Abstract
Following primary infection Herpes simplex virus-1 (HSV-1) establishes lifelong latency in the neurons of human sensory ganglia. Upon reactivation HSV-1 can cause neurological diseases such as facial palsy, vestibular neuritis or encephalitis. Certain populations of sensory neurons have been shown to be more susceptible to latent infection in the animal model, but this has not been addressed in human tissue. In the present study, trigeminal ganglion (TG) neurons expressing six neuronal marker proteins were characterized, based on staining with antibodies against the GDNF family ligand receptor Ret, the high-affinity nerve growth factor receptor TrkA, neuronal nitric oxide synthase (nNOS), the antibody RT97 against 200kDa neurofilament, calcitonin gene-related peptide and peripherin. The frequencies of marker-positive neurons and their average neuronal sizes were assessed, with TrkA-positive (61.82%) neurons being the most abundant, and Ret-positive (26.93%) the least prevalent. Neurons positive with the antibody RT97 (1253 µm2) were the largest, and those stained against peripherin (884 µm2) were the smallest. Dual immunofluorescence revealed at least a 4.5% overlap for every tested marker combination, with overlap for the combinations TrkA/Ret, TrkA/RT97 and Ret/nNOS lower, and the overlap between Ret/CGRP being higher than would be expected by chance. With respect to latent HSV-1 infection, latency associated transcripts (LAT) were detected using in situ hybridization (ISH) in neurons expressing each of the marker proteins. In contrast to the mouse model, co-localization with neuronal markers Ret or CGRP mirrored the magnitude of these neuron populations, whereas for the other four neuronal markers fewer marker-positive cells were also LAT-ISH+. Ret and CGRP are both known to label neurons related to pain signaling.
Introduction
Herpes simplex virus-1 (HSV-1) is a human neurotropic DNA virus of the herpesviridae family [1]. After an initial oral infection, the virus can travel along axons innervating the affected region, to reach the trigeminal ganglia (TG) where it establishes a lifelong latency. The sensory neurons of the TG are the principal site for HSV-1 latency in humans, although the vestibular, geniculate, spiral, and sacral ganglia can also harbor latent virus as summarized in [2], [3]. During latency HSV-1 viral activity is restricted, with only the latency associated transcript (LAT) being abundantly expressed [4]. LAT is engaged in establishing latency [5], [6], in facilitating the process of reactivation [7], [8], and at the same time promoting neuronal survival after HSV-1 infection by reducing apoptosis [9]. Hence, it is reasonable to consider LAT a surrogate marker of HSV-1 latency. Reactivation can occur spontaneously or be induced by various stimuli, leading to diseases including herpes labialis, facial palsy, vestibular neuritis or encephalitis [10], [11]. The mechanisms underlying establishment and maintenance of latency, as well as viral reactivation, are still under investigation. Several viral factors have been identified which may play a role [2], [11], [12], but host factors which influence latency and reactivation are less well understood: the host´s immune system definitely plays an important role [13]–[16], but data from the animal model suggest that neuronal characteristics might also be important.
Sensory neurons in the TG form a diverse population of cells that can be classified according to cellular morphology, neurochemistry and functional characteristics [17], [18]. Classically, studies of the peripheral nervous system have subdivided the neurons into three major classes on the basis of their sizes: A (also known as large light), B (also known as small dark) and C [19]. Neurochemical classification of neurons is based on their protein expression profile, including the presence of growth factor receptors, neuropeptides and neurotransmitters [20], but a full neurochemical classification of neurons in the human TG has not been performed.
Different neuronal populations have been shown to display variable susceptibilities for HSV-1 infection in the mouse model. Although all TG neuronal populations in the mouse model are capable of supporting an HSV-1 infection, neurons positive with the monoclonal antibody KH10 are more permissive for a productive infection [21], while those expressing the globoseries glycoconjugate stage specific embryonic antigen 3 (SSEA3) are productively infected less than might be expected from the magnitude of their neuronal population [22]. Similarly, during latent infections in the mouse model, trigeminal and dorsal root neuronal populations reactive for the monoclonal antibody A5 are more susceptible [21]–[23] than neurons positive for KH10 [18].
In the current study, we characterized the neuronal composition of the human TG using six different marker proteins: the GDNF family ligand receptor Ret, the high-affinity nerve growth factor receptor TrkA, neuronal nitric oxide synthase (nNOS), the antibody RT97 against 200 kDa neurofilament, calcitonin gene-related peptide (CGRP) and peripherin (for details see Table 1). Secondly, we assessed the susceptibility of neurons expressing these markers towards latent HSV-1 infection via the expression of HSV-1 LAT using in situ hybridization (ISH).
Table 1. Characteristics and frequency of neuronal markers analyzed.
| Antigen recognized by antibody | Expected cellular localizationa | Proposed characteristics of labeled neurons | Data from animal models | % labeled neurons (number of counted neurons (present study)) | |
| Ret | GDNF-family ligand receptor tyrosine kinase | Membrane protein, also seen as cytoplasmic and Golgi hn [34] | Small diameter neurons, especially nociceptors [51] | Labels neurons equivalent to KH10 antibody in mouse [23] | 26.93 (2306) |
| TrkA | High-affinity nerve growth factor receptor tyrosine kinase | Cytoplasmic and Golgi hn [52] Plasma membrane ho [53] | Small diameter neurons, especially nociceptors [51] | Labels neurons equivalent to A5 antibody in mouse [23] | 61.82 (2499) |
| nNOS | Neuronal nitric oxide synthase | Cytoplasmic hn [54] | Prevalence in ophthalmic branch of TG [20] | NO can inhibit HSV-1 replication [21] | 33.13 (2792) |
| RT97 | Phosphorylated 200kDa neurofilament | Cytoplasmic an [19], [55] | Larger A-type neurons [45] | 39.26 (2127) | |
| CGRP | Calcitonin gene-related peptide, a vasodilator and pain mediator | Cytoplasmic hn [56] | Smaller sized neurons [20] | Found throughout the TG of mice [20], and innervate the gingiva in rats [33] | 27.16 (2003) |
| Peripherin | Type III intermediate filament | Cytoplasmic hn [57] | Neurons of the peripheral nervous system and small nerve fibers of the sensory ganglia [58] | Upregulated in response to nerve injury and proinflammatory cytokines [59] | 40.87 (2109) |
a: References are marked according to whether the experiments were performed in humans (h) or animals (a), and neurons (n) or other cell types (o).
Materials and Methods
Ethics statement
The use of autopsy samples for the present study was approved by the Ethics Committee of the Medical Faculty of the Ludwig-Maximilians University, Munich, Germany (project number: 017-11). The tissue samples were collected from the cadavers as part of the routine autopsy procedure and were irreversibly anonymized, so that according to the national ethics guidelines obtaining consent is neither possible nor required. The Ethics Committee approved this consent procedure.
Human tissue samples
Fourteen human trigeminal ganglia (5 LAT-negative, 9 LAT-positive; Table 2) from 14 individuals were obtained at autopsy at Ludwig-Maximilians University (Munich, Germany). LAT negativity or positivity was determined using ISH at different levels within each ganglion (see protocol below) and RT-PCR analysis [24] which gave identical results. All LAT+ samples were also positive using nested HSV-1 DNA PCR [25], while all LAT- samples were negative using this method. Nested HSV-2 DNA PCR [26] was negative in all samples. Samples 1–10 (Table 2) were used for dual LAT ISH and marker protein staining, and samples 11–14 (Table 2) for double marker protein fluorescence staining. The average age at death was 38yr±8 for the LAT– and 55yr±5 for the LAT+ samples, and the average post-mortem delay was 15hr±2.4 for LAT– and 23hr±2.3 for the LAT+ samples. No donors had evidence of active herpesvirus infection at death. Ganglia were embedded directly in Jung Tissue Freezing Medium (Leica Microsystems, Nussloch, Germany) and stored at –80°C for further use.
Table 2. Information on trigeminal ganglion donors.
| ID | LAT status | Sexa | Age at death (years) | Cause of death |
| 1 | – | M | 35 | Cardiac |
| 2 | – | M | 29 | Drug overdose |
| 3 | – | F | 36 | Cardiac |
| 4 | – | F | 20 | Cardiac |
| 5 | – | F | 70 | Cardiac |
| 6 | + | F | 78 | Cardiac |
| 7 | + | M | 57 | Trauma |
| 8 | + | F | 41 | Cranial aneurism |
| 9 | + | M | 69 | Cardiac |
| 10 | + | M | 47 | Cardiac |
| 11 | + | M | 71 | Cardiac |
| 12 | + | M | 44 | Trauma |
| 13 | + | M | 40 | Cardiac |
| 14 | + | M | 50 | Trauma |
a: M = Male, F = Female.
In situ analyses
It is still a matter of debate whether LAT can be visualized in all latently infected neurons using ISH or if some neurons do not express LAT to a level which allows for detection using this method [27], [28]. We thus refer to the neurons in this manuscript as “LAT-ISH+” or “LAT-ISH– “ to indicate this fact. In situ hybridization was performed in combination with immunohistochemistry as described previously [27]. In brief, defrosted tissue sections were paraformaldehyde-fixed, then endogenous peroxidase-blocked using 1.5% H2O2 in methanol, acetylated with 0.25% acetic anhydride in triethanolamine, then prehybridized for 1hr in PHO buffer (1xDenhard’s solution, 0.1 µg/ml herring sperm DNA, 5 mg/ml sodium pyrophosphate, 5 mM Tris-HCl, in 4x sodium citrate buffer SSC). Samples were incubated overnight at 37°C with 4 ng/µl of an oligonucleotide probe against HSV-1 LAT (not recognizing HSV-2 LAT), labeled with a 3′-DIG tag (5′-CAT AGA GAG CCA GGC ACA AAA ACA C-3′; nucleotides 119.783 – 119.759; Eurofins MWG Operon, Ebersberg, Germany). After further washing and blocking, samples were incubated for 2 hours with an anti-DIG-AP conjugated antibody (Roche Diagnostics, Mannheim, Germany), and reactivity was visualized using NBT/BCIP (Roche Diagnostics). Samples were again fixed in paraformaldehyde, then incubated overnight with primary antibodies at pre-determined optimal dilutions, before this reactivity was visualized using biotin-conjugated secondary antibodies (Dako, Hamburg, Germany; Abcam, Cambridge, UK), HRP-conjugated streptavidin (BioLegend, Fell, Germany), and 3-3′-diaminobenzidine (DAB; Dako). Samples were counterstained with haematoxylin to help visualize cells and nuclei.
Primary antibodies used in this study: mouse monoclonal to Ret (Ret01), ab1840, Abcam, Cambridge, UK, used at 1∶10; goat polyclonal to TrkA, AF175, R&D Systems, Wiesbaden, Germany, used at 1∶50; rabbit polyclonal to nNOS, AB5380, Merck Millipore, Molsheim, France, used at 1∶2000; mouse monoclonal to NEFH (RT97), MA1-34345, Thermo Scientific/Pierce Biotechnology, Rockford, IL, USA, used at 1∶100; sheep polyclonal to CGRP, CA1137, Enzo Life Sciences, Lörrach, Germany, used at 1∶1500; rabbit polyclonal to peripherin, ab4666, Abcam, Cambridge, UK, used at 1∶750. Controls without primary or secondary antibodies as well as with isotype control antibodies were performed (see Figures S1 and S2). No mouse ascites derived antibodies were used in the current study [29]. All experiments were performed in duplicate or triplicate on non-consecutive sections. Staining was visualized using an Olympus BX41 microscope and pictures were taken with an Olympus C4040 Zoom digital camera. Images were adjusted in CorelDraw to reflect the brightness and contrast as seen through the microscope.
Fluorescence immunohistochemistry for dual visualization
For fluorescence IHC experiments, sections from LAT+ donors were paraformaldehyde fixed, blocked in 2% bovine serum albumin (BSA, Sigma Aldrich, Steinheim, Germany), then exposed to the first primary antibody for 2 hours at room temperature. Antibodies were used at the same concentrations as given above. After washing in PBS, slides were incubated with an appropriate fluorophore-conjugated secondary antibody, as listed below, for 30 minutes. After further washing and blocking with BSA, slides were then incubated with the second primary antibody overnight, at 4°C, then washed again, and incubated with a secondary antibody of alternative color. After washing in both PBS and 75% ethanol, slides were incubated in a solution of 1% Sudan Black (Sigma Aldrich) in 70% ethanol for 10 minutes to reduce lipofuscin autofluorescence. Slides were then washed in 75% ethanol, before being mounted with Fluoro-Gel II, with DAPI (Science Services, Munich, Germany). Controls omitting primary or secondary antibodies as well as using isotype control antibodies were performed (see Figure S3).
All secondary antibodies were purchased from Life Technologies Molecular Probes (Darmstadt, Germany), and used at a 1∶200 dilution. These included AlexaFluor568 conjugated donkey anti-goat, donkey anti-rabbit and donkey anti-mouse, and AlexaFluor488 conjugated goat anti-mouse and goat anti-rabbit. All experiments were performed in duplicate. Slides were visualized using an All-in-one Type Fluorescence Microscope (BZ-8100E, Keyence) and images were obtained with an integrated camera and software. Images were adjusted in CorelDraw to reflect the brightness and contrast as seen through the microscope.
Neuron assessment and measurement
Following staining, neurons were assessed and counted based on their staining pattern. Only neurons with a visible nucleus were counted to ensure potential LAT staining was not missed. Neurons were assigned as singly or doubly positive or negative.
For size analysis, only neurons with a nucleus were measured, as this suggested that the neuron was cut through a central plane. Additionally, neurons were only included in the analysis where the surrounding tissue was intact to confirm that the neuron had not shrunk post-mortem or during the experimental procedure. Due to these strict inclusion criteria many neurons could not be assessed for the current study, and thus a bias towards more “robust” neurons cannot be excluded. The cross-sectional area of neurons inµm2 was measured using Image J (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2012.). The measurements were calibrated by imaging a calibration grid using the same microscope and camera settings, and calculating the distance per pixel.
Statistical analysis
Statistical analyses were performed in Microsoft Excel and SPSS, p<0.05 was regarded as significant for all analyses. For data with a Gaussian distribution mean ± SEM are given, for those with non-Gaussian distribution, values for the median, 25th and 75th percentiles are presented. Differences in the percentage of Marker+ neurons between LAT-ISH+ and LAT-ISH– samples, and between DAB marker staining and fluorescence staining, were calculated using a two-tailed Mann-Whitney U test. Correlations between the age or post-mortem delay of the samples, and the percentage of positive staining were calculated using the Spearman Correlation coefficient and 2-tailed T-test. Differences in the sizes between Marker+ and Marker– neurons from the same experiments were calculated using a Mann-Whitney U-test. For analysis of the co-localization of two marker proteins, the Pearson Chi-squared test was used to calculate p-values. For analysis of the co-localization of LAT with marker proteins, the two-tailed Fisher exact Chi-squared test was used to calculate p-values, due to the low incidence of LAT-ISH+ neurons.
Results
Distribution of neuronal marker proteins in TG neurons
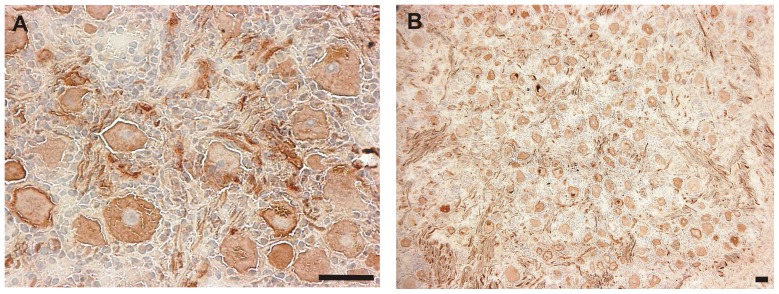
Sections from 5 LAT+ donors and 5 LAT− donors (donors 1-10, Table 2) were stained immunohistochemically for each of six neuronal marker proteins (Table 1). Neurons positive for the high-affinity growth factor receptor TrkA were most abundant with 61.82% of all assessed neurons staining positive, peripherin was detected in 40.87%, the neurofilament antibody RT97 in 39.26%, the neuronal nitric oxide synthase nNOS in 33.13%, calcitonin gene-related peptide (CGRP) in 27.16% and the GDNF family ligand receptor Ret in 26.93% of all neurons. The percentage of Marker+ neurons was compared between the LAT+ and LAT− donors. No differences in frequency between LAT+ and LAT− donors were found for any neuronal marker proteins (p>0.05 in all cases, Mann-Whitney U test, 2-tailed). Correlations between the number of Marker+ neurons and either the age of the donor, or the post-mortem delay between death and autopsy, were also evaluated. No correlations were seen, except for a negative correlation between age and the percentage of neurons positive for CGRP (Spearman correlation coefficient –0.73, T-test p = 0.027; data not shown). Experiments performed using all six marker protein antibodies gave complete coverage of all TG neurons (Figure 1).
Figure 1. Labeling of a TG with a marker mix of all markers used in the current study (Ret, TrkA, nNOS, RT97, CGRP and peripherin).
The micrograph on the left (A) was taken at 400x magnification for a more detailed view, that on the right (B) at 100x for an overview. Marker proteins are labeled with DAB (brown) resulting in complete labeling of all detected neurons. The tissue was counterstained with haematoxylin. Scale bars represent 50 µm in all cases.
Size of differential neuronal populations in TG
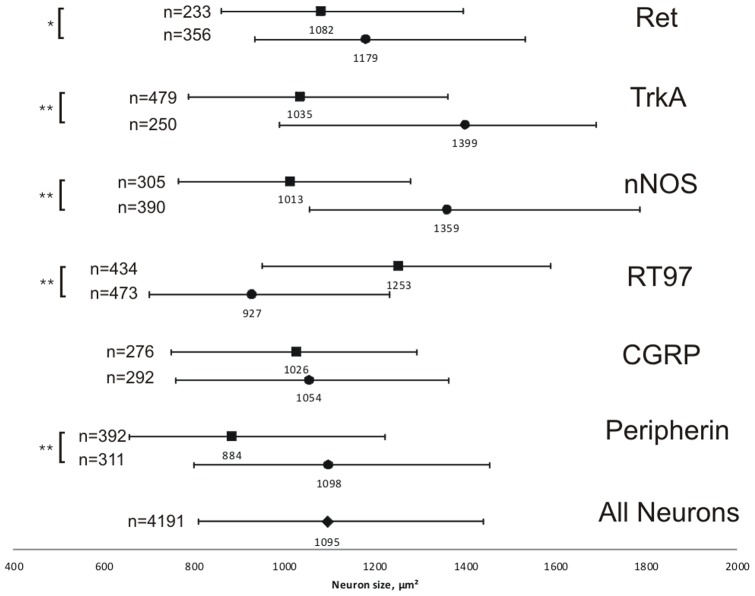
To further characterize the neurons expressing each marker protein, the cross-sectional area of neurons was measured for both neuronal Marker+ and Marker− cells from the same experiments (Figure 2). The median for all neurons, both labeled and unlabeled from all experiments, was 1095 µm2. In the case of Ret, TrkA, nNOS and peripherin, those neurons expressing the protein were significantly smaller than those negative for staining on the same slides (p = 0.013 (Ret) and p<0.001 (TrkA, nNOS, peripherin), Mann-Whitney U test, 2-tailed), whilst for RT97, positive neurons were significantly larger (p<0.001 Mann-Whitney U test, 2-tailed). Cells stained with antibody against CGRP showed no significant difference in cross-sectional area compared with negative neurons. All of the markers stained neurons from a wide range of sizes with a large overlap between the different neuronal marker proteins (Figure 2).
Figure 2. Median cross-sectional areas of cells labeled with different marker antibodies.
Central values represent the median values inµm2, whilst the bars indicate the 25th–75th percentile ranges. Filled squares = marker positive neurons, filled circles = marker negative neurons from the same experiment. n represents the number of counted neurons. Asterisks represent significant differences (* = p<0.01, ** = p<0.001, Mann-Whitney U-test).
Co-localization of marker proteins in neurons
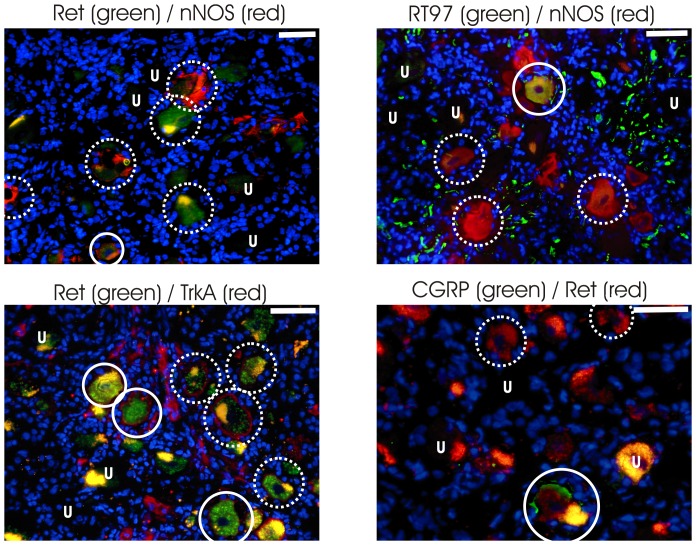
The high percentage of neurons expressing each of the six marker proteins investigated suggested that there was some overlap in the expression of these proteins. To investigate this co-expression, we performed dual immunofluorescence experiments for a subset of antibody combinations in LAT+ donors (samples 11-14, Table 2, Figure 3). The percentage of cells labeled with TrkA and nNOS using fluorescence secondary antibodies was significantly lower (p<0.05, Mann-Whitney U test, 2-tailed) compared to DAB staining, possibly reflecting inter-donor variability or increased sensitivity due to the extra streptavidin amplification in the DAB protocol.
Figure 3. Fluorescence co-labeling of marker combinations.
. The markers are labeled with Alexa568 (red), Alexa488 (green) and DAPI (blue). Marker combinations are indicated above each pictogram. Dotted-line circles indicate examples of single labeled neurons, solid-line circles indicate examples of double labeled neurons. The letter “U” indicates examples of unlabeled neurons, some with lipofuscin. Scale bars represent 50 µm.
With all of the tested combinations of antibodies, a subset of neurons was observed to be double labeled (Marker 1+/Marker 2+ in Table 3). The greatest overlap was between TrkA and RT97 (20.13%), whilst the least was between Ret and nNOS (4.5%). These results were analyzed using a Chi-squared test, to confirm if this overlap reflected the distribution of the individual markers in the population of neurons as a whole. The co-localizations of TrkA/nNOS, and nNOS/RT97 were as expected by chance, based on the frequency of these markers in the population. However, the Chi-squared results for TrkA/Ret, TrkA/RT97, Ret/nNOS and Ret/CGRP were all significant (p<0.05, see Table 3 for exact values and Table S1 for median and interquartile ranges). Comparison of the expected and observed frequencies shows that the co-localization of Ret/CGRP was more frequent than expected by chance, while it was less than expected by chance for the combinations TrkA/Ret, TrkA/RT97 and Ret/nNOS.
Table 3. Co-localization of neuronal marker proteins using immunofluorescencea,b.
| Marker 1 | Marker 2 | Marker 1+/ Marker 2+ | Marker 1+/ Marker 2– | Marker 1–/ Marker 2+ | Marker 1–/ Marker 2– | pc |
| Ret | nNOS | 4.5% | 23% | 16.63% | 55.88% | 0.042* |
| Ret | CGRP | 15.75% | 19.13% | 16.88% | 48.25% | <0.01* |
| TrkA | Ret | 8.38% | 34.75% | 15% | 41.88% | 0.021* |
| TrkA | nNOS | 10% | 35.88% | 14.5% | 39.63% | 0.102 |
| TrkA | RT97 | 20.13% | 25.25% | 19.63% | 35% | 0.015* |
| nNOS | RT97 | 9.25% | 17.75% | 27% | 46% | 0.476 |
a: see Supplementary Table S4 for median and interquartile ranges
b: 800 neurons were counted per marker pair.
c: p-value as calculated by the Pearson Chi-squared test, with p<0.05 taken as significant, shown by an asterisk.
Frequency and size of LAT-ISH+ neurons in LAT+ donors
Using samples from 5 LAT+ donors (samples 6–10 in Table 2), individual neurons were assessed to be LAT-ISH+ or LAT-ISH−. Overall, 4.72%±0.39 (mean ± SEM) of neurons were LAT-ISH+, indicating latent HSV-1 infection. To further characterize these infected neurons, the cross-sectional area of LAT-ISH+ neurons was measured and compared to LAT-ISH- neurons from the same donors. The median cross-sectional area of LAT-ISH+ neurons was 1106 µm2 (821 µm2–1439 µm2; 25th percentile - 75th percentile) and of LAT-ISH- neurons was 1070 µm2 (786 µm2–1429 µm2). There was no significant difference between the sizes of LAT-ISH+ and LAT-ISH– neurons (p = 0.502, Mann-Whitney U test, 2-tailed). The median cross-sectional area of 1106 µm2 for LAT-ISH+ neurons is closest in size to the median of Ret-positive neurons (Figure 2).
Co-localization of LAT and neuronal markers in LAT+ ganglia
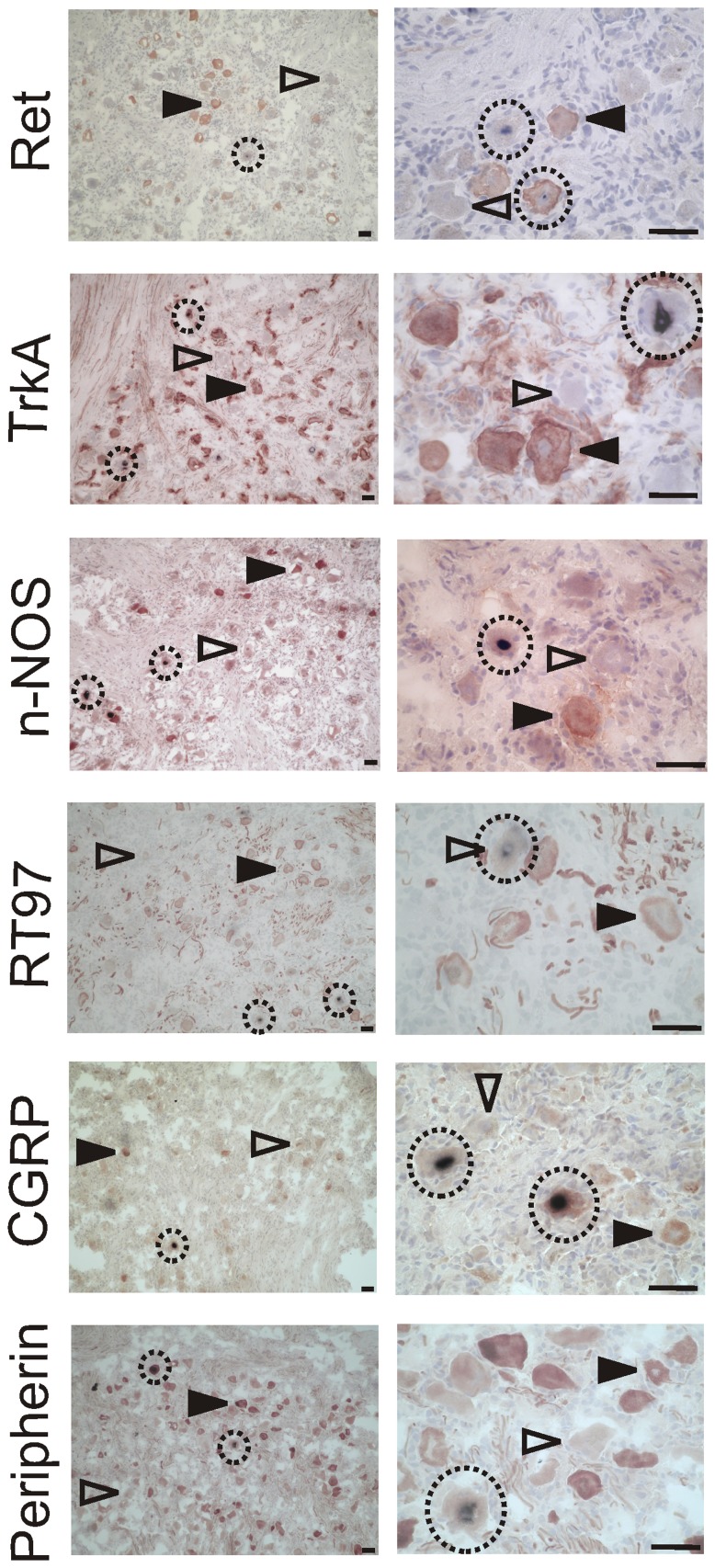
Slides from LAT+ donors (samples 6-10, Table 2) were co-stained for LAT and one of each of the six markers investigated in this study, and assigned to one of four categories. These categories were: LAT-ISH+, and either Marker+ or Marker–, or LAT-ISH– and either Marker+ or Marker– (Table 4, Figure 4 and Table S2 for median and interquartile ranges).
Table 4. Percentage of neurons labeled for neuronal markers and LATa.
| Marker | LAT+ / Marker+ | LAT+ / Marker– | LAT– / Marker+ | LAT– / Marker– | No. counted neurons | Pb |
| Ret | 0.95% | 3.39% | 25.50% | 70.15% | 1149 | 0.516 |
| TrkA | 1.87% | 3.82% | 54.91% | 39.40% | 1231 | <0.001* |
| nNOS | 0.98% | 4.29% | 32.33% | 62.40% | 1423 | 0.0054* |
| RT97 | 0.63% | 3.59% | 39.37% | 56.41% | 1115 | <0.001* |
| CGRP | 0.6% | 3% | 24.8% | 71.6% | 1000 | 0.249 |
| Peripherin | 0.73% | 3.09% | 40.20% | 55.99% | 1102 | 0.0035* |
a: see Table S2 for median and interquartile ranges.
b: p-value calculated using Fisher’s exact Chi-squared test, with p<0.05 taken as significant, shown by an asterisk.
Figure 4. Detection of HSV-1 LAT in neurons expressing different maker proteins in the human TG.
The micrographs in the left column are taken at 100x magnification for an overview, those in the right column at 400x for a more detailed view. The name of the marker used is indicated in each row. Marker proteins are labeled with DAB (brown), LAT using NBT (purple), and the tissue was counterstained with haematoxylin. Open arrowheads indicate marker negative cells, closed arrowheads marker positive cells. LAT positive cells are circled. Scale bars represent 50 µm in all cases.
The distribution of LAT-ISH+ neurons with and without the marker protein was compared to the distribution of the marker protein in the LAT-ISH- neurons (Table 4, Fisher’s exact Chi-squared test). For the marker proteins Ret and CGRP, there was no significant difference in the percentage of Marker+ neurons for LAT-ISH+ cells and LAT-ISH- cells. In the case of TrkA, nNOS, RT97 and peripherin, there was a significant difference. Comparison of the expected and observed frequencies in each case suggests that fewer of the LAT-ISH+ cells are also Marker+ than would be expected based on the distribution of the marker in LAT-ISH- cells, and thus that LAT is preferentially expressed in Marker- cells.
Discussion
The major findings of this study are: 1) neuronal markers can be used to characterize neuronal populations within the human TG, 2) a subgroup of neurons was observed to display an overlap with all tested combinations of markers, with the marker combination Ret/CGRP occurring more frequently than expected by chance, and 3) HSV-1 LAT associates proportionally with the neuronal marker proteins Ret and CGRP, in a divergence from the animal model.
The most prevalent of the six neuronal marker proteins investigated was TrkA, with 61.82% of neurons staining positive. This is higher than the 2.60%, 6.22% and 34.61% of Marker+ neurons seen in three adult human TG from two studies [30], [31], but lower than the 70% reported in rat DRG [32]. The median cross-sectional area of TrkA+ neurons in this study (1035 µm2) matches well with previous reports of TrkA neurons in rat and human TG [31], [33]. The presence of nNOS in 33.13% of neurons was also higher than previous work on brain NOS in mouse TG (3% of neurons) [21], suggesting possible differences between species. No data are available in the literature on nNOS frequency in human TG. The percentage of neurons positive for Ret (26.93%) matched previously published results of 23.23% positive neurons from a single adult human TG [34] though this was lower than the 61.9% of positive mouse DRG neurons [35], again highlighting potential differences when comparing species and sensory ganglia. The 39.26% of neurons stained with antibody RT97 are within the range of the 23–41% of positive neurons in mouse TG [21], [36]. No data on staining frequency in the human TG for the RT97 antibody is available. The larger cross-sectional area of these neurons compared to Marker- neurons reflects the association of this antibody with larger A-type neurons [19]. Results with the CGRP antibody were slightly lower than those seen in other work of 30–50% positive TG neurons in mice, rats and humans [20], [37], [38]. Interestingly, the percentage of neurons positive for CGRP also seemed to decrease with increasing donor age, though this was based on a small sample number. Peripherin was found in neurons with a smaller cross-sectional area, reflecting previous work on this marker in rats [39], [40]. Little data is available on frequency in the TG – a recent paper found 72.3% of TG neurons traced from rat upper molars along dental primary afferents to the maxillary region of the TG were peripherin+ [39], but no data are available on frequencies in human TG.
The total percentages of the six neuronal marker protein frequencies suggested that these proteins were overlapping in some neurons. This was confirmed for a subset of antibodies using dual immunofluorescence. Comparison of the dual staining to the proportion of neurons labeled by each antibody in the neuron population as a whole revealed that TrkA/Ret, TrkA/RT97, and Ret/nNOS dual staining occurred less frequently than would be expected. This reflects previous work in the literature that TrkA and Ret are present in two distinct populations of nociceptive neurons [35], and that RT97 is labeling larger A-type neurons in comparison to the smaller B-type neurons that contain TrkA [19]. All of the markers used in the present study stained neurons from a wide range of sizes with a large overlap between the different neuronal markers. The Ret/CGRP dually labeled neurons occurred more frequently than expected by chance. Lectin BSL IB4 (which labels a roughly comparable neuron population to the marker Ret [18]) has been shown to co-label neurons with CGRP in the mouse and more so in the TG of rats [41], [42]. Ret and CGRP both label neurons related to pain signaling, albeit they are commonly considered to be located in different classes of neurons [43].
LAT was found using ISH in neurons expressing all of the six marker proteins studied, reflecting the work in animal models that all neurons are susceptible to HSV-1 infection [21]. It is still a matter of debate whether ISH can detect LAT in all latently infected neurons in the human TG or if some neurons do not express LAT to a level that allows for detection using this method, and thus neurons expressing LAT at a very low level could have been missed in the present study [27], [28], [44]. The distribution of LAT in Ret+ or CGRP+ neurons reflected the proportion of these neurons in the population, whilst LAT was found less often in neurons labeled with antibodies against TrkA, nNOS, RT97 and peripherin. None of the marker proteins investigated showed a positive correlation between LAT and the marker. This suggests either that LAT has a preference for a certain neuron type that was not identified by one of the six markers in this study, or alternatively that there is a neuron population characterized by a combination of two or more of the marker proteins from this study. The statistically significant co-localization of Ret and CGRP in a subset of neurons suggests that these might be a target, a finding that warrants further investigation.
The reduced association of LAT with RT97+ and peripherin+ neurons could reflect a preference for intermediate sized B-type neurons, as RT97 in this study and the literature [45] labels larger neurons, whilst peripherin was found in neurons with a smaller cross-sectional area. In fact, the average size of LAT-ISH+ neurons best fitted with the size of neurons stained with antibody against Ret, which did not show this negative distribution. The reduced association of LAT with nNOS+ neurons in this study could also reflect the potential anti-viral activity of NO seen in animal models [21], or the association of nNOS+ neurons with the ophthalmic branch of the TG, which shows less latent infection in humans [20], [46]. LAT was found in CGRP neurons as would be expected based on the magnitude of this population. If these neurons are associated with innervation of the gingiva as is seen in rats [33] then this could be important based on the oral route of human HSV-1 infection, but the distribution of CGRP in the branches of the human TG has not so far been investigated.
Conclusions
In the present study co-localization of LAT using ISH occurred most frequently with neuronal markers Ret or CGRP, two markers which also preferably co-localize within the same neuron population. The distribution of LAT in the human TG in this study contrasts with the mouse model, where A5+ neurons, which have a high co-localization with TrkA+ neurons, are more permissive for HSV-1 latent infection, whilst KH10+ neurons, which have a high co-localization with Ret+ neurons, are less permissive [18], [21], [23]. The disparity between the mouse and human results is also intriguing in light of the failure of the mouse model to show full spontaneous reactivation similar to that seen in humans and rabbits [47]–[49]. This difference could be purely due to inherent differences in the distribution of these neuronal markers between species or related to other host factors such as immunological factors. It could also reflect the ocular route of infection used in the mouse model, as compared to the typical route of human infection via the oral mucosa, as different neuronal populations could innervate these tissues differentially [50]. However, in the rabbit model, the ocular route of infection is also often used [49]. Since the clinical presentation of HSV-1 reactivation can result in differential diseases, among them herpes labialis, facial palsy, vestibular neuritis or encephalitis, one might speculate that from the clinical localization and distribution of HSV-1 reactivation, one could infer the affected neuronal subpopulation.
Supporting Information
Labeling of a TG with isotypes of all markers used in the current study. Isotype control for mouse primary antibody (08-6599), Life Technologies, Darmstadt, Germany; Isotype control for rabbit primary antibody (086199), Life Technologies; Isotype control for goat primary antibody (02-6202), Life Technologies; Isotype control for sheep primary antibody (013-000-002), Jackson Immuno Research Europe Ltd., Suffolk, UK. The micrograph on the left was taken for an overview, that on the right for a more detailed view. The tissue was counterstained with haematoxylin. Scale bars represent 50 µm in all cases.
(TIF)
Labeling of TG with isotypes for each marker used in the current study. Isotype control for mouse primary antibody (08-6599), Life Technologies, Darmstadt, Germany; Isotype control for rabbit primary antibody (086199), Life Technologies; Isotype control for goat primary antibody (02-6202), Life Technologies; Isotype control for sheep primary antibody (013-000-002), Jackson Immuno Research Europe Ltd., Suffolk, UK. The name of the isotype used is indicated in each row. The micrograph on the left was taken for an overview, that on the right for a more detailed view. The tissue was counterstained with haematoxylin. Scale bars represent 50 µm in all cases.
(TIF)
Fluorescence co-labeling of isotype controls. The isotypes are labeled with Alexa568 (red), Alexa488 (green) and DAPI (blue). Isotype combinations are indicated above each pictogram. The letter “U” indicates examples of unlabeled neurons, some with lipofuscin. Scale bars represent 50 µm.
(TIF)
The median and interquartile ranges of Marker+ neurons using immunofluorescence.
(DOCX)
The median and interquartile ranges of LAT-ISH+ and Marker+ neurons.
(DOCX)
Acknowledgments
We thank Christine Adrion for help with statistical analysis.
Funding Statement
This study was funded by the German Ministry for Education and Research (IFB 01EO0901) and the Hertie Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Griffin BD, Verweij MC, Wiertz EJ (2010) Herpesviruses and immunity: the art of evasion. VetMicrobiol 143: 89–100. [DOI] [PubMed] [Google Scholar]
- 2. Wilson AC, Mohr I (2012) A cultured affair: HSV latency and reactivation in neurons. Trends Microbiol 20: 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Held K, Derfuss T (2011) Control of HSV-1 latency in human trigeminal ganglia-current overview. JNeurovirol 17: 518–527. [DOI] [PubMed] [Google Scholar]
- 4. Stevens JG, Wagner EK, vi-Rao GB, Cook ML, Feldman LT (1987) RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235: 1056–1059. [DOI] [PubMed] [Google Scholar]
- 5. Thompson RL, Sawtell NM (1997) The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. JVirol 71: 5432–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perng GC, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, et al. (2000) The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J Virol 74: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hill JM, Sedarati F, Javier RT, Wagner EK, Stevens JG (1990) Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174: 117–125. [DOI] [PubMed] [Google Scholar]
- 8. Perng GC, Dunkel EC, Geary PA, Slanina SM, Ghiasi H, et al. (1994) The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol 68: 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, et al. (2000) Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287: 1500–1503. [DOI] [PubMed] [Google Scholar]
- 10. Steiner I, Kennedy PG, Pachner AR (2007) The neurotropic herpes viruses: herpes simplex and varicella-zoster. Lancet Neurol 6: 1015–1028. [DOI] [PubMed] [Google Scholar]
- 11. Steiner I, Kennedy PG (1995) Herpes simplex virus latent infection in the nervous system. JNeurovirol 1: 19–29. [DOI] [PubMed] [Google Scholar]
- 12. Thompson RL, Preston CM, Sawtell NM (2009) De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoSPathog 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sheridan BS, Knickelbein JE, Hendricks RL (2007) CD8 T cells and latent herpes simplex virus type 1: keeping the peace in sensory ganglia. ExpertOpinBiolTher 7: 1323–1331. [DOI] [PubMed] [Google Scholar]
- 14. Mark KE, Wald A, Magaret AS, Selke S, Kuntz S, et al. (2010) Rapidly cleared episodes of oral and anogenital herpes simplex virus shedding in HIV-infected adults. JAcquirImmuneDeficSyndr 54: 482–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wauters O, Lebas E, Nikkels AF (2012) Chronic mucocutaneous herpes simplex virus and varicella zoster virus infections. JAm AcadDermatol 66: e217–e227. [DOI] [PubMed] [Google Scholar]
- 16. Theil D, Derfuss T, Paripovic I, Herberger S, Meinl E, et al. (2003) Latent herpesvirus infection in human trigeminal ganglia causes chronic immune response. AmJ Pathol 163: 2179–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sommer EW, Kazimierczak J, Droz B (1985) Neuronal subpopulations in the dorsal root ganglion of the mouse as characterized by combination of ultrastructural and cytochemical features. Brain Res 346: 310–326. [DOI] [PubMed] [Google Scholar]
- 18. Margolis TP, Imai Y, Yang L, Vallas V, Krause PR (2007) Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J Virol 81: 1872–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kai-Kai MA (1989) Cytochemistry of the trigeminal and dorsal root ganglia and spinal cord of the rat. Comp BiochemPhysiol A Comp Physiol 93: 183–193. [DOI] [PubMed] [Google Scholar]
- 20. Lazarov NE (2002) Comparative analysis of the chemical neuroanatomy of the mammalian trigeminal ganglion and mesencephalic trigeminal nucleus. ProgNeurobiol 66: 19–59. [DOI] [PubMed] [Google Scholar]
- 21. Yang L, Voytek CC, Margolis TP (2000) Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. JVirol 74: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Margolis TP, Dawson CR, LaVail JH (1992) Herpes simplex viral infection of the mouse trigeminal ganglion. Immunohistochemical analysis of cell populations. Invest OphthalmolVisSci 33: 259–267. [PubMed] [Google Scholar]
- 23. Bertke AS, Swanson SM, Chen J, Imai Y, Kinchington PR, et al. (2011) A5-positive primary sensory neurons are nonpermissive for productive infection with herpes simplex virus 1 in vitro. JVirol 85: 6669–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Theil D, Arbusow V, Derfuss T, Strupp M, Pfeiffer M, et al. (2001) Prevalence of HSV-1 LAT in human trigeminal, geniculate, and vestibular ganglia and its implication for cranial nerve syndromes. Brain Pathol 11: 408–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aurelius E, Johansson B, Skoldenberg B, Staland A, Forsgren M (1991) Rapid diagnosis of herpes simplex encephalitis by nested polymerase chain reaction assay of cerebrospinal fluid. Lancet 337: 189–192. [DOI] [PubMed] [Google Scholar]
- 26. Aurelius E, Johansson B, Skoldenberg B, Forsgren M (1993) Encephalitis in immunocompetent patients due to herpes simplex virus type 1 or 2 as determined by type-specific polymerase chain reaction and antibody assays of cerebrospinal fluid. JMedVirol 39: 179–186. [DOI] [PubMed] [Google Scholar]
- 27. Held K, Junker A, Dornmair K, Meinl E, Sinicina I, et al. (2011) Expression of herpes simplex virus 1-encoded microRNAs in human trigeminal ganglia and their relation to local T-cell infiltrates. JVirol 85: 9680–9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang K, Lau TY, Morales M, Mont EK, Straus SE (2005) Laser-capture microdissection: refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella-zoster virus DNA in human trigeminal Ganglia at the single-cell level. JVirol 79: 14079–14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zerboni L, Sobel RA, Lai M, Triglia R, Steain M, et al. (2012) Apparent expression of varicella-zoster virus proteins in latency resulting from reactivity of murine and rabbit antibodies with human blood group a determinants in sensory neurons. JVirol 86: 578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quartu M, Setzu MD, Del FM (1996) trk-like immunoreactivity in the human trigeminal ganglion and subnucleus caudalis. Neuroreport 7: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 31. Quartu M, Geic M, Del FM (1997) Neurotrophin-like immunoreactivity in the human trigeminal ganglion. Neuroreport 8: 3611–3617. [DOI] [PubMed] [Google Scholar]
- 32. Fang X, Djouhri L, McMullan S, Berry C, Okuse K, et al. (2005) trkA is expressed in nociceptive neurons and influences electrophysiological properties via Nav1.8 expression in rapidly conducting nociceptors. JNeurosci 25: 4868–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gaspersic R, Kovacic U, Cor A, Skaleric U (2007) Expression of TrkA receptor for neurotrophins in trigeminal neurons innervating the rat gingivomucosal tissue. NeurosciLett 418: 253–256. [DOI] [PubMed] [Google Scholar]
- 34. Quartu M, Serra MP, Mascia F, Boi M, Lai ML, et al. (2006) GDNF family ligand receptor components Ret and GFRalpha-1 in the human trigeminal ganglion and sensory nuclei. Brain ResBull 69: 393–403. [DOI] [PubMed] [Google Scholar]
- 35. Molliver DC, Wright DE, Leitner ML, Parsadanian AS, Doster K, et al. (1997) IB4-binding DRG neurons switch from NGF to GDNF dependence in early postnatal life. Neuron 19: 849–861. [DOI] [PubMed] [Google Scholar]
- 36. Felipe CD, Gonzalez GG, Gallar J, Belmonte C (1999) Quantification and immunocytochemical characteristics of trigeminal ganglion neurons projecting to the cornea: effect of corneal wounding. EurJPain 3: 31–39. [DOI] [PubMed] [Google Scholar]
- 37. Quartu M, Diaz G, Floris A, Lai ML, Priestley JV, et al. (1992) Calcitonin gene-related peptide in the human trigeminal sensory system at developmental and adult life stages: immunohistochemistry, neuronal morphometry and coexistence with substance P. JChemNeuroanat. 5: 143–157. [DOI] [PubMed] [Google Scholar]
- 38. Alvarez FJ, Morris HR, Priestley JV (1991) Sub-populations of smaller diameter trigeminal primary afferent neurons defined by expression of calcitonin gene-related peptide and the cell surface oligosaccharide recognized by monoclonal antibody LA4. JNeurocytol 20: 716–731. [DOI] [PubMed] [Google Scholar]
- 39. Vang H, Chung G, Kim HY, Park SB, Jung SJ, et al. (2012) Neurochemical properties of dental primary afferent neurons. ExpNeurobiol 21: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flores CM, DeCamp RM, Kilo S, Rogers SW, Hargreaves KM (1996) Neuronal nicotinic receptor expression in sensory neurons of the rat trigeminal ganglion: demonstration of alpha3beta4, a novel subtype in the mammalian nervous system. JNeurosci 16: 7892–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price TJ, Flores CM (2007) Critical evaluation of the colocalization between calcitonin gene-related peptide, substance P, transient receptor potential vanilloid subfamily type 1 immunoreactivities, and isolectin B4 binding in primary afferent neurons of the rat and mouse. J Pain 8: 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bergman E, Carlsson K, Liljeborg A, Manders E, Hokfelt T, et al. (1999) Neuropeptides, nitric oxide synthase and GAP-43 in B4-binding and RT97 immunoreactive primary sensory neurons: normal distribution pattern and changes after peripheral nerve transection and aging. Brain Res 832: 63–83. [DOI] [PubMed] [Google Scholar]
- 43. Zylka MJ (2005) Nonpeptidergic circuits feel your pain. Neuron 47: 771–772. [DOI] [PubMed] [Google Scholar]
- 44. Arbusow V, Derfuss T, Held K, Himmelein S, Strupp M, et al. (2010) Latency of herpes simplex virus type-1 in human geniculate and vestibular ganglia is associated with infiltration of CD8+ T cells. JMedVirol 82: 1917–1920. [DOI] [PubMed] [Google Scholar]
- 45. Rusu MC, Pop F, Hostiuc S, Dermengiu D, Lala AI, et al. (2011) The human trigeminal ganglion: c-kit positive neurons and interstitial cells. AnnAnat 193: 403–411. [DOI] [PubMed] [Google Scholar]
- 46. Hufner K, Horn A, Derfuss T, Glon C, Sinicina I, et al. (2009) Fewer latent herpes simplex virus type 1 and cytotoxic T cells occur in the ophthalmic division than in the maxillary and mandibular divisions of the human trigeminal ganglion and nerve. JVirol 83: 3696–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harbour DA, Hill TJ, Blyth WA (1983) Recurrent herpes simplex in the mouse: inflammation in the skin and activation of virus in the ganglia following peripheral stimulation. JGenVirol 64 (Pt 7): 1491–1498. [DOI] [PubMed] [Google Scholar]
- 48. Feldman LT, Ellison AR, Voytek CC, Yang L, Krause P, et al. (2002) Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. ProcNatlAcadSciUSA 99: 978–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perng GC, Slanina SM, Yukht A, Ghiasi H, Nesburn AB, et al. (1999) Herpes simplex virus type 1 serum neutralizing antibody titers increase during latency in rabbits latently infected with latency-associated transcript (LAT)-positive but not LAT-negative viruses. J Virol 73: 9669–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hilliges M, Astback J, Wang L, Arvidson K, Johansson O (1996) Protein gene product 9.5-immunoreactive nerves and cells in human oral mucosa. AnatRec 245: 621–632. [DOI] [PubMed] [Google Scholar]
- 51. Golden JP, Hoshi M, Nassar MA, Enomoto H, Wood JN, et al. (2010) RET signaling is required for survival and normal function of nonpeptidergic nociceptors. JNeurosci 30: 3983–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quartu M, Setzu MD, Del Fiacco M (1996) trk-like immunoreactivity in the human trigeminal ganglion and subnucleus caudalis. Neuroreport 7: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 53. Sniderhan LF, Garcia-Bates TM, Burgart M, Bernstein SH, Phipps RP, et al. (2009) Neurotrophin signaling through tropomyosin receptor kinases contributes to survival and proliferation of non-Hodgkin lymphoma. Exp Hematol 37: 1295–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Borsani E, Giovannozzi S, Boninsegna R, Rezzani R, Labanca M, et al. (2010) Nitroxidergic system in human trigeminal ganglia neurons: a quantitative evaluation. Acta Histochem 112: 444–451. [DOI] [PubMed] [Google Scholar]
- 55. Price J (1985) An immunohistochemical and quantitative examination of dorsal root ganglion neuronal subpopulations. JNeurosci 5: 2051–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eftekhari S, Salvatore CA, Calamari A, Kane SA, Tajti J, et al. (2010) Differential distribution of calcitonin gene-related peptide and its receptor components in the human trigeminal ganglion. Neuroscience 169: 683–696. [DOI] [PubMed] [Google Scholar]
- 57. Xiao S, McLean J, Robertson J (2006) Neuronal intermediate filaments and ALS: a new look at an old question. Biochim Biophys Acta 1762: 1001–1012. [DOI] [PubMed] [Google Scholar]
- 58. Lariviere RC, Nguyen MD, Ribeiro-da-Silva A, Julien JP (2002) Reduced number of unmyelinated sensory axons in peripherin null mice. JNeurochem 81: 525–532. [DOI] [PubMed] [Google Scholar]
- 59. Xiao S, McLean J, Robertson J (2006) Neuronal intermediate filaments and ALS: a new look at an old question. BiochimBiophysActa 1762: 1001–1012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Labeling of a TG with isotypes of all markers used in the current study. Isotype control for mouse primary antibody (08-6599), Life Technologies, Darmstadt, Germany; Isotype control for rabbit primary antibody (086199), Life Technologies; Isotype control for goat primary antibody (02-6202), Life Technologies; Isotype control for sheep primary antibody (013-000-002), Jackson Immuno Research Europe Ltd., Suffolk, UK. The micrograph on the left was taken for an overview, that on the right for a more detailed view. The tissue was counterstained with haematoxylin. Scale bars represent 50 µm in all cases.
(TIF)
Labeling of TG with isotypes for each marker used in the current study. Isotype control for mouse primary antibody (08-6599), Life Technologies, Darmstadt, Germany; Isotype control for rabbit primary antibody (086199), Life Technologies; Isotype control for goat primary antibody (02-6202), Life Technologies; Isotype control for sheep primary antibody (013-000-002), Jackson Immuno Research Europe Ltd., Suffolk, UK. The name of the isotype used is indicated in each row. The micrograph on the left was taken for an overview, that on the right for a more detailed view. The tissue was counterstained with haematoxylin. Scale bars represent 50 µm in all cases.
(TIF)
Fluorescence co-labeling of isotype controls. The isotypes are labeled with Alexa568 (red), Alexa488 (green) and DAPI (blue). Isotype combinations are indicated above each pictogram. The letter “U” indicates examples of unlabeled neurons, some with lipofuscin. Scale bars represent 50 µm.
(TIF)
The median and interquartile ranges of Marker+ neurons using immunofluorescence.
(DOCX)
The median and interquartile ranges of LAT-ISH+ and Marker+ neurons.
(DOCX)