Abstract
Solid tumors often exhibit simultaneously inflammatory and hypoxic microenvironments. The ‘signal transducer and activator of transcription-3’ (STAT3)-mediated inflammatory response and the hypoxia-inducible factor (HIF)-mediated hypoxia response have been independently shown to promote tumorigenesis through the activation of HIF or STAT3 target genes and to be indicative of a poor prognosis in a variety of tumors. We report here for the first time that STAT3 is involved in the HIF1, but not HIF2-mediated hypoxic transcriptional response. We show that inhibiting STAT3 activity in MDA-MB-231 and RCC4 cells by a STAT3 inhibitor or STAT3 small interfering RNA significantly reduces the levels of HIF1, but not HIF2 target genes in spite of normal levels of hypoxia-inducible transcription factor 1α (HIF1α) and HIF2α protein. Mechanistically, STAT3 activates HIF1 target genes by binding to HIF1 target gene promoters, interacting with HIF1α protein and recruiting coactivators CREB binding protein (CBP) and p300, and RNA polymerase II (Pol II) to form enhanceosome complexes that contain HIF1α, STAT3, CBP, p300 and RNA Pol II on HIF1 target gene promoters. Functionally, the effect of STAT3 knockdown on proliferation, motility and clonogenic survival of tumor cells in vitro is phenocopied by HIF1α knockdown in hypoxic cells, whereas STAT3 knockdown in normoxic cells also reduces cell proliferation, motility and clonogenic survival. This indicates that STAT3 works with HIF1 to activate HIF1 target genes and to drive HIF1-depedent tumorigenesis under hypoxic conditions, but also has HIF-independent activity in normoxic and hypoxic cells. Identifying the role of STAT3 in the hypoxia response provides further data supporting the effectiveness of STAT3 inhibitors in solid tumor treatment owing to their usefulness in inhibiting both the STAT3 and HIF1 pro-tumorigenic signaling pathways in some cancer types.
Keywords: cotranscriptional activation, HIF, hypoxia, STAT3, transcription
INTRODUCTION
A hypoxic microenvironment is common in solid tumors. The hypoxic microenvironment stabilizes hypoxia-inducible transcription factor 1α (HIF1α) and 2α (HIF2α) that are normally degraded under normoxia. The stabilized HIF1α and HIF2α proteins translocate to the nucleus, where they dimerize with aryl hydrocarbon receptor nuclear translocator (ARNT) to form HIF1α/ARNT (HIF1) and HIF2α/ARNT (HIF2) complexes. HIF1 and HIF2 bind to HIF-binding sites and activate genes involved in neovascularization, glycolysis, cellular proliferation and metastasis. Thus, the HIF-mediated hypoxia response is critical for tumor progression by allowing cancer cells to adapt to a low oxygen environment.1–4
In addition to HIF activity, many types of solid tumors such as breast, kidney, lung, prostate and head/neck tumors also exhibit constitutive ‘signal transducer and activator of transcription-3’ (STAT3) activity.5–9 STAT3 is a latent cytoplasmic transcription factor, whose activity is induced by interleukin (IL)-6 and IL-10 family cytokines and growth factors such as epidermal growth factor and platelet-derived growth factor. The binding of cytokines to their receptors activates receptor-associated tyrosine kinases that are often members of the Janus kinase family or cytoplasmic SRC family kinases, whereas the binding of growth factors to their receptors activates intrinsic receptor tyrosine kinases. These activated tyrosine kinases phosphorylate the STAT3 protein at the tyrosine-705 (Y705) residue, which facilitates homodimerization and nuclear translocation of the STAT3 dimer. Nuclear STAT3 dimer then binds directly to STAT3-binding elements on target gene promoters to activate transcription. In normal cells, the activation of STAT3 is tightly controlled by the availability of cytokines or growth factors. In tumor cells, however, STAT3 activators such as IL-6, IL-10 and IL-11 are commonly overexpressed by cancer cells as well as by stromal cells in the solid tumor microenvironment. In addition, some cancers exhibit constitutive tyrosine kinase activity because of activating mutations, gene amplification or fusions of the GF receptors, GF or the cytoplasmic tyrosine kinases. A vast number of studies have established the role of STAT3 as an important oncogene in multiple cancer types by activating genes such as VEGF, MMP2, MMP9, BCL-2, BCL-XL, survivin (also called BIRC5), cyclin D1 and MYC that promote angiogenesis, cell proliferation, survival and metastasis.5–9
Recently hypoxia has been shown to activate STAT3 in several tumor cell lines and animal models.10–14 In addition, STAT3 has been shown to work with HIF1 in activating vascular endothelial growth factor (VEGF) and haptoglobin gene transcription during hypoxia.13,15–19 Given the fact that the hypoxic and inflammatory microenvironments often coexist in solid tumors and that hypoxia activates STAT3 activity, we hypothesized that STAT3 is a general cotranscription factor of the HIF-mediated hypoxia response. Here, the role of STAT3 in the general hypoxia response and the mechanism of STAT3 in contributing to the HIF-mediated hypoxia response were probed.
RESULTS
Hypoxia promotes STAT3 activity in a HIF-independent manner in cancer cell lines
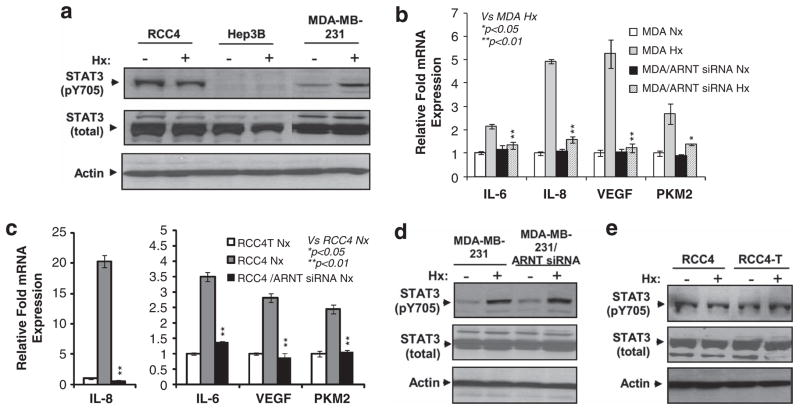
A number of papers report activation of STAT3 by hypoxia in several cell types.10–14 To confirm these results, we investigated STAT3 activity in RCC4 renal clear cell carcinoma cells, MDA-MB-231 breast cancer cells and Hep3B hepatoma cells. These cell lines express both HIF1α and HIF2α and have been extensively used in hypoxia studies.20–25 Both Hep3B and MDA-MB-231 cells have functional pVHL, whereas RCC4 cells are deficient in functional pVHL and exhibit HIF activity even under normoxia.20,26,27 Although all three cell lines expressed similar levels of total STAT3 protein under normoxia and hypoxia (Figure 1a), levels of Y705 phosphorylated STAT3 (pY705STAT3), the transcriptionally active form of STAT3 protein, varied significantly among the cell lines (Figure 1a). RCC4 cells exhibited high levels of pY705STAT3 regardless of oxygen concentration; MDA-MB-231 cells displayed low levels of pY705STAT3 under normoxia while hypoxia increased STAT3 phosphorylation. However, no pY705STAT3 was detected even in hypoxic Hep3B cells (Figure 1a). Although STAT3 activation by hypoxia is well established, different pathways have been reported to activate STAT3 during hypoxia.11,12,28,29 IL-6, IL-8, VEGF and pyruvate kinase, muscle, isoform 2 (PKM2) are reported STAT3 activators8,30–33 and hypoxia-inducible genes.21,34 The correlation between HIF activity and STAT3 activity in RCC4 and MDA-MB-231 cells promoted us to test whether HIF activity is responsible for STAT3 activation in these cell lines. VEGF and PKM2 are well-known HIF target genes; however, it is still not clear whether hypoxic induction of IL-6 and IL-8 is ARNT-dependent. Interestingly, like VEGF and PKM2, IL-6 and IL-8 were HIF target genes in MDA-MB-231 cells (Figure 1b). Furthermore, high levels of these genes in normoxic RCC4 cells were also HIF-/ARNT-dependent, as the levels of these STAT3 activators were significantly reduced in normoxic RCC4T cells or RCC4 cells targeted with ARNT small interfering RNA (siRNA; Figure 1c). In comparison with hypoxic MDA-MB-231 cells, hypoxic MDA-MB-231/ARNT siRNA cells exhibited similar pY705STAT3 levels according to densitometry scanning (Figure 1d). Furthermore, the levels of pY705STAT3 were found to be similar between normoxic RCC4 and normoxic RCC4T cells, and between normoxic RCC4T and hypoxic RCC4T cells (Figure 1e). These data suggested that the HIF-mediated hypoxia response did not have a significant role in hypoxia-mediated STAT3 activation in RCC4 and MDA-MB-231 cells.
Figure 1.
Hypoxia activates STAT3 activity in breast cancer cell line MDA-MB-231. (a) Western blot analysis of total and phosphorylated (Y705) STAT3 protein levels in normoxic and hypoxic (Hx) RCC4 (pVHL-null), Hep3B and MDA-MB-231 cancer cell lines. Beta-actin serves as loading control for this and other western blot analysis in the paper. (b) qPCR analysis of mRNA levels of reported STAT3 activators in normoxic and hypoxic MDA-MB-231 cells or cells targeted with ARNT siRNA. qPCR results were normalized to 18S rRNA expression and calibrated to normoxia control cells; error bars are ±1 s.d. from at least three independent experiments in this and other figures. Two-tail t-tests were performed for this and other studies in the paper with ‘*’ indicating P<0.05 and ‘**’ indicating P<0.01. Controls for the t-tests are indicated in the figures. (c) qPCR analysis of mRNA levels of reported STAT3 activators in normoxic RCC4 cells, RCC4 cells targeted with ARNT siRNA or normoxic pVHL-tranfected RCC4-T cells. (d) Western blot analysis of total and pY705STAT3 protein in normoxic and hypoxic MDA-MB-231 cells or MDA-MB-231 cells targeted with ARNT siRNA. (e) Western blot analysis of total and pY705STAT3 protein in normoxic or hypoxic RCC4 and RCC4 cells reconstituted with pVHL (RCC4-T cells).
STAT3 activity corresponds with maximal hypoxia-inducible gene expression in MDA-MB-231 cells
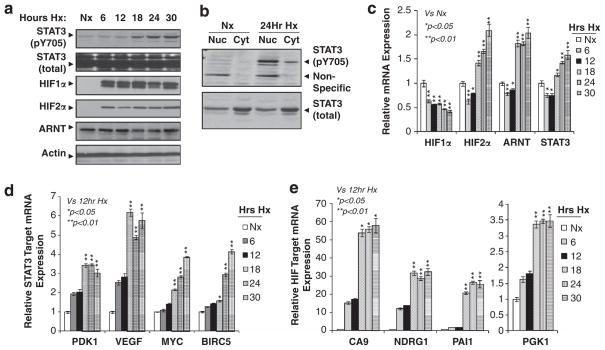
To uncover the role of STAT3 in the hypoxia response of MDA-MB-231 cells, we determined the kinetics of hypoxia-mediated STAT3 activation and hypoxia-mediated gene expression by conducting a time-course experiment. The levels of pY705STAT3 began to increase in cells exposed to hypoxia for 18 h, and this pY705STAT3 level was maintained during the 18–30-h hypoxia treatment (Figure 2a). Importantly, exposure of cells to hypoxia for 24 h dramatically increased the nuclear levels of pY705STAT3 protein (Figure 2b) in spite of similar levels of total nuclear STAT3 between normoxic cells and hypoxic cells (Figure 2b). STAT3 was reported to activate HIF1α gene transcription in several other cell types;35–37 we found that levels of HIF1α mRNA were actually reduced about 50% in 6–30 h hypoxic MDA-MB-231 cells, whereas HIF2α, ARNT and STAT3 mRNA levels were induced in cells treated with 18–30 h hypoxia (Figure 2c). However, HIF1α, HIF2α, ARNT and total STAT3 protein levels were not changed during 6–30 h hypoxia treatment (Figure 2a). Indeed, known STAT3 and HIF common target genes PDK1 and VEGF exhibited upregulation in 6 and 12 h hypoxic MDA-MB-231 cells, and further increased induction in 18–30 h hypoxic cells in which STAT3 was also further activated (Figure 2d), whereas known STAT3 targets MYC and BIRC5 were mainly induced in 18–30 h hypoxic cells in which STAT3 was activated by hypoxia (Figure 2d).16,38–43 CA9, NDRG1, PGK1 and PAI1 genes are known HIF target genes,20,21,34 but these genes have never been reported to be regulated by STAT3. Interestingly, these genes also exhibited higher levels of expression in MDA-MB-231 cells containing higher STAT3 activity (18–30 h hypoxia) than MDA-MB-231 cells having lower STAT3 activity (6–12 h hypoxia) (Figure 2e), demonstrating a possible role of STAT3 in enhancement of the hypoxia response in MDA-MB-231 cells.
Figure 2.
Hypoxia-mediated STAT3 activation corresponds with maximal HIF target gene expression in MDA-MB-231 cells. (a) Western blot analysis of pY705STAT3, total STAT3, HIF1α, HIF2α and ARNT protein in MDA-MB-231 cells cultured under normoxia (Nx) or 6–30 h hypoxia (Hx). (b) Western blot analysis of pY705STAT3 and total STAT3 in nuclear (Nuc) and cytosolic (Cyt) fractions of the MDA-MB-231 cells cultured under Nx or 24 h Hx. (c) qPCR analysis of mRNA levels of HIF1α, HIF2α, ARNT and STAT3 in MDA-MB-231 cells cultured under Nx or 6–30 h of Hx. (d) qPCR analysis of mRNA levels of known STAT3/HIF common targets (PDK1 and VEGF) or STAT3 targets (MYC and BIRC5) in MDA-MB-231 cells under Nx and 6–30 h Hx. (e) qPCR analysis of mRNA levels of known HIF target genes in MDA-MB-231 cells under Nx and 6–30 h Hx.
Optimal hypoxia activation of HIF1 target genes in MDA-MB-231 cells requires STAT3 activity
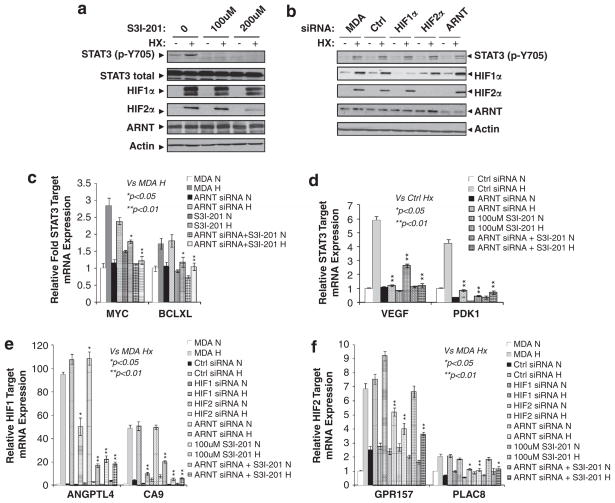
After having observed a correlation between STAT3 activity and optimal hypoxic induction of HIF target genes in MDA-MB-231 cells (Figure 2), we next attempted to determine the role of STAT3 in the HIF-mediated hypoxia response. Both 100 and 200 μm STAT3 inhibitor S3I-201 significantly and similarly reduced the levels of pY705STAT3 in normoxic or 24 h hypoxic MDA-MB-231 cells (Figure 3a). In addition, 200 μM, but not 100 μM STAT3 inhibitor reduced the levels of HIF1α and HIF2α, but not ARNT protein (Figure 3a). Thus, 100 μm STAT3 inhibitor was used to assess the role of STAT3 in the hypoxia response in MDA-MB-231 cells. To determine the role of STAT3 specifically in the HIF/ARNT-mediated hypoxia response, MDA-MB-231 cells were targeted with ARNT siRNA (Figure 3b). To assess the possible specific requirement of STAT3 for HIF1 and/or HIF2 target genes, we used siRNA to reduce HIF1α or HIF2α mRNA levels in MDA-MB-231 cells (Figure 3b).
Figure 3.
Optimal hypoxic induction of HIF1 target genes in MDA-MB-231 cells requires STAT3 activity. (a) Western blot analysis of levels of pY705STAT3, total STAT3, HIF1α, HIF2α and ARNT protein in MDA-MB-231 cells cultured under normoxia (Nx) or 24 h hypoxia (Hx) in the absence or presence of 100 or 200 μM of the STAT3 inhibitor S3I-201. (b) Western blot analysis of indicated protein in Nx or 24 h Hx MDA-MB-231 cells (MDA), or MDA-MB-231 cells targeted with control siRNA or siRNA against HIF1α, HIF2α, or ARNT mRNA. (c) qPCR analysis of mRNA levels of known STAT3 target genes MYC and BCL-XL in Nx or 24 h Hx MDA cells, or cells targeted with ARNT siRNA, or with 100 μM STAT3 inhibitor S3I-201 or both ARNT siRNA and STAT3 inhibitor. (d) qPCR analysis of mRNA levels of known STAT3/HIF common target genes VEGF and PDK1 in Nx or 24 h Hx MDA cells as for Figures 3c, e and f) qPCR analysis of mRNA levels of (e) HIF1- and (f) HIF2-specific target genes in Nx or 24 h Hx MDA cells, siRNA-targeted MDA cells, 100 μM S3I-201-treated MDA cells or ARNT siRNA with STAT3 inhibitor.
Hypoxic induction of MYC and BCL-XL was reduced by STAT3 inhibitor, but not by ARNT siRNA, confirming MYC and BCL-XL as STAT3, but not HIF target genes (Figure 3c). Hypoxic induction of the known STAT3/HIF common target genes VEGF and PDK1 was significantly reduced by either ARNT siRNA or STAT3 inhibitor (Figure 3d), confirming both STAT3 and HIF/ARNT activate these genes under hypoxia. Interestingly, STAT3 inhibition reduced hypoxic induction of HIF1-specifc genes ANGPTL4 and CA9 (Figure 3e), but not HIF2-specific target genes GPR157 and PLAC8 (Figure 3f), indicating a possible role of STAT3 in HIF1 target gene activation during hypoxia. As expected, combined inhibition of STAT3 and ARNT did not further reduce the levels of HIF2 target genes GPR157 and PLAC8 relative to their levels in MDA/ARNT siRNA cells (Figure 3f). Interestingly, combined inhibition of STAT3 and ARNT also exhibited no additional reduction of HIF1 target genes ANGPTL4 and CA9 (Figure 3e) and known STAT3/HIF common targets (Figure 3d), suggesting that STAT3 and ARNT activated these HIF target genes in the same, and not parallel pathway. STAT3 siRNA also significantly reduced hypoxic induction of HIF1-specific target genes PDK1, ANGPTL4 and CA9, and HIF1/2 common target VEGF, but not HIF2-specific targets GPR157 and PLAC8 (data not shown). In summary, these data support a role of STAT3 in the HIF/ARNT-mediated hypoxia response, particularly in the induction of HIF1 target genes in MDA-MB-231 cells.
STAT3 activity is required for hypoxia-induced HIF1 target gene expression in RCC4 cells
To extend the role of STAT3 in the hypoxia response to another cell type, and most importantly to assess the role of STAT3 in global HIF-mediated target gene activation, RCC4 cells were targeted with siRNA against HIF1α, HIF2α, HIF1α + 2α or STAT3 mRNA. Interestingly, knockdown of STAT3 exhibited 30% reduction of HIF1α and HIF2α mRNA (Figure 4a), but not HIFα protein (Figure 4b). HIF1-, HIF2-specific or HIF1/HIF2 common target genes were first identified using Affymetrix Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA, USA). Table 1 lists the top 10 most likely HIF1-specifc, HIF2-specifc and HIF1/HIF2 common target genes based on levels of reduction by HIF1α, HIF2α or HIF1α + 2α siRNAs, as determined by microarray (Table 1, microarray). Quantitative reverse transcription-PCR (qPCR) analysis of the levels of HIF target genes in RNA prepared from normoxic parental, HIF1α, HIF2α, ARNT or STAT3 siRNA-transfected RCC4 cells validated the microarray-identified top 10 HIF target genes. Importantly, STAT3 was found to be required for higher expression of all 20 HIF1 (including HIF1-specific and HIF1/2 common genes), but not the majority of top 10 HIF2-specific target genes (Table 1, qPCR). These results demonstrated for the first time that the STAT3 is specifically important for hypoxic activation of HIF1 target genes in RCC4 cells. Therefore, we focused specifically on STAT3’s role in HIF1 target gene activation.
Figure 4.
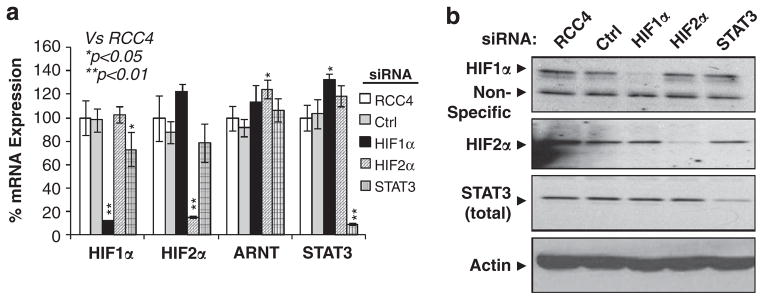
HIF1α, HIF2α or STAT3 siRNA efficiently reduced mRNA and protein expression of HIF1α, HIF2α and STAT3 in RCC4 cells. (a) qPCR analysis of mRNA levels of HIF1α, HIF2α, ARNT and STAT3 in normoxic RCC4 cells and RCC4 cells targeted with HIF1α, HIF2α or STAT3 siRNA. (b) Western blot analysis of HIF1α, HIF2α or STAT3 protein levels in parental or siRNA-transfected RCC4 cells.
Table 1.
Efficient expression of HIF1 target genes in RCC4 cells requires STAT3 activity
| Probe ID | Accession | Gene symbol | Microarray
|
qPCR
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| HIF1 siRNA | HIF2 siRNA | HIF1 + HIF2 siRNA | HIF1 siRNA | HIF2 siRNA | ARNT siRNA | STAT3 siRNA | |||
| HIF1-specific targets | |||||||||
| 213943_at | NM_000474 | TWIST1 | 0.06 | 0.82 | 0.06 | 0.43** | 1.39 | 0.21** | 0.29** |
| 1553036_at | NM_153839 | GPR111 | 0.08 | 0.89 | 0.11 | 0.23** | 0.92 | 0.28** | 0.55* |
| 218995_s_at | NM_001168319 | EDN1 | 0.16 | 0.83 | 0.06 | 0.61** | 0.94 | 0.35** | 0.6** |
| 205199_at | NM_001216 | CA9 | 0.25 | 0.94 | 0.31 | 0.25** | 0.86 | 0.22** | 0.38** |
| 203685_at | NM_000633 | BCL2 | 0.31 | 0.62 | 0.47 | 0.38* | 0.67 | 0.14** | 0.42* |
| 206367_at | NM_001002914 | KCTD11 | 0.39 | 0.93 | 0.54 | 0.31** | 1.28 | 0.3** | 0.37** |
| 200650_s_at | NM_001135239 | LDHA | 0.44 | 1.23 | 0.31 | 0.36** | 0.7 | 0.17** | 0.24** |
| 202095_s_at | NM_001012270 | BIRC5 | 0.46 | 2.14 | 0.47 | 0.51** | 0.92 | 0.09** | 0.64* |
| 202364_at | NM_001008541 | MXI1 | 0.47 | 0.87 | 0.44 | 0.43** | 0.89 | 0.14** | 0.52** |
| 200737_at | NM_000291 | PGK1 | 0.5 | 1.41 | 0.66 | 0.49** | 1.02 | 0.27** | 0.51** |
| HIF2-Specific Targets | |||||||||
| 217287_s_at | NM_004621 | TRPC6 | 0.88 | 0.06 | 0.04 | 0.84 | 0.2** | 0.2** | 0.85 |
| 202627_s_at | NM_000602 | PAI1 | 2.46 | 0.37 | 0.28 | 0.78 | 0.28* | 0.16** | 0.85 |
| 1552846_s_at | NM_152304 | RAB42 | 0.93 | 0.38 | 0.27 | 1.65* | 0.61* | 0.44** | 1.08 |
| 205670_at | NM_004861 | CST | 1.15 | 0.41 | 0.41 | 0.77* | 0.34** | 0.33** | 0.63** |
| 209652_s_at | NM_001207012 | PGF | 2 | 0.41 | 0.35 | 0.62* | 0.09** | 0.08** | 0.54** |
| 230250_at | NM_001109754 | PTPRB | 0.93 | 0.47 | 0.35 | 1.37 | 0.47** | 0.28** | 1.43* |
| 219888_at | NM_003116 | SPAG4 | 1.05 | 0.5 | 0.13 | 1.37* | 0.64** | 0.05** | 1.04 |
| 227970_at | NM_024980 | GPR157 | 0.81 | 0.52 | 0.41 | 0.88 | 0.29** | 0.28** | 1.22 |
| 208286_x_at | NM_001173531 | POU5F1 | 0.95 | 0.63 | 0.62 | 0.52** | 0.2** | 0.26** | 0.55* |
| 205015_s_at | NM_001099691 | TGFA | 2.46 | 0.71 | 0.71 | 1 | 0.38* | 0.23** | 1.12 |
| HIF1/2 common targets | |||||||||
| 202912_at | NM_001124 | ADM | 0.5 | 0.5 | 0.16 | 0.65* | 0.63* | 0.28** | 0.75* |
| 204298_s_at | NM_001178102 | LOX | 0.35 | 0.76 | 0.2 | 0.41** | 0.3** | 0.34** | 0.42** |
| 200632_s_at | NM_001135242 | NDRG1 | 0.7 | 0.62 | 0.29 | 0.54** | 0.31** | 0.16** | 0.34** |
| 221478_at | NM_004331 | BNIP3L | 0.57 | 0.5 | 0.33 | 0.26** | 0.59* | 0.28** | 0.29** |
| 227353_at | NM_152468 | TMC8 | 0.44 | 0.66 | 0.35 | 0.54* | 0.47** | 0.42** | 0.36** |
| 206686_at | NM_002610 | PDK1 | 0.47 | 0.47 | 0.36 | 0.31** | 0.53** | 0.16** | 0.43** |
| 201250_s_at | NM_006516 | GLUT1 | 0.71 | 0.62 | 0.38 | 0.68* | 0.37** | 0.26** | 0.79* |
| 221748_s_at | NM_022648 | TNS1 | 0.35 | 0.66 | 0.41 | 0.48** | 0.39** | 0.42** | 0.44** |
| 208329_at | NM_021635 | PBOV1 | 0.71 | 0.66 | 0.5 | 0.25** | 0.4** | 0.19** | 0.3** |
| 210512_s_at | NM_001025366 | VEGFA | 0.63 | 0.57 | 0.71 | 0.69* | 0.52* | 0.27** | 0.7* |
Abbreviations: ARNT, aryl hydrocarbon receptor nuclear translocator; HIF1, hypoxia-inducible factor-1; STAT3, signal transducer and activator of transcription-3. Vs RCC4.
P<0.05.
P<0.01. DNA microarray was performed to determine global HIF1-, HIF2-specific and HIF1/2 common target genes in RCC4 cells or RCC4 cells targeted with HIF1α, HIF2α or HIF1α + HIF2α siRNA. Listed are the top most likely HIF1-, HIF2-specific and HIF1/HIF2 common target genes (Microarray). HIF target gene specificity was confirmed by qPCR using RNAs from RCC4 cells targeted with HIF1α, HIF2α or ARNT siRNA. In addition, the role of STAT3 in HIF target gene regulation was determined by qPCR using complementary DNA generated from RCC4 cells targeted with STAT3 siRNA (qPCR).
STAT3, not HIF/ARNT, is primarily responsible for the hypoxia-mediated increased recruitment of CBP and p300 coactivators to endogenous HIF1 target promoters
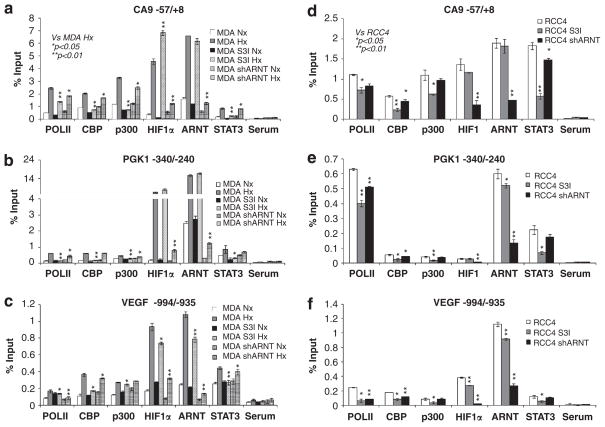
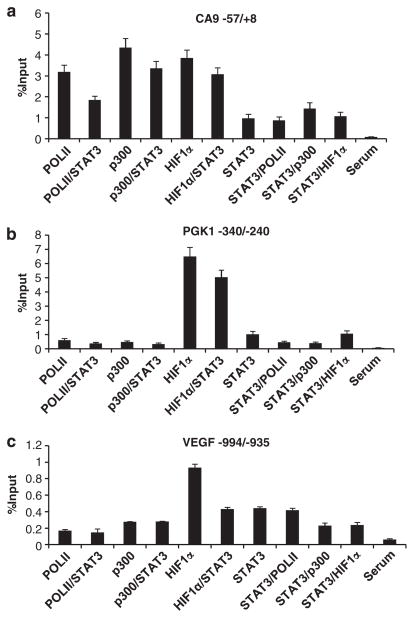
Having showed that STAT3 is required for maximal hypoxic induction of HIF1 target genes, we wanted to determine the molecular mechanism(s) behind STAT3’s contribution to HIF1 target gene activation during hypoxia. STAT3 activates gene transcription by recruiting histone acetylases CBP and p300, and basal components of the RNA polymerase II (Pol II) transcription complex.44–46 Thus, we performed chromatin immunoprecipitation (ChIP) experiments to assess the binding of CBP, p300, the largest subunit of RNA Pol II, HIF1α and STAT3 to HIF1 target gene promoters in 24 h normoxic or hypoxic parental MDA-MB-231 cells with or without 100 μM S3I-201. We selected the VEGF promoter as a positive control, as VEGF is regulated by both HIF1 and STAT3.13,15–18 CA9 and PGK1 genes were chosen as they are established HIF1 target genes, but they are newly identified STAT3 targets (Figure 2 and Table 1). As expected, hypoxia increased binding of HIF1α, ARNT, STAT3, CBP, p300 and Pol II on CA9, PGK1 and VEGF promoter regions around the reported HREs at −6 for CA9, at −233 and −224 for PGK1 and at −975 for VEGF in MDA-MB-231 cells (Figures 5a–c). However, no significant binding of these factors to control regions of exon 8 for PGK1 and exon 3 for VEGF was detected (data not shown). S3I-201 significantly decreased STAT3 binding to all three promoters in hypoxic cells (Figures 5a–c). HIF1α and ARNT binding to the VEGF (Figure 5c), but not CA9 and PGK1 promoters (Figures 5a and b) was reduced slightly in S3I-201-treated hypoxic MDA-MB-231 cells (Figure 5c). The most significant and consistent change in response to treatment with S3I-201 was the reduction or complete blockage of hypoxia-mediated increased binding of CBP, p300 and Pol II to all three promoters (Figures 5a–c), providing evidence for a critical role of STAT3 in hypoxic recruitment of CBP, p300 and Pol II to these genes. HIF/ARNT has been reported to activate hypoxia-inducible genes by recruiting CBP and p300 via N-terminal and C-terminal transactivation domains.47–49 However, hypoxic MDA-MB-231/ARNT small hairpin RNA (shRNA) cells exhibited only a slight reduction in CBP, p300 and Pol II binding to CA9, PGK1 and VEGF promoters in spite of significant reduction of HIF1α and ARNT binding (Figures 5a–c), demonstrating that STAT3, not HIF/ARNT, is the primary recruiter of CBP and p300 to HIF1 target genes CA9, PGK1 and VEGF promoters, whereas both STAT3 and HIF/ARNT contribute to Pol II recruitment. To confirm these results in a different cell type, RCC4 cells were targeted with ARNT shRNA or S3I-201 (Figures 5d–f). Again, STAT3 inhibitor was more effective in reducing CBP and p300 binding to CA9, PGK1 and VEGF promoters than ARNT shRNA, whereas both STAT3 and ARNT were equally important for Pol II recruitment (Figures 5d–f). In summary, these results demonstrate that STAT3 binds to the endogenous HIF1 target genes in an ARNT-independent fashion and participates in the HIF1-mediated hypoxia response by recruiting histone acetylases CBP and p300 and Pol II to HIF1 target gene promoters.
Figure 5.
STAT3, but not HIF1α/ARNT is primarily responsible for recruiting CBP and p300 coactivators to the endogenous HIF1 target promoters. ChIP analysis of Pol II, CBP, p300, HIF1α, ARNT and STAT3 binding to the HIF1 target promoters around the reported HREs of the CA9 (a), PGK1 (b) and VEGF genes (c) in MDA-MB-231 cells under normoxic (Nx) or 24 h hypoxic (Hx) conditions or MDA cells treated with STAT3 inhibitor, or with ARNT stably knocked down. Co-precipitated DNA is presented as percent DNA compared with input controls and background level is quantified by DNA pulled down using non-reactive serum (Serum). Similar ChIP was performed in normoxic RCC4 cells, or RCC4 cells targeted with STAT3 inhibitor or ARNT stably knocked down for CA9 promoter (d), PGK1 promoter (e) and VEGF promoter (f).
STAT3 physically interacts with HIF1α, but not with HIF2α
The above ChIP data demonstrated the binding of HIF1α, STAT3, CBP, p300 and Pol II to HIF1 target gene promoters (Figure 5). In addition, HIF1α (but not HIF2α) and STAT3 exhibited functional cooperation in activating HIF1 target genes (Figure 3 and Table 1). These data support a hypothesis that HIF1α and STAT3 are components of the HIF1-specific target gene ‘enhanceosome’ complex. To test this hypothesis, we first assessed the physical interaction between STAT3 and HIFα proteins. STAT3 interaction with HIF1α or HIF2α was first tested using an overexpression system in which Flag-tagged HIF1α but not Flag-tagged HIF2α protein co-precipitated HA-tagged STAT3 protein in HEK293T cells transfected with STAT3C-HA (constitutively active STAT3 with HA-tag) alone for control, STAT3C-HA with human HIF1αTM-Flag (triple mutation of HIF1α with the Flag tag), or STAT3C-HA with human HIF2αTM-Flag (Figure 6a). To confirm STAT3/HIF1α interaction under physiological conditions, endogenous STAT3 protein in normoxic RCC4 cells was precipitated by STAT3 antibodies and protein-A/protein-G beads (Figure 6b, STAT3-IP lane); interestingly, STAT3 only co-precipitated HIF1α, but not HIF2α protein (Figure 6b, STAT3-IP lane).
Figure 6.
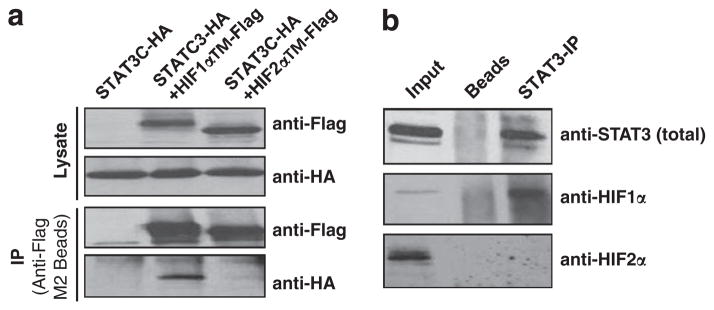
STAT3 protein specifically interacts HIF1α, but not HIF2α protein. (a) Western blot analysis of Flag-tagged HIF1α and HIF2α proteins (anti-Flag) or HA-tagged STAT3 (anti-HA) in the lysates (Lysate) or anti-Flag M2 bead immunoprecipitated materials (IP) from 293T cells transfected with STAT3C-HA (HA-tagged constitutively active STAT3), STAT3C-HA +HIF1αTM-Flag (Flag tagged HIF1α with triple mutation to make HIF1α stable and active under normoxia), or STAT3C-HA +HIF2αTM-Flag. (b) Western blot analysis of STAT3, HIF1α or HIF2α protein in the RCC4 lysates (input) or IP materials from the protein-A/protein-G beads (beads) or IP materials using anti-STAT3 pY705 antibody and protein-A/protein-G beads.
STAT3, HIF1α, p300 and Pol II form a transcriptional complex on endogenous HIF1 target promoters
We next tested whether there were physical interactions among STAT3, HIF1α, p300 and Pol II on endogenous HIF1 target gene promoters in 24 h hypoxia-treated MDA-MB-231 cells using ChIP/Re-chromatin Immunoprecipitation (Re-ChIP). Analysis of ChIP/ReChIP precipitated DNA by qPCR for CA9, PGK1 and VEGF promoters showed a high percentage of Pol II bound to the same physical promoter in proximity to STAT3, as was p300 with STAT3 and HIF1α with STAT3 (Figure 7). These results were confirmed by the reciprocal ChIP/ReChIP of STAT3 with Pol II, STAT3 with p300 and STAT3 with HIF1α (Figure 7). These results provided strong evidence that STAT3, HIF1α, p300 and Pol II are components of a multifactorial transcription complex binding to HIF1 target gene promoters during hypoxia.
Figure 7.
STAT3, HIF1α, p300 and Pol II form a transcriptional complex on the endogenous HIF1 target promoters. ChIP/ReChIP analysis of STAT3, HIF1α, Pol II and P300 binding to the endogenous HIF1 target gene promoters CA9 (a), PGK1 (b) and VEGF (c) in 24 h hypoxic MDA-MB-231 cells. Antibodies used for the primary and secondary immunoprecipitation are listed as first/second. The co-precipitated DNA is presented in comparison with pull-down of each individual factor and non-reactive serum controls (Serum).
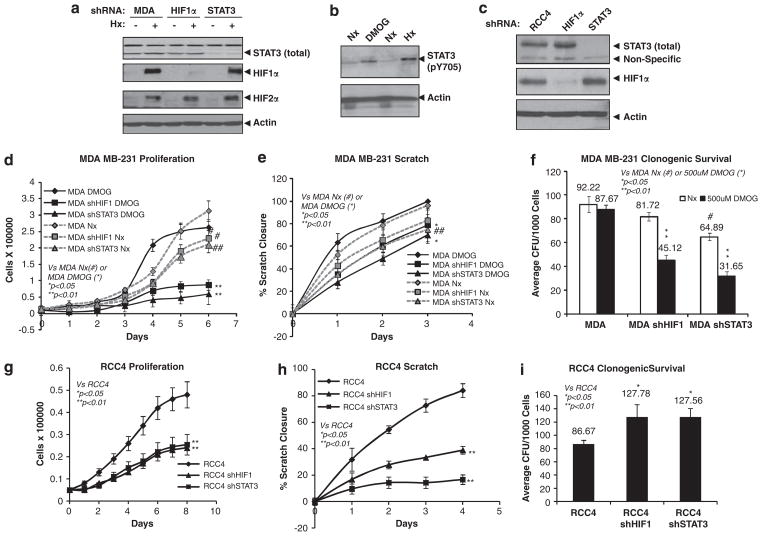
STAT3 knockdown phenocopies HIF1α knockdown in tumorigenic properties of MDA-MB-231 and RCC4 cells in vitro
Next we investigated whether STAT3 inhibition would phenocopy HIF1 inhibition in functional assays. To test this hypothesis, MDA-MB-231 cells were targeted with STAT3 or HIF1α shRNAs to reduce STAT3 or HIF1α protein levels (Figure 8a). To activate HIF and STAT3 activity for functional assays, parental, MDA-MB-231/HIF1α shRNA or MDA-MB-231/STAT3 shRNA cell lines were cultured in media with 500 μM dimethyloxaloylglycine (DMOG). Importantly, like hypoxia, the hypoxia mimic DMOG induced STAT3 activation (Figure 8b). In comparison with parental MDA-MB-231 cells without DMOG, MDA-MB-231/HIF1α shRNA cells exhibited slight reduction in proliferation, migration and clonogenic survival (Figures 8d–e), consistent with the fact that normoxic MDA-MB-231 cells did not express HIF1α protein (Figure 3a). Interestingly, reduction of STAT3 also weakly reduced cell proliferation and migration, but strongly decreased clonogenic survival (Figure 8e). Compared with MDA-MB-231 cells without DMOG treatment, DMOG-treated parental MDA-MB-231 cells exhibited quicker proliferation during day 3–5 (Figure 8d), but had no significant changes in wound healing and clonogenic survival (Figures 8e and f). Furthermore, identical to MDA-MB-231/HIF1α shRNA cells, DMOG-treated MDA-MB-231/STAT3 shRNA cells exhibited significantly decreased cell proliferation (Figure 8d), wound healing (Figure 8e) and clonogenic survival (Figure 8f), demonstrating that the HIF1/STAT3-mediated hypoxia response is important for maintaining cell proliferation, migration and survival of these cells under hypoxia.
Figure 8.
STAT3 knockdown phenocopies HIF1α knockdown in tumorigenic properties of MDA-MB-231 and RCC4 cells in vitro. (a) Western blot analysis of total STAT3, HIF1α and HIF2α in normoxic (Nx) or hypoxic (Hx) MDA-MB-231 cells or MDA-MB-231 cells with stably knocked down HIF1α or STAT3 protein. (b) Western blot analysis of pY705STAT3 in Nx or 24 h Hx MDA-MB-231 cells or Nx cells treated with or without 500 μM DMOG. (c) Western blot analysis of HIF1α or total STAT3 protein in RCC4 cells or RCC4 cells with HIF1α or STAT3 shRNA. (d) Proliferation of MDA-MB-231 cells, MDA/shHIF1α and MDA/shSTAT3 cells treated with or without 500 μM DMOG. (e) Motility of MDA-MB-231, MB-231/shHIF1 and MDA-MB-231/shSTAT3 cells grown with or without 500 μM DMOG was measured and presented as percent closure of a scratch assay. (f) Percent survival of colony-forming units of MDA-MB-231, MDA-MB-231/shHIF1 and MDA-MB-231/shSTAT3 cells grown in the presence or absence of 500 μM DMOG in a clonogenic survival assay. (g) Proliferation of RCC4, RCC4/shHIF1 and RCC4/shSTAT3 cells under Nx. (h) Motility of Nx RCC4, RCC4/shHIF1 and RCC4/shSTAT3 cells was measured and presented as percent closure in a scratch assay. (i) Percent survival of colony-forming units of RCC4, RCC4/shHIF1 and RCC4/shSTAT3 cells in a clonogenic survival assay.
These functional assays were also conducted in normoxic RCC4 cells that exhibited constitutive HIF1α and STAT3 activity. Reduction of STAT3 and HIF1α activity was achieved by shRNA targeting HIF1α or STAT3 mRNA (Figure 8c). Again, HIF1α or STAT3 shRNAs greatly and similarly reduced RCC4 cell proliferation (Figure 8g) and motility (Figure 8h). However, clonogenic survival of RCC4 cells was similarly enhanced by HIF1α or STAT3 shRNA (Figure 8i), demonstrating HIF1α or STAT3 activity in RCC4 cells actually decreases clonogeneic survival, consistent with HIF1α’s tumor suppressor role in the VHL-deficient RCC4 cells.50
DISCUSSION
Cytokines and growth factors are well-known activators of the inflammatory response mediator STAT3.5–9,38 Recent studies show that hypoxia also activates STAT3 via different mechanisms, including activation of SRC kinase in pancreatic carcinoma BxPC3 and PANC1 cells,28 activation of Janus kinase 2 in lung carcinoma IGR-Heu cells,11 activation of nuclear factor-κβ and activator protein-1 (AP1) and IL-6 expression in several head and neck carcinoma cells,29 or increased levels of reactive oxygen species in the human ovarian A2780 cancer cells.12 However, it is not clear whether all or some of these pathways are activated in specific cancer cell types. We found that hypoxia activates STAT3 in MDA-MB-231 cells, but not in Hep3B cells. In addition, we show that hypoxic activation of STAT3 in MDA-MB-231 cells and constitutive STAT3 activity in RCC4 cells is ARNT independent (Figure 1). However, it is still possible that STAT3 activators induced by hypoxia could activate STAT3 in the absence of SRC or other STAT3 activators.
Several previous reports indicate that STAT3 functions as an activator of HIF1α gene transcription and/or a promoter of HIF1α protein stability.35–37,51 However, other studies reported that inhibition of STAT3 activity has no effect on the levels of HIF1α mRNA and/or protein. For example, the STAT3 inhibitor STATTIC inhibits STAT3 phosphorylation, but not the levels of HIF1α protein in hypoxic head and neck cancer UM-SCC-17B and SC-19 cells;52 hypoxia activates Janus kinase 2 and STAT3 in pulmonary arterial smooth muscle cells, but STAT3 siRNA does not change the levels of HIF1α protein in the hypoxic PASM cells.53 Furthermore, c-Src inhibitor saracatinib significantly reduces p-STAT3, but has no effects on the HIF1α protein levels in hypoxic kidney cancer Caki-1 cells.54 We also show that Hep3B cells express high levels of HIF1α mRNA20 although STAT3 is not active in Hep3B cells (Figure 1a). In addition, we found that STAT3 shRNA (Figure 8a) or 100 μM STAT3 inhibitor S3I-201 (Figure 3a) does not reduce the levels of HIF1α mRNA and/or protein in MDA-MB-231 cells. In addition, STAT3 target genes STAT3, MYC and BIRC5, but not HIF1α (Figures 2c and d) were induced in 18–30 h hypoxia-treated MDA-MB-231 cells. In contrast, we found that STAT3 is a weak activator of the HIF1α gene in RCC4 cells. Furthermore, we found that STAT3 activates the HIF1α and HIF2α promoter reporters in HEK293T cells (Supplementary Figure S1). Thus, these different findings are likely due to cell-type differences whereby reduction of STAT3 activity is not enough to reduce HIF1α gene expression in some cell types in which HIF1α gene expression could be maintained by other HIF1α gene activators such as nuclear factor-κβ, PI3K/AKT and activator protein-1.55
VEGF and haptoglobin are known STAT3 target genes16,56,57 and hypoxia inducible genes.58–60 It makes sense that STAT3 and HIF1α cooperatively activate VEGF and haptoglobin genes during hypoxia.13,15,17–19 We confirmed the activation of VEGF by HIF1α and STAT3 in MDA-MB-231 and RCC4 cells (Figure 2 and Table 1). More importantly, for the first time, we establish the role of STAT3 as a general cotranscriptional activator to a larger set of HIF target genes that have never been reported as STAT3 targets (Table 1).
Although the role of STAT3 in the hypoxia response was also probed previously,11,14 these studies used IGR-Heu lung and U87 glioblastoma cells in which knockdown of STAT3 also significantly reduced HIF1α mRNA and protein.11,14 Thus, these studies did not separate STAT3’s role in regulating HIF1α gene expression from its role as a HIF coactivator during hypoxia. In contrast, we studied the role of STAT3 in the hypoxia response by using cell lines that have reduced STAT3 activity, but normal levels of HIF1α, HIF2α and ARNT. In addition, by using siRNAs targeting HIF1α, HIF2α or both HIFα mRNAs, we specifically studied the role of STAT3 in the HIF-mediated hypoxia response. Most importantly, we found for the first time that STAT3 activity is required for HIF1, but not HIF2 target gene activation in RCC4 and MDA-MB-231 cells.
Although increased binding of STAT3, HIF1α, p300 and RNA Pol II to the VEGF promoter during hypoxia has been reported,13,15 we for the first time show that STAT3 is primarily responsible for p300 and CBP recruitment, whereas both STAT3 and HIF1α/ARNT recruit RNA Pol II to the VEGF promoter. In addition, we demonstrate the role of STAT3 in recruiting CBP, p300 and RNA Pol II to two novel STAT3 target genes CA9 and PGK1 (Figures 6 and 7).
Although STAT3 was found to be involved in the hypoxia response, STAT3 clearly has HIF-independent activity. For example, STAT3 specifically activates MYC and BCL-XL (Figures 2c and 3c), HIF2α, ARNT and STAT3 in MDA-MB-231 cells (Figure 2b). This is consistent with functional assays demonstrating that inhibition of STAT3 in normoxic MDA-MB-231 cells reduces cell survival (Figure 8).
Our target gene and functional studies demonstrate integration between the inflammatory STAT3 pathway and the hypoxic HIF1 pathway under hypoxia. Our findings suggest that pharmacological inhibition of STAT3 activity could prove anti-tumorigenic and especially effective in some solid tumors in which the HIF1-mediated hypoxia response has a critical role in cancer progression, whereas anti-STAT3 treatment could be less effective in cells such as RCC4 in which HIF1 could function as a tumor suppressor.
MATERIALS AND METHODS
Cell culture
HEK293T, RCC4, RCC4T, Hep3B and MDA-MB-231 cells were grown in high-glucose Dulbecco’s modified Eagle medium (Hyclone, Logan, UT, USA) with 10% fetal bovine serum. For hypoxia, 25 mM hydroxyethyl piperazineethanesulfonic acid was added to growth media and cells were incubated at 21 or 1.2% O2 for 24 h (or otherwise noted). Phosphorylation of Y705-STAT3 was inhibited by adding 100 μM S3I-201 (sc-204304; Santa Cruz, Dallas, TX, USA) to media 1 h before hypoxia treatment.
Knockdown of HIFα, ARNT and STAT3 mRNAs using siRNAs or shRNAs
Control or siRNAs specific for human HIF1α, HIF2α and ARNT mRNAs were described previously.61 STAT3 siRNA (siSGENOME M-003544-02) was purchased from Dharmacon (Walthan, MA, USA). Transient knockdown of mRNA in RCC4 or MDA-MB-231 cells were performed as described.61 Stable knockdown of HIF1α, ARNT or STAT3 mRNA in RCC4 and MDA-MB-231 cells were achieved by pLKO.1 lentiviruses expressing shRNAs targeting mRNAs of HIF1α (TRCN0000003810), ARNT (TRCN0000003816) or STAT3 (TRCN0000020840).
DNA constructs
pcDNA3.1hHIF1αTM-Flag (triple mutations of P402A/P577A/N813A of human HIF1α protein with Flag-tag) and pcDNA3.1hHIF2αTM-Flag have been described.61 The pRc/CMV/STAT3C-Flag construct was purchased from Addgene (Cambridge, MA, USA; plasmid 8722, deposited by Jim Darnell) and used as a template to make pcDNA3.1 STAT3C-HA (HA-tagged at STAT3 C-terminus).
RNA preparation, microarray and qPCR
RNA was isolated from cells using the RNeasy Plus mini kit (Qiagen, Germantown, MD, USA). RNA was reverse-transcribed using the iSCRIPT Advanced complementary DNA synthesis kit (Bio-Rad, Hercules, CA, USA). Levels of mRNA were quantified by Sybr Green qPCR using the CFX384 Real-Time System (Bio-Rad). All primer sets designed to measure target gene mRNA levels or used in ChIP were validated for their specificity and amplification efficiency (85–110%) using melt curve analysis, qPCR product sequencing and standard dilution analysis. qPCR results were normalized using 18S rRNA. Results were the average of a minimum of three independent experiments performed in triplicate. Microarray analysis of HIF target gene specificity in RCC4 cells was basically identical to HIF target gene analysis in Hep3B cells, as described.61
Protein analysis
Typically total cell lysates were used for western blot analysis. To detect nuclear and cytoplasmic protein, the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo, Walthan, MA, USA, 78833) was used. Protein concentration was determined by BCA protein assay kit (Pierce, Walthan, MA, USA, 23223 and 23224) and the same amount of protein was loaded in western blot procedures, as described below. Western blot analysis was performed using standard protocols with the following primary antibodies: anti-HIF1α monoclonal antibodies (NB 100–105; Novus Biologicals, Littleton, CO, USA), anti-HIF2α polyclonal antibodies (NB100–122; Novus Biological), anti-ARNT mAb (NB 100–124; Novus Biological), anti-STAT3 (total) pAb (K-15, sc-483; Santa Cruz), anti-Flag M2 mAb (F-3165; Sigma-Aldrich, St Louis, MO, USA), anti-HA (MMS-101P, Covance, Alice, TX, USA) and anti-beta Actin (SC-1616; Santa Cruz). Anti-STAT3 western blot was blocked in 5% bovine serum albumin in Tris buffered saline with Tween 20 (TBST) and probed using anti-pSTAT3 (pY705) mAb (B-7, sc-8059; Santa Cruz).
STAT3/HIFα protein interaction
For in vitro STAT3 and HIF1α (or HIF2α) protein interaction, HEK293T cells were transfected with HA-tagged constitutively active STAT3 alone, or STAT3C-HA with Flagged constitutively active HIF1α or HIF2α. Lysates from transfected 293T cells were immunoprecipitated with anti-Flag M2 beads (Sigma-Aldrich) to pull down Flag-tagged HIF1α or HIF2α, in which HA-tagged STAT3 was detected. For endogenous HIF/STAT3 protein interaction, nuclear extracts from RCC4 cells were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit in which STAT3 antibody and protein-A/protein-G beads were added.
ChIP and ReChIP
ChIP assays were performed as described.62 Anti-HIF1α (NB 100-134B3; Novus Biologicals), anti-ARNT (NB 100–110; Novus Biologicals), anti-CBP (A-22, sc-369; Santa Cruz), anti-p300 (N-15, sc-584; Santa Cruz), anti-STAT3 (K-15, sc-483; Santa Cruz) and anti-Pol II (H-224, SC-9001, Santa Cruz) antibodies were used for protein–DNA complex precipitation, whereas rabbit preimmune serum served as a control. DNA from input or immunoprecipitated samples was assayed using Sybr Green-based quantitative PCR with specific primers designed to amplify the CA9, PGK1 or VEGF promoter around the reported HREs or in exons for negative controls. ChIP/ReChIP: ChIP was performed as above, binding complexes from the first immunoprecipitation were eluted from the sepharose beads using Re-ChIP buffer as described.61 The eluted protein–DNA complexes were diluted in radioimmunoprecipitation buffer and resubjected to ChIP using a different antibody.
In vitro tumorigenic assays
Proliferation assays, scratch assays and clonogenic survival assays were performed for MDA-MB-231 cells or MDA/HIF1α shRNA or MDA/STAT3 shRNA cells with or without 500 μM DMOG under normoxia. The same experiments were carried out in RCC4 cells or RCC4/HIF1α shRNA or RCC4/STAT3 shRNA cells under normoxia, as described.61
Supplementary Material
Acknowledgments
This work was supported by grants from the National Cancer Institute (RO1CA134687, Hu) and Cancer Leagues of Colorado (Hu).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Finger EC, Giaccia AJ. Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev. 2010;29:285–293. doi: 10.1007/s10555-010-9224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelin WG., Jr Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 7.Horiguchi A, Oya M, Shimada T, Uchida A, Marumo K, Murai M. Activation of signal transducer and activator of transcription 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J Urol. 2002;168:762–765. [PubMed] [Google Scholar]
- 8.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 9.Horvath CM. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci. 2000;25:496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 10.Lee MY, Joung YH, Lim EJ, Park JH, Ye SK, Park T, et al. Phosphorylation and activation of STAT proteins by hypoxia in breast cancer cells. Breast. 2006;15:187–195. doi: 10.1016/j.breast.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Noman MZ, Buart S, Van Pelt J, Richon C, Hasmim M, Leleu N, et al. The cooperative induction of hypoxia-inducible factor-1 alpha and STAT3 during hypoxia induced an impairment of tumor susceptibility to CTL-mediated cell lysis. J Immunol. 2009;182:3510–3521. doi: 10.4049/jimmunol.0800854. [DOI] [PubMed] [Google Scholar]
- 12.Selvendiran K, Bratasz A, Kuppusamy ML, Tazi MF, Rivera BK, Kuppusamy P. Hypoxia induces chemoresistance in ovarian cancer cells by activation of signal transducer and activator of transcription 3. Int J Cancer. 2009;125:2198–2204. doi: 10.1002/ijc.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung JE, Lee HG, Cho IH, Chung DH, Yoon SH, Yang YM, et al. STAT3 is a potential modulator of HIF-1-mediated VEGF expression in human renal carcinoma cells. Faseb J. 2005;19:1296–1298. doi: 10.1096/fj.04-3099fje. [DOI] [PubMed] [Google Scholar]
- 14.Kang SH, Yu MO, Park KJ, Chi SG, Park DH, Chung YG. Activated STAT3 regulates hypoxia-induced angiogenesis and cell migration in human glioblastoma. Neurosurgery. 2010;67:1386–1395. doi: 10.1227/NEU.0b013e3181f1c0cd. discussion 95. [DOI] [PubMed] [Google Scholar]
- 15.Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, et al. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 16.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 17.Rathinavelu A, Narasimhan M, Muthumani P. A novel regulation of VEGF expression by HIF-1alpha and STAT3 in HDM2 transfected prostate cancer cells. J Cell Mol Med. 2012;16:1750–1757. doi: 10.1111/j.1582-4934.2011.01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santra M, Santra S, Zhang J, Chopp M. Ectopic decorin expression up-regulates VEGF expression in mouse cerebral endothelial cells via activation of the transcription factors Sp1, HIF1alpha, and Stat3. J Neurochem. 2008;105:324–337. doi: 10.1111/j.1471-4159.2007.05134.x. [DOI] [PubMed] [Google Scholar]
- 19.Oh MK, Park HJ, Kim NH, Park SJ, Park IY, Kim IS. Hypoxia-inducible factor-1alpha enhances haptoglobin gene expression by improving binding of STAT3 to the promoter. J Biol Chem. 2011;286:8857–8865. doi: 10.1074/jbc.M110.150557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu CJ, Sataur A, Wang L, Chen H, Simon MC. The N-terminal transactivation domain confers target gene specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha. Mol Biol Cell. 2007;18:4528–4542. doi: 10.1091/mbc.E06-05-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, et al. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. Faseb J. 2004;18:1462–1464. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 24.Bando H, Toi M, Kitada K, Koike M. Genes commonly upregulated by hypoxia in human breast cancer cells MCF-7 and MDA-MB-231. Biomed Pharmacother. 2003;57:333–340. doi: 10.1016/s0753-3322(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 25.Indelicato M, Pucci B, Schito L, Reali V, Aventaggiato M, Mazzarino MC, et al. Role of hypoxia and autophagy in MDA-MB-231 invasiveness. J Cell Physiol. 2010;223:359–368. doi: 10.1002/jcp.22041. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 27.Zia MK, Rmali KA, Watkins G, Mansel RE, Jiang WG. The expression of the von Hippel-Lindau gene product and its impact on invasiveness of human breast cancer cells. Int J Mol Med. 2007;20:605–611. [PubMed] [Google Scholar]
- 28.Pham NA, Magalhaes JM, Do T, Schwock J, Dhani N, Cao PJ, et al. Activation of Src and Src-associated signaling pathways in relation to hypoxia in human cancer xenograft models. Int J Cancer. 2009;124:280–286. doi: 10.1002/ijc.23912. [DOI] [PubMed] [Google Scholar]
- 29.Squarize CH, Castilho RM, Sriuranpong V, Pinto DS, Jr, Gutkind JS. Molecular cross-talk between the NFkappaB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia. 2006;8:733–746. doi: 10.1593/neo.06274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartoli M, Platt D, Lemtalsi T, Gu X, Brooks SE, Marrero MB, et al. VEGF differentially activates STAT3 in microvascular endothelial cells. FASEB J. 2003;17:1562–1564. doi: 10.1096/fj.02-1084fje. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Byfield G, Jiang Y, Smith GW, McCloskey M, Hartnett ME. VEGF-mediated STAT3 activation inhibits retinal vascularization by down-regulating local erythropoietin expression. Am J Pathol. 2012;180:1243–1253. doi: 10.1016/j.ajpath.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piperi C, Samaras V, Levidou G, Kavantzas N, Boviatsis E, Petraki K, et al. Prognostic significance of IL-8-STAT-3 pathway in astrocytomas: correlation with IL-6, VEGF and microvessel morphometry. Cytokine. 2011;55:387–395. doi: 10.1016/j.cyto.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Hu CJ, Iyer S, Sataur A, Covello KL, Chodosh LA, Simon MC. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol Cell Biol. 2006;26:3514–3526. doi: 10.1128/MCB.26.9.3514-3526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niu G, Briggs J, Deng J, Ma Y, Lee H, Kortylewski M, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1alpha RNA expression in both tumor cells and tumor-associated myeloid cells. Mol Cancer Res. 2008;6:1099–1105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 2007;21:1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D’Angeli L, Bartoli A, et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging. 2010;2:823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Grad JM, Zeng XR, Boise LH. Regulation of Bcl-xL: a little bit of this and a little bit of STAT. Curr Opin Oncol. 2000;12:543–549. doi: 10.1097/00001622-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Horiguchi A, Asano T, Kuroda K, Sato A, Asakuma J, Ito K, et al. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. Br J Cancer. 2010;102:1592–1599. doi: 10.1038/sj.bjc.6605691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carraro F, Pucci A, Pellegrini M, Pelicci PG, Baldari CT, Naldini A. p66Shc is involved in promoting HIF-1alpha accumulation and cell death in hypoxic T cells. J Cell Physiol. 2007;211:439–447. doi: 10.1002/jcp.20951. [DOI] [PubMed] [Google Scholar]
- 44.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 45.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 46.Schuringa JJ, Schepers H, Vellenga E, Kruijer W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 2001;495:71–76. doi: 10.1016/s0014-5793(01)02354-7. [DOI] [PubMed] [Google Scholar]
- 47.Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ebert BL, Bunn HF. Regulation of transcription by hypoxia requires a multiprotein complex that includes hypoxia-inducible factor 1, an adjacent transcription factor, and p300/CREB binding protein. Mol Cell Biol. 1998;18:4089–4096. doi: 10.1128/mcb.18.7.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruas JL, Berchner-Pfannschmidt U, Malik S, Gradin K, Fandrey J, Roeder RG, et al. Complex regulation of the transactivation function of hypoxia-inducible factor-1 alpha by direct interaction with two distinct domains of the CREB-binding protein/p300. J Biol Chem. 2010;285:2601–2609. doi: 10.1074/jbc.M109.021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jung JE, Kim HS, Lee CS, Shin YJ, Kim YN, Kang GH, et al. STAT3 inhibits the degradation of HIF-1alpha by pVHL-mediated ubiquitination. Exp Mol Med. 2008;40:479–485. doi: 10.3858/emm.2008.40.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adachi M, Cui C, Dodge CT, Bhayani MK, Lai SY. Targeting STAT3 inhibits growth and enhances radiosensitivity in head and neck squamous cell carcinoma. Oral Oncol. 2012;48:1220–1226. doi: 10.1016/j.oraloncology.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu T, Li Y, Lin K, Yin H, He B, Zheng M, et al. Regulation of S100A4 expression via the JAK2-STAT3 pathway in rhomboid-phenotype pulmonary arterial smooth muscle cells exposure to hypoxia. Int J Biochem Cell Biol. 2012;44:1337–1345. doi: 10.1016/j.biocel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Bai L, Yang JC, Ok JH, Mack PC, Kung HJ, Evans CP. Simultaneous targeting of Src kinase and receptor tyrosine kinase results in synergistic inhibition of renal cell carcinoma proliferation and migration. Int J Cancer. 2012;130:2693–2702. doi: 10.1002/ijc.26303. [DOI] [PubMed] [Google Scholar]
- 55.Belaiba RS, Bonello S, Zahringer C, Schmidt S, Hess J, Kietzmann T, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 57.Kim H, Baumann H. The carboxyl-terminal region of STAT3 controls gene induction by the mouse haptoglobin promoter. J Biol Chem. 1997;272:14571–14579. doi: 10.1074/jbc.272.23.14571. [DOI] [PubMed] [Google Scholar]
- 58.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gleadle JM, Ratcliffe PJ. Induction of hypoxia-inducible factor-1, erythropoietin, vascular endothelial growth factor, and glucose transporter-1 by hypoxia: evidence against a regulatory role for Src kinase. Blood. 1997;89:503–509. [PubMed] [Google Scholar]
- 60.Wenger RH, Rolfs A, Marti HH, Bauer C, Gassmann M. Hypoxia, a novel inducer of acute phase gene expression in a human hepatoma cell line. J Biol Chem. 1995;270:27865–27870. doi: 10.1074/jbc.270.46.27865. [DOI] [PubMed] [Google Scholar]
- 61.Pawlus MR, Wang L, Ware K, Hu CJ. USF2 and HIF2alpha cooperatively activate HIF2 target genes during hypoxia. Mol Cell Biol. 2012;32:4595–4610. doi: 10.1128/MCB.00724-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.