating enzymes that could target ISG15 conjugates identified USP2, USP5, USP13, and USP14 as potential candidates, although their significance remains to be determined in vivo (Catic and others 2007). Interestingly, like ISG15 itself, all members of the ISG15 conjugation cascade are upregulated by type I IFNs. In addition to ISG15 conjugates, free unconjugated ISG15 can be detected within IFN-treated cells and can be released from cells (Fig. 1) (Knight and Cordova 1991; Recht and others 1991; D'Cunha and others 1996b). Recent efforts in the field have focused on understanding the biological function of ISG15.
FIG. 1.
Schematic of the 3 forms of ISG15. The IFN-induced E1/E2/E3 enzymatic cascade catalyzes ISGylation of substrate proteins. The activating E1 enzyme UbE1L forms a thioester bond with free ISG15 in an ATP-dependent manner (1). ISG15 is then transferred to the E2 conjugating enzyme Ubc8 (2). Interaction with the E3 ligase HERC5 (3) then catalyzes the conjugation of ISG15 to lysine residues of target substrate proteins (4). The deconjugating enzyme Ubp43 reverses conjugation by removing ISG15 from target proteins (5). Free intracellular ISG15 may bind noncovalently to unknown intracellular proteins (6) and impact downstream signaling. Secretion of free intracellular ISG15 (7) results in extracellular ISG15 that may bind to its unknown receptor(s) (8) to initiate unknown signaling pathways. IFN, interferon; ISG, IFN-stimulated gene.
Antiviral Activity of ISG15
Despite being identified in 1979, for many years the functional significance of ISG15 was unclear. Since ISG15 and its conjugation cascade are strongly induced by type I IFN and IFNs play such a critical role during viral infection, early studies focused on its potential role during viral infection. Multiple approaches, including gene knockdown, overexpression, and genetic deletion of various components of the ISG15 cascade, have been taken to determine whether ISG15 participates in the host response in a variety of viral models. We will briefly summarize these results here, but refer the reader to past reviews for a more comprehensive analysis of the various antiviral effects of ISG15 (Harty and others 2009; Lenschow 2010; Skaug and Chen 2010).
Several studies have shown that ISG15 regulates viral growth in cell culture. Influenza virus, vaccinia virus, vesicular stomatitis virus (VSV), Sendai virus, Japanese encephalitis virus, Newcastle disease virus, avian sarcoma leucosis virus, human papilloma virus (HPV), HIV-1, Ebola virus-like particles (VLPs), megalocytivirus, dengue virus, and West Nile virus (WNV) are all modestly inhibited by ISG15 in vitro (Lenschow and others 2005; Kim and others 2006; Okumura and others 2006; Lenschow and others 2007; Guerra and others 2008; Malakhova and Zhang 2008; Okumura and others 2008; Giannakopoulos and others 2009; Hsiang and others 2009; Lai and others 2009; Broering and others 2010; Durfee and others 2010; Hsiao and others 2010; Pincetic and others 2010; Shi and others 2010; Dai and others 2011; Tang and others 2012; Wang and others 2012). It is, however, important to note that in many cases these antiviral phenotypes have not been confirmed in an in vivo setting. In addition, there are examples where the in vitro phenotypes are not necessarily reflected in vivo. For example, neither ISG15−/− nor UbE1L−/− mice display a phenotypic difference compared to wildtype animals during VSV infection, despite the observation in vitro that ISG15 overexpression lowers VSV titers (Osiak and others 2005; Kim and others 2006; Okumura and others 2008). We have also not detected a phenotype in ISG15−/− mice after WNV infection (Lenschow and others, unpublished observations). The reasons for this lack of correlation are unclear but could reflect compensation by other ISGs in vivo, which are not evident in vitro, when only a single cell type is being evaluated.
Clear evidence that ISG15 contributes significantly to the host's antiviral response was shown with the analysis of ISG15−/− mice. While phenotypes have not been seen in every viral model evaluated, including both VSV and lymphocytic choriomeningitis virus, increased lethality was observed in ISG15 deficient mice after infection with Sindbis, influenza A and B, Chikungunya, HSV-1, and vaccinia virus (Osiak and others 2005; Lenschow and others 2007; Guerra and others 2008; Werneke and others 2011). Efforts to determine the mechanism by which ISG15 is functioning in these models are ongoing. To date, there are 2 mechanisms of action that have been defined for ISG15. The first are the conjugation dependent actions of ISG15 that inhibit viral replication. The second are the biological properties of ISG15 that appear to be mediated by free ISG15. We will briefly discuss the conjugation-dependent antiviral properties of ISG15, but refer the readers to several outstanding reviews that have been written on this topic (Knobeloch 2010; Zhang and Zhang 2011). We will then focus the remainder of this review on the more recent observations that have described the immunomodulatory properties of free ISG15 and discuss its implications on disease pathogenesis.
Conjugation-Dependent Actions of ISG15
The utilization of UbE1L−/− mice, which lack ISG15 conjugates due to the absence of the E1 but still express free ISG15, was vital in distinguishing whether ISGylation contributed to the ISG15-dependent protection during viral infection. UbE1L−/− mice displayed a similar increase in lethality as seen in ISG15−/− mice during both Sindbis and influenza B viral infections (Giannakopoulos and others 2009; Lai and others 2009). These results indicated that ISG15 conjugates were required for the ISG15-mediated resistance to these viruses. Further support for the importance of ISGylation came from studies utilizing recombinant Sindbis viruses. The infection of ISG15−/− mice with a Sindbis virus encoding wildtype ISG15 rescued the ISG15−/− mice from death. In contrast, a recombinant virus expressing an ISG15 mutant in which the C-terminal LRLRGG was mutated such that it could not undergo conjugation failed to rescue the knockout mice, indicating that ISGylation is necessary for the ISG15-dependent control of Sindbis virus infection (Lenschow and others 2007).
The distinct immune evasion strategies developed by numerous viruses has provided further evidence supporting the importance of ISG15 conjugation during the antiviral response. The influenza NS1 protein was the first immunoevasin to be described that antagonizes the ISG15 conjugation system (Yuan and Krug 2001). NS1B, but not NS1A, binds noncovalently to ISG15, in a species-specific manner, to prevent it from interacting with UbE1L, effectively inhibiting conjugate formation (Yuan and Krug 2001). Another viral protein, the vaccinia virus E3L, also inhibits the generation of ISG15 conjugates, although the mechanism by which this occurs is not yet understood (Guerra and others 2008). Targeting the ISGylation system appears to be a successful immune evasion strategy for vaccinia virus since wildtype and ISG15−/− mice respond similarly to infection with wildtype vaccinia virus, but ISG15−/− mice displayed increased mortality when infected with an E3L-deficient virus (Guerra and others 2008). These 2 viruses provide examples of evasion strategies that inhibit ISG15 conjugation.
In addition to influenza and vaccinia, which prevent ISG15 conjugate formation, several viruses encode proteins with ISG15 deconjugating activity. The L proteins from Crimean Congo hemorrhagic fever virus (CCHFV) and Dugbe virus and the nsP2 proteins from equine arteritis virus and porcine respiratory and reproductive syndrome virus, all contain ovarian tumor (OTU) domains (Frias-Staheli and others 2007). These viral OTU-containing proteins are cysteine proteases with both deubiquitinating and de-ISGylating activity. The antiviral activity of ISG15 is lost in transgenic mice expressing the CCHFV L protein during Sindbis infection, as these mice display enhanced susceptibility. Finally, the papain-like proteases of the coronaviruses SARS and NL63 contain both deubiquitinating and de-ISGylating activity (Lindner and others 2005; Clementz and others 2010). The contribution of these ISG15 antagonists to immune evasion in vivo is still unknown but the targeting of ISG15 conjugates by multiple unrelated viruses suggests that ISGylation plays an important role in host antiviral responses.
Functions of ISGylated Proteins
To understand how ISG15 and UbE1L mediate protection from viral infection, it is necessary to define the functional effects of ISGylation upon modified target proteins. Numerous screens and individual studies have identified several hundred candidate ISG15 targets which span a diverse array of biological processes and include both host and viral proteins (Hamerman and others 2002; Kim and others 2004; Giannakopoulos and others 2005; Takeuchi and others 2005; Zhao and others 2005; Zou and others 2005; Takeuchi and Yokosawa 2005; Lu and others 2006; Takeuchi and others 2006a, 2006b; Wong and others 2006; Okumura and others 2007; Zou and others 2007; Feng and others 2008; Kim and others 2008; Shah and others 2008; Jeon and others 2009; Durfee and others 2010; Pincetic and others 2010; Shi and others 2010; Tang and others 2010; Zhao and others 2010; Kuang and others 2011; Jeon and others 2012; Okumura and others 2012). Only a limited number of these proteins have been validated. There is currently no known universal fate for all ISGylated proteins. Unlike K48-linked ubiquitination, for example, ISGylation does not appear to directly target proteins for proteasome-mediated degradation (Liu and others 2003). To further complicate this matter, numerous studies have reported that only a small fraction of a given target protein pool is ISGylated (Hamerman and others 2002; Zhao and others 2005; Jeon and others 2009). While it is still largely unknown what impact ISG15 modification has on a specific protein, the field has gained some insight from the analysis of the limited number of conjugated targets that have been studied (Table 1). In these cases, ISG15 has been shown to function by either disrupting the activity of a target protein, enhancing its activity, and/or by altering its localization within the cell. There are examples of both host and viral proteins that are modified by ISG15 that impact viral replication. For example, ISGylation induces the autophosphorylation and activation of the host protein dsRNA-dependent protein kinase (PKR) even in the absence of viral RNA (Okumura and others 2012). PKR activation leads to eIF2α phosphorylation and translation suppression which impacts upon viral replication. ISGylation of viral proteins has also been shown to inhibit viral replication. Two different research groups found that the NS1 protein from influenza A virus is ISGylated. In 1 study the modification was localized predominantly to the lysine at position 41 of influenza A/WSN NS1 (Zhao and others 2010). This modification disrupts the association between NS1 and importin-α, which is critical for nuclear translocation and replication of influenza. Infection with a recombinant virus in which lysine 41 was mutated to arginine (K41R) resulted in reduced NS1 ISGylation. The K41R virus was less sensitive to the antiviral effects of IFN-β pretreatment of A549 cells compared to the wildtype virus, demonstrating that ISGylation of NS1 inhibits replication. Another group studying NS1 from the PR8 strain of influenza A identified lysines 126 and 127 as the critical amino acids required for conjugation in vivo (Tang and others 2010). Recombinant viruses encoding mutations at either residue exhibited increased viral growth in vitro and increased virulence in vivo. The fate of ISGylated proteins is still an area of active research. As this body of research continues to evolve we should gain further insight into how ISGylation mediates antiviral activity.
Table 1.
ISGylated Target Proteins with Known Consequences of Conjugation
| Protein | Effect of ISGylation | Modified lysine | Reference |
|---|---|---|---|
| Ubc13 |
Suppresses thioester bond formation to ubiquitin |
92 |
Takeuchi and Yokosawa (2005) |
| UbcH6 |
Suppresses thioester bond formation to ubiquitin |
136 |
Takeuchi and others (2005) |
| PP2Cβ |
Suppresses NF-κB activation |
12/142 |
Takeuchi and others (2006) |
| IRF3 |
Inhibits ubiquitin-mediated degradation |
193/360/366 |
Lu and others (2006); Shi and others (2010) |
| EFP |
Inhibits E3 ligase activity |
117 |
Zou and others (2007) |
| 4EHP |
Enhances m7GTP cap structure-binding activity |
121/130/134/222 |
Okumura and others (2007) |
| RIG-I |
Negatively regulates RIG-I-mediated signaling |
ND |
Kim and others (2008) |
| Cyclin D1 |
Suppresses cyclin D1 protein levels |
ND |
Feng and others (2008) |
| PML/RARα |
Represses PML domain |
ND |
Shah and others (2008) |
| Filamin B |
Inhibits ability to scaffold RAC1, MEKK1, and MKK4 |
2467 |
Jeon and others (2009) |
| Influenza A NS1 |
Inhibits nuclear import; inhibits dimerization |
41;20/31/108/110/126/217/219 |
Tang and others (2010); Zhao and others (2010) |
| CHMP5 |
Inhibits CHMP5-LIP5 interaction |
ND |
Pincetic and others (2010) |
| CHMP2A |
Inhibits CHMP2A-LIP5 interaction |
ND |
Kuang and others (2011) |
| CHMP4B |
Inhibits binding to Vps4 |
ND |
Kuang and others (2011) |
| CHMP6 |
Inhibits binding to Vps4 |
ND |
Kuang and others (2011) |
| ΔNp63α |
Becomes susceptible to caspase-2 cleavage |
139/324 |
Jeon and others (2012) |
| PKR | Activates PKR in absence of viral RNA | 69/159 | Okumura and others (2012) |
ND, not determined.
Conjugation-Independent Actions of ISG15
While much of the early research in the ISG15 field has focused on ISGylation and the identification of the members of the ISG15 conjugation cascade, the identification of target proteins, and analysis of the impact of ISGylation on protein function—there has recently been increasing evidence that free ISG15 can also play a critical role during infection.
Free ISG15 Is Present Both Intra- and Extracellularly
In addition to the conjugated form of ISG15 that is detected in cell lysates, free ISG15 exists both intra- and extracellularly. ISG15 lacks a hydrophobic leader peptide and its release is not affected by inhibitors of the conventional ER-Golgi secretory pathway, so it is unclear how ISG15 traffics outside the cell (D'Cunha and others 1996b). It is possible that intracellular free ISG15 is released passively into the extracellular space from dead or damaged cells in a manner that is distinct from active secretion, but since the trafficking mechanism of ISG15 is undefined we will refer to extracellular ISG15 as secreted ISG15 in this review. Despite its lack of a leader peptide, ISG15 can be detected in the supernatants of type I IFN-treated leukocytes and cell lines and in the serum from IFN-β-treated patients (Knight and Cordova 1991; Recht and others 1991; D'Cunha and others 1996b). The existence of a secreted form of ISG15 suggested that it might bind to a cell surface receptor to modulate intracellular signaling events during an immune response. Although the identity of a cell surface receptor(s) for ISG15 is currently unknown, multiple studies have indicated that secreted ISG15 has cytokine-like activity. In contrast, much less is known about the function of unconjugated ISG15 that exists within the cell. However, recent studies have suggested that both forms of free ISG15 may be functional.
Models of Positive Regulation by Free ISG15
The initial investigation into whether secreted ISG15 has biological activity reported that recombinant soluble ISG15 could induce T cells, but not natural killer (NK) cells, to secrete IFN-γ (Recht and others 1991). It was further shown that incubation with ISG15 enhanced the LPS-induced cytolytic activity of peripheral blood mononuclear cell (PBMC) cultures. This was attributed to the ISG15-induced production of IFN-γ by T cells, which then activated monocytes. The ability of ISG15 to induce IFN-γ secretion by T cells was confirmed in a subsequent study demonstrating that recombinant ISG15 could stimulate significant NK cell proliferation when added to PBMC cultures (D'Cunha and others 1996a). This effect was indirect because ISG15 did not stimulate proliferation of purified NK cells alone, but required total PBMCs. The results from these 2 early articles suggested a model in which ISG15 release from IFN-stimulated cells induced the production of IFN-γ by T cells with subsequent monocyte and NK cell activation. For these reasons, it was suggested that secreted ISG15 could regulate the immune response after type I IFN production.
After these early reports, another group examined the effect of ISG15 on dendritic cells (Padovan and others 2002). Using microarray analysis to identify melanoma cell-derived soluble factors that could impair DC migration, it was shown that ISG15 upregulated E-cadherin, CD15, and CD86 expression on monocyte-derived DCs. Soon after it was shown that ISG15 was localized to the extracellular surface of erythrocytes that were either uninfected or undergoing an early stage infection with Plasmodium yoelii, while erythrocytes containing the later schizont stage parasites lacked surface ISG15 staining (Owhashi and others 2003). ISG15 derived from the uninfected or early stage-infected erythrocytes was shown to act as a chemoattractant for and activator of neutrophils.
Taken together, these 4 initial studies suggested that secreted ISG15 acts as a positive regulator of multiple cell types. A recent study by Casanova and colleagues of patients with Mendelian susceptibility to mycobacterial disease (MSMD) has provided further evidence that ISG15 can function as an immunomodulatory cytokine (Bogunovic and others 2012). Through the use of whole-exome sequencing and genome-wide linkage analysis they identified 3 patients, including 2 siblings, encoding homozygous mutations in ISG15. The mutations were different between the 2 families, but both resulted in a loss of ISG15 protein expression. Cells derived from these patients lacked free and conjugated ISG15, although both could be reconstituted after transfection of wildtype ISG15, indicating that the ISGylation machinery was intact. After stimulation with Bacillus Calmette-Guérin (BCG), a vaccine strain of Mycobacterium bovis, both PBMCs and neutrophils from control but not ISG15−/− patients secreted ISG15. These data provided the first evidence in humans that inherited mutations in ISG15 can lead to the loss of protein expression.
The characterized germline mutations associated with MSMD are located in genes whose protein products are involved in IFN-γ signaling (Cottle 2011). Given this, the authors revisited the idea that ISG15 functions as a cytokine to regulate IFN-γ production and compared ISG15-mediated IFN-γ production in cells from control and ISG15−/− patients. In accordance with earlier studies, the addition of recombinant ISG15 to the culture medium of control PBMCs induced IFN-γ production. Interestingly, coincubation of PBMCs with ISG15 and IL-12 greatly augmented IFN-γ secretion compared to ISG15 alone, suggesting that a synergistic relationship exists between these 2 cytokines. In this study NK cells, not T cells, were the main IFN-γ producer. The reason for this is unknown but may be explained by the difference in IL-12 costimulation, which was not performed in the earlier studies. After stimulation with BCG alone or BCG and IL-12, ISG15−/− PBMCs were found to secrete less IFN-γ than control PBMCs, despite normal expression of both IL-12p40 and IL-12p70. Importantly, in the presence of a monoclonal antibody against ISG15, IFN-γ production was greatly reduced when control PBMCs were stimulated with BCG or BCG plus IL-12. Flow cytometric analysis revealed CD3+ cells as the main population to lose IFN-γ production during ISG15 blockade, while CD3− cells exhibited a less severe reduction. Exogenous ISG15 was able to partially reconstitute the phenotype. Recombinant ISG15 did not rescue IFN-γ production by ISG15−/− PBMCs stimulated with BCG alone, but it did increase IFN-γ secretion when the cells were stimulated with BCG plus IL-12. In addition, while exogenous ISG15 increased the number of CD56+IFN-γ+ cells in BCG plus IL-12-stimulated ISG15−/− cultures, it was not effective in restoring the CD56−IFN-γ+ population. While it is clear that cells derived from ISG15−/− patients produce less IFN-γ upon BCG challenge, the inability of recombinant soluble ISG15 to fully restore both the population of IFN-γ+ cells and the total amount of secreted IFN-γ raises the possibility that other defects may also contribute to this phenotype. Sequence analysis identified 2 other homozygous coding variants located in the chromosomal regions linked to MSMD in one ISG15−/− patient and 9 in another. It is possible that these mutations also contribute to susceptibility to MSMD and perhaps to the regulation of IFN-γ by ISG15.
Despite this, the defective IFN-γ production from ISG15−/− cells and the critical role of IFN-γ in mycobacterial immune responses suggest a model in which BCG stimulates secretion of ISG15, which then synergizes with IL-12 to promote IFN-γ production from CD56+ and CD3+ cells. In this model, the ISG15-dependent production of IFN-γ is abrogated in patients with ISG15 deficiency and is responsible for the improper control of mycobacteria. The authors also reported that ISG15−/− mice were more susceptible than wildtype mice to infection with Mycobacterium tuberculosis infection, although whether this increased susceptibility was independent of conjugation was not assessed. This in vivo model will be a useful tool in further dissecting the role of ISG15 in mycobacterial pathogenesis.
Finally, a recent study has shown that secreted ISG15 exerts immunoregulatory effects not only in mammals but also in teleost (fish) species (Wang and others 2012). When added to the media of macrophages from the tongue sole Cynoglossus semilaevis, recombinant soluble ISG15 induced respiratory burst and nitric oxide production. Soluble ISG15 also enhanced the expression of several immune genes in C. semilaevis lymphocyte cultures, including TLR9, IL-8, and IL-1β. Interestingly, while the effects on macrophages were dependent on the C-terminal LRGG residues of ISG15, the phenotypes in lymphocytes had mixed dependence on this motif. The wildtype C-terminus was required for IL-8 induction, but TLR9 and IL-1β were still induced by a mutant ISG15 in which the C-terminus was mutated to LAAG. This was the first study to suggest a role for the LRGG motif of ISG15 for something other than conjugation. This report also highlights the significance of ISG15 in nonmammalian species and again suggests species-specificity for various effects of ISG15.
There are several important observations that come from these studies and should help to guide future work on the cytokine activity of ISG15. First, several of these studies support the ability of ISG15 to function as an immunomodulatory molecule, with the strongest evidence being its ability to regulate IFN-γ production. Whether ISG15 regulates other cytokines or chemokines in these models is unclear. In the study of C. semilaevis ISG15, both IL-8 and IL-1β were also induced by ISG15, raising the possibility that in the correct setting or by stimulating different cell types, ISG15 may regulate additional pathways (Wang and others 2012). Second, these studies indicated that ISG15 can be released from different cell types, although the mechanisms may be distinct. Neutrophils were found to contain ISG15 in gelatinase and secretory granules even in an unstimulated state (Bogunovic and others 2012). However, ISG15 was also found to be on the surface of erythrocytes during malaria infection and released from lymphocytes and monocyte during type I IFN treatment (Knight and Cordova 1991; D'Cunha and others 1996b; Owhashi and others 2003). The mechanisms by which it gets released from these different cell types will need to be investigated further. Finally, these studies may provide important clues that will lead to the identification of the ISG15 receptor. The synergy noted between ISG15 and IL-12 in the Casanova study could indicate that the receptor is induced by IL-12 or works in concert with another IL-12-induced protein to stimulate IFN-γ production. The study in C. semilaevis which revealed that the LRGG motif was important for this stimulatory activity may also provide important insight into the structure of released ISG15, which will be critical in moving forward. Finally, in all of these studies, different cell types were found to respond to ISG15, providing insight into which cell types may express an ISG15 receptor.
Model of Negative Regulation by Free ISG15
All of the studies evaluating the cytokine–like activity of ISG15 were performed in vitro, and early studies using the knockout mouse models had indicated the antiviral activities of ISG15 were conjugation-dependent, with the ISG15−/− and UbE1L−/− mice phenocopying each other. However, a recent study using a neonatal model of Chikungunya virus (CHIKV) provided the first in vivo evidence that free ISG15 contributes to the host response during viral infection (Werneke and others 2011). CHIKV is an arthropod-borne alphavirus that causes an acute febrile arthritis of the peripheral joints in affected patients. Using a murine model of CHIKV, Werneke and others were able to demonstrate a significant increase in lethality in mice lacking ISG15, with 100% of ISG15−/− mice succumbing to infection within 3–4 days as compared to only 40% lethality in wildtype mice at later times postinfection. Unlike previously described influenza and Sindbis virus models, where the antiviral activity of ISG15 was dependent on conjugation, during CHIKV infection the action of ISG15 was independent of UbE1L-mediated conjugation. UbE1L−/− mice displayed no increase in lethality and instead phenocopied the wildtype mice. Since these mice lack ISG15 conjugates but still contain free ISG15, this study implied that the protection afforded by ISG15 was not dependent on its conjugated form but rather on free ISG15. It should be noted that there is the possibility that UbE1L is not the only E1 enzyme for ISG15, and that an alternative E1 could generate low levels of ISG15 conjugates that protect these mice from CHIKV infection. However, no other E1 enzyme has been identified for the ISG15 pathway and ISG15 conjugates were not detected by immunoblot in UbE1L−/− mice or cells. It also possible that UbE1L has an ISG15-independent function that is crucial for resistance to CHIKV—but aside from its role as an E1 no other functions are currently known. Therefore, it is likely that free ISG15 mediates the resistance to CHIKV-induced lethality.
Although this study indicated that free ISG15 was required for protection during CHIKV infection, its role was not directly antiviral. In contrast to the conjugation-dependent influenza and Sindbis virus models that resulted in dramatically higher viral loads in both ISG15−/− and UbE1L−/− mice, ISG15 deficiency did not lead to increased CHIKV titers in any organ tested. Instead, mice lacking ISG15 were found to have elevated levels of several proinflammatory cytokine and chemokines. TNF-α, IL-1β, IL-6, CCL2, CCL3, and CCL5 levels in ISG15−/− mice were as high or higher than those seen in IFNAR−/− mice, which die from CHIKV infection even more rapidly than ISG15−/− mice. Mice lacking ISG15 displayed clinical signs of shock, including elevated liver enzymes and blood urea nitrogen levels, and the blockade of proinflammatory cytokines was able to prolong survival of these mice, suggesting that cytokine storm was contributing to the increased lethality (Lenschow and others, unpublished observations). These data indicate that free ISG15 functions not as an antiviral factor, but as an immunomodulatory protein that negatively regulates the expression of proinflammatory cytokines and chemokines.
While this study provided the first in vivo evidence that free ISG15 contributed to the host response during viral infection, it does not distinguish between the actions of extracellular or intracellular free ISG15. The actions of extracellular ISG15 have been most clearly tied to the induction of IFN-γ secretion by cytotoxic lymphocytes. However, in the CHIKV model, wildtype, ISG15−/−, and UbE1L−/− mice produced equivalent IFN-γ levels during the course of the infection (Lenschow and others, unpublished data), implying that IFN-γ production is sufficient or unnecessary in CHIKV-infected ISG15−/− mice. Several other cytokines and chemokines were upregulated in CHIKV-infected ISG15−/− mice. Therefore, it is possible that secreted ISG15 may bind to a cell surface receptor that downregulates these cytokines. The absence of ISG15 then would result in their elevation. Since the receptor(s) for ISG15 has not been identified, it is difficult to know how free ISG15 is acting in this system. Blocking antibodies and/or a mutated ISG15 that lacks receptor binding would be essential tools in defining whether the protection mediated by ISG15 in the CHIKV model is through the action of extracellular, free ISG15. However, both of these approaches require knowledge of the receptor's identity.
An alternative explanation to the ISG15-dependent, UbE1L-independent protection against CHIKV infection is that free intracellular ISG15 is responsible for the effect by binding noncovalently to other intracellular proteins and modulating their functions. This noncovalent, intracellular interaction appears to control the ability of Ebola VLPs to egress from cells in vitro. Ebola VLP budding requires ubiquitination of the VP40 viral matrix protein through its interaction with Nedd4, a host E3 ubiquitin ligase. Overexpression of ISG15, independent of the conjugation cascade, was shown to inhibit Ebola VP40 VLP budding from 293T cells by binding to the Nedd4 ligase and disrupting the interaction between Nedd4 and the E2 enzyme UbcH6, which is required for ubiquitin transfer (Malakhova and others 2008; Okumura and others 2008). This resulted in reduced VP40 ubiquitination and less efficient VLP budding. Although these studies have yet to be confirmed in vivo, they suggest that free intracellular ISG15 can modulate intracellular biochemical events. With respect to CHIKV infection, it is unlikely that free intracellular ISG15 alters a binding event that affects viral production, since titers are similar between wildtype, ISG15−/−, and UbE1L−/− mice. However, it is possible that ISG15 impacts intracellular host protein interactions that regulate cytokine and chemokine production. Further examination of intracellular, noncovalent binding partners of ISG15 will shed further light on this matter. Ongoing work will help define whether free ISG15 is required in its intracellular and/or extracellular form during CHIKV-infection and in other model systems.
Defining the Mechanisms of ISG15-Dependent Immunomodulation
It is now clear that certain conditions exist in which free ISG15 acts as a positive regulator of cytokine production, particularly with regard to IFN-γ (Fig. 2). In contrast, other systems implicate free ISG15 as a negative regulator of cytokine production. How can these seemingly opposite scenarios be reconciled, especially when the molecular mechanisms by which they are each mediated are unknown? One explanation may be that secreted ISG15 binds to a cell surface receptor that stimulates IFN-γ production. The 3 studies using recombinant soluble ISG15 as an inducer of IFN-γ support this hypothesis. In contrast, intracellular free ISG15 may interact within cytoplasmic signaling networks to downmodulate proinflammatory cytokine production. Discovery of the extracellular and intracellular proteins with which ISG15 interacts will be necessary to confirm these hypotheses.
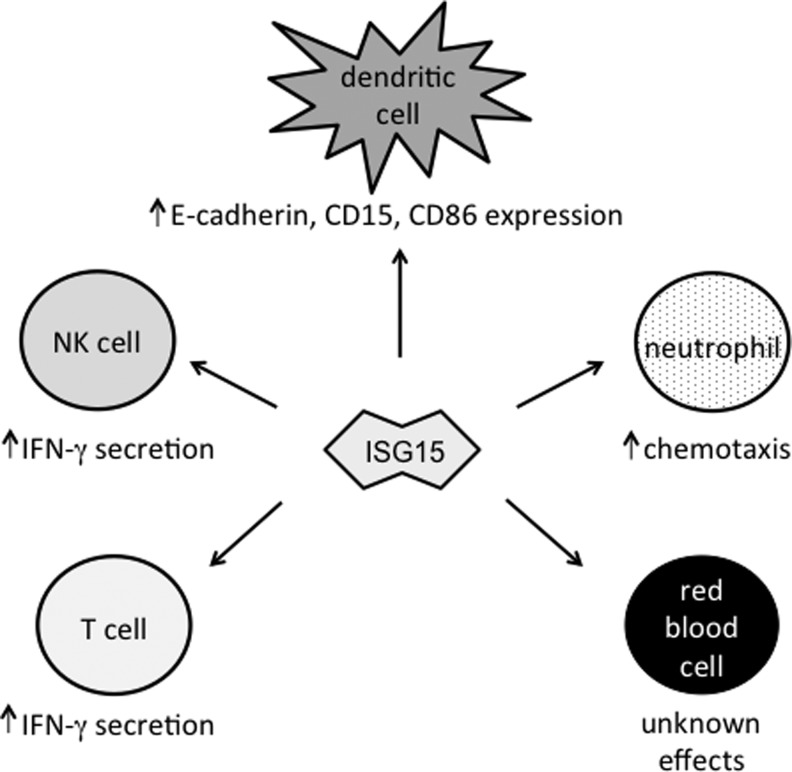
FIG. 2.
Secreted ISG15 modulates the activities of multiple mammalian cell types. ISG15 has been shown to alter the phenotypes of various cells. T and NK cells secrete IFN-γ upon ISG15 stimulation, while dendritic cells upregulate expression of E-cadherin, CD15, and CD86 in response to ISG15. ISG15 enhances the chemotactic activity of neutrophils. Although ISG15 has been detected on the surface of red blood cells, it is not known what the functional impact of this is.
The identity of the receptor to which secreted ISG15 binds will be fundamental to our understanding of ISG15 biology. It is possible that more than 1 receptor exists for ISG15, especially since it has been shown to directly modulate the functions of or bind to diverse cell types, including T cells, NK cells, neutrophils, DCs, macrophages, and red blood cells. The receptor may be localized on the cell surface, where it could bind ISG15 in the extracellular space, or it may be localized inside cytoplasmic vesicles, in which case endocytosis could deliver ISG15 to its receptor. Two other Ubls, SUMO-3 and MNSF-β, along with ubiquitin itself, are present extracellularly (Nakamura and others 1996; Hosono and Yokosawa 2008; Majetschak 2011). The receptors for SUMO-3 and MNSF-β are unknown, but ubiquitin binds to the chemokine receptor CXCR4 (Saini and others 2010). Since ubiquitin and Ubls adopt a similar 3-dimensional structure, it will be interesting to learn whether they also bind receptors with structural similarity. The novel finding that IL-12 synergizes with ISG15 to induce IFN-γ expression raises the possibility that IL-12 may induce expression of the ISG15 receptor, a situation that should be examined.
The work on Ebola VLPs demonstrated that free intracellular ISG15 can bind noncovalently to cytoplasmic proteins to alter their functions. It is conceivable that ISG15 interacts with other unidentified intracellular proteins independent of conjugation. Recent work in the ubiquitin field has demonstrated that unanchored ubiquitin chains can bind to innate signaling molecules, such as RIG-I and members of the NF-κB signaling cascade to influence signaling through these important pathways (Xia and others 2009; Zeng and others 2010). This raises the possibility that other Ubls could function in a similar manner. The body of research on noncovalent binding partners of ISG15 is quite small compared to the conjugation-dependent interactions, but these interactions may influence important biological processes. Uncovering these binding partners for ISG15 will be instrumental in understanding its molecular mechanism.
Concluding Remarks
Significant progress has been made over the past several years with respect to the identification of the ISG15 conjugation machinery, the identities of proteins that undergo ISGylation, and the role of ISG15 as an antiviral factor with both conjugation-dependent and-independent modes of action. Despite these achievements, many questions still remain for all 3 forms of ISG15. With respect to the conjugation-dependent aspects of ISG15, a very small proportion of the identified ISGylated host target proteins have been examined. Additionally, with the exception of influenza and HPV proteins, it is unknown whether other viral or bacterial proteins are modified by ISG15. The elucidation of the functional consequences of ISGylation of both host and microbial proteins will be important in understanding the conjugation-dependent role of ISG15 during infection. The recognition that several viruses encode immune evasion proteins aimed at disrupting conjugation suggests that ISGylation is an important post-translational modification during infection. Mutant viruses that lack this evasion tactic may be useful tools in the identification of ISGylated proteins that are vital to viral control.
Important advances have also been made in the study of free ISG15. Identification of the proteins that bind noncovalently to free intracellular ISG15 is just beginning, but will shed light on the mechanism by which ISG15 may directly participate in intracellular signaling pathways. Similarly, identification of the receptor for secreted ISG15 will clarify how ISG15 can induce diverse phenotypes on various cell types.
Addressing these remaining questions will undoubtedly provide interesting insights into the basic biology of ISG15. At the same time, a better understanding of the antiviral and immunomodulatory activities of ISG15 may aid the development of therapeutics targeted at ISG15. For example, since ISGylation is important to the host response to multiple viruses, inhibition of the deconjugating enzyme Ubp43 may be a useful strategy to augment the antiviral activity of ISG15 conjugation. Other circumstances may benefit from the manipulation of free ISG15. Since secreted ISG15 induces IFN-γ secretion, recombinant ISG15 may be a valuable tool when greater IFN-γ levels would be beneficial, such as during insufficient responses to mycobacteria. Conversely, neutralization of secreted ISG15 may contribute to the dampening of IFN-γ-mediated immunopathology. Finally, depending on the nature of the interactions between free intracellular ISG15 and its noncovalent binding partners, small molecule inhibitors and/or antagonists may allow us to modify the inflammatory cytokines that ISG15 negatively regulates. Further examination into the mechanisms of all 3 forms of ISG15 will help us progress toward these exciting possibilities.
Acknowledgments
This work is supported by grants from National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MCRE), NIH R01 AI080672, and the Children's Discovery Institute of Washington University and St. Louis Children's Hospital.
Author Disclosure Statement
J.A.C. and D.J.L. have no competing financial interests.
References
- Blomstrom DC, Fahey D, Kutny R, Korant BD, Knight E., Jr.1986. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J Biol Chem 261(19):8811–8816 [PubMed] [Google Scholar]
- Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, Mansouri N, Okada S, Bryant VL, Kong XF, Kreins A, Velez MM, Boisson B, Khalilzadeh S, Ozcelik U, Darazam IA, Schoggins JW, Rice CM, Al-Muhsen S, Behr M, Vogt G, Puel A, Bustamante J, Gros P, Huibregtse JM, Abel L, Boisson-Dupuis S, Casanova JL. 2012. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 337(6102):1684–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering R, Zhang X, Kottilil S, Trippler M, Jiang M, Lu M, Gerken G, Schlaak JF. 2010. The interferon stimulated gene 15 functions as a proviral factor for the hepatitis C virus and as a regulator of the IFN response. Gut 59(8):1111–1119 [DOI] [PubMed] [Google Scholar]
- Catic A, Fiebiger E, Korbel GA, Blom D, Galardy PJ, Ploegh HL. 2007. Screen for ISG15-crossreactive deubiquitinases. PLoS One 2(7):e679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chairatvit K, Wongnoppavich A, Choonate S. 2012. Up-regulation of interferon-stimulated gene15 and its conjugates by tumor necrosis factor-alpha via type I interferon-dependent and -independent pathways. Mol Cell Biochem 368(1–2):195–201 [DOI] [PubMed] [Google Scholar]
- Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84(9):4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottle LE. 2011. Mendelian susceptibility to mycobacterial disease. Clin Genet 79(1):17–22 [DOI] [PubMed] [Google Scholar]
- D'Cunha J, Knight E., Jr.Haas AL, Truitt RL, Borden EC. 1996a. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci U S A 93(1):211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight E., Jr.Borden EC. 1996b. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol 157(9):4100–4108 [PubMed] [Google Scholar]
- Dai J, Pan W, Wang P. 2011. ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol J 8:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem 281(7):4334–4338 [DOI] [PubMed] [Google Scholar]
- Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95(26):15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee LA, Lyon N, Seo K, Huibregtse JM. 2010. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell 38(5):722–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ, Broeze RJ, Lengyel P. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279(5713):523–525 [DOI] [PubMed] [Google Scholar]
- Feng Q, Sekula D, Guo Y, Liu X, Black CC, Galimberti F, Shah SJ, Sempere LF, Memoli V, Andersen JB, Hassel BA, Dragnev K, Dmitrovsky E. 2008. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol Cancer Ther 7(12):3780–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, Garcia-Sastre A, Virgin HWt. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2(6):404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulos NV, Arutyunova E, Lai C, Lenschow DJ, Haas AL, Virgin HW. 2009. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J Virol 83(4):1602–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulos NV, Luo JK, Papov V, Zou W, Lenschow DJ, Jacobs BS, Borden EC, Li J, Virgin HW, Zhang DE. 2005. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun 336(2):496–506 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Navajas JM, Lee J, David M, Raz E. 2012. Immunomodulatory functions of type I interferons. Nat Rev Immunol 12(2):125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog 4(7):e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AL, Ahrens P, Bright PM, Ankel H. 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem 262(23):11315–11323 [PubMed] [Google Scholar]
- Hamerman JA, Hayashi F, Schroeder LA, Gygi SP, Haas AL, Hampson L, Coughlin P, Aebersold R, Aderem A. 2002. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J Immunol 168(5):2415–2423 [DOI] [PubMed] [Google Scholar]
- Harty RN, Pitha PM, Okumura A. 2009. Antiviral activity of innate immune protein ISG15. J Innate Immun 1(5):397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono H, Yokosawa H. 2008. Small ubiquitin-related modifier is secreted and shows cytokine-like activity. Biol Pharm Bull 31(5):834–837 [DOI] [PubMed] [Google Scholar]
- Hsiang TY, Zhao C, Krug RM. 2009. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol 83(12):5971–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao NW, Chen JW, Yang TC, Orloff GM, Wu YY, Lai CH, Lan YC, Lin CW. 2010. ISG15 over-expression inhibits replication of the Japanese encephalitis virus in human medulloblastoma cells. Antiviral Res 85(3):504–511 [DOI] [PubMed] [Google Scholar]
- Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, Oh KH, Kim KI, Zhang DE, Bang OS, Chung CH. 2009. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep 10(4):374–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon YJ, Jo MG, Yoo HM, Hong SH, Park JM, Ka SH, Oh KH, Seol JH, Jung YK, Chung CH. 2012. Chemosensitivity is controlled by p63 modification with ubiquitin-like protein ISG15. J Clin Invest 122(7):2622–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketscher L, Basters A, Prinz M, Knobeloch KP. 2012. mHERC6 is the essential ISG15 E3 ligase in the murine system. Biochem Biophys Res Commun 417(1):135–140 [DOI] [PubMed] [Google Scholar]
- Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. 2004. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol Cell Biol 24(21):9592–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KI, Yan M, Malakhova O, Luo JK, Shen MF, Zou W, de la Torre JC, Zhang DE. 2006. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol Cell Biol 26(2):472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KI, Zhang DE. 2005. UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays. Methods Enzymol 398:491–499 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Hwang SY, Imaizumi T, Yoo JY. 2008. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol 82(3):1474–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. 2010. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A 107(34):15181–15186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight E., Jr.Cordova B. 1991. IFN-induced 15-kDa protein is released from human lymphocytes and monocytes. J Immunol 146(7):2280–2284 [PubMed] [Google Scholar]
- Knight E., Jr.Fahey D, Cordova B, Hillman M, Kutny R, Reich N, Blomstrom D. 1988. A 15-kDa interferon-induced protein is derived by COOH-terminal processing of a 17-kDa precursor. J Biol Chem 263(10):4520–4522 [PubMed] [Google Scholar]
- Knobeloch KP. 2010. In vivo functions of isgylation. Subcell Biochem 54:215–227 [DOI] [PubMed] [Google Scholar]
- Kuang Z, Seo EJ, Leis J. 2011. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol 85(14):7153–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, Garcia-Sastre A, Zhang DE, Lenschow DJ. 2009. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol 83(2):1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ. 2010. Antiviral properties of ISG15. Viruses 2(10):2154–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HWt. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol 79(22):13974–13983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcia-Sastre A, Leib DA, Pekosz A, Knobeloch KP, Horak I, Virgin HW. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104(4):1371–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79(24):15199–15208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Li XL, Hassel BA. 2003. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J Biol Chem 278(3):1594–1602 [DOI] [PubMed] [Google Scholar]
- Loeb KR, Haas AL. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem 267(11):7806–7813 [PubMed] [Google Scholar]
- Lu G, Reinert JT, Pitha-Rowe I, Okumura A, Kellum M, Knobeloch KP, Hassel B, Pitha PM. 2006. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol Biol (Noisy-le-grand) 52(1):29–41 [PubMed] [Google Scholar]
- Majetschak M. 2011. Extracellular ubiquitin: immune modulator and endogenous opponent of damage-associated molecular pattern molecules. J Leukoc Biol 89(2):205–219 [DOI] [PubMed] [Google Scholar]
- Malakhova O, Malakhov M, Hetherington C, Zhang DE. 2002. Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J Biol Chem 277(17):14703–14711 [DOI] [PubMed] [Google Scholar]
- Malakhova OA, Zhang DE. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem 283(14):8783–8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Nagata T, Xavier M, Tanigawa Y. 1996. Ubiquitin-like polypeptide inhibits the IgE response of lipopolysaccharide-activated B cells. Int Immunol 8(11):1659–1665 [DOI] [PubMed] [Google Scholar]
- Nakasato N, Ikeda K, Urano T, Horie-Inoue K, Takeda S, Inoue S. 2006. A ubiquitin E3 ligase Efp is up-regulated by interferons and conjugated with ISG15. Biochem Biophys Res Commun 351(2):540–546 [DOI] [PubMed] [Google Scholar]
- Narasimhan J, Wang M, Fu Z, Klein JM, Haas AL, Kim JJ. 2005. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem 280(29):27356–27365 [DOI] [PubMed] [Google Scholar]
- Okumura A, Lu G, Pitha-Rowe I, Pitha PM. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A 103(5):1440–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Pitha PM, Harty RN. 2008. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A 105(10):3974–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Okumura AJ, Uematsu K, Hatakeyama S, Zhang DE, Kamura T. 2012. Activation of double-stranded RNA-activated protein kinase (PKR) by interferon stimulated gene 15 (ISG15) modification down-regulates protein translation. J Biol Chem 288(4):2839–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura F, Zou W, Zhang DE. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev 21(3):255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiak A, Utermohlen O, Niendorf S, Horak I, Knobeloch KP. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol 25(15):6338–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn D, van Boheemen S, Sanchez-Aparicio MT, Rajsbaum R, Garcia-Sastre A, Versteeg GA. 2012. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS One 7(1):e29870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owhashi M, Taoka Y, Ishii K, Nakazawa S, Uemura H, Kambara H. 2003. Identification of a ubiquitin family protein as a novel neutrophil chemotactic factor. Biochem Biophys Res Commun 309(3):533–539 [DOI] [PubMed] [Google Scholar]
- Padovan E, Terracciano L, Certa U, Jacobs B, Reschner A, Bolli M, Spagnoli GC, Borden EC, Heberer M. 2002. Interferon stimulated gene 15 constitutively produced by melanoma cells induces e-cadherin expression on human dendritic cells. Cancer Res 62(12):3453–3458 [PubMed] [Google Scholar]
- Pincetic A, Kuang Z, Seo EJ, Leis J. 2010. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol 84(9):4725–4736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter JL, Narasimhan J, Mende-Mueller L, Haas AL. 1999. Precursor processing of pro-ISG15/UCRP, an interferon-beta-induced ubiquitin-like protein. J Biol Chem 274(35):25061–25068 [DOI] [PubMed] [Google Scholar]
- Recht M, Borden EC, Knight E., Jr.1991. A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J Immunol 147(8):2617–2623 [PubMed] [Google Scholar]
- Saini V, Marchese A, Majetschak M. 2010. CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin. J Biol Chem 285(20):15566–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SJ, Blumen S, Pitha-Rowe I, Kitareewan S, Freemantle SJ, Feng Q, Dmitrovsky E. 2008. UBE1L represses PML/RAR{alpha} by targeting the PML domain for ISG15ylation. Mol Cancer Ther 7(4):905–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, Zhu LH, Wang C. 2010. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol 30(10):2424–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143(2):187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Inoue S, Yokosawa H. 2006a. Identification and Herc5-mediated ISGylation of novel target proteins. Biochem Biophys Res Commun 348(2):473–477 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Iwahara S, Saeki Y, Sasajima H, Yokosawa H. 2005. Link between the ubiquitin conjugation system and the ISG15 conjugation system: ISG15 conjugation to the UbcH6 ubiquitin E2 enzyme. J Biochem 138(6):711–719 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kobayashi T, Tamura S, Yokosawa H. 2006b. Negative regulation of protein phosphatase 2Cbeta by ISG15 conjugation. FEBS Lett 580(18):4521–4526 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yokosawa H. 2005. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun 336(1):9–13 [DOI] [PubMed] [Google Scholar]
- Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. 2012. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev 249(1):158–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhong G, Zhu L, Liu X, Shan Y, Feng H, Bu Z, Chen H, Wang C. 2010. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J Immunol 184(10):5777–5790 [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11(11):997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Hale BG, van Boheemen S, Wolff T, Lenschow DJ, Garcia-Sastre A. 2010. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J Virol 84(10):5423–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang M, Xiao ZZ, Sun L. 2012. Cynoglossus semilaevis ISG15: a secreted cytokine-like protein that stimulates antiviral immune response in a LRGG motif-dependent manner. PLoS One 7(9):e44884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneke SW, Schilte C, Rohatgi A, Monte KJ, Michault A, Arenzana-Seisdedos F, Vanlandingham DL, Higgs S, Fontanet A, Albert ML, Lenschow DJ. 2011. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog 7(10):e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Pung YF, Sze NS, Chin KC. 2006. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc Natl Acad Sci U S A 103(28):10735–10740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461(7260):114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Krug RM. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J 20(3):362–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141(2):315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 31(1):119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. 2004. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc Natl Acad Sci U S A 101(20):7578–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci U S A 102(29):10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Hsiang TY, Kuo RL, Krug RM. 2010. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc Natl Acad Sci U S A 107(5):2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Papov V, Malakhova O, Kim KI, Dao C, Li J, Zhang DE. 2005. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun 336(1):61–68 [DOI] [PubMed] [Google Scholar]
- Zou W, Wang J, Zhang DE. 2007. Negative regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem Biophys Res Commun 354(1):321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]