Abstract
Objective
Three common X-ray repair cross-complementing groups 1 (XRCC1) polymorphisms, Arg399Gln, Arg194Trp, and Arg280His, have been reported to be implicated in the development of leukemia. However, previous results from different studies were inconsistent. Consequently, we performed a meta-analysis in order to accurately evaluate the association between XRCC1 Arg399Gln, Arg194Trp, and Arg280His polymorphisms and leukemia risk.
Methods
Through computerized searching of PubMed, ISI Web of Knowledge, Cochrane, EBSCO, and OpenGrey databases, and manually searching relevant references, a total of 19 studies with 3387 cases and 6168 controls for Arg399Gln (G>A) polymorphism, 12 studies with 2043 cases and 4550 controls for Arg194Trp (C>T), and 6 studies with 1445 cases and 1905 controls for Arg280His (G>A) were collected to perform meta-analysis and stratified analysis to explore the associations between these variants and leukemia susceptibility. Based on three genetic models, the codominant model, dominant model and recessive model, odds ratios (ORs) as well as their 95% confidence intervals (CIs) were used to evaluate the association strength between XRCC1 genotypes and leukemia risk.
Results
With respect to overall leukemia susceptibility, no association was detected. In stratified analyses by tumor type, Arg399Gln was associated with higher acute lymphoblastic leukemia (ALL) risk (AA vs. GG, OR = 1.50, 95% CI: 1.11-2.02; AA+GA vs. GG, OR = 1.35, 95% CI: 1.02-1.78). Additionally, Arg399Gln, Arg194Trp, and Arg280His may influence the susceptibilities of some leukemia type and race populations.
Conclusion
This meta-analysis indicates these three polymorphisms of XRCC1 do not associate with overall leukemia risks but could be associated with the risks for some specific subgroups.
Introduction
Leukemia is one of the most common human cancers, with an estimated 48610 new cases and 23720 deaths expected in the US in 2013 [1]. According to the cell type and growth rate, leukemia can be classified into four groups: acute myeloid leukemia (AML), acute lymphocytic leukemia (ALL), chronic myeloid leukemia (CML), and chronic lymphocytic leukemia (CLL). Although studies for leukemogenesis have been conducted for many years, the mechanisms underlying the development of this hemotologic malignancy remains unclear.
Impaired DNA repair may be associated with increased susceptibility to human cancers [2]. X-ray repair cross-complementing groups 1 (XRCC1) binds to DNA repair related proteins and takes part in the DNA repair process [3], [4], [5]. In the past decade, a number of studies have been performed to explore the relationship between three common XRCC1 single nucleotide polymorphisms (SNPs)—Arg399Gln (base G to A polymorphism), Arg194Trp (base C to T polymorphism), and Arg280His (base G to A polymorphism)—and leukemia risk. However, the conclusions of these studies are inconsistent. Therefore, a meta-analysis followed by stratified analysis of 19 published studies was performed to estimate the association between XRCC1 Arg399Gln, Arg194Trp, and Arg280His polymorphisms and leukemia risk.
Materials and Methods
Study identification
Computer bibliographic searches through PubMed, ISI Web of Knowledge, Cochrane, EBSCO, and grey literature database OpenGrey were conducted using the keywords: “leukemia,” “leukaemia” and “polymorphisms,” “genotypes,” “variants,” and “XRCC1,” “X-ray repair cross-complementing groups 1,” with the final search completed in May 2013. Studies from the references of the related reports were checked. Articles in all languages were searched to ensure the relevant studies were not missed. The following inclusion criteria were applied: (1) case-control studies or nested case-control studies within cohort studies, if any (2) studies evaluating association between XRCC1 polymorphism and leukemia risk, (3) full text reports which including enough data to calculate odds ratios (ORs) and 95% confidence intervals (CIs). The exclusion criteria were as follows: (1) duplicated reports, (2) reviews or meta-analyses, if they were performed without additional eligible studies; otherwise, the additional eligible study was included in our meta-analysis, (3) if the same population was used in multiple studies, only the most complete or latest study was selected for further analysis.
Data extraction procedure
According to the study identification criteria, the available studies were reviewed, selected, and the following information from the eligible studies was extracted: first author’s last name, publication year, country, leukemia type, number of case and control subjects, ethnicity, control subjects source population, and genotype numbers of cases and controls.
Study quality assessment
Critical quality assessment of the included studies was performed by Effective Public Health Practice Project Quality Assessment Tool (EPHPP). With this tool, assessments of the risk of bias or methodological quality were made separately for six individual domains: selection bias, study design, confounders, blinding, data collection method, and withdrawals and drop-outs. The comprehensive dictionary for the assessment tool was used to guide the rating of the studies. Each domain was rated as strong, moderate, or weak. The study quality was then evaluated as strong, moderate, or weak if there were no, one, or two or more in total weak ratings for all the domains, respectively [6]. Two reviewers (Haijun Zhang and Hang Liu) independently reviewed the studies and they resolved discrepancies through discussion.
Statistical analysis
Statistical analyses were performed as described previously [7], [8]. Briefly, for individual studies, Hardy-Weinberg equilibrium of control subjects was tested by Pearson’s goodness-of-fit χ 2 test. The strength of association between XRCC1 Arg399Gln, Arg194Trp, and Arg280His polymorphisms and leukemia risk was measured by odds ratios (ORs) and the corresponding 95% confidence intervals (CIs). We used codominant, dominant, and recessive genetic models to assess the pooled ORs. For both Arg399Gln and Arg280His (G>A) polymorphisms, the codominant model included homozygous comparison of AA vs. GG and heterozygous comparison of GA vs. GG, the dominant model was AA+GA vs. GG, and the recessive model was AA vs. GA+GG. For Arg194Trp (C>T) polymorphism, the codominant model included homozygous comparison of TT vs. CC and heterozygous comparison of CT vs. CC, the dominant model was TT+CT vs. CC, and the recessive model was TT vs. CT+CC. Stratified analyses were performed by race, control source, and tumor type. Due to the high heterogeneity across the studies, the random effects model based on the DerSimonian and Laird method was applied [9]. For calculating the OR of a subgroup containing a single stud, the inverse variance method was used instead. Publication bias was evaluated by Begg’s rank correlation test [10] and Egger’s linear regression test [11]. If the publication bias tests indicated bias existed, the Duval and Tweedie “trim and fill” method was used to adjust the bias [12]. The statistical power for XRCC1 polymorphisms and leukemia risk in three genetic models was calculated by OpenEpi program (Version 2.3.1, www.OpenEpi.com). Statistical analysis was conducted in R 2.15.2 using “meta” package [13].
Results
Study identification and characteristics of studies
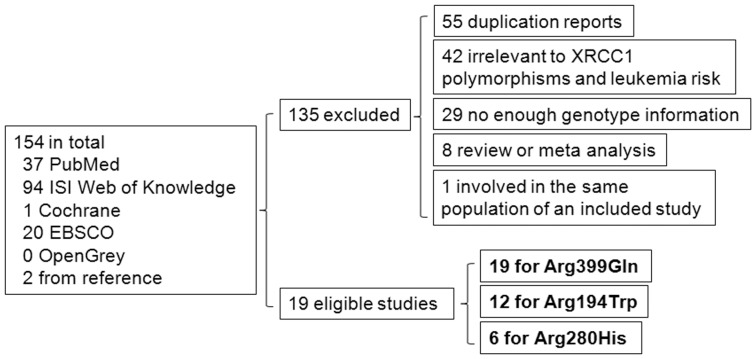
Searches of PubMed, ISI Web of Knowledge, Cochrane, EBSCO, and OpenGrey databases and manually searching references returned 154 studies. Among them, 135 reports were excluded for the following reasons: 55 were duplications, 42 were irrelevant to XRCC1 polymorphisms and leukemia risk, 29 did not provide enough genotype information, and 8 were reviews or meta-analyses., In addition, studies that involve the same population as another eligible study were excluded to avoid bias [14]. Also of note was Annamaneni et al. report [15], in which the minor allele frequencies of Arg194Trp polymorphism in both case and control groups were much higher than major allele frequencies, which indicated the genotype numbers of the Arg194Trp polymorphism may include errors in this study. We therefore excluded this study from Arg194Trp analysis. After removing these ineligible studies, a total 19 studies with 3387 cases and 6168 controls for Arg399Gln (G>A) polymorphism [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], 12 studies with 2043 cases and 4550 controls for Arg194Trp (C>T) [17], [18], [20], [21], [22], [23], [24], [25], [26], [28], [29], [32], and 6 studies with 1445 cases and 1905 controls for Arg280His (G>A) [15], [22], [23], [26], [28], [30] were included for further analysis (Figure 1). A database was established to display the study characteristics of each eligible study (Table 1). The distribution of genotypes in the control group of each study was in agreement with Hardy-Weinberg equilibrium except for 4 studies for Arg399Gln, 3 studies for Arg194Trp and 1 study for Arg280His (Table 1). All the included studies were rated as strong or moderate by EPHPP Quality Assessment Tool, indicating that the synthesis of results in a meta-analysis should be reliable (Table 1).
Figure 1. Flow diagram of study identification process.
Table 1. Study characteristics of the meta-analysis.
| Polymorphism | Author | Year | Country | Racial descent | Tumor type(# of cases) | Control source | Case | Control | HWE | QA | |||||
| Arg399Gln | GG | GA | AA | GG | GA | AA | |||||||||
| (G>A) | Seedhouse | 2002 | UK | Caucasian | AML (167) | Population | 70 | 69 | 28 | 55 | 76 | 47 | 0.05 | Strong | |
| Joseph | 2005 | India | Asian | ALL (117) | Hospital | 55 | 46 | 16 | 75 | 33 | 9 | 0.06 | Strong | ||
| Matullo | 2006 | Multiple | Unknown | Unknown (169) | Population | 67 | 74 | 28 | 484 | 482 | 128 | 0.63 | Moderate | ||
| Deligezer | 2007 | Turkey | Caucasian | AML (72), CML (182) | Population | 103 | 121 | 30 | 96 | 101 | 29 | 0.76 | Moderate | ||
| Pakakasama | 2007 | Thailand | Asian | ALL (108) | Population | 39 | 60 | 9 | 175 | 124 | 18 | 0.51 | Strong | ||
| Batar | 2009 | Turkey | Caucasian | ALL (70) | Population | 24 | 37 | 9 | 24 | 37 | 14 | 0.97 | Strong | ||
| Ganster | 2009 | Austria | Unknown | CLL (429) | Population | 173 | 192 | 64 | 184 | 193 | 52 | 0.90 | Moderate | ||
| Meza-Espinoza | 2009 | Mexico | Unknown | ALL (120) | Population | 57 | 51 | 12 | 65 | 47 | 8 | 0.90 | Moderate | ||
| Tumer | 2010 | Turkey | Caucasian | ALL (167) | Population | 63 | 77 | 27 | 92 | 78 | 20 | 0.57 | Strong | ||
| Shi | 2011 | China | Asian | AML (306) | Population | 173 | 114 | 19 | 316 | 213 | 29 | 0.37 | Strong | ||
| Stanczyk | 2011 | Poland | Caucasian | ALL (97) | Population | 34 | 45 | 18 | 50 | 57 | 24 | 0.28 | Moderate | ||
| Canalle | 2011 | Brazil | Caucasian,unknown | ALL (201) | Hospital | 112 | 72 | 17 | 186 | 152 | 23 | 0.27 | Strong | ||
| Özcan | 2011 | Turkey | Caucasian | AML (36), ALL (9) | Population | 22 | 22 | 1 | 42 | 43 | 15 | 0.47 | Strong | ||
| Duman | 2012 | Turkey | Caucasian | CLL (73) | Population | 7 | 50 | 16 | 19 | 26 | 5 | 0.36 | Moderate | ||
| Kim | 2012 | Korea | Asian | AML (415) | Population | 234 | 155 | 26 | 914 | 693 | 91 | 0.01 | Strong | ||
| Abramenko | 2012 | Ukraine | Caucasian | CLL (169) | Population | 67 | 82 | 20 | 38 | 41 | 15 | 0.48 | Strong | ||
| El-Din | 2012 | Egypt | Caucasian | AML (40) | Population | 20 | 16 | 4 | 16 | 2 | 2 | 0.01 | Strong | ||
| Annamaneni | 2012 | India | Asian | CML (350) | Population | 79 | 191 | 80 | 61 | 235 | 54 | <0.01 | Strong | ||
| Sorour | 2013 | Egypt | Caucasian | AML (90) | Population | 54 | 27 | 9 | 33 | 27 | 0 | 0.02 | Strong | ||
| Arg194Trp | CC | CT | TT | CC | CT | TT | |||||||||
| (C>T) | Seedhouse | 2002 | UK | Caucasian | AML (126) | Population | 112 | 14 | 0 | 78 | 7 | 2 | <0.01 | Strong | |
| Joseph | 2005 | India | Asian | ALL (117) | Hospital | 77 | 32 | 8 | 91 | 22 | 4 | 0.09 | Strong | ||
| Matullo | 2006 | Multiple | Unknown | Unknown (169) | Population | 145 | 23 | 1 | 951 | 141 | 2 | 0.17 | Moderate | ||
| Pakakasama | 2007 | Thailand | Asian | ALL (108) | Population | 62 | 44 | 2 | 150 | 145 | 22 | 0.10 | Strong | ||
| Batar | 2009 | Turkey | Caucasian | ALL (70) | Population | 52 | 16 | 2 | 64 | 11 | 0 | 0.49 | Strong | ||
| Ganster | 2009 | Austria | Unknown | CLL (439) | Population | 371 | 63 | 5 | 389 | 45 | 5 | 0.01 | Moderate | ||
| Meza-Espinoza | 2009 | Mexico | Unknown | ALL (120) | Population | 80 | 34 | 6 | 86 | 31 | 3 | 0.92 | Moderate | ||
| Tumer | 2010 | Turkey | Caucasian | ALL (167) | Population | 140 | 27 | 0 | 159 | 26 | 5 | 0.01 | Strong | ||
| Canalle | 2011 | Brazil | Caucasian,unknown | ALL (201) | Hospital | 168 | 32 | 1 | 298 | 59 | 4 | 0.58 | Strong | ||
| Duman | 2012 | Turkey | Caucasian | CLL (73) | Population | 64 | 8 | 1 | 41 | 9 | 0 | 0.48 | Moderate | ||
| Kim | 2012 | Korea | Asian | AML (413) | Population | 167 | 208 | 38 | 775 | 741 | 164 | 0.50 | Strong | ||
| El-Din | 2012 | Egypt | Caucasian | AML (40) | Population | 11 | 14 | 15 | 14 | 4 | 2 | 0.09 | Strong | ||
| Arg280His | GG | GA | AA | GG | GA | AA | |||||||||
| (G>A) | Joseph | 2005 | India | Asian | ALL (117) | Hospital | 76 | 38 | 3 | 85 | 30 | 2 | 0.73 | Strong | |
| Pakakasama | 2007 | Thailand | Asian | ALL (108) | Population | 94 | 14 | 0 | 272 | 42 | 3 | 0.34 | Strong | ||
| Ganster | 2009 | Austria | Unknown | CLL (443) | Population | 396 | 47 | 0 | 388 | 53 | 2 | 0.90 | Moderate | ||
| Meza-Espinoza | 2009 | Mexico | Unknown | ALL (120) | Population | 87 | 31 | 2 | 88 | 31 | 1 | 0.33 | Moderate | ||
| Shi | 2011 | China | Asian | AML (307) | Population | 236 | 66 | 5 | 445 | 109 | 4 | 0.34 | Strong | ||
| Annamaneni | 2012 | India | Asian | CML (350) | Population | 346 | 4 | 0 | 338 | 11 | 1 | 0.01 | Strong | ||
AML, acute myeloid leukemia; ALL, acute lymphocytic leukemia; CML, chronic myeloid leukaemia; CLL, chronic lymphocytic leukemia; HWE, P value of Pearson’s goodness-of-fit χ2 test for Hardy-Weinberg equilibrium; QA, quality assessment; Unknown, including study populations in which the race was mixed/unclear or tumor type was not described.
Meta-analysis results
Meta-analysis and relevant subgroups analysis by tumor type, race and control sources were conducted to examine the association between XRCC1 Arg399Gln (G>A), Arg194Trp (C>T), and Arg280His (G>A) polymorphisms and leukemia risk in three genetic models. Stratified analysis by race and control sources in each leukemia type and by etiology in AML was further performed to explore the possible associations. The analyses results were shown in Table 2. The statistical power for XRCC1 polymorphisms and leukemia risk in three genetic models was shown in Table 3.
Table 2. Pooled ORs and 95% CIs for XRCC1 Arg399Gln, Arg194Trp and Arg280His meta-analysis.
| Polymorphism | Study group | n | Codominant | Dominant | Recessive | |||||
| Arg399Gln | AA vs. GG | GA vs. GG | AA + GA vs. GG | AA vs. GA + GG | ||||||
| (G>A) | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| Total | 19 | 1.25 (0.98–1.59) | 0.08 | 1.10 (0.93–1.31) | 0.27 | 1.13 (0.96–1.34) | 0.15 | 1.19 (0.98–1.45) | 0.08 | |
| Racial decent | ||||||||||
| Asian | 5 | 1.31 (1.00–1.71) | 0.05 | 1.12 (0.77–1.64) | 0.55 | 1.18 (0.83–1.69) | 0.35 | 1.42 (1.12–1.81) | <0.01 | |
| Caucasian | 11 | 1.10 (0.68–1.78) | 0.69 | 1.12 (0.86–1.47) | 0.40 | 1.12 (0.85–1.48) | 0.44 | 0.97 (0.68–1.38) | 0.87 | |
| Unknown | 4 | 1.44 (1.07–1.93) | 0.02 | 1.04 (0.80–1.35) | 0.77 | 1.12 (0.90–1.39) | 0.32 | 1.39 (1.06–1.84) | 0.02 | |
| Control source | ||||||||||
| Population | 17 | 1.21 (0.93–1.57) | 0.16 | 1.10 (0.92–1.31) | 0.32 | 1.12 (0.94–1.33) | 0.21 | 1.16 (0.94–1.43) | 0.18 | |
| Hospital | 2 | 1.62 (0.84–3.11) | 0.15 | 1.19 (0.50–2.82) | 0.69 | 1.27 (0.54–2.98) | 0.58 | 1.54 (0.91–2.58) | 0.11 | |
| Tumor type | ||||||||||
| AML | 7 | 0.94 (0.56–1.56) | 0.80 | 0.92 (0.73–1.15) | 0.46 | 0.92 (0.73–1.16) | 0.46 | 0.94 (0.60–1.49) | 0.80 | |
| Etiology | ||||||||||
| De novo | 4 | 1.12 (0.48–2.59) | 0.79 | 0.96 (0.59–1.56) | 0.87 | 1.00 (0.65–1.54) | 0.99 | 1.07 (0.50–2.29) | 0.86 | |
| Secondary | 2 | 0.30 (0.11–0.82) | 0.02 | 0.60 (0.35–1.03) | 0.06 | 0.53 (0.31–0.89) | 0.02 | 0.40 (0.15–1.06) | 0.07 | |
| Racial decent | ||||||||||
| Asian | 2 | 1.14 (0.79–1.65) | 0.47 | 0.91 (0.76–1.09) | 0.31 | 0.94 (0.79–1.11) | 0.47 | 1.19 (0.83–1.70) | 0.34 | |
| Caucasian | 5 | 0.83 (0.32–2.16) | 0.71 | 0.97 (0.60–1.57) | 0.90 | 0.94 (0.59–1.51) | 0.80 | 0.79 (0.35–1.76) | 0.56 | |
| Control source | ||||||||||
| Population | 7 | 0.94 (0.56–1.56) | 0.80 | 0.92 (0.73–1.15) | 0.46 | 0.92 (0.73–1.16) | 0.46 | 0.94 (0.60–1.49) | 0.80 | |
| ALL | 8 | 1.50 (1.11–2.02) | 0.01 | 1.32 (0.99–1.75) | 0.06 | 1.35 (1.02–1.78) | 0.03 | 1.31 (0.99–1.74) | 0.06 | |
| Racial decent | ||||||||||
| Asian | 2 | 2.33 (1.25–4.34) | 0.01 | 2.06 (1.44–2.95) | < 0.01 | 2.11 (1.50–2.97) | < 0.01 | 1.69 (0.93–3.07) | 0.09 | |
| Caucasian | 5 | 1.27 (0.86–1.86) | 0.23 | 1.10 (0.86–1.41) | 0.45 | 1.13 (0.89–1.43) | 0.31 | 1.16 (0.81–1.66) | 0.41 | |
| Unknown | 2 | 1.55 (0.70–3.43) | 0.28 | 0.76 (0.25–2.28) | 0.62 | 0.88 (0.36–2.16) | 0.78 | 1.61 (0.74–3.47) | 0.23 | |
| Control source | ||||||||||
| Population | 6 | 1.47 (1.02–2.11) | 0.04 | 1.45 (1.15–1.83) | <0.01 | 1.45 (1.13–1.85) | <0.01 | 1.23 (0.88–1.72) | 0.23 | |
| Hospital | 2 | 1.62 (0.84–3.11) | 0.15 | 1.19 (0.50–2.82) | 0.69 | 1.27 (0.54–2.98) | 0.58 | 1.54 (0.91–2.58) | 0.11 | |
| CML | 2 | 1.05 (0.72–1.55) | 0.78 | 0.81 (0.48–1.36) | 0.43 | 0.86 (0.61–1.21) | 0.38 | 1.26 (0.70–2.26) | 0.45 | |
| Racial decent | ||||||||||
| Asian | 1 | 1.14 (0.71–1.85) | 0.58 | 0.63 (0.43–0.92) | 0.02 | 0.72 (0.50–1.05) | 0.09 | 1.62 (1.11–2.38) | 0.01 | |
| Caucasian | 1 | 0.91 (0.48–1.73) | 0.78 | 1.06 (0.70–1.61) | 0.77 | 1.03 (0.69–1.53) | 0.88 | 0.89 (0.49–1.61) | 0.69 | |
| Control source | ||||||||||
| Population | 2 | 1.05 (0.72–1.55) | 0.78 | 0.81 (0.48–1.36) | 0.43 | 0.86 (0.61–1.21) | 0.38 | 1.26 (0.70–2.26) | 0.45 | |
| CLL | 3 | 1.73 (0.64–4.67) | 0.28 | 1.58 (0.79–3.14) | 0.20 | 1.61 (0.79–3.27) | 0.19 | 1.21 (0.69–2.10) | 0.51 | |
| Racial decent | ||||||||||
| Caucasian | 2 | 2.41 (0.22–26.39) | 0.47 | 2.30 (0.52–10.22) | 0.27 | 2.32 (0.43–12.54) | 0.33 | 1.25 (0.36–4.35) | 0.72 | |
| Unknown | 1 | 1.31 (0.86–1.99) | 0.21 | 1.06 (0.79–1.41) | 0.70 | 1.11 (0.85–1.46) | 0.45 | 1.27 (0.86–1.88) | 0.23 | |
| Control source | ||||||||||
| Population | 3 | 1.73 (0.64–4.67) | 0.28 | 1.58 (0.79–3.14) | 0.20 | 1.61 (0.79–3.27) | 0.19 | 1.21 (0.69–2.10) | 0.51 | |
| Arg194Trp | TT vs. CC | CT vs. CC | TT + CT vs. CC | TT vs. CT + CC | ||||||
| (C>T) | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| Total | 12 | 1.21 (0.65–2.27) | 0.55 | 1.20 (1.00–1.43) | 0.05 | 1.20 (0.96–1.48) | 0.10 | 1.11 (0.63–1.94) | 0.72 | |
| Racial decent | ||||||||||
| Asian | 3 | 0.92 (0.33–2.53) | 0.87 | 1.16 (0.75–1.78) | 0.50 | 1.13 (0.69–1.85) | 0.62 | 0.87 (0.36–2.08) | 0.75 | |
| Caucasian | 6 | 1.06 (0.20–5.68) | 0.94 | 1.22 (0.81–1.83) | 0.34 | 1.27 (0.77–2.08) | 0.35 | 0.97 (0.22–4.31) | 0.97 | |
| Unknown | 4 | 1.61 (0.69–3.74) | 0.27 | 1.28 (0.98–1.66) | 0.07 | 1.29 (1.00–1.67) | 0.05 | 1.54 (0.66–3.57) | 0.31 | |
| Control source | ||||||||||
| Population | 10 | 1.18 (0.56–2.47) | 0.66 | 1.19 (0.97–1.47) | 0.09 | 1.19 (0.93–1.52) | 0.17 | 1.08 (0.56–2.08) | 0.82 | |
| Hospital | 2 | 1.32 (0.28–6.33) | 0.73 | 1.24 (0.70–2.18) | 0.46 | 1.27 (0.66–2.45) | 0.48 | 1.27 (0.31–5.19) | 0.74 | |
| Tumor type | ||||||||||
| AML | 3 | 1.48 (0.24–9.08) | 0.67 | 1.53 (0.92–2.55) | 0.10 | 1.74 (0.79–3.86) | 0.17 | 1.21 (0.27–5.42) | 0.80 | |
| Etiology | ||||||||||
| De novo | 2 | 1.49 (0.03–87.31) | 0.85 | 2.21 (0.69–7.09) | 0.18 | 2.43 (0.43–13.83) | 0.32 | 1.16 (0.04–37.87) | 0.94 | |
| Secondary | 1 | 1.26 (0.06–27.73) | 0.89 | 1.86 (0.34–10.01) | 0.47 | 1.44 (0.28–7.51) | 0.66 | 1.18 (0.05–25.84) | 0.92 | |
| Racial decent | ||||||||||
| Asian | 1 | 1.08 (0.73–1.59) | 0.72 | 1.30 (1.04–1.63) | 0.02 | 1.26 (1.01–1.57) | 0.04 | 0.94 (0.65–1.36) | 0.73 | |
| Caucasian | 2 | 1.40 (0.02–92.08) | 0.87 | 2.24 (0.73–6.86) | 0.16 | 2.47 (0.45–13.49) | 0.30 | 1.08 (0.03–39.94) | 0.97 | |
| Control source | ||||||||||
| Population | 3 | 1.48 (0.24–9.08) | 0.67 | 1.35 (1.09–1.68) | 0.10 | 1.74 (0.79–3.86) | 0.17 | 1.21 (0.27–5.42) | 0.80 | |
| ALL | 6 | 0.88 (0.28–2.77) | 0.82 | 1.11 (0.85–1.45) | 0.46 | 1.12 (0.80–1.55) | 0.51 | 0.87 (0.30–2.52) | 0.79 | |
| Racial decent | ||||||||||
| Asian | 2 | 0.75 (0.07–7.97) | 0.81 | 1.09 (0.48–2.51) | 0.84 | 1.08 (0.40–2.89) | 0.88 | 0.75 (0.09–6.12) | 0.79 | |
| Caucasian | 3 | 0.60 (0.07–5.11) | 0.64 | 1.14 (0.79–1.64) | 0.48 | 1.10 (0.71–1.68) | 0.68 | 0.58 (0.07–4.68) | 0.61 | |
| Unknown | 2 | 2.08 (0.57–7.63) | 0.27 | 1.27 (0.78–2.08) | 0.34 | 1.32 (0.82–2.13) | 0.25 | 1.97 (0.54–7.18) | 0.30 | |
| Control source | ||||||||||
| Population | 4 | 0.71 (0.13–3.97) | 0.70 | 1.06 (0.75–1.50) | 0.75 | 1.05 (0.69–1.61) | 0.82 | 0.72 (0.14–3.64) | 0.69 | |
| Hospital | 2 | 1.32 (0.28–6.33) | 0.73 | 1.24 (0.70–2.18) | 0.46 | 1.27 (0.66–2.45) | 0.48 | 1.27 (0.31–5.19) | 0.74 | |
| CLL | 2 | 1.14 (0.35–3.63) | 0.83 | 1.03 (0.42–2.53) | 0.94 | 1.10 (0.53–2.29) | 0.80 | 1.10 (0.34–3.52) | 0.87 | |
| Racial decent | ||||||||||
| Caucasian | 1 | 1.93 (0.08–48.52) | 0.69 | 0.57 (0.20–1.59) | 0.28 | 0.64 (0.23–1.75) | 0.38 | 2.09 (0.08–52.34) | 0.65 | |
| Unknown | 1 | 1.05 (0.30–3.65) | 0.94 | 1.47 (0.98–2.21) | 0.07 | 1.43 (0.96–2.11) | 0.08 | 1.00 (0.29–3.48) | 1.00 | |
| Control source | ||||||||||
| Population | 2 | 1.14 (0.35–3.63) | 0.83 | 1.03 (0.42–2.53) | 0.94 | 1.10 (0.53–2.29) | 0.80 | 1.10 (0.34–3.52) | 0.87 | |
| Arg280His | AA vs. GG | GA vs. GG | AA + GA vs GG | AA vs GA + GG | ||||||
| (G>A) | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| Total | 6 | 1.32 (0.56–3.11) | 0.52 | 1.02 (0.82–1.27) | 0.87 | 1.01 (0.78–1.30) | 0.97 | 1.28 (0.55–3.01) | 0.57 | |
| Racial decent | ||||||||||
| Asian | 4 | 1.50 (0.57–3.91) | 0.41 | 1.05 (0.73–1.51) | 0.79 | 1.02 (0.68–1.54) | 0.91 | 1.44 (0.55–3.75) | 0.46 | |
| Unknown | 2 | 0.76 (0.08–7.39) | 0.81 | 0.92 (0.65–1.28) | 0.61 | 0.90 (0.65–1.26) | 0.55 | 0.77 (0.08–7.31) | 0.82 | |
| Control source | ||||||||||
| Population | 5 | 1.23 (0.47–3.26) | 0.67 | 0.97 (0.78–1.22) | 0.81 | 0.94 (0.72–1.24) | 0.68 | 1.22 (0.46–3.22) | 0.69 | |
| Hospital | 1 | 1.68 (0.27–10.31) | 0.58 | 1.42 (0.80–2.50) | 0.23 | 1.43 (0.82–2.50) | 0.21 | 1.51 (0.25–9.23) | 0.65 | |
| Tumor type | ||||||||||
| AML | 1 | 2.36 (0.63–8.86) | 0.20 | 1.14 (0.81–1.61) | 0.45 | 1.18 (0.85–1.66) | 0.32 | 2.29 (0.61–8.60) | 0.22 | |
| Etiology | ||||||||||
| De novo | 1 | 1.58 (0.35–7.13) | 0.55 | 1.10 (0.77–1.58) | 0.59 | 1.12 (0.79–1.59) | 0.53 | 1.55 (0.34–6.98) | 0.57 | |
| Secondary | 1 | 8.90 (1.56–50.94) | 0.01 | 1.47 (0.67–3.24) | 0.34 | 1.73 (0.83–3.63) | 0.14 | 8.15 (1.44–46.06) | 0.02 | |
| Racial decent | ||||||||||
| Asian | 1 | 2.36 (0.63–8.86) | 0.20 | 1.14 (0.81–1.61) | 0.45 | 1.185 (0.85–1.66) | 0.32 | 2.29 (0.61–8.60) | 0.22 | |
| Control source | ||||||||||
| Population | 1 | 2.36 (0.63–8.86) | 0.20 | 1.14 (0.81–1.61) | 0.45 | 1.185 (0.85–1.66) | 0.32 | 2.29 (0.61–8.60) | 0.22 | |
| ALL | 3 | 1.35 (0.37–4.98) | 0.65 | 1.13 (0.80–1.59) | 0.49 | 1.13 (0.80–1.58) | 0.49 | 1.28 (0.35–4.71) | 0.71 | |
| Racial decent | ||||||||||
| Asian | 2 | 1.15 (0.24–5.39) | 0.86 | 1.20 (0.78–1.84) | 0.41 | 1.17 (0.74–1.84) | 0.50 | 1.07 (0.23–5.00) | 0.94 | |
| Unknown | 1 | 2.02 (0.18–22.72) | 0.57 | 1.01 (0.57–1.81) | 0.97 | 1.04 (0.59–1.84) | 0.88 | 2.02 (0.18–22.54) | 0.57 | |
| Control source | ||||||||||
| Population | 2 | 1.07 (0.16–7.01) | 0.94 | 0.99 (0.64–1.53) | 0.97 | 0.98 (0.64–1.50) | 0.92 | 1.07 (0.17–7.00) | 0.94 | |
| Hospital | 1 | 1.68 (0.27–10.31) | 0.58 | 1.42 (0.80–2.51) | 0.23 | 1.43 (0.82–2.50) | 0.21 | 1.51 (0.25–9.23) | 0.65 | |
| CML | 1 | 0.33 (0.01–8.02) | 0.49 | 0.36 (0.11–1.13) | 0.08 | 0.33 (0.10–1.02) | 0.05 | 0.33 (0.01–8.19) | 0.50 | |
| Racial decent | ||||||||||
| Asian | 1 | 0.33 (0.01–8.02) | 0.49 | 0.36 (0.11–1.13) | 0.08 | 0.33 (0.10–1.02) | 0.05 | 0.33 (0.01–8.19) | 0.50 | |
| Control source | ||||||||||
| Population | 1 | 0.33 (0.01–8.02) | 0.49 | 0.36 (0.11–1.13) | 0.08 | 0.33 (0.10–1.02) | 0.05 | 0.33 (0.01–8.19) | 0.50 | |
| CLL | 1 | 0.20 (0.01–4.10) | 0.29 | 0.87 (0.57–1.32) | 0.51 | 0.84 (0.55–1.27) | 0.40 | 0.20 (0.01–4.16) | 0.30 | |
| Racial decent | ||||||||||
| Unknown | 1 | 0.20 (0.01–4.10) | 0.29 | 0.87 (0.57–1.32) | 0.51 | 0.84 (0.55–1.27) | 0.40 | 0.20 (0.01–4.16) | 0.30 | |
| Control source | ||||||||||
| Population | 1 | 0.20 (0.01–4.10) | 0.29 | 0.87 (0.57–1.32) | 0.51 | 0.84 (0.55–1.27) | 0.40 | 0.20 (0.01–4.16) | 0.30 | |
n, number of studies; unknown, including study populations in which the race was mixed or unclear; secondary, including secondary and therapy-related AML; OR, odds ratio; CI, confidence interval; P, P value.
Table 3. Statistical power (%) for XRCC1 polymorphisms and leukemia risk.
| Polymorphism | Homozygous codominant | Heterozygous codominant | Dominant | Recessive | |
| Arg399Gln (G>A) | AA vs. GG | GA vs. GG | AA+GA vs. GG | AA vs. GA+GG | |
| 100 | 80.4 | 98.8 | 99.8 | ||
| Arg194Trp (C>T) | TT vs. CC | TC vs. CC | TT+TC vs. CC | TT vs. TC+CC | |
| 36.7 | 50.4 | 65.4 | 30.1 | ||
| Arg280His (G>A) | AA vs. GG | GA vs. GG | AA+GA vs. GG | AA vs. GA+GG | |
| 1.1 | 8.0 | 7.9 | 1.1 | ||
XRCC1 Arg399Gln (G>A) polymorphism
In the overall analysis, no statistically significant association between XRCC1 Arg399Gln polymorphism and leukemia susceptibility was observed in three genetic models. A very mild publication bias was detected by Egger’s test (P = 0.04; Begg’s test, P = 0.12) in the codominant heterozygous comparison (GA vs. GG) (other data not shown). We repeated meta-analysis with the “trim and fill” method to adjust publication bias in this genetic model. The conclusion was not influenced, which indicated the robustness our conclusions (data not shown). In the stratified analysis by racial descent, increased risk of leukemia was found among Asians (AA vs. GA+GG, OR = 1.42, 95% CI: 1.12-1.81), and the unknown or mixed race subgroup (AA vs. GG, OR = 1.44, 95% CI: 1.07-1.93; AA vs. GA+GG, OR = 1.39, 95% CI: 1.06-1.84).
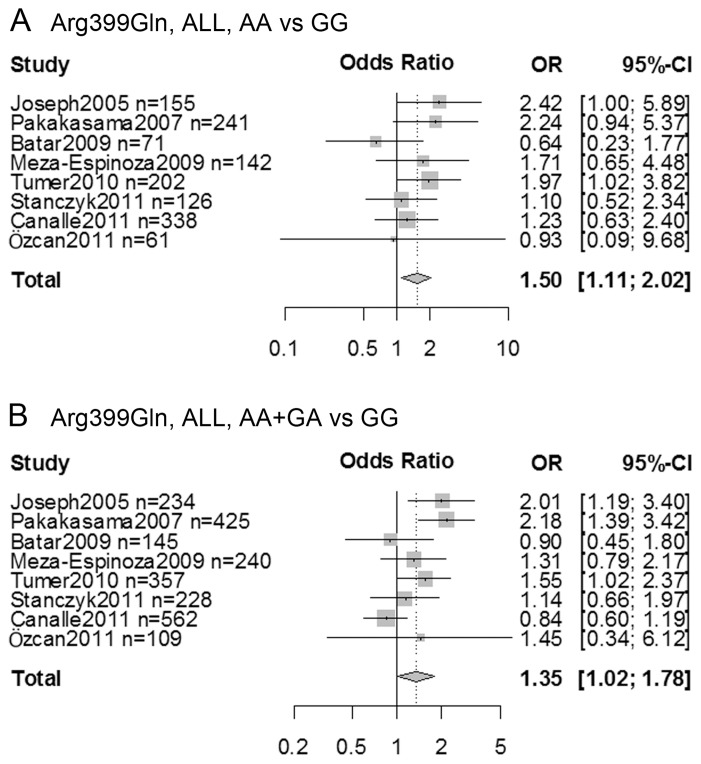
When performing meta-analysis by tumor type, higher risk can be detect in ALL (AA vs. GG, OR = 1.50, 95% CI: 1.11-2.02; AA+GA vs. GG, OR = 1.35, 95% CI: 1.02-1.78) (Figure 2A and 2B) and among ALL Asian (AA vs. GG, OR = 2.33, 95% CI: 1.25-4.34; GA vs. GG, OR = 2.06, 95% CI: 1.44-2.95; AA+GA vs. GG, OR = 2.11, 95% CI: 1.50-2.97) and population-based control subgroups (AA vs. GG, OR = 1.47, 95% CI: 1.02-2.11; GA vs. GG, OR = 1.45, 95% CI: 1.15-1.83; AA+GA vs. GG, OR = 1.45, 95% CI: 1.13-1.85). Although significant association was not found in AML, CML and CLL and in most subgroups by race and control sources of these leukemia types, a protective effect was exhibited among the secondary and therapy-related AML (AA vs. GG, OR = 0.30, 95% CI: 0.11-0.82; AA+GA vs. GG, OR = 0.53, 95% CI: 0.31-0.89) and the CML Asian subjects (GA vs. GG, OR = 0.63, 95% CI: 0.43-0.92; AA vs. GA+GG, OR = 1.62, 95% CI: 1.11-2.38).
Figure 2. Forest plots showed meta-analysis of XRCC1 Arg399Gln polymorphism and acute lymphocytic leukemia (ALL) risk in (A) homozygous codominant (AA vs. GG), and (B) dominant (AA+GA vs. GG) models.
OR, Odds ratio; CI, confidence interval.
XRCC1 Arg194Trp (C>T) polymorphism
XRCC1 Arg194Trp was not associated with the leukemia susceptibility in the overall population or in different race and control source subgroups. Publication bias does not exist across the studies (data not shown). Arg194Trp worked as a risk factor in the AML Asians (CT vs. CC, OR = 1.30, 95% CI: 1.04-1.63; TT+CT vs. CC, OR = 1.26, 95% CI: 1.01-1.57) albeit there was only one study for this race subgroup.
XRCC1 Arg280His (G>A) polymorphism
As for Arg280His polymorphism, no statistically significant association was present in any genetic model or subgroup except for among the secondary and therapy-related AML codominant model homozygous comparison (AA vs. GG, OR = 8.90, 95% CI: 1.56-50.94), and the recessive model (AA vs. GA+GG, OR = 8.15, 95% CI: 1.44-46.06). Begg’s test and Egger’s test showed publication bias in the main meta-analysis in the codominant homozygous comparison (AA vs. GG, Begg’s test P = 0.04, Egger’s test P = 0.02), and the recessive model (AA vs. GA+GG, Begg’s test P = 0.04, Egger’s test P = 0.02) (other data not shown). Adjusting these two models by “trim and fill” method did not influence the conclusion (data not shown).
Discussion
Nonsynonymous XRCC1 polymorphisms Arg399Gln, Arg194Trp, and Arg280His have been implicated in the risk of various cancers [7], [8], [34]. The relationship between these XRCC1 polymorphisms and leukemia risk has been examined in some case-control studies, but the results of these studies were contradictory and inconclusive. Although the association between XRCC1 polymorphisms and risk of some types of leukemia was recognized by a number of studies [15], [17], [20], [21], [22], [23], [24], [25], [27], [28], [29], [32], [33], other reports did not take the XRCC1 genetic variants as risk or protective factors for leukemia [16], [18], [19], [26], [30], [31]. We conducted a meta-analysis that includes 19 studies for Arg399Gln (G>A) polymorphism, 12 studies for Arg194Trp (C>T), and 6 studies for Arg280His (G>A) to evaluate XRCC1 genotype-leukemia association (Figure 1).
Although associations between Arg399Gln, Arg194Trp, and Arg280His and overall leukemia risks were lacking, higher leukemia susceptibility was detected for Arg399Gln among Asians (AA vs. GA+GG, OR = 1.42, 95% CI: 1.12-1.81), and the unknown or mixed race subgroup (AA vs. GG, OR = 1.44, 95% CI: 1.07-1.93; AA vs. GA+GG, OR = 1.39, 95% CI: 1.06-1.84) while no such effect was found among Caucasians (Table 2). The finding in Asians is consistent with previous studies that the Arg399Gln polymorphism increases glioma risk among Asians [8] but does not alter glioma [8] or skin cancer [7] risks among Caucasians, which indicates a race-specific effect of this polymorphism in some tumors. The analysis for unknown or mixed population implies that, excepting Caucasians, other races in these populations may be sensitive to Arg399Gln associated leukemia risk. Collecting more samples from races other than Asian and Caucasian will be necessary to verify the conclusion in future studies.
The predisposition of each leukemia type could be differentially influenced by genetic factors. We performed stratified analysis by leukemia type in order to clarify the role of XRCC1 polymorphisms in the development of individual types of leukemia (Table 2). In the secondary and therapy-related AML, Arg399Gln is a protective factor (AA vs. GG, OR = 0.30, 95% CI: 0.11-0.82; AA+GA vs. GG, OR = 0.53, 95% CI: 0.31-0.89) whereas Arg280His is a risk factor (AA vs. GG, OR = 8.90, 95% CI: 1.56-50.94; AA vs. GA+GG, OR = 8.15, 95% CI: 1.44-46.06). The occurrence of secondary and therapy-related AML is correlated with prior chemotherapy and/or radiation therapy, which probably involves XRCC1 mediated DNA repair. XRCC1 polymorphisms could alter the susceptibility of this AML category by changing XRCC1 DNA repair capacity. However, only one or two studies were included in these subgroups, which compromises the reliability of these findings. Arg399Gln increases CML risk (GA vs. GG, OR = 0.63, 95% CI: 0.43-0.92; AA vs. GA+GG, OR = 1.62, 95% CI: 1.11-2.38), and Arg194Trp increases AML risk (CT vs. CC, OR = 1.30, 95% CI: 1.04-1.63; TT+CT vs. CC, OR = 1.26, 95% CI: 1.01-1.57) among Asians. The conclusions are both derived from one single study and need to be interpreted carefully. Arg399Gln is associated with higher risk in ALL (AA vs. GG, OR = 1.50, 95% CI: 1.11-2.02; AA+GA vs. GG, OR = 1.35, 95% CI: 1.02-1.78) (Figure 2A and 2B) and among ALL Asian (AA vs. GG, OR = 2.33, 95% CI: 1.25-4.34; GA vs. GG, OR = 2.06, 95% CI: 1.44-2.95; AA+GA vs. GG, OR = 2.11, 95% CI: 1.50-2.97) and ALL population-based control subgroups (AA vs. GG, OR = 1.47, 95% CI: 1.02-2.11; GA vs. GG, OR = 1.45, 95% CI: 1.15-1.83; AA+GA vs. GG, OR = 1.45, 95% CI: 1.13-1.85). The results from population-based and hospital-based studies are different. The former may be more reliable since patient controls usually carry other disease conditions, which might potentially influence leukemia risk.
The advantages of this meta-analysis are that it is the most complete and the information from the eligible studies is utilized as much as possible through genetic model and stratified analysis. However, there are several limitations in this study. First, expression of specific genes is highly regulated by a transcriptional control mechanism requiring transcription factors-gene promoter interaction [35], [36], [37], [38]. XRCC1 -77 T>C polymorphism might change the binding capacity of the transcription factor SP1 to the XRCC1 promoter, and then downregulate XRCC1 expression [39]. This polymorphism contributes to the development of lung cancer [40] and breast cancer [39]. However, studies addressing the association between XRCC1 -77 T>C polymorphism and leukemia risk are absent, and cannot be analyzed by our meta-analysis. In addition to the promoter control, the regulation of 3’ untranslated region (3’UTR), by regulators for example microRNAs, influences gene expression in cancer and developmental process [41], [42]. However, whether the XRCC1 polymorphisms in 3’UTR alter leukemia risk was not fully studied. Second, although the statistical power for Arg399Gln was greater than 80% in all three genetic models, the power for Arg194Trp and Arg280His was not as high as that in Arg399Gln (Table 3). The lower power for these two polymorphisms indicates that the weak associations between Arg194Trp and Arg280His and leukemia risk might not be detected while the associations that are detected in our meta-analysis still remain interesting. Statistical power in the genetic association study is primarily affected by the participant numbers and the effect size (OR) when choosing conventional level of α significance criterion 0.05 in the target population. Because the effects of the Arg194Trp and Arg280His polymorphisms on leukemia risk are weak (Table 2, ORs are close to 1), further study with larger sample numbers will be helpful to improve the power for detecting the positive effects. To the current meta-analysis, the negative association between these two polymorphisms and leukemia susceptibility should be cautiously interpreted. Third, the eligible study number for some individual subgroups is small, which restricts the application of the conclusions drawn from those subgroups. Fourth, four eligible studies contained an "unknown" race population. Specific race information was not collect by two of them [22], [25]. The other two studies included non-Caucasian mestizos or mulattos [18], [26]. Since the ethnicities of mestizos and mulattos were difficult to determine, we then described the non-Caucasian population in these two studies as "unknown" race. We have discussed the necessity of performing study with different races in the future studies according to the findings from the unknown population. In the report of Matullo et al. [25], information about the leukemia subtype was not presented. The purpose of the original study was not to conduct a detailed analysis of the subtypes of leukemia. In addition to leukemia, multiple types of cancer as well as emphysema and chronic obstructive pulmonary disease were evaluated based on the data collected by 23 centers from 10 countries. It would be difficult for the researchers to extract the detailed subtype information for a single cancer type, for example leukemia, after the study have been completed. Moreover, the case numbers of this study were not very large; thus the study would not strongly affect the outcome of our subgroup analysis by tumor type of leukemia. Accordingly, we did not use the leukemia tumor type information of this study in our meta-analysis. Finally, African populations have not been well studied previously regarding XRCC1 polymorphisms and leukemia susceptibility. Data from this population will be useful to establish a better overview of polymorphism-leukemia association.
Taken together, results from this meta-analysis demonstrate that XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms might not be associated with overall leukemia risk. However, these polymorphisms might potentially be protective factors or risk factors in specific leukemia types or among particular ethnicities.
Acknowledgments
We thank Dr. Tao Sun at Weill Medical College of Cornell University Department of Cell and Developmental Biology for comments and suggestions on the manuscript, and Jennifer L. Knauss and Aisha I. Abdullah for language-editing of the manuscript.
Funding Statement
This work was partly supported by the National Natural Science Foundation of China (No. 81172462). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1.Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, et al. SEER Cancer Statistics Review, 1975-2010. National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2010/. Based on November 2012 SEER data submission, posted to the SEER web site, 2013.
- 2. Berwick M, Vineis P (2000) Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst 92: 874–897. [DOI] [PubMed] [Google Scholar]
- 3. Caldecott KW, Aoufouchi S, Johnson P, Shall S (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids Res 24: 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dianov GL, Prasad R, Wilson SH, Bohr VA (1999) Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. J Biol Chem 274: 13741–13743. [DOI] [PubMed] [Google Scholar]
- 5. Thompson LH, West MG (2000) XRCC1 keeps DNA from getting stranded. Mutat Res 459: 1–18. [DOI] [PubMed] [Google Scholar]
- 6. Armijo-Olivo S, Stiles CR, Hagen NA, Biondo PD, Cummings GG (2012) Assessment of study quality for systematic reviews: a comparison of the Cochrane Collaboration Risk of Bias Tool and the Effective Public Health Practice Project Quality Assessment Tool: methodological research. J Eval Clin Pract 18: 12–18. [DOI] [PubMed] [Google Scholar]
- 7. Zhang H, Li W, Franklin MJ, Dudek AZ (2011) Polymorphisms in DNA repair gene XRCC1 and skin cancer risk: a meta-analysis. Anticancer Res 31: 3945–3952. [PubMed] [Google Scholar]
- 8. Zhang H, Liu H, Knauss JL (2013) Associations between three XRCC1 polymorphisms and glioma risk: a meta-analysis. Tumor Biol 34: 3003–3013. [DOI] [PubMed] [Google Scholar]
- 9. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 10. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 11. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duval S, Tweedie R (2000) Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56: 455–463. [DOI] [PubMed] [Google Scholar]
- 13.R Development Core Team (2010) R: A language and environment for statistical computing.
- 14. Celkan T, Guven M, Batar B, Alhaj S (2008) The difference between pre-B cell acute lymphoblastic leukemia and Burkitt lymphoma in relation to DNA damage repair gene polymorphisms in childhood. Leuk Lymphoma 49: 1638–1640. [DOI] [PubMed] [Google Scholar]
- 15.Annamaneni S, Gorre M, Kagita S, Addepalli K, Digumarti RR, et al.. (2012) Association of XRCC1 gene polymorphisms with chronic myeloid leukemia in the population of Andhra Pradesh, India. Hematology. [DOI] [PubMed]
- 16. Abramenko I, Bilous N, Chumak A, Kostin A, Martina Z, et al. (2012) DNA repair polymorphisms in B-cell chronic lymphocytic leukemia in sufferers of Chernobyl Nuclear Power Plant accident. J Radiat Res 53: 497–503. [DOI] [PubMed] [Google Scholar]
- 17. Batar B, Güven M, Bariş S, Celkan T, Yildiz I (2009) DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res 33: 759–763. [DOI] [PubMed] [Google Scholar]
- 18. Canalle R, Silveira VS, Alberto Scrideli C, Queiroz RGP, Fernando Lopes L, et al. (2011) Impact of thymidylate synthase promoter and DNA repair gene polymorphisms on susceptibility to childhood acute lymphoblastic leukemia. Leukemia & Lymphoma 52: 1118–1126. [DOI] [PubMed] [Google Scholar]
- 19. Deligezer U, Akisik EE, Dalay N (2007) Lack of association of XRCC1 codon 399Gln polymorphism with chronic myelogenous leukemia. Anticancer Res 27: 2453–2456. [PubMed] [Google Scholar]
- 20. Duman N, Aktan M, Ozturk S, Palanduz S, Cakiris A, et al. (2012) Investigation of Arg399Gln and Arg194Trp Polymorphisms of the XRCC1 (X-Ray Cross-Complementing Group 1) Gene and Its Correlation to Sister Chromatid Exchange Frequency in Patients with Chronic Lymphocytic Leukemia. Genetic Testing and Molecular Biomarkers 16: 287–291. [DOI] [PubMed] [Google Scholar]
- 21. El-Din M, Raslan H, Abdel-Hamid S, Makhlouf M (2012) Detection of XRCC1 gene polymorphisms in Egyptian patients with acute myeloid leukemia. Comparative Clinical Pathology 21: 505–513. [Google Scholar]
- 22. Ganster C, Neesen J, Zehetmayer S, Jäger U, Esterbauer H, et al. (2009) DNA repair polymorphisms associated with cytogenetic subgroups in B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer 48: 760–767. [DOI] [PubMed] [Google Scholar]
- 23. Joseph T, Kusumakumary P, Chacko P, Abraham A, Pillai MR (2005) DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett 217: 17–24. [DOI] [PubMed] [Google Scholar]
- 24. Kim HN, Kim NY, Yu L, Huong Thi Thanh T, Kim YK, et al. (2012) Association of GSTT1 polymorphism with acute myeloid leukemia risk is dependent on smoking status. Leukemia & Lymphoma 53: 681–687. [DOI] [PubMed] [Google Scholar]
- 25. Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, et al. (2006) DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis 27: 997–1007. [DOI] [PubMed] [Google Scholar]
- 26. Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, Macias-Gomez N, Ayala-Madrigal ML, et al. (2009) XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res 8: 1451–1458. [DOI] [PubMed] [Google Scholar]
- 27. Özcan A, Pehlivan M, Tomatir AG, Karaca E, Özkinay C, et al. (2011) Polymorphisms of the DNA repair gene XPD (751) and XRCC1 (399) correlates with risk of hematological malignancies in Turkish population. African Journal of Biotechnology 10: 8860–8870. [Google Scholar]
- 28. Pakakasama S, Sirirat T, Kanchanachumpol S, Udomsubpayakul U, Mahasirimongkol S, et al. (2007) Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer 48: 16–20. [DOI] [PubMed] [Google Scholar]
- 29. Seedhouse C, Bainton R, Lewis M, Harding A, Russell N, et al. (2002) The genotype distribution of the XRCC1 gene indicates a role for base excision repair in the development of therapy-related acute myeloblastic leukemia. Blood 100: 3761–3766. [DOI] [PubMed] [Google Scholar]
- 30. Shi JY, Ren ZH, Jiao B, Xiao R, Yun HY, et al. (2011) Genetic variations of DNA repair genes and their prognostic significance in patients with acute myeloid leukemia. International Journal of Cancer 128: 233–238. [DOI] [PubMed] [Google Scholar]
- 31. Stanczyk M, Sliwinski T, Cuchra M, Zubowska M, Bielecka-Kowalska A, et al. (2011) The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep 38: 445–451. [DOI] [PubMed] [Google Scholar]
- 32. Tumer TB, Yilmaz D, Tanrikut C, Sahin G, Ulusoy G, et al. (2010) DNA repair XRCC1 Arg399Gln polymorphism alone, and in combination with CYP2E1 polymorphisms significantly contribute to the risk of development of childhood acute lymphoblastic leukemia. Leuk Res 34: 1275–1281. [DOI] [PubMed] [Google Scholar]
- 33. Sorour A, Ayad MW, Kassem H (2013) The genotype distribution of the XRCC1, XRCC3, and XPD DNA repair genes and their role for the development of acute myeloblastic leukemia. Genet Test Mol Biomarkers 17: 195–201. [DOI] [PubMed] [Google Scholar]
- 34. Hu Z, Ma H, Chen F, Wei Q, Shen H (2005) XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiol Biomarkers Prev 14: 1810–1818. [DOI] [PubMed] [Google Scholar]
- 35. Zhang H, Pan Y, Zheng L, Choe C, Lindgren B, et al. (2011) FOXO1 inhibits Runx2 transcriptional activity and prostate cancer cell migration and invasion. Cancer Res 71: 3257–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang HJ, Li WJ, Gu YY, Li SY, An GS, et al. (2010) p14ARF interacts with E2F factors to form p14ARF-E2F/partner-DNA complexes repressing E2F-dependent transcription. J Cell Biochem 109: 693–701. [DOI] [PubMed] [Google Scholar]
- 37. Zhang HJ, Li WJ, Yang SY, Li SY, Ni JH, et al. (2009) 8-Chloro-adenosine-induced E2F1 promotes p14ARF gene activation in H1299 cells through displacing Sp1 from multiple overlapping E2F1/Sp1 sites. J Cell Biochem 106: 464–472. [DOI] [PubMed] [Google Scholar]
- 38. Li WJ, Gu YY, Zhang HJ, Zhou J, Jia HT (2009) Induction of p14ARF by E2F1 contributes to 8-chloro-adenosine-induced apoptosis in human lung cancer H1299 cells. Chemotherapy 55: 335–343. [DOI] [PubMed] [Google Scholar]
- 39. Ginsberg G, Angle K, Guyton K, Sonawane B (2011) Polymorphism in the DNA repair enzyme XRCC1: utility of current database and implications for human health risk assessment. Mutat Res 727: 1–15. [DOI] [PubMed] [Google Scholar]
- 40. Dai L, Duan F, Wang P, Song C, Wang K, et al. (2012) XRCC1 gene polymorphisms and lung cancer susceptibility: a meta-analysis of 44 case-control studies. Mol Biol Rep 39: 9535–9547. [DOI] [PubMed] [Google Scholar]
- 41. Wang D, Qiu C, Zhang H, Wang J, Cui Q, et al. (2010) Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One 5: e13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang H, Shykind B, Sun T (2012) Approaches to manipulating microRNAs in neurogenesis. Front Neurosci 6: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]