Abstract
Breast cancer initiation, invasion and metastasis span multiple length and time scales. Molecular events at short length scales lead to an initial tumorigenic population, which left unchecked by immune action, acts at increasingly longer length scales until eventually the cancer cells escape from the primary tumor site. This series of events is highly complex, involving multiple cell types interacting with (and shaping) the microenvironment. Multiscale mathematical models have emerged as a powerful tool to quantitatively integrate the convective-diffusion-reaction processes occurring on the systemic scale, with the molecular signaling processes occurring on the cellular and subcellular scales. In this study, we reviewed the current state of the art in cancer modeling across multiple length scales, with an emphasis on the integration of intracellular signal transduction models with pro-tumorigenic chemical and mechanical microenvironmental cues. First, we reviewed the underlying biomolecular origin of breast cancer, with a special emphasis on angiogenesis. Then, we summarized the development of tissue engineering platforms which could provide highfidelity ex vivo experimental models to identify and validate multiscale simulations. Lastly, we reviewed top-down and bottom-up multiscale strategies that integrate subcellular networks with the microenvironment. We present models of a variety of cancers, in addition to breast cancer specific models. Taken together, we expect as the sophistication of the simulations increase, that multiscale modeling and bottom-up agent-based models in particular will become an increasingly important platform technology for basic scientific discovery, as well as the identification and validation of potentially novel therapeutic targets.
Keywords: Multiscale modeling, Breast cancer, Hypoxia, Angiogenesis
INTRODUCTION
Breast cancer is one of the predominant cancers diagnosed among women, and the second leading cause of cancer death.66 In the past, most experimental cancer research has focused on the genetic and molecular scale malfunctions which deregulate cell growth.10 Understanding the deregulation of the wiring which controls central molecular programs is a daunting and multifaceted problem. These molecular pathways are large, and contain complex architectural features such as redundancy, feedback and crosstalk. 148 While this complexity ensures robustness and efficiency, it also complicates the reprogramming of signal flow and the interpretation of experimental findings. For example, Jones et al. showed in a study of pancreatic cancer patients that on average each patient had 63 genetic alterations spread throughout 12 core signaling pathways.69 Thus, there was not a single dominant malfunction or pathway. Rather a combinatorial interplay of malfunctions acting in concert deregulated cellular function. This integration underscores the realization that cancer is a systems disease, even at the subcellular length scale.
Unfortunately, tumorigenesis involves far more than just malfunctions in signal transduction pathways in homogenous cell populations. Breast tumors are highly heterogenous, involving the simultaneous transmission and processing of many chemical and mechanical signals between multiple cell types within a time- and spatially-varying microenvironment. Furthermore, this cellular variety often includes diverse genetic populations within the same cell type. For example, Navin et al.104 sequenced single cells in highgrade (III) ductal carcinomas and found a complex polygenomic population containing approximately 63% normal and 37% tumor cells, with a large fraction of leukocytes. Interestingly, within the same tumor, they identified four major genetically diverse tumorigenic cell subpopulations. Thus, understanding and ultimately reprogramming the integration of central programs such as proliferation, differentiation or death within multiple cell types, or genetic variants of the same cell type, in concert with the chemical and mechanical cues of the microenvironment is a grand challenge.
To attack a complex disease like breast cancer, we must build comprehensive experimental and computational tools which integrate intracellular signaling architectures with the extracellular microenvironment. Multiscale simulation methods, in combination with novel in vitro tissue engineering platforms, are rapidly evolving to meet this critical challenge. In this study, we review the current state of the art in cancer modeling across multiple length scales, with an emphasis on the integration of intracellular signal transduction models with pro-tumorigenic chemical and mechanical microenvironmental cues. First, we review the underlying biomolecular origins of breast cancer with a special emphasis on angiogenesis. Next, we summarize the development of tissue engineering platforms which could provide high-fidelity ex vivo experimental models to identify and validate multiscale simulations. Following that, we review top-down and bottom-up multiscale computational strategies that integrate subcellular networks with the microenvironment and tumorigenesis. We present models of a variety of cancers, in addition to breast cancer specific models. Thus, as our understanding of the complexity of these processes evolves, multiscale simulation could be a critical tool which provides fundamental biological understanding and potentially important clinical insight.
THE BIOMOLECULAR ORIGINS OF BREAST CANCER
Breast cancer is a highly heterogenous disease which can be broadly subdivided into three major subtypes: hormone receptor-positive tumors, ERBB2-amplified tumors and a third category collectively referred to as triple-negative tumors. The molecular understanding of each subtype, along with the possible treatments for each,58 continues to evolve. High-throughput analytical technologies, such as gene expression profiling or rapid whole-genome sequencing, have been used to great effect to characterize the tumor type and microenvironment, 2,114 and specific gene signatures associated with stages of the disease.25,104,115,147 The traditional tumor initiation hypothesis posits that genetic transforming events in single cells, e.g., TP53 mutations110 or epigenetic changes,64 leads to clonal expansion and the accumulation of additional genetic changes. However, mutations in genes classically associated with breast cancer, e.g., BRCA1, BRCA2 and TP53, account for less than 25% of the excess risk associated with family history.115 Thus, there are likely other transformation pathways that initiate the disease. For example, this traditional view has recently been challenged by the cancer stem cell (CSC) hypothesis in which differentiated cancer cells, which are unable to self-renew, are the progeny of a population of self-renewing CSCs.16,106,115. These tumorigenic cells can then recruit (or phenotypically transform) many other cell types which collectively form the microenvironment of the growing tumor (Fig. 1). Interactions between the tumorigenic cells and the microenvironment, and even the cellular composition of the microenvironment, is a complex function of many factors.73 It is thought that autocrine and bidirectional paracrine signaling regulates the tumorigenic cell population (including CSCs), and these cells in turn secrete factors which influence the makeup and behavior of the microenvironment.82,116 However, soluble signals are likely not the only important cues. Physical changes, solid stresses, matrix stiffness, fluid pressure and other biomechanical forces have also been implicated in tumorigenesis and may influence the recruitment of other cell types including circulating tumor cells (CTCs), fibroblasts and immune cells (recently reviewed by Shieh et al.127 and Lu et al.86). The complexity of the tumor microenvironment may even play a critical role in drug resistance (see Correia et al.29). However, CSCs/CTCs and their respective role in driving tumorigenesis remains controversial. Yet another hypothesis, which builds upon an older idea, is that tumorigenesis is actually a malfunctioning wound-healing process.96 Whatever the initiation events and source of heterogeneity, it is agreed that the complexity of breast tumorigenesis complicates our understanding of the disease, and ultimately limits the development of effective targeted treatment options.
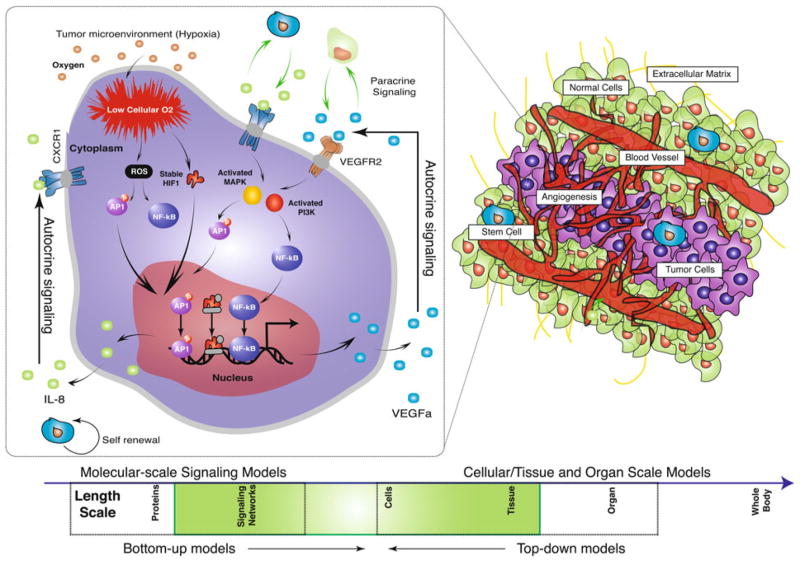
FIGURE 1.
Schematic of the tumor microenvironment. Breast cancer initiation, invasion and metastasis span multiple length and time scales. Molecular events at short length scales lead to an initial tumorigenic population, which left unchecked by immune action, acts at increasingly longer length scales until eventually the cancer cells escape from the primary tumor site. One of the central programs associated with this transition is angiogenesis. Tumor angiogenesis is stimulated by reduced oxygen tension (i.e., hypoxia) which up-regulates the secretion of pro-angiogenic signaling molecules, e.g., VEGF, Interleukin-6 (IL-6) and Interleukin- 8 (IL-8) by tumorigenic cells and other cell types in the tumor microenvironment. These signals then initiate autocrine and paracrine programs which shape the chemical, mechanical and cellular composition of the microenvironment.
The transition from localized ductal carcinoma to invasive and ultimately metastatic breast cancer is a critical milestone impacting the clinical management and outcome of the disease. One of the central programs associated with this transition is angiogenesis (Fig. 1). Tumor angiogenesis is stimulated by many factors, including reduced oxygen tension (i.e., hypoxia) which up-regulates the secretion of proangiogenic signaling molecules by tumorigenic cells.56 Vascular endothelial growth factor (VEGF) is the most potent angiogenic factor secreted by tumor cells in response to hypoxia.39,43 Anti-angiogenic therapies aimed at disrupting the molecular coupling between hypoxia, VEGF signaling, and tumor angiogenesis initially showed great promise.40 Several of the regulatory axes which control VEGF expression in the microenvironment, for example the role of oxygen tension, are relatively well understood. Oxygen in the microenvironment is sensed by hypoxia inducible factor 1α (HIF1α) and the generation of reactive oxygen species (ROS).79,100 HIF1α mediates the initial phase of the angiogenic program by forming a transcriptionally active complex with HIF1β and co-activators such as p300. The stability of the HIF1α subunit is oxygendependent. 117 In normoxic conditions, hydroxylation at two prolyl residues (P402 and P564) by PHD proteins promotes the association of HIF1α with the Von Hippel- Lindau (VHL) E3 ubiquitin ligase and subsequently leads to degradation. An additional hydroxylation site at N803 near the C-terminus of HIF1α is regulated by the asparaginyl hydroxylase FIH. Hydroxylation at N803 does not influence stability; rather, it blocks the interaction of the HIF1α C-terminal domain with transcriptional co-activators such as p300. Activated HIF1 up-regulates the expression of many factors including VEGF and Interleukin-8 (IL-8).146 On the other hand, ROSpromotes nuclear factor κB (NF-κB) activation.100 NF-κB also regulates both VEGF and IL-8 expression. 100,146 The exact relationship between ROS and NF-κB activation is unclear; ROS has been hypothesized to activate serine kinases which in-turn phopshorylate the N-terminal serine residues (S32/S36) on IKK.44 Unfortunately, the initial success of the anti- VEGF-A monoclonal antibody bevacizumab has been reexamined in light of clinical evidence suggesting anti- VEGF therapy often prolongs patient survival by only months, without offering an enduring cure.72 Studies have also emerged questioning the overall survival advantage of bevacizumab in combination with chemotherapeutics, 98 while other studies suggested that even short-term exposure to potent anti-angiogenic therapies might actually induce invasiveness.36,108 These studies, in combination with potential safety concerns,27 led the US Food and Drug Administration (USFDA) to remove the breast cancer indication from the bevacizumab label.
The finding that anti-VEGF therapy may induce invasiveness seemingly contradicts years of dogma suggesting VEGF-induced vessel recruitment is essential for cancer progression. Carmeliet and coworkers recently reviewed two of the leading hypotheses explaining this apparent contradiction.84 The first has suggested that reduced angiogenesis selects for a hypoxia-tolerant tumorigenic population that is better adapted to the low-oxygen microenvironment.18 These hyper-tolerant tumorigenic cells thrive in the noxious microenvironment by adapting their metabolism or escape by inducing invasive programs such the epithelial mesenchymal transition (EMT).18 The role of EMT in cancer progression and metastasis has long been recognized.55,140 However, this population of hyper-tolerant tumorigenic cells may also recruit other vascular precursor cell types, for example angiocompetent bone marrow-derived cells,52 or co-opt existing vasculature that is not inhibited by anti-VEGF therapy. 15 Ebos and coworkers suggested a second hypothesis where VEGF inhibitors induce a chronically inflamed state characterized by the expression of several factors including stromal cell-derived factors 1-alpha (SDF1α), placenta growth factor (PlGF), interleukin-6 (IL-6), erythropoietin, osteopontin, and other cytokines.35 These cytokines may then recruit angiogenic bone marrow-derived endothelial and myeloid progenitors,70 many of which express vascular endothelial growth factor receptor 1 (VEGFR1), thus their recruitment is not blocked by VEGF inhibitors.70 Both of these hypotheses involve the recruitment of immunomodulatory cell types by the secretion of cytokines and other factors. The integration between the immune system, inflammation and cancer progression (including the modulation of the CSC population) is an emerging area with classical roots.34 The immune system can both inhibit and stimulate tumorigenesis, where these influences are mediated by complex mechanisms.93 Inflammatory signals, such as Interleukin-6 (IL-6) and IL-8, are secreted by many cell types in the microenvironment.147 IL-6 is known to promote breast cancer progression,125,126 and serum levels of both IL-6 and IL-8 correlate with patient outcome.14,124 Interestingly, both IL-6 (via the GP130 receptor) and IL-8 (via the CXCR1 receptor) have also been shown to directly regulator breast cancer stem cell (BCSC) self-renewal.64 The expression of both of these cytokines is regulated by NF-κB,12 thereby potentially linking this critical signaling axis with ROS formation in hypoxic environments.
Ex Vivo Experimental Models
The development of effective anti-angiogenic therapies depends critically upon a comprehensive understanding of proliferation and vascularization programs and the interaction of these programs with the microenvironment. Multiscale simulation tools in combination with high-fidelity ex-vivo experimental models can help unravel this complexity.111 However, multiscale models require fine-grained training and validation data to be successful. Unfortunately, wideranging but fine-tuned experimental control of the receptor signaling cascades involved in angiogenesis or other tumorigenic processes is not possible with current in vitro and in vivo approaches. For example, conventional angiogenesis models (e.g., tube formation on Matrigel) fail to capture: (i) the intrinsic, threedimensional morphology and diffusion-limited formation of intratumor niches, (ii) microscale integration of multiple cell types within physiologically relevant architectures, and (iii) coupling to a vascular interface that provides systemic convective transfer of endocrine signals and other cellular nutrients. Tissue engineering approaches to model tumor physiology have recapitulated the reaction-diffusion processes of solid tumors and begun to elucidate the microphysiological details of the angiogenic and other tumorigenic processes. These advancements have been enabled by new synthetic materials,88 development of microfluidic lab on a chip technologies33,63 as well as a new appreciation for the significant role played by the microenvironment in shaping tumor progression.86 The integration of microfluidic and three dimensional tissue engineering technologies permits control over and monitoring of the soluble microenvironment experienced by cells.21–26 Nelson et al.105 developed one of the first three-dimensional patterning techniques to construct multicellular epithelial tissues in three-dimensional gels composed of extracellular matrix (ECM) proteins. Using this patterning technology, they later explored the signaling forces driving cell organization in engineered three dimensional mammary ducts101 as well as how complex interactions between mammary progenitor cells and the microenvironment drive cell fate decisions.75 Using three-dimensional polymeric scaffolds to mimic the tumor ECM, we recently showed that dimensionality (i.e., two-dimensional vs. threedimensional), hypoxia, and integrin engagement play a critical role in VEGF and IL-8 up-regulation.42,43 Zheng et al.153 created ex-vivo microvascular networks using human umbilical vein endothelial cells (HUVECs) seeded into microfluidic circuits formed via soft lithography in a type I collagen gel. They quantified sprout formation following exogenous administration of vasculogenic medium throughout the device. Seok and coworkers used a similar three-dimensional microfluidic strategy to explore sprouting in the presence of angiopoietin 1 (ANG-1) and VEGF gradients. 68,128 Engineered culture systems could advance studies of tumor vascularization by faithfully replicating the in vivo microenvironment, while providing highly quantifiable, and controlled conditions. Microfluidic devices have also been used to reconstruct realistic microenvironmental mimics to study other processes important in breast tumorigenesis e.g., differentiation and migration.62 These experimental tools and others, such as bead-based methods,103 when combined with mathematical models of signaling driving the evolution of the microenvironment, could unravel the complexity of tumor vascularization and perhaps identify molecular targets for improved proangiogenic therapies.
MULTISCALE MODELING METHODS IN CANCER
Many factors act in concert to drive tumor formation. These forces act across multiple length and time scales, involve heterogenous cell populations and involve both biophysical and biochemical cues. To understand how these disparate forces drive tumor formation generally, and breast cancer tumorigenesis in particular, we need to develop predictive multiscale models. Multiscale models of tumorigenic processes e.g., growth-factor induced proliferation or angiogenesis dynamics are not new. Mathematical models exploring this space of problems have been developed since the 1970s (see Quatub et al.119 for a review angiogenesis models). A wonderful compilation of recent work in multiscale modeling has been organized in a book edited by Deisboeck and Stamatakos.31 The individual chapters (authored by several groups) describe the application of agent based and continuum modeling strategies to study several cancer types, including breast cancer. Moreover, several journals have dedicated special issues to multiscale simulation methods and their application to cancer modeling.121 While the objectives of multiscale simulation studies have not changed in several decades, current models are significantly more sophisticated. This increased sophistication has largely been driven by increased biological understanding and the rapid increase in computing power.
Multiscale strategies can broadly be organized into continuous, discrete and hybrid approaches. Continuous approaches use continuum mechanical principles encoded in partial differential equations (PDEs) or integral partial differential equations (IPDEs) to describe the variation of population-averaged phenomena, e.g., tumor cell density as a function of space and time. Continuum models offer the advantage of easily describing whole tumor dynamics, including complex physical phenomena such as interstitial pressure gradients and convective transport from the tumor.65 For example, Murray and coworkers used continuum approaches to model prostate cancer137 and many aspects of glioma formation,136 including the response to treatment.135 More recently, Swanson and colleagues used continuum approaches to model glial progenitor cell recruitment.94 Continuous approaches have also been used to explore therapeutic antibody distribution in tumors,142 as well as the design of therapeutic antibodies.120 While these and other continuum studies have generated nontrivial insights, continuum models are limited to a population-averaged picture of the tumor. This is an issue if you are interested in population distributed behavior at the cellular and subcellular length scales, or the behavior of your system is strongly stochastic. On the other hand, discrete approaches such fully stochastic simulations, can predict emergent properties generated by interactions between individual cells.23 Fully stochastic methods, such as the next subvolume method (NSM), naturally integrate stochastic reaction dynamics with physical models.57 Unfortunately, stochastic methods such as NSM typically scale poorly with problem size.
Agent Based Models (ABMs)
In between continuum and fully discrete approaches are hybrid strategies. Perhaps the best known hybrid strategy in the cancer and complex systems community is agent based modeling (ABM).17 ABMs are a class of simulation in which combinations of autonomous actors or agents are embedded in a spatially and temporally varying computational universe. Both the agents and universe may have state, meaning variables or variable combinations which describe the current configuration of the system. The stateful agents individually interact with the universe (and each other) using predefined rules. These interactions can be twoway, i.e., the state of the agents can be informed by the universe (often governed by continuum mechanics), and conversely the state of the universe can inform the agents (Fig. 2). Integration between the behavior of the agents and the microenvironment occurs naturally by making the behavior rules functions of spatially or temporally distributed microenvironmental variables. Arguably, ABMs have had the largest impact simulating morphogen-induced developmental programs 50,141 as well as immunological processes such as cell trafficking.9 However, ABMs have also proven useful in modeling tumorigenesis,1,134 subprocesses such as normal and pathological angiogenesis30,81 and microvascular patterning.112 ABMs have also been used extensively in ecology,51 epidemiology,22 crowd behavior102 as well as non-biological fields such as transportation management.17 Thus, ABM is a powerful technique with wide applicability to a broad spectrum of problems, not just modeling cancer progression.
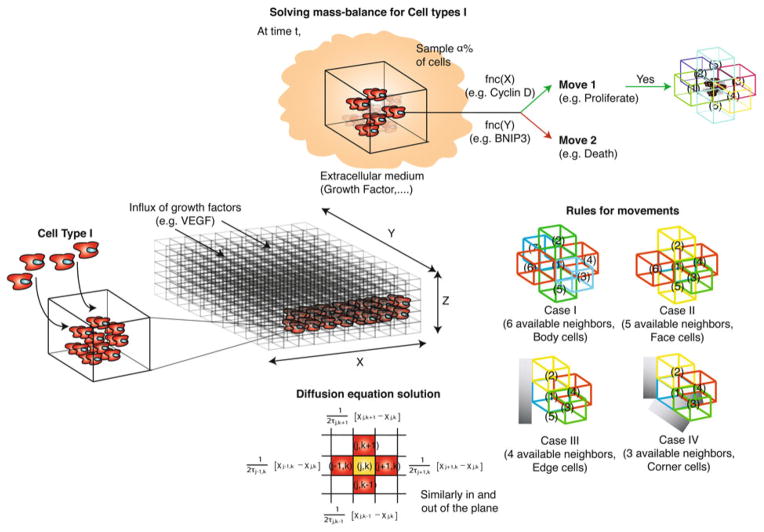
FIGURE 2.
Schematic of a generic bottom-up ABM strategy. A three-dimensional computational domain representing the microenvironment is discretized into well-mixed microcompartments. The extracellular state e.g., the concentration of pO2 or VEGF in each of the microcompartments is governed by the solution of continuum mass balances equations (partial differential equations). Agents representing different cell-types, each equipped with perhaps many signal processing networks, are embedded into the computational microenvironment and allowed to evolve according to rules that are functions of the output of the signaling networks. The agents make decisions about possible actions e.g., move, proliferate, differentiate etc. by evaluating the network models. Thus, the decisions of the agents depend upon both the position and temporal state of the agent.
There are two schools of thought governing ABM rule formulation. Top-down approaches, which have traditionally been the most popular strategy, encode system attributes as coarse-grained empirical rules which describe global control mechanisms. Often these rules are based on experimental observations, thus topdown ABMs can predict sophisticated cellular behavior without mechanistic information.23 Furthermore, software packages, e.g., NetLogo129 or CompuCell4 facilitate ABM formulation and simulation, making this strategy easy to implement. On the other hand, bottom-up approaches use mechanistic signal transduction pathway models to inform the behavior of agents. Each agent in the simulation is equipped with these signaling networks. Thus, the signaling profile of each agent can vary as a function of time and position, within the microenvironment. This integration allows agents to make complex decisions which vary as the extracellular microenvironment varies. Moreover, if these signaling programs result in secretion, the agents can transform the local extracellular matrix or initiate autocrine or paracrine signaling programs. The advantage of a bottom-up strategy is the direct coupling of agent behavior with cellular or subcellular signaling programs. Arguably one of the most advanced examples of a bottom-up biophysical tumor simulation is the recent vascularized tumor growth model of Perfahl et al.113. In the Perfahl et al. study, a comprehensive simulation that integrated several biophysical and biochemical facets of tumorigenesis e.g., blood flow, angiogenesis, vascular remodeling, extracellular transport and nutrient-dependent cell cycle dynamics was used to explore three-dimensional tumor formation. While the solid tumors simulated were not breast tumors, the strategy used in the Perfahl et al. study could be easily adopted to model breast tumorigenesis. Of course the Perfahl et al. study was build upon or extended several other important previous multiscale studies.91,107 The integration of subcellular networks with macroscopic tumor formation, also potentially allows bottom-up ABMs to be useful as in silico surrogates for therapeutic target identification, or ultimately to understand the morphological outcome of cellular mutations. For example, Rejniak et al. used the IBCell framework to simulate the formation of epithelial structures e.g., hollow acinar structures which were qualitatively consistent with three dimensional MCF10A cell culture studies.122 This study was important in two ways. First, it an excellent case study of the integration of experimental tools with multiscale simulation. Second, the authors performed a parameter sampling calculation that identified regions of distinct epithelial morphologies. These possible configurations were then validated with engineered MCF10A cell lines. The latter aspect of this study firmly established that bottom- up multiscale models could be used as predictive tools. However, including mechanistic information can also be a disadvantage; intracellular signal transduction models are difficult to formulate, identify and validate. This is especially true in breast cancer because of the multiple cytokine and growth-factor signaling axes involved in the disease.76 Thus, one of the central challenges to using bottom-up ABMs is the identification of the signal transduction models used in the rule sets. Typically, these models are formulated as a coupled system of nonlinear ordinary differential equations (ODEs), however many other model formulations could be possible.71
To formulate and solve ODE signaling models requires a deep understanding of both network structure and model parameters. The rates of biochemical or biophysical transformations within ODE models can be described using a variety of kinetic formulations, e.g., mass-action kinetics.24 These various kinetic forms have a variety of parameter types that must be estimated or measured. The parameter estimation problem is often very difficult, given the underdetermined and noisy nature of most training data sets. Moreover, signal transduction models typically exhibit complex behavior with respect to inputs and their parameters. For example, models of growth factor, hormone signaling, differentiation and MAPK signaling all showed threshold or switch-like behavior.7–138. Thus, it is often impossible to uniquely identify parameters in signaling models, even with extensive training data.46 Despite identification standards47 and the integration of model identification with experimental design,11 parameter estimation remains challenging. Towards this issue, a number of groups have turned to ensemble methods. Instead of identifying a single (but uncertain) model, the goal of an ensemble approach is to identify a family of models consistent with, and constrained by, the available experimental data. This strategy has been used in systems biology and other fields like weather prediction to identify parameter rich models using incomplete or sparse data.74–109 Their central value is the ability to constrain model predictions despite uncertainty in the model parameters (and sometimes structure). For example, Sethna and coworkers showed that an ensemble of growth factor signaling models gave good predictions despite incomplete parameter information (sometimes only order of magnitude estimates).20 They further showed that model ensembles were predictive using many different mathematical model formulations.54 Model ensembles have also been hypothesized as a general coarse-grained means to capture population distributed phenomena when stochastic simulation is too expensive. For example, the population specific translation regulation77 or the response of a patient population to treatment.87 There are several numerical techniques to generate model ensembles. Battogtokh et al. introduced a Metropolis-type random walk strategy97 to estimate an ensemble of models describing the quinic acid gene cluster of Neurospora crassa.13 This Monte Carlo strategy was later modified by Tasseff et al. to control for ensemble correlation in models of prostate cancer,138 and later Retinoic Acid (RA) induced differentiation of hematopoietic precursors cells.139 A similar Markov-Chain Monte Carlo technique was developed by Song et al. to generate a family of models describing Neutrophil trafficking in Sepsis.132 Other strategies such as the Pareto optimal ensemble technique (POET), a multiobjective optimization strategy which uses simulated annealing to sample parameter space, have also been proposed. 130,131 POETs has been used, in combination with cross-validation, to generate predictive ensembles for several networks including fundamental programs such as translation initiation.77 Taken together, identification of intracellular signal transduction models that are parameter rich will continue to be a challenge. However, ensembles are one strategy to develop reasonably predictive models which could be useful for ABM simulations, despite uncertainty.
Perhaps the more fundamental challenge to developing predictive signal transduction models is representing the signaling network architecture. Yeast Two- Hybrid (Y2H),41, Fluorescence Resonance Energy Transfer (FRET)150 or Chromatin Immunoprecipitation (ChIP) combined with DNA-microarrays (ChIP-chip) or high-throughput DNA-sequencing (ChIP-seq) techniques92 have all been used to estimate protein–protein or protein–DNA interactions. These techniques when combined with low-throughput immunoprecipitation have been the basis for most experimental network discovery. Computational motif discovery,99 network discovery and reconstruction using high-throughput data sources145,149 or text mining, 3,38 have also contributed significantly to network identification. These studies and many others have led to comprehensive on-line network databases such as STRING,67 NetworKIN,80 PhosphoSitePlus60 or KEGG6 which continue to evolve as new information is made available. On-line model repositories such as the BioModels database78 have also been created to archive published signal transduction models. Thus, as more network architecture information becomes available, and model development continues to evolve perhaps the challenge of developing comprehensive signal transduction simulation models will decrease. However, for the foreseeable future, biologically realistic network models are likely to be parameter rich and data poor, even with the advent of advanced analytical techniques.
Despite identification challenges, there are several examples where bottom-up strategies have been used to integrate subcellular data with the microenvironment. 32 Deisboeck and coworkers developed a number of ABM simulations exploring growth-factor signaling within brain and non-small cell lung cancer (NSCLC) tumors. An ODE-based epidermal growth factor (EGF) signaling model was embedded within two- and three-dimensional computational domains, where the spatial-temporal dynamics of the microenvironment domain was governed by PDEs. This framework was then used to explore a number of complex questions: the role of epidermal growth factor receptor (EGFR) density in tumor progression,8 the influence of genetic instability in tumor heterogeneity,152 the components that control the proliferation-to-migration switch for brain tumors,151 and the role of EGF and TGFβ signal integration in non-small cell lung cancer.144 Macklin et al. used an ABM approach to investigate breast ductal carcinoma in situ (DCIS),90 using patient-specific molecular and cellular measurements to calibrate their model. Likewise, Frieboes et al. used multiscale modeling to identify specific functional relationships linking tumor growth and regression to the underlying phenotype of breast cancer following chemotherapy.45 Simulations of the factors controlling tumor shape and morphology is another area where ABMs have made an impact. For example, Engler et al. used multiscale modeling to investigate how emergent properties of adhesion-directed multicellular structures sculpt the tissue, promote its functionality, and maintain its homeostasis.37 While the majority of bottom-up decision networks are mechanistic, non-mechanistic network models have also been used to guide agent behavior. For example, Gerlee and Anderson used a neural network formulation where extracellular variables formed the input layer, intracellular variables were the hidden layer and phenotype was the output layer.48 The neural-network agent framework was used in several studies of the general properties of invasion and tumorogenesis,5,49 including the response of the tumor to treatment.59 Potentially, other hybrid mechanistic models, such as discrete logic models, could also be used in the ABM rules.123
There is growing enthusiasm for using signaling assisted multiscale models as tools for therapeutic target discovery and validation.143 However, the signaling models used to date have not been comprehensive, typically containing perhaps two abstracted pathway architectures at most. Thus, while the concept of using bottom-up ABMs for drug discovery is intriguing, the description of the biology must be significantly expanded to capture the intricate responses of signaling architectures to perturbation. Increasing the level of detail of the signaling architectures used by agents brings several challenges. We have already mentioned the challenges of network model identification. Assuming we already have identified network models, the next big challenge is then the scaling performance of the simulation. While the exact performance of bottom-up ABM simulations is problemspecific, in the worst case we would expect exponential scaling with the number of agents. Thus, detailed simulations using many bottom-up agents, each equipped with multiple decision networks, is not tractable on single processor machines. However, this is a common issue faced in many multiscale modeling application domains. For example, combustion applications often have chemical reaction networks with hundreds or thousands of species which are coupled to turbulent flow models. For these problems, Pope and coworkers developed the in situ adaptive tabulation (ISAT) algorithm which minimizes expensive function updates. Interestingly, the ISAT strategy has resulted in speed-ups of up to a thousand-fold for complex combustion calculations.85 While ISAT has not been applied to bottom-up ABMs, this and other highperformance computing strategies could be adapted to facilitate increasingly detailed multiscale simulations.
CONCLUSIONS
Breast cancer initiation, invasion and metastasis span multiple length and time scales. Molecular events at short length scales lead to an initial tumorigenic population, which left unchecked by immune action, acts at increasingly longer length scales until eventually these cells escape from the primary tumor site. This series of events is highly complex, involving multiple cell types interacting with (and shaping) the microenvironment. Multiscale mathematical models have emerged as a powerful tool to quantitatively integrate the convective-diffusion-reaction processes occurring on the systemic scale, with the molecular signaling processes occurring on the cellular and subcellular scales. In this study, we reviewed the current state of the art in cancer modeling across multiple length scales, with an emphasis on the integration of intracellular signal transduction models with pro-tumorigenic chemical and mechanical microenvironmental cues. First, we reviewed the underlying biomolecular origin of breast cancer, with a special emphasis on angiogenesis. Then we summarized the development of tissue engineering platforms which could provide highfidelity ex vivo experimental models to identify and validate multiscale simulations. Lastly, we reviewed top-down and bottom-up multiscale strategies that integrate subcellular networks with the microenvironment. Taken together, we expect as the sophistication of the simulations increase, that multiscale modeling and bottom-up agent-based models in particular will become an increasingly important platform technology for basic scientific discovery.
Acknowledgments
I apologize to the investigators whose work I was unable to discuss in this focused review. This study was supported by Award Number #U54CA143876 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Abbott RG, Forrest S, Pienta KJ. Simulating the hallmarks of cancer. Artif Life. 2006;12:617–634. doi: 10.1162/artl.2006.12.4.617. [DOI] [PubMed] [Google Scholar]
- 2.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Ananiadou S, Kell DB, Tsujii JI. Text mining and its potential applications in systems biology. Trends Biotechnol. 2006;24:571–579. doi: 10.1016/j.tibtech.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Andasari V, Roper RT, Swat MH, Chaplain MAJ. Integrating intracellular dynamics using CompuCell3D and Bionetsolver: applications to multiscale modelling of cancer cell growth and invasion. PLoS One. 2012;7:e33726. doi: 10.1371/journal.pone.0033726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson ARA, Rejniak KA, Gerlee P, Quaranta V. Microenvironment driven invasion: a multiscale multimodel investigation. J Math Biol. 2009;58:579–624. doi: 10.1007/s00285-008-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki-Kinoshita KF, Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol Biol. 2007;396:71–91. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
- 7.Asthagiri AR, Lauffenburger DA. A computational study of feedback effects on signal dynamics in a mitogen-activated protein kinase (MAPK) pathway model. Biotechnol Prog. 2001;17:227–239. doi: 10.1021/bp010009k. [DOI] [PubMed] [Google Scholar]
- 8.Athale CA, Deisboeck TS. The effects of EGFreceptor density on multiscale tumor growth patterns. J Theor Biol. 2006;238:771–779. doi: 10.1016/j.jtbi.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 9.Bailey AM, Thorne BC, Peirce SM. Multi-cell agent-based simulation of the microvasculature to study the dynamics of circulating inflammatory cell trafficking. Ann Biomed Eng. 2007;35:916–936. doi: 10.1007/s10439-007-9266-1. [DOI] [PubMed] [Google Scholar]
- 10.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33(Suppl):238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 11.Bandara S, Schlöder JP, Eils R, Bock HG, Meyer T. Optimal experimental design for parameter estimation of a cell signaling model. PLoS Comput Biol. 2009;5:e1000558. doi: 10.1371/journal.pcbi.1000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 13.Battogtokh D, Asch DK, Case ME, Arnold J, Schuttler HB. An ensemble method for identifying regulatory circuits with special reference to the QA gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 2002;99:16904–16909. doi: 10.1073/pnas.262658899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, et al. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10:7157–7162. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 15.Bergers G, Hanahan D. Modes of resistance to antiangiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertos NR, Park M. Breast cancer—one term, many entities. J Clin Invest. 2011;121:3789–3796. doi: 10.1172/JCI57100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonabeau E. Agent-based modeling: methods and techniques for simulating human systems. Proc Natl Acad Sci U S A. 2002;99(Suppl 3):7280–7287. doi: 10.1073/pnas.082080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brahimi-Horn MC, Chiche J, Pouysségur J. Hypoxia and cancer. J Mol Med (Berl) 2007;85:1301–1307. doi: 10.1007/s00109-007-0281-3. [DOI] [PubMed] [Google Scholar]
- 19.Brown KS, Sethna JP. Statistical mechanical approaches to models with many poorly known parameters. Phys Rev E Stat Nonlin Soft Matter Phys. 2003;68:021904. doi: 10.1103/PhysRevE.68.021904. [DOI] [PubMed] [Google Scholar]
- 20.Brown KS, Hill CC, Calero GA, Myers CR, Lee KH, et al. The statistical mechanics of complex signaling networks: nerve growth factor signaling. Phys Biol. 2004;1:184– 195. doi: 10.1088/1478-3967/1/3/006. [DOI] [PubMed] [Google Scholar]
- 21.Cabodi M, Choi NW, Gleghorn JP, Lee CSD, Bonassar LJ, et al. A microfluidic biomaterial. J Am Chem Soc. 2005;127:13788–13789. doi: 10.1021/ja054820t. [DOI] [PubMed] [Google Scholar]
- 22.Chao DL, Halloran ME, Obenchain VJ, Longini IM., Jr Flute, a publicly available stochastic influenza epidemic simulation model. PLoS Comput Biol. 2010;6:e1000656. doi: 10.1371/journal.pcbi.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chavali AK, Gianchandani EP, Tung KS, Lawrence MB, Peirce SM, et al. Characterizing emergent properties of immunological systems with multi-cellular rule-based computational modeling. Trends Immunol. 2008;29:589–599. doi: 10.1016/j.it.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Chen WW, Schoeberl B, Jasper PJ, Niepel M, Nielsen UB, et al. Input–output behavior of ERBB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol. 2009;5:239. doi: 10.1038/msb.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 26.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, et al. Microfluidic scaffolds for tissue engineering. Nat Mater. 2007;6:908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 27.Choueiri TK, Mayer EL, Je Y, Rosenberg JE, Nguyen PL, et al. Congestive heart failure risk in patients with breast cancer treated with bevacizumab. J Clin Oncol. 2011;29:632–638. doi: 10.1200/JCO.2010.31.9129. [DOI] [PubMed] [Google Scholar]
- 28.Chrobak KM, Potter DR, Tien J. Formation of perfused, functional microvascular tubes in vitro. Microvasc Res. 2006;71:185–196. doi: 10.1016/j.mvr.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Correia AL, Bissell MJ. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist Updat. 2012;15:39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das A, Lauffenburger D, Asada H, Kamm RD. A hybrid continuum-discrete modelling approach to predict and control angiogenesis: analysis of combinatorial growth factor and matrix effects on vessel-sprouting morphology. Philos Trans A Math Phys Eng Sci. 2010;368:2937–2960. doi: 10.1098/rsta.2010.0085. [DOI] [PubMed] [Google Scholar]
- 31.Deisboeck TS, Stamatakos GS, editors. Multiscale Cancer Modeling. Boca Raton, FL: CRC Press; 2010. [Google Scholar]
- 32.Deisboeck TS, Wang Z, Macklin P, Cristini V. Multiscale cancer modeling. Annu Rev Biomed Eng. 2011;13:127–155. doi: 10.1146/annurev-bioeng-071910-124729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 34.Dvorak HF. Tumors: wounds that do not heal. similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 35.Ebos JML, Lee CR, Christensen JG, Mutsaers AJ, Kerbel RS. Multiple circulating proangiogenic factors induced by sunitinib malate are tumorindependent and correlate with antitumor efficacy. Proc Natl Acad Sci U S A. 2007;104:17069–17074. doi: 10.1073/pnas.0708148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebos JML, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, et al. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–239. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engler AJ, Humbert PO, Wehrle-Haller B, Weaver VM. Multiscale modeling of form and function. Science. 2009;324:208–212. doi: 10.1126/science.1170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faro A, Giordano D, Spampinato C. Combining literature text mining with microarray data: advances for system biology modeling. Brief Bioinform. 2012;13:61–82. doi: 10.1093/bib/bbr018. [DOI] [PubMed] [Google Scholar]
- 39.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 40.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti- VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 41.Fields S, Sternglanz R. The two-hybrid system: an assay for protein–protein interactions. Trends Genet. 1994;10:282–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 42.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 43.Fischbach C, Kong HJ, Hsiong SX, Evangelista MB, Yuen W, et al. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc Natl Acad Sci U S A. 2009;106:399–404. doi: 10.1073/pnas.0808932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flohé L, Brigelius-Flohé R, Saliou C, Traber MG, Packer L. Redox regulation of NF-kappa B activation. Free Radic Biol Med. 1997;22:1115–1126. doi: 10.1016/s0891-5849(96)00501-1. [DOI] [PubMed] [Google Scholar]
- 45.Frieboes HB, Edgerton ME, Fruehauf JP, Rose FRAJ, Worrall LK, et al. Prediction of drug response in breast cancer using integrative experimental/computational modeling. Cancer Res. 2009;69:4484–4492. doi: 10.1158/0008-5472.CAN-08-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gadkar KG, Varner J, Doyle FJ. Model identification of signal transduction networks from data using a state regulator problem. Syst Biol (Stevenage) 2005;2:17–30. doi: 10.1049/sb:20045029. [DOI] [PubMed] [Google Scholar]
- 47.Gennemark P, Wedelin D. Benchmarks for identification of ordinary differential equations from time series data. Bioinformatics. 2009;25:780–786. doi: 10.1093/bioinformatics/btp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerlee P, Anderson ARA. Modelling evolutionary cell behaviour using neural networks: application to tumour growth. Biosystems. 2009;95:166–174. doi: 10.1016/j.biosystems.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerlee P, Anderson ARA. Evolution of cell motility in an individual-based model of tumour growth. J Theor Biol. 2009;259:67–83. doi: 10.1016/j.jtbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant MR, Mostov KE, Tlsty TD, Hunt CA. Simulating properties of in vitro epithelial cell morphogenesis. PLoS Comput Biol. 2006;2:e129. doi: 10.1371/journal.pcbi.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grimm V, Revilla E, Berger U, Jeltsch F, Mooij WM, et al. Pattern-oriented modeling of agent-based complex systems: lessons from ecology. Science. 2005;310:987– 991. doi: 10.1126/science.1116681. [DOI] [PubMed] [Google Scholar]
- 52.Grunewald M, Avraham I, Dor Y, Bachar-Lustig E, Itin A, et al. VEGF-induced adult neovascularization: recruitment, retention, and role of accessory cells. Cell. 2006;24:175–189. doi: 10.1016/j.cell.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 53.Gupta A, Varner J, Maranas C. Large-scale inference of the transcriptional regulation of Bacillus subtilis. Comput Chem Eng. 2005;29:565–576. [Google Scholar]
- 54.Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, et al. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol. 2007;3:1871–1878. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 56.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 57.Hattne J, Fange D, Elf J. Stochastic reaction-diffusion simulation with MesoRD. Bioinformatics. 2005;21:2923–2924. doi: 10.1093/bioinformatics/bti431. [DOI] [PubMed] [Google Scholar]
- 58.Higgins MJ, Baselga J. Targeted therapies for breast cancer. J Clin Invest. 2011;121:3797–3803. doi: 10.1172/JCI57152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinow P, Gerlee P, McCawley LJ, Quaranta V, Ciobanu M, et al. A spatial model of tumor-host interaction: application of chemotherapy. Math Biosci Eng. 2009;6:521–546. doi: 10.3934/mbe.2009.6.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, et al. Phosphositeplus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D70. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hornberg JJ, Binder B, Bruggeman FJ, Schoeberl B, Heinrich R, et al. Control of MAPK signalling: from complexity to what really matters. Oncogene. 2005;24:5533– 5542. doi: 10.1038/sj.onc.1208817. [DOI] [PubMed] [Google Scholar]
- 62.Huang Y, Agrawal B, Sun D, Kuo JS, Williams JC. Microfluidics-based devices: new tools for studying cancer and cancer stem cell migration. Biomicrofluidics. 2011;5:13412. doi: 10.1063/1.3555195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huh D, Torisawa YS, Hamilton GA, Kim HJ, Ingber DE. Microengineered physiological biomimicry: organs-on-chips. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 64.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, Signaling Assisted Multiscale Modeling (SAMM) 2497 and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain RK, Baxter LT. Mechanisms of heterogeneous distribution of monoclonal antibodies and other macromolecules in tumors: significance of elevated interstitial pressure. Cancer Res. 1988;48:7022–7032. [PubMed] [Google Scholar]
- 66.Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69– 90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 67.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, et al. String 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong GS, Han S, Shin Y, Kwon GH, Kamm RD, et al. Sprouting angiogenesis under a chemical gradient regulated by interactions with an endothelial monolayer in a microfluidic platform. Anal Chem. 2011;83:8454–8459. doi: 10.1021/ac202170e. [DOI] [PubMed] [Google Scholar]
- 69.Jones S, Zhang X, Parsons DW, Lin JCH, Leary RJ, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karlebach G, Shamir R. Modelling and analysis of gene regulatory networks. Nat Rev Mol Cell Biol. 2008;9:770– 780. doi: 10.1038/nrm2503. [DOI] [PubMed] [Google Scholar]
- 72.Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuepfer L, Peter M, Sauer U, Stelling J. Ensemble modeling for analysis of cell signaling dynamics. Nat Biotechnol. 2007;25:1001–1006. doi: 10.1038/nbt1330. [DOI] [PubMed] [Google Scholar]
- 75.LaBarge MA, Nelson CM, Villadsen R, Fridriksdottir A, Ruth JR, et al. Human mammary progenitor cell fate decisions are products of interactions with combinatorial microenvironments. Integr Biol (Camb) 2009;1:70– 79. doi: 10.1039/b816472j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lazzara MJ, Lauffenburger DA. Quantitative modeling perspectives on the ERBB system of cell regulatory processes. Exp Cell Res. 2009;315:717–725. doi: 10.1016/j.yexcr.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 77.Lequieu J, Chakrabarti A, Nayak S, Varner JD. Computational modeling and analysis of insulin induced eukaryotic translation initiation. PLoS Comput Biol. 2011;7:e1002263. doi: 10.1371/journal.pcbi.1002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, et al. Biomodels database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst Biol. 2010;4:92. doi: 10.1186/1752-0509-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao D, Corle C, Seagroves TN, Johnson RS. Hypoxia-inducible factor-1alpha is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 2007;67:563–72. doi: 10.1158/0008-5472.CAN-06-2701. [DOI] [PubMed] [Google Scholar]
- 80.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jørgensen C, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu G, Qutub AA, Vempati P, Mac Gabhann F, Popel AS. Module-based multiscale simulation of angiogenesis in skeletal muscle. Theor Biol Med Model. 2011;8:6. doi: 10.1186/1742-4682-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Locasale JW, Wolf-Yadlin A. Maximum entropy reconstructions of dynamic signaling networks from quantitative proteomics data. PLoS One. 2009;4:e6522. doi: 10.1371/journal.pone.0006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loges S, Schmidt T, Carmeliet P. Antimyeloangiogenic” therapy for cancer by inhibiting PLGF. Clin Cancer Res. 2009;15:3648–3653. doi: 10.1158/1078-0432.CCR-08-2276. [DOI] [PubMed] [Google Scholar]
- 85.Lu L, Pope S. An improved algorithm for in situ adaptive tabula tion. J Comput Phys. 2009;228:361–386. [Google Scholar]
- 86.Lu P, V, Weaver M, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luan D, Szlam F, Tanaka KA, Barie PS, Varner JD. Ensembles of uncertain mathematical models can identify network response to therapeutic interventions. Mol Biosyst. 2010;6:2272–2286. doi: 10.1039/b920693k. [DOI] [PubMed] [Google Scholar]
- 88.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 89.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macklin P, Kim J, Tomaiuolo G, Edgerton M, Cristini V. Agent-based modeling of ductal carcinoma in situ: application to patient-specific breast cancer modeling. In: Pharm TD, editor. Computational Biology Issues and Applications in Oncology. New York: Springer; 2010. pp. 77–111. [Google Scholar]
- 91.Macklin P, McDougall S, Anderson ARA, Chaplain MAJ, Cristini V, et al. Multiscale modelling and nonlinear simulation of vascular tumour growth. J Math Biol. 2009;58:765–798. doi: 10.1007/s00285-008-0216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 2011;27:141–148. doi: 10.1016/j.tig.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 94.Massey SC, Assanah MC, Lopez KA, Canoll P, Swanson KR. Glial progenitor cell recruitment drives aggressive glioma growth: mathematical and experimental modelling. J R Soc Interface. 2012;9(73):1757–1766. doi: 10.1098/rsif.2012.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mayawala K, Gelmi CA, Edwards JS. MAPK cascade possesses decoupled controllability of signal amplification and duration. Biophys J. 2004;87:L01–L02. doi: 10.1529/biophysj.104.051888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meng X, Zhong J, Liu S, Murray M, Gonzalez-Angulo AM. A new hypothesis for the cancer mechanism. Cancer Metastasis Rev. 2011;31(1–2):247–268. doi: 10.1007/s10555-011-9342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Metropolis N, Rosenbluth A, Rosenbluth M, Teller A, Teller E. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1093. [Google Scholar]
- 98.Miles DW, Chan A, Dirix LY, Cortés J, Pivot X, et al. Phase III study of bevacizumab plus docetaxel compared with placebo plus docetaxel for the first-line treatment of human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol. 2010;28:3239–3247. doi: 10.1200/JCO.2008.21.6457. [DOI] [PubMed] [Google Scholar]
- 99.Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 100.Mizukami Y, Kohgo Y, Chung DC. Hypoxia inducible factor-1 independent pathways in tumor angiogenesis. Clin Cancer Res. 2007;13:5670–5674. doi: 10.1158/1078-0432.CCR-07-0111. [DOI] [PubMed] [Google Scholar]
- 101.Mori H, Gjorevski N, Inman JL, Bissell MJ, Nelson CM. Self-organization of engineered epithelial tubules by differential cellular motility. Proc Natl Acad Sci U S A. 2009;106:14890–14895. doi: 10.1073/pnas.0901269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moussaïd M, Guillot EG, Moreau M, Fehrenbach J, Chabiron O, et al. Traffic instabilities in self-organized pedestrian crowds. PLoS Comput Biol. 2012;8:e1002442. doi: 10.1371/journal.pcbi.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakatsu MN, Davis J, Hughes CCW. Optimized fibrin gel bead assay for the study of angiogenesis. J Vis Exp. 2007;3:186. doi: 10.3791/186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nelson CM, Inman JL, Bissell MJ. Threedimensional lithographically defined organotypic tissue arrays for quantitative analysis of morphogenesis and neoplastic progression. Nat Protoc. 2008;3:674–678. doi: 10.1038/nprot.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 107.Owen MR, Alarcón T, Maini PK, Byrne HM. Angiogenesis and vascular remodelling in normal and cancerous tissues. J Math Biol. 2009;58:689–721. doi: 10.1007/s00285-008-0213-z. [DOI] [PubMed] [Google Scholar]
- 108.Pààez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palmer T, Shutts G, Hagedorn R, Doblas-Reyes F, Jung Y, et al. Representing model uncertainty in weather and climate prediction. Annu Rev Earth Planetary Sci. 2005;33:163–193. [Google Scholar]
- 110.Patocs A, Zhang L, Xu Y, Weber F, Caldes T, et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–2551. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- 111.Peirce SM, Gabhann FM, Bautch VL. Integration of experimental and computational approaches to sprouting angiogenesis. Curr Opin Hematol. 2012;19(3):184– 191. doi: 10.1097/MOH.0b013e3283523ea6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peirce SM, Van Gieson EJ, Skalak TC. Multicellular simulation predicts microvascular patterning and in silico tissue assembly. FASEB J. 2004;18:731–733. doi: 10.1096/fj.03-0933fje. [DOI] [PubMed] [Google Scholar]
- 113.Perfahl H, Byrne HM, Chen T, Estrella V, Alarcón T, et al. Multiscale modelling of vascular tumour growth in 3D: the roles of domain size and boundary conditions. PLoS One. 2011;6:e14790. doi: 10.1371/journal.pone.0014790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 115.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–38. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 117.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 118.Quo CF, Kaddi C, Phan JH, Zollanvari A, Xu M, et al. Reverse engineering biomolecular systems using -omic data: challenges, progress and opportunities. Brief Bioinform. 2012;13:430–445. doi: 10.1093/bib/bbs026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qutub AA, Mac Gabhann F, Karagiannis ED, Vempati P, Popel AS. Multiscale models of angiogenesis. IEEE Eng Med Biol Mag. 2009;28:14–31. doi: 10.1109/MEMB.2009.931791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rao BM, Lauffenburger DA, Wittrup KD. Integrating cell-level kinetic modeling into the design of engineered protein therapeutics. Nat Biotechnol. 2005;23:191– 194. doi: 10.1038/nbt1064. [DOI] [PubMed] [Google Scholar]
- 121.Rejniak KA, Anderson ARA. State of the art in computational modelling of cancer. Math Med Biol. 2012;29:1–2. doi: 10.1093/imammb/dqr029. [DOI] [PubMed] [Google Scholar]
- 122.Rejniak KA, Wang SE, Bryce NS, Chang H, Parvin B, et al. Linking changes in epithelial morphogenesis to cancer mutations using computational modeling. PLoS Comput Biol. 2010;6:e1000900. doi: 10.1371/journal.pcbi.1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saez-Rodriguez J, Alexopoulos LG, Epperlein J, Samaga R, Lauffenburger DA, et al. Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol Syst Biol. 2009;5:331. doi: 10.1038/msb.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Salgado R, Junius S, Benoy I, Van Dam P, Vermeulen P, et al. Circulating interleukin-6 predicts survival in patients with metastatic breast cancer. Int J Cancer. 2003;103:642–646. doi: 10.1002/ijc.10833. [DOI] [PubMed] [Google Scholar]
- 125.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shieh AC. Biomechanical forces shape the tumor microenvironment. Ann Biomed Eng. 2011;39:1379–1389. doi: 10.1007/s10439-011-0252-2. [DOI] [PubMed] [Google Scholar]
- 128.Shin Y, Jeon JS, Han S, Jung GS, Shin S, et al. In vitro 3D collective sprouting angiogenesis under orchestrated ANG-1 and VEGF gradients. Lab Chip. 2011;11:2175–2181. doi: 10.1039/c1lc20039a. [DOI] [PubMed] [Google Scholar]
- 129.Sklar E. Netlogo, a multi-agent simulation environment. Artif Life. 2007;13:303–311. doi: 10.1162/artl.2007.13.3.303. [DOI] [PubMed] [Google Scholar]
- 130.Song SO, Chakrabarti A, Varner JD. Ensembles of signal transduction models using pareto optimal ensemble techniques (POETs) Biotechnol J. 2010;5:768–780. doi: 10.1002/biot.201000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Song SO, Varner J. Modeling and analysis of the molecular basis of pain in sensory neurons. PLoS One. 2009;4:e6758. doi: 10.1371/journal.pone.0006758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Song SOK, Hogg J, Peng ZY, Parker R, Kellum JA, et al. Ensemble models of neutrophil trafficking in severe sepsis. PLoS Comput Biol. 2012;8:e1002422. doi: 10.1371/journal.pcbi.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spencer SL, Gerety RA, Pienta KJ, Forrest S. Modeling somatic evolution in tumorigenesis. PLoS Comput Biol. 2006;2:e108. doi: 10.1371/journal.pcbi.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Swanson KR, Alvord EC, Jr, Murray JD. Quantifying efficacy of chemotherapy of brain tumors with homogeneous and heterogeneous drug delivery. Acta Biotheor. 2002;50:223–237. doi: 10.1023/a:1022644031905. [DOI] [PubMed] [Google Scholar]
- 136.Swanson KR, Bridge C, Murray JD, Alvord EC., Jr Virtual and real brain tumors: using mathe- Signaling Assisted Multiscale Modeling (SAMM) 2499 matical modeling to quantify glioma growth and invasion. J Neurol Sci. 2003;216:1–10. doi: 10.1016/j.jns.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 137.Swanson KR, True LD, Murray JD. On the use of quantitative modeling to help understand prostatespecific antigen dynamics and other medical problems. Am J Clin Pathol. 2003;119:14–17. doi: 10.1309/AR06-82JH-8B1B-G058. [DOI] [PubMed] [Google Scholar]
- 138.Tasseff R, Nayak S, Salim S, Kaushik P, Rizvi N, et al. Analysis of the molecular networks in androgen dependent and independent prostate cancer revealed fragile and robust subsystems. PLoS One. 2010;5:e8864. doi: 10.1371/journal.pone.0008864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tasseff R, Nayak S, Song SO, Yen A, Varner D. Modeling and analysis of retinoic acid induced differentiation of uncommitted precursor cells. Integr Biol (Camb) 2011;3:578–591. doi: 10.1039/c0ib00141d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 141.Thorne BC, Bailey AM, Peirce SM. Combining experiments with multi-cell agent-based modeling to study biological tissue patterning. Brief Bioinform. 2007;8:245–257. doi: 10.1093/bib/bbm024. [DOI] [PubMed] [Google Scholar]
- 142.Varner JD. Systems biology and the mathematical modelling of antibody-directed enzyme prodrug therapy (ADEPT) Syst Biol (Stevenage) 2005;152:291–302. doi: 10.1049/ip-syb:20050047. [DOI] [PubMed] [Google Scholar]
- 143.Wang Z, Bordas V, Deisboeck TS. Discovering molecular targets in cancer with multiscale modeling. Drug Dev Res. 2011;72:45–52. doi: 10.1002/ddr.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Z, Bordas V, Sagotsky J, Deisboeck TS. Identifying therapeutic targets in a combined EGFRTGFBR signalling cascade using a multiscale agent-based cancer model. Math Med Biol. 2012;29:95–108. doi: 10.1093/imammb/dqq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang J, Zhang Y, Marian C, Ressom HW. Identification of aberrant pathways and network activities from high-throughput data. Brief Bioinform. 2012;13:406–419. doi: 10.1093/bib/bbs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Waugh DJJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 147.Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, et al. Combined CDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 148.Yarden Y, Sliwkowski MX. Untangling the erbb signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 149.Yeung MKS, Tegnér J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc Natl Acad Sci U S A. 2002;99:6163–6168. doi: 10.1073/pnas.092576199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.You X, Nguyen AW, Jabaiah A, Sheff MA, Thorn KS, et al. Intracellular protein interaction mapping with FRET hybrids. Proc Natl Acad Sci USA. 2006;103:18458–18463. doi: 10.1073/pnas.0605422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang L, Athale CA, Deisboeck TS. Development of a three-dimensional multiscale agent-based tumor model: simulating gene-protein interaction profiles, cell phenotypes and multicellular patterns in brain cancer. J Theor Biol. 2007;244:96–107. doi: 10.1016/j.jtbi.2006.06.034. [DOI] [PubMed] [Google Scholar]
- 152.Zhang L, Strouthos CG, Wang Z, Deisboeck TS. Simulating brain tumor heterogeneity with a multiscale agent-based model: Linking molecular signatures, phenotypes and expansion rate. Math Comput Model. 2009;49:307–319. doi: 10.1016/j.mcm.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci U S A. 2012;109:9342– 9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]