Background: Proteins from the malaria parasite Plasmodium contain extensive asparagine repeats.
Results: Plasmodium tRNAAsn concentration and in vitro asparaginylation catalytic parameters were analyzed to explain the frequency of asparagine repeats.
Conclusion: The requirement for high levels of asparaginylated tRNAs hinders efficient protein translation.
Significance: Limiting the availability of asparaginylated tRNAs represents a plausible regulatory mechanism for the production of properly folded proteins.
Keywords: Aminoacyl tRNA Synthesis, Plasmodium, Protein Folding, Transfer RNA (tRNA), Translation, Asparagine Repeats, Low Complexity Regions
Abstract
Genome sequencing revealed an extreme AT-rich genome and a profusion of asparagine repeats associated with low complexity regions (LCRs) in proteins of the malarial parasite Plasmodium falciparum. Despite their abundance, the function of these LCRs remains unclear. Because they occur in almost all families of plasmodial proteins, the occurrence of LCRs cannot be associated with any specific metabolic pathway; yet their accumulation must have given selective advantages to the parasite. Translation of these asparagine-rich LCRs demands extraordinarily high amounts of asparaginylated tRNAAsn. However, unlike other organisms, Plasmodium codon bias is not correlated to tRNA gene copy number. Here, we studied tRNAAsn accumulation as well as the catalytic capacities of the asparaginyl-tRNA synthetase of the parasite in vitro. We observed that asparaginylation in this parasite can be considered standard, which is expected to limit the availability of asparaginylated tRNAAsn in the cell and, in turn, slow down the ribosomal translation rate when decoding asparagine repeats. This observation strengthens our earlier hypothesis considering that asparagine rich sequences act as “tRNA sponges” and help cotranslational folding of parasite proteins. However, it also raises many questions about the mechanistic aspects of the synthesis of asparagine repeats and about their implications in the global control of protein expression throughout Plasmodium life cycle.
Introduction
Malaria is one of the main menaces to human health in developing countries. There were ∼220 million cases of malaria (between 154–289 million) and 660,000 deaths estimated (between 610,000–971,000) in 2010 (according to the World Malaria Report of 2012 by the World Health Organization). Most deaths were caused by Plasmodium falciparum, which has developed resistance to nearly all anti-malarial treatments.
The sequence of the P. falciparum genome was completed in 2002 (1) and led to many fundamental observations about the molecular features of this parasite. Since then, sequences of other Plasmodium genomes (Plasmodium berghei, Plasmodium chabaudi, Plasmodium yoelii, Plasmodium knowlesi, and Plasmodium vivax) have also been reported. Plasmodium genomes are AT-rich, except for a divergent group, including P. knowlesi and P. vivax, which are characterized by a higher GC content (2). In AT-rich genomes, lysine, asparagine, phenylalanine, and isoleucine should occur in greater numbers than other amino acids. This is indeed the case in P. falciparum, P. berghei, P. chabaudi, and P. yoelii, where asparagine, lysine, and isoleucine are predominant in the proteomes of the parasite. These residues comprise 11.3, 10.4, and 10.5% of the P. falciparum proteome, respectively; only phenylalanine does not follow this probability trend, occurring at a frequency of only 3.9% (3). Moreover, asparagine residues are characterized by a particular distribution in these proteomes. They concentrate in long single amino acid repeats that are typically associated with regions of low complexity (LCR)3 (4). LCRs and asparagine repeats are found in 87% of the encoded proteins (5), even in metabolic enzymes and heat shock proteins that are usually resistant to these types of insertions (6). The presence of LCRs makes malarial proteins longer than their homologues in other species and it has been proposed that LCRs will form bulging domains from the core of each protein so that they would not impair structure or function (6). Numerous hypotheses have tried to address the function of plasmodial LCRs. It has been proposed that they would promote mRNA stability (7), protein-protein interactions (8), or may be involved in antigen diversification (9–12). It has also been proposed that LCRs are only neutral spacers between protein structural modules (13) or that they may be excised by some unknown mechanism (6). Yet, experimental evidence suggesting that LCRs play a functional role is rare. In addition to these hypotheses, we proposed that LCRs in general and asparagine-rich LCRs in particular modulate the rate of translating ribosomes and thus participate in cotranslational folding of plasmodial proteins (14). This proposed process would be driven by limiting access to asparaginylated transfer RNAAsn (tRNA) in the parasite during translation of LCRs.
It is generally accepted that variations in ribosome decoding rates are a consequence of the abundance of tRNAs in the cell; translation is optimal at codons decoded by highly abundant tRNAs and limited at codons read by rare tRNAs, as the ribosome pauses until the cognate tRNA becomes available. Still, it has been shown that tRNA abundance is not the only determinant of elongation rates. Interactions between the mRNA and the 16S ribosomal RNA as well as the presence of positively charged amino acids in the nascent peptide decelerate the ribosomes along transcripts in bacteria and yeast, respectively (15, 16). Among prokaryotic and eukaryotic genomes, genes encoding a given tRNA are usually present in multiple copies. Plasmodia are exceptions to this rule: the plasmodial nuclear genomes display 45 tRNA genes, with only one gene copy per tRNA isoacceptor (1). Thus, this minimal set of tRNA genes leads to the appealing assessment that the product of the unique tRNAAsn gene would decode >10% of the Plasmodium proteome. Translation specificity and efficiency depend on the accuracy of tRNA aminoacylation catalyzed by their cognate aminoacyl-tRNA synthetases (17) and on the availability of charged tRNAs, respectively. In the present study, P. falciparum cytosolic asparaginyl-tRNA synthetase (AsnRS) and tRNAAsn were cloned and their kinetic parameters were determined in vitro to estimate asparaginylation efficiency in this parasite. Moreover, the relative concentrations of each molecular partners involved in P. falciparum asparaginylation were compared with other plasmodial aminoacylation systems to deduce the relative accumulation of asparaginyl-tRNAAsn in Plasmodium and eventually its impact on the rate of synthesis of asparagine-rich LCRs.
EXPERIMENTAL PROCEDURES
Materials
Pyrococcus abyssi asparagine synthetase A was a generous gift from D. Kern. Radioactive l-[14C]tyrosine (483 mCi/mmol) was from Amersham Biosciences (Velizy-Villacoublay, France) and l-[14C]aspartate (200 mCi/mmol) was from PerkinElmer Life Sciences.
Database Screening for AsnRS Sequences
The P. falciparum genome encodes two copies of AsnRS, a cytoplasmic version (PlasmoDB ID PF3D7_0211800) and an apicoplastic one (PlasmoDB ID PF3D7_0509600) (14). The cytoplasmic P. falciparum AsnRS gene was identified by blasting the sequence of yeast cytoplasmic AsnRS (NP_011883.1) against the PlasmoDB database. Sequence alignments were computed with Tcoffee software.
RNA Seq Data of aaRS mRNA Expression in Sexual and Asexual Stages
The study was performed in the laboratory of Xin-zhuan Su (18), and data were retrieved from PlasmoDB for each P. falciparum cytosolic aminoacyl-tRNA synthetase gene.
Proteomic Data of P. falciparum Life Cycle Stages
Data were retrieved from Florens et al. (19). We considered proteins (i) expressed specifically in a single developmental stage and (ii) with sequences covered by at least three peptides. Thus, we used 27, 18, 31, and 66 protein sequences, corresponding to 50,027, 9629, 24,152, and 55,394 amino acids, identified in the sporozoite, merozoite, trophozoite, and gametocyte stages, respectively.
Cloning and Expression of Recombinant P. falciparum and H. sapiens AsnRSs
Genes coding for both the P. falciparum and human AsnRSs were amplifed by PCR from either P. falciparum (provided by H. Vial, Montpellier, France) or human liver cDNA libraries, respectively. Primers for PCR were designed according to the sequences of the AsnRS genes. In the case of the parasite AsnRS, the PCR reaction was performed with a customized dNTP mixture adapted to the P. falciparum genome composition (80% AT-rich): 240 μm dATP, 240 μm dTTP, 80 μm dCTP, 80 μm dGTP, and elongation was performed at 65 °C. Amplified DNA fragments were entirely sequenced and cloned into pQE30 (Qiagen, Courtaboeuf, France) to yield N-terminal His6-tagged recombinant proteins. Escherichia coli Top10 strains, separately expressing each construct, were grown at 37 °C in Luria broth with 2% glucose and 50 μg/ml ampicillin until they reached an absorbance of 2.0 (1-cm path length). The medium was then substituted with the same medium lacking glucose and AsnRS expression was induced for 3 h at 37 °C with 1 mm isopropyl-β-d-thiogalactopyranoside. Recombinant proteins were purified on nickel-nitrilotriacetic acid resin according to the manufacturer's instructions (Qiagen). Fractions containing AsnRS were pooled and dialyzed overnight at 4 °C against 50 mm KH2PO4/K2HPO4, pH 7.4, 10 mm β-mercaptoethanol, 150 mm KCl, and 50% glycerol. AsnRS proteins were quantified by Bradford assay (Bio-Rad) and stored at −20 °C. Preparations yielded one band on SDS-polyacrylamide gel and were estimated to be at least 95% pure.
Cloning and Expression of Recombinant P. falciparum AspRS and TyrRS
Purification of recombinant P. falciparum AspRS was performed as described in Ref. 20. The gene coding for P. falciparum TyrRS was amplified by PCR from P. falciparum cDNA, cloned into pQE30, and purified as described for P. falciparum AsnRS.
Dynamic Light Scattering
P. falciparum and H. sapiens AsnRS samples (20 μm) were incubated in HEPES-KOH, pH 7.0, 50 mm NaCl, 10% glycerol and were analyzed with a Zetasizer Nano S (Malvern, Orsay, France). Five intensity measurements were recorded at 20 °C, and the data were processed with the manufacturer's software, assuming that the enzyme particles were spherical. This method measures distributions of particle diffusion coefficients (D) that were transformed via the Einstein equation (D = kT/6pη0Rh) into hydrodynamic radii (Rh) and thus apparent particle sizes. Corrections for solvent refractive index and viscosity were applied, and the contribution of the solvent components was subtracted.
Preparation of l-[14C]Asparagine
This procedure was adapted from Roy and collaborators (21). The reaction mixture (200 μl), containing 50 mm HEPES-NaOH, pH 7.2, 10 mm MgCl2,2 mm NH4Cl, 2 mm KF, 240 μm l-[14C]Asp (300 cpm/pmol), and 10 mm ATP, was incubated at 70 °C for 2 h in the presence of 2 μm Pyrococcus abyssi asparagine synthetase A. Under these conditions, 95% of the initial aspartic acid was amidated into asparagine (as analyzed by thin layer chromatography). The reaction was stopped by phenol extraction, the l-[14C]Asn was further purified on an Illustra NAP-5 column (GE Healthcare Life Sciences) and concentrated by cryo-desiccation. The pellet was recovered in H2O to reach a final concentration of 200 μm.
Preparation of tRNA Samples
Transfer RNA transcripts were produced in vitro using either the classical method for P. falciparum tRNAAsn (22) or the “transzyme” system for H. sapiens tRNAAsn (23). Briefly, synthetic genes made of overlapping oligonucleotides reconstructed the T7 RNA polymerase promoter, the optional hammerhead ribozyme, the tRNA, and a BstNI site matching the CCA 3′-end of the tRNA sequence. When transcribed, the hammerhead ribozyme self-cleaved and released the mature tRNA that was further purified on a denaturing polyacrylamide gel. Before any assay, tRNA transcripts were heated at 65 °C for 2 min in water and refolded at 37 °C in the presence of MgCl2 (10 mm).
Total RNA from P. falciparum 3D7 strain parasites were cultured at 37 °C in 1% hematocrit as described previously (24), and cultures were harvested with 12–15% parasitemia. Infected erythrocytes were washed with incomplete growth medium and lysed with 0.15% saponin, and the parasites were used immediately for total RNA isolation by the Tri®-reagent BD (Sigma) extraction method according to the manufacturer's instructions.
Northern Blots
Samples of P. falciparum RNA were resolved on an 8% polyacrylamide gel, with 8 m urea in Tris-borate-EDTA buffer. Molecules were electrotransferred to an N+ nylon membrane (Amersham Biosciences) for 3 h at 250 mA at 4 °C and cross-linked to the membrane by UV irradiation. Specific 32P-labeled oligodeoxyribonucleotide probes were incubated with the membrane at 60 °C in ultra-sensitive hybridization buffer (Ambion, Saint Aubin, France), and the membrane was washed twice for 10 min at room temperature in 2× SSC, 0.1% SDS. Ribosomal 5S RNA was used as an internal loading control when quantifying tRNAs. Two amounts (5 μg and 15 μg) of three different P. falciparum RNA samples were tested, and probes were designed to detect tRNAAsnGUU (5′-CCGCACGCTCTAACCAACTGAGCTACGGAACC-3′), tRNAAspGUC (5′-GCGTGACAGGCGGAAATACTTGCACTATACTATCTCGGA-3′), tRNATyrGUA (5′-ATTTCAACTACAGTCTGCCGCTCTACCAACTGAGCTATCATCGG-3′), tRNALysCUU/UUU (5′-CCAGATTAAGAGTCTGGCGCTCTACCGACTGAGCTAAGGCAGC), and 5S rRNA (5′-CGAGCGGTCTCCCACCTCAGTATCT-3′) with melting temperatures ranging between 80 and 83 °C.
ATP/PPi Exchange and tRNA Aminoacylation Assays
ATP/PPi exchange assays were performed at 37 °C in 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1.5 mm [32P]PPi (3 cpm/pmol), 2 mm KF, and 250 nm AsnRS. The concentrations of either l-Asn or ATP varied from 0.05 to 1.5 mm. The synthesized [32P]ATP was quantified as described by Campanacci and collaborators (25).
Aminoacylation assays were conducted in 50 mm HEPES-NaOH, pH 7.2, 10 mm MgCl2, 2 mm ATP, 30 mm KCl, and appropriate amounts of tRNAAsn and AsnRSs (diluted in 100 mm HEPES-NaOH, pH 7.4, 1 mm DTT, and 10% glycerol for the plasmodial enzyme and the same buffer containing 5 mg/ml bovine serum albumin for the human enzyme). The mixture was incubated for 15 min at 37 °C and the reaction was initiated by adding 20 versus 10 μm l-[14C]asparagine for the plasmodial and the human enzymes, respectively. Apparent kinetic parameters were determined from Lineweaver-Burk plots. Data were expressed as averages of at least three independent experiments.
Aminoacylation plateaus of Plasmodium total RNA were performed in the presence of 0.5 μm of the relevant aminoacyl-tRNA synthetases (aaRS; P. falciparum recombinant AspRS, TyrRS, and AsnRS). Values were measured after 60-min incubations at 37 °C.
RESULTS
In Silico Identification of Plasmodial AsnRSs
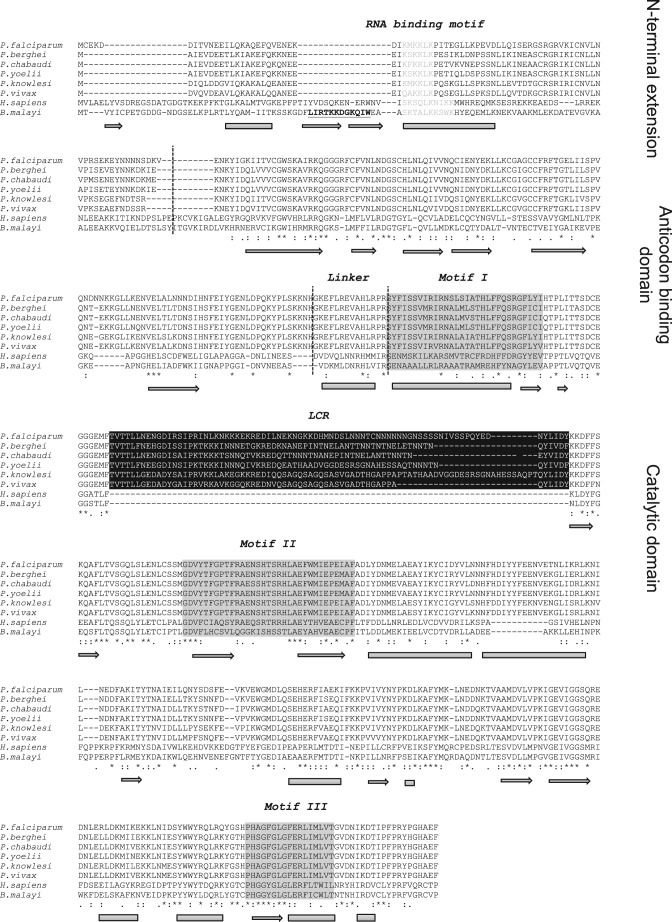
Six plasmodial genomes were screened for cytosolic AsnRS genes. Complete AsnRS genes were found in primate-infecting (P. falciparum, P. vivax, and P. knowlesi) and rodent-infecting (P. yoelii, P. berghei, and P. chabaudi) plasmodia (Fig. 1). The gene coding for the cytosolic P. falciparum AsnRS (PF3D7_0211800) is 1833 base pairs, is located on chromosome 2, and codes for a 610-amino acid protein. AsnRS is classified with both the aspartyl- and lysyl-tRNA synthetases as a class IIb aaRSs (26). Plasmodia AsnRSs were compared with homologues from H. sapiens, the vertebrate host of P. falciparum, and from Brugia malayi, for comparison with the crystal structure determined by Crepin and collaborators (27). Structure-based sequence alignments show characteristic eukaryotic AsnRS features, in particular in the C-terminal catalytic domain that contains the three class II aaRS motifs. All AsnRSs exhibit N-terminal appendices, which are predicted to fold as helical modules. The plasmodial modules are shorter (75 to 82 amino acids), and their sequences diverge from the N-terminal appendices of B. malayi AsnRS (110 amino acids) (28) and H. sapiens AsnRS (103 amino acids) (29). Yet, they all contain a helical lysine-rich motif (KMKKLK) that is similar to the RNA-binding motif identified in eukaryotic AspRSs and LysRSs (30–32). Lastly, the N-terminal module of plasmodia AsnRSs does not contain the amino acid sequence involved in the cytokine activity of B. malayi AsnRS (28).
FIGURE 1.
Multiple alignments between plasmodia, H. sapiens, and B. malayi AsnRS sequences. Sequence alignments between P. falciparum (PF3D7_0211800), P. berghei (PBANKA_030860), P. chabaudi (PCHAS_031080), P. yoelii (PY05639), P. knowlesi (PKH_040840), P. vivax (PVX_002940), H. sapiens (NP_004530.1), and B. malayi (XP_001898693) AsnRSs were performed with Tcoffee and were further refined by comparisons between the crystal structure of the B. malayi AsnRS (27), and the secondary structure prediction of P. falciparum AsnRS (on the PredictProtein website) (data not shown). Secondary structures related to the B. malayi AsnRS crystal structure are indicated below the alignment. Cylinders correspond to α-helices, and arrows indicate to β-strands. Dashed lines delineate the N-terminal extensions containing the putative RNA-binding motif, the tRNA anticodon recognition domain, the linker, and the C-terminal catalytic domains. The class II-specific motifs are boxed in gray, and the plasmodia-specific insertions are boxed in black. The chemokine-like activity of B. malayi AsnRS involves the underlined and boldface residues located in its 80-amino acid N-terminal domain (28).
All plasmodial AsnRSs are characterized by the presence of an insertion in the first half of the catalytic domain, between motifs 1 and 2 (Fig. 1), which likely does not disturb the active site architecture. This insertion corresponds to an LCR and has not been previously identified in AsnRSs of other phylogenetic origins. The insertion site of this LCR is strictly conserved among plasmodial AsnRSs, but the size and sequence vary from one species to another. In P. falciparum, P. berghei, P. chabaudi, and P. yoelii, this insertion contains several asparagine stretches.
To test the contribution of both the N-terminal extension and the LCR, P. falciparum AsnRS variants missing these domains were designed and constructed. However, they were not tested because recombinant mutant proteins were not soluble (data not shown).
Physicochemical Analysis of P. falciparum and H. sapiens AsnRSs by Dynamic Light Scattering
Dynamic light scattering spectroscopy is a convenient and non-invasive method for protein sizing (33) and has been applied to measure the aggregation states of aaRSs (e.g. Ref. 20). Measured diffusion coefficients of P. falciparum and H. sapiens AsnRS samples showed that both recombinant enzymes are dispersed at the concentrations used (20 μm monomeric protein). Only a small portion of the human AsnRS (13%) was aggregated (particle diameter, 110 nm; data not shown). Analysis of dynamic light scattering data allowed the determination of the size of AsnRS particles. Distributions of scattered intensities indicated that the average diameter of the P. falciparum and the H. sapiens AsnRSs were 13.8 and 12.5 nm, respectively, which correspond to spherical particles of 295 and 210 kDa (versus 142 and 127 kDa, theoretically). Because the crystallographic structure of the B. malayi AsnRS is more elongated than spherical (27), these data indicate that, as expected, both AsnRSs organize as dimers.
Catalytic Constants for the ATP/PPi Exchange Activity
The apparent Km and kcat values for asparagine activation were determined under identical conditions for the P. falciparum and H. sapiens AsnRSs (Table 1). The kinetic parameters for both enzyme orthologues are similar to the values determined for the AsnRSs from E. coli and CHO cells (34, 35). However, the Km for asparagine is 11-fold higher for the human AsnRS (460 μm) than for the other enzymes (40 μm for P. falciparum and E. coli AsnRS and 60 μm for CHO AsnRS).
TABLE 1.
Kinetic parameters of P. falciparum and H. sapiens AsnRSs the ATP/PPi exchange reaction
Results represent averages of at least three independent experiments.
Kinetic Analyses of tRNA Aminoacylation by P. falciparum and H. sapiens AsnRSs
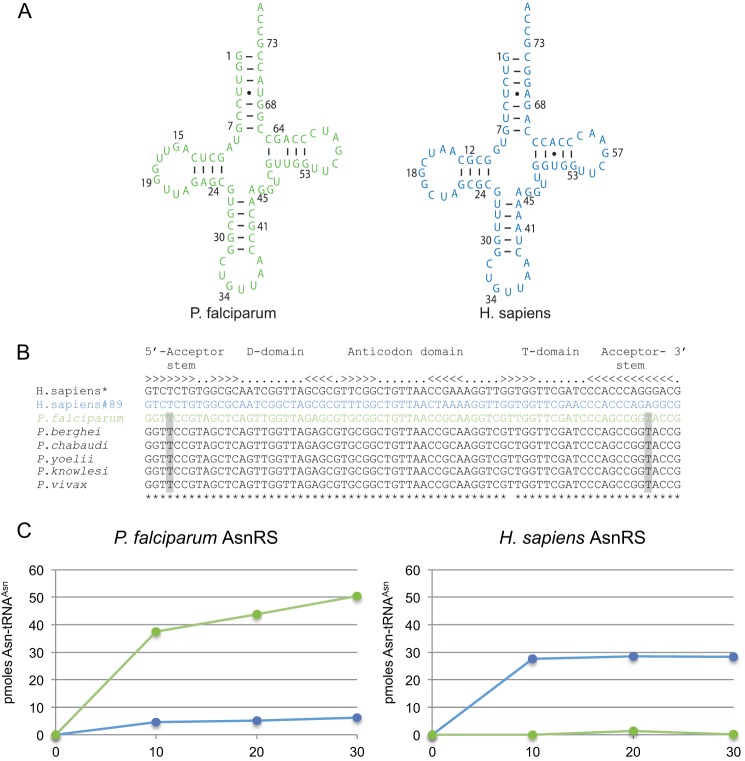
Both recombinant P. falciparum and H. sapiens AsnRSs were tested for tRNA charging under the same conditions (Fig. 2). They aminoacylated their cognate tRNAAsn transcripts at plateau levels of 65 and 35%, respectively (Fig. 2C). The apparent Km for their cognate tRNA transcripts were the same, but the velocity (apparent kcat) was 13-fold lower for P. falciparum AsnRS than for H. sapiens AsnRS, significantly decreasing (13-fold) the catalytic efficiency of the parasite AsnRS (Table 2).
FIGURE 2.
Sequence, secondary structure, and aminoacylation plateaus of Plasmodium and H. sapiens tRNAAsn transcripts. A, cloverleaf structure of tRNAAsn transcripts: the motifs involved in tRNA folding are present in both the plasmodial (green) and the human (blue) tRNAAsn sequences. The few differences are spread across their sequences and do not involve residues strategic to the structure and are presumed to be harmless for folding and aminoacylation. B, sequence alignments of human and plasmodial tRNAAsn sequences: the human genome codes for 35 tRNAAsn genes partitioned into two isoaccepting families either with an AUU anticodon (two genes coding for a unique isodecoder tRNA sequence) or a GUU anticodon (GtRNAdb, Genomic tRNA Database). The GUU isoacceptor tRNA family contains 14 different isodecoders encoded by the 33 other genes. Among them, the sequence of the major tRNAAsn isodecoder (asterisk) is represented 11 times. This sequence was first chosen to produce the human tRNAAsn transcript in vitro but was a poor aminoacylation substrate. Another sequence was thus chosen, tRNAAsn-89, whose transcript is a better substrate for the human AsnRS. The sequences of plasmodial tRNAAsn are highly conserved, except for a U-G base pair that becomes the C-G in the T-stem of P. chabaudi and P. knowlesi tRNAAsn. The U4·U69 mismatch conserved in the acceptor stem is highlighted in gray. C, aminoacylation plateaus of tRNAAsn transcripts. Reactions were performed in the presence of 80 pmol of transcript and 25 pmol pf of P. falciparum AsnRS or 3.5 pmol of H. sapiens AsnRS. The color code is the same as described in A.
TABLE 2.
Kinetic parameters of P. falciparum and H. sapiens AsnRSs for tRNA transcript aminoacylation
Results represent averages of at least three independent experiments. nd indicates not detectable.
|
P. falciparum AsnRS |
H. sapiens AsnRS |
|||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | min−1 | min−1 μm−1 10−3 | μm | min−1 | min−1 μm−1 10−3 | |
| tRNAAsn transcripts | ||||||
| P. falciparum WT | 2.0 ± 0.82 | 0.5 ± 0.17 | 252 | nd | nd | nd |
| H. sapiens WT | 6.0 ± 0.28 | 0.2 ± 0.06 | 31 | 2.0 ± 1.12 | 6.7 ± 3.73 | 3350 |
Despite the similarity between the P. falciparum and H. sapiens tRNAAsn sequences (Fig. 2, A and B), cross-species aminoacylation was not efficient (Fig. 2C). The human enzyme did not recognize the plasmodial tRNAAsn transcript, and the extent of asparaginylation of the human tRNAAsn transcript by the P. falciparum AsnRS was reduced 8-fold compared with its cognate tRNAAsn. The only remarkable feature distinguishing these two tRNAs is the presence of a UU mismatch at base pairs 4–69 in the acceptor arm of the P. falciparum tRNAAsn; it is conserved in all plasmodial tRNAAsn sequences (Fig. 2B), which is an indication that the acceptor stem might play a role in this species-specific barrier. However, a human tRNAAsn mutant, where the UG base pair was replaced by UU did not show any improvement in its aminoacylation capacity with P. falciparum AsnRS (data not shown).
Expression of Asparaginylation Molecular Partners and Comparison with Other Aminoacylation Systems in P. falciparum
tRNA Concentrations
We investigated the expression levels of tRNAAsn, tRNATyr, tRNAAsp, and tRNALys in non-synchronized in vitro cultures of P. falciparum. Transfer RNATyr and tRNAAsp are unique isoacceptors, which are each encoded by a single tRNA gene similar to tRNAAsn, but they decode fewer codons (4.4 and 6.4%, respectively) than tRNAAsn (11.2%) (Codon Usage Database). On the contrary, frequencies of asparagine and lysine codons are comparable (11.2 and 10.4%, respectively), but tRNALys is encoded by two tRNA genes (tRNALysAAA and tRNALysAAG). Northern blots and aminoacylation detection showed that two of the four selected tRNAs, tRNAAsn and tRNATyr, are globally expressed at the same level, independent of their use in translation (Fig. 3). Logically, the two tRNALys genes result in twice as much tRNALys compared with either tRNAAsn or tRNATyr. Surprisingly, tRNAAsp appeared ∼3-fold more concentrated than the others, despite only one copy of the tRNAAsp gene.
FIGURE 3.
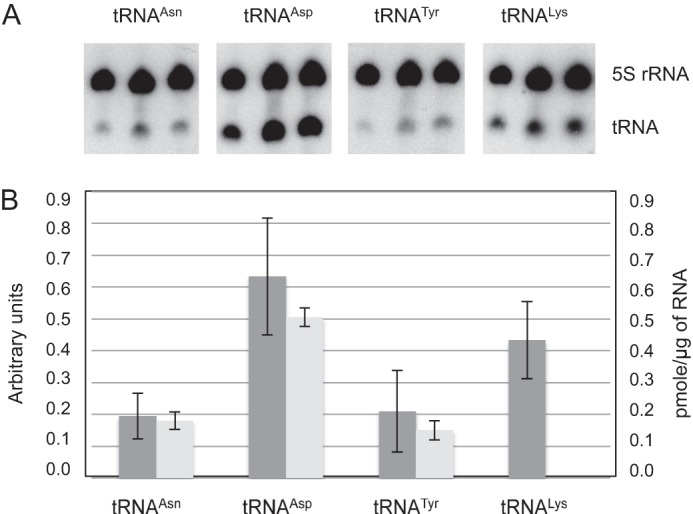
Quantification of P. falciparum selected tRNAs. A, total RNA was purified from blood stage P. falciparum and tRNAAsn (one isoacceptor), tRNAAsp (one isoacceptor), tRNATyr (one isoacceptor), and tRNALys (two isoacceptors) were detected by Northern blot. B, their relative concentrations were estimated by (i) Northern blot quantification (dark gray) and (ii) by determining aminoacylation plateaus in presence of the corresponding recombinant AsnRS, TyrRS, and AspRS (light gray).
aaRS Expression
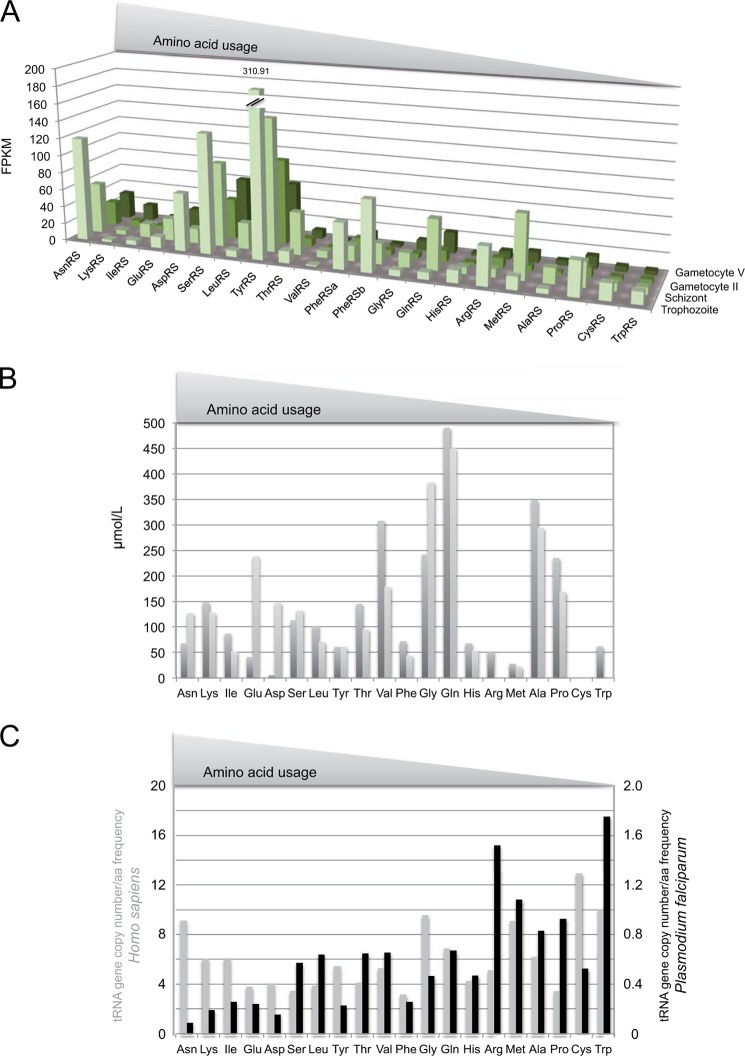
RNA-Seq coverages from different P. falciparum development stages (18) were used to estimate the relative expression levels of AsnRS compared with other aaRSs in P. falciparum (Fig. 4A). Concentrations of mRNAs varied from one aaRS to another. For example, trophozoites had 300-fold less ValRS mRNA than TyrRS mRNA. They also differed from one developmental stage to another: the mRNA encoding AsnRS was 4-fold higher in trophozoites than it was in gametocytes. This mRNA belongs to a group of three aaRS mRNAs (AsnRS, SerRS, and TyrRS) that are, on average, more concentrated than the remaining 18 aaRS mRNAs (Fig. 4A). Thus, the AsnRS mRNA has a propensity to accumulate noticeably; however, it is not the most abundant mRNA at any stage growth.
FIGURE 4.
Theoretical availability of aaRS mRNAs, amino acids, and tRNAs in P. falciparum. A, RNA Seq data of aaRSs mRNA expression in sexual and asexual stages. Sequencing data of strand-specific cDNA libraries are from the laboratory of Xin-zhuan Su (18). The y axis corresponds to fragments per kilobase of exon model per million mapped reads (FPKM). Data were retrieved from the PlasmoDB website and map four different developmental stages: late trophozoite, schizont, gametocyte II, and gametocyte V. B, amino acid availability in the host blood. During the blood stage, the Plasmodium translational machinery uses a small portion of blood cell amino acids produced by the degradation of the hemoglobin of erythrocyte (light gray) in particular; most of the exogenous amino acids come from the host plasma (dark gray). Amino acids concentrations in human blood are derived from Ref. 57. C, comparison of tRNA availability in H. sapiens (gray) and P. falciparum (black). Bars (arbitrary units) represent the ratio between the number of tRNA gene copies encoded by each genome (tRNA isoacceptor genes were combined, GtRNAdb, Genomic tRNA Database) and the corresponding amino acid frequency (Codon Usage Database). Lower bars correspond to the rarest tRNAs. Note that scales on the y axes are different for H. sapiens (left) and P. falciparum (right). L, liter.
Amino Acid Availability
Based on genome analysis, Plasmodium possesses biosynthetic pathways for Asn, Glu, Asp, Gly, Pro, and Gln (36). However, its growth depends on a supply of a complete set of exogenous amino acids. Thus, during the blood stage, Plasmodium development relies mostly on hemoglobin catabolism and circulating amino acids (37). Based on the amino acid concentrations measured in human blood, the amount of asparagine available for translation in Plasmodium corresponds to 3.5% of all amino acids (Fig. 4B).
Asparagine Content of P. falciparum Stage-specifically Expressed Proteins
Data from stage-specific proteomes were retrieved from Ref. 19. They relate to four distinct stages of the parasite: sporozoites (the infectious form injected by the mosquito), merozoites (the invasive stage of the erythrocytes), trophozoites (the form multiplying in erythrocytes), and gametocytes (the sexual stage). For each stage, amino acid contents and amino acid repeats were determined. Global proteome asparagine content is not strictly conserved among the different stages; for example, there is 1.8-fold more asparagine in sporozoite proteins than in trophozoites proteins (Fig. 5A). This observation is strengthened when looking at the frequency of asparagine repeats (Fig. 5B): asparagine repeats are more abundant in sporozoites (2.7- and 4.2-fold) and in gametocytes (2- and 3-fold) than in merozoites and trophozoites, respectively, and longer. Glutamate and valine contents are also stage-specific, but unlike asparagine, these amino acids do not cluster in long repeats (data not shown).
FIGURE 5.
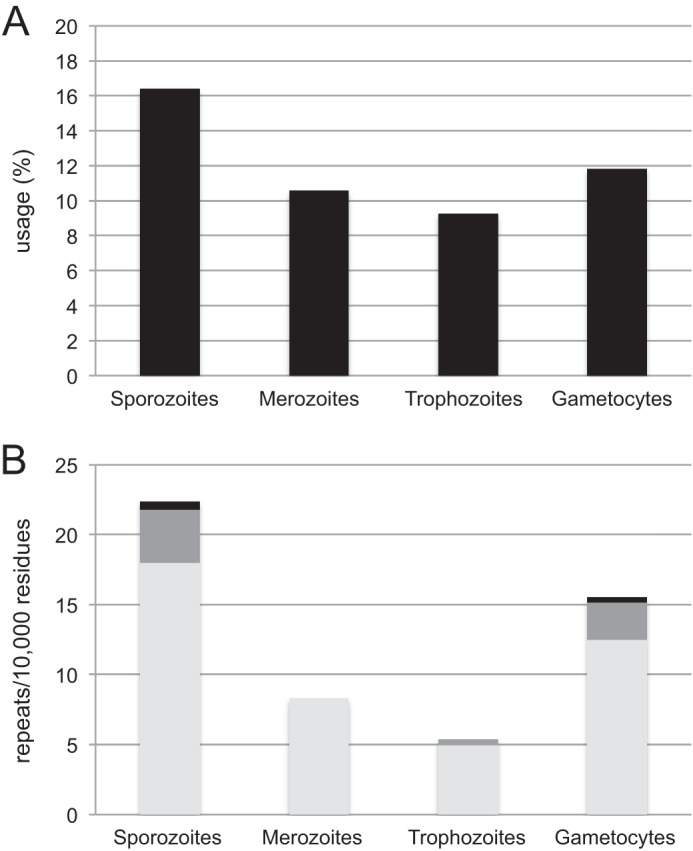
Asparagine content of P. falciparum stage-specific proteins. Stage-specific proteome data were retrieved from the study by Florens and collaborators (19). They correspond to four distinct stages of the parasite: sporozoites, merozoites, trophozoites, and gametocytes. A, asparagine frequencies were compared in stage specific proteins; B, asparagine repeats were sorted according to their length: five to nine consecutive residues (light gray), 10 to 19 residues (dark gray), and >20 residues (black).
DISCUSSION
AsnRS Activity
aaRSs are diverse in sequence, size, and structure but perform the same function: they catalyze specific tRNA aminoacylation (17). In turn, the efficiency and accuracy of these reactions govern the efficiency and accuracy of protein synthesis. Crystal structures of Thermus thermophilus and B. malayi AsnRSs have been reported (27, 38). Yet, biochemical and enzymatic studies are rare. The only exception relates to the T. thermophilus asparaginylation system (39).
In the present work, we studied P. falciparum AsnRS and its host H. sapiens AsnRSs to directly compare both enzymes. Neither the plasmodial nor the human enzymes are optimal in charging their cognate tRNAAsn transcripts in vitro, as compared with the asparaginylation kinetics measured for the T. thermophilus system with native tRNAAsn. The kinetic parameters determined for the prokaryotic system are 2000- and 150-fold more efficient than the parasite and the human systems, respectively. Similarly, the Km of the CHO AsnRS for its cognate tRNAAsn (35) is ∼30-fold lower (0.06 μm) than the apparent Km values determined in the present study (2 μm). These differences could be explained by the use of in vitro transcripts in this study, which are deprived of post-transcriptional modifications. Occasionally, these modifications have been shown to be involved in tRNA aminoacylation (i.e. Ref. 40). In particular, the absence of a queuosine at position 34 in both the Plasmodium and the human tRNAAsn could explain the reduced Km and kcat values. The presence of two genes coding for putative queuosine tRNA ribosyltransferases in the P. falciparum genome (PF3D7_1242200, probably targeted to the apicoplast, and PF3D7_0717400, the putative cytosolic enzyme) support the existence of this particular post-transcriptional modification in P. falciparum tRNAAsn, as it is the case in all organisms except yeast (41).
tRNAAsn Availability
The genomes of most organisms display multiple copies of tRNA genes; the number of copies for each tRNA isoacceptor diverges from species to species (42). It is accepted that (i) tRNA gene content determines the relative amount of each tRNA in the cell, (ii) the most abundant codons are decoded by the most abundant tRNAs, and (iii) translation efficiency is governed by the relative abundance of each tRNA (reviewed in Ref. 43). In this context, the fact that P. falciparum but also all other plasmodia genomes encodes only one gene copy per tRNA isoacceptor is confusing. Assuming that each gene is transcribed to similar extents, one can calculate the theoretical availability of Plasmodium tRNA species compared with their codon usage in P. falciparum protein synthesis (Fig. 4C). Strikingly, unlike other organisms, there is no correlation between the number of tRNA genes and its codon usage in this parasite (or the related amino acid frequency). For example, the ratios of tRNA gene copy number to amino acid frequency varies at most by 4-fold among all H. sapiens tRNA/amino acid pairs. In contrast, in P. falciparum, these ratios vary over a 20-fold range, with the lowest value assigned to tRNAAsn and asparagine (Fig. 4C). Thus, tRNAAsn is the most limiting tRNA in the translation of P. falciparum. This hypothesis was confirmed by Northern blots and aminoacylation tests on total RNA isolated from in vitro cultures of different P. falciparum blood stages (Fig. 3). In these experiments, tRNAAsn did not accumulate more than any of the three other tRNAs tested (tRNAAsp, tRNALys, and tRNATyr). Because the codons that are decoded by tRNAAsn are highly represented in the Plasmodium genome, translation efficiency from that particular tRNA is expected to be compromised. This scenario might be partially compensated for by an increased local concentration of this specific tRNA near the translation site of asparagine stretches, as has been suggested for other organisms (44).
Asn-tRNAAsn Availability
The regulating action of tRNAAsn depends above all on the proportion of asparagine-charged tRNAAsn available for translation. Based on the RNA seq data, the mRNA encoding AsnRS is highly expressed in the cell, suggesting that, in the absence of specific post-transcriptional inhibitory mechanism, AsnRS would substantially accumulate in the parasite. Moreover, P. falciparum AsnRS shows an optimal affinity for asparagine (Table 1), indicating that even if the asparagine concentration is getting low, it should not affect significantly the rate of the first step of the asparaginylation reaction. These observations favor efficient asparaginylation of the tRNAAsn pool in Plasmodium.
Open Questions
Slippery Sequences?
The relative scarcity of tRNAAsn in plasmodia could sequentially decrease the rate of Asn-tRNAAsn selection at the ribosome A-site, trigger ribosomal frameshifting, and finally lead to a reading frameshift when translating repeated asparagine codons. Moreover, one can wonder whether the post-transcriptionally added queuosine modification in the tRNAAsn anticodon would also influence the fidelity of asparagine insertion in asparagine repeats. It was previously shown that queuosine does not influence frameshifting in bacteria (45) or when decoding a specific sequence of the infectious bronchitis virus (46). However, this hypothesis has not been tested on the long AU-rich sequences that characterize Plasmodium mRNAs or in the presence of limiting concentrations of Asn-tRNAAsn.
Asparagine Repeats and Protein Aggregation?
In P. falciparum, the presence of asparagine repeats could be deleterious because it leads to the expression of a large number of prion-like domains (∼25% of the proteome) (47), which in turn make the corresponding proteins suitable for aggregation. Recently, it was shown that P. falciparum Hsp110c, with other chaperones, plays an essential role in overcoming the aggregation tendencies of asparagine-rich proteins (48). Furthermore, we propose that accumulation of such prion-like sequences could be permitted through the presence of charged amino acids, lysine, glutamate, and aspartate, which also cover >25% of the proteome. The presence of so many charged amino acids could, in theory, improve the production of more soluble proteins (49, 50).
Potential Regulatory Activities?
According to the models discussed above, the tRNA pool should remain the same not only in different cell types but also throughout their life cycle. As a matter of fact, recent measurements in yeast and human tissues show that the abundance of different tRNAs deviates from the copy number of the corresponding genes (51, 52). As a consequence, variations in tRNA levels during the parasite life cycle would offer an opportunity to regulate and induce wide variations in translation efficiency and could modulate the expression of stage-specific proteins depending on their content in asparagine repeats. Our analysis of P. falciparum stage-specific proteomes (19) supports this idea by revealing an increase in asparagine repeats in proteins expressed exclusively in sporozoites and gametocytes compared with erythrocytic stages. These differences suggest that translation in sporozoites and to a lesser extent in gametocytes requires more asparaginylated-tRNAAsn. When looking at trophozoite and gametocyte stages, there is no apparent correlation between the concentrations of mRNA for AsnRS (Fig. 4A) and the presence of long single amino acid repeats (Fig. 5). However, one should be aware that mRNA concentrations do not consider post-transcriptional regulatory mechanisms, which control the final cellular concentrations of these enzymes, at least in other organisms (53, 54).
tRNAAsn Limitation and Cotranslational Folding of Proteins?
Our favorite hypothesis is that LCRs and asparagine repeats are intrinsic chaperones. Indeed, the low availability of Asn-tRNAAsn should decrease the rate of translating ribosomes when decoding asparagine-rich LCRs and, in consequence, govern cotranslational folding of the proteins of the parasite: ribosome pauses would give enough time to the newly synthesized N-terminal domain to fold correctly, independently from the protein C-terminal sequence not yet translated. This scenario is consistent with our previous observation that LCRs exist in boundaries between independent folding modules in the aaRSs themselves (14). LCRs are inserted in almost all Plasmodium proteins. Their own translation spatially separates the synthesis of defined portions in these proteins (Fig. 6), allowing sequential folding of individual protein domains with translation pauses or slow downs in between. Furthermore, the presence of asparagine repeats in LCRs would increase local Asn-tRNAAsn demand and further postpone translating ribosomes.
FIGURE 6.
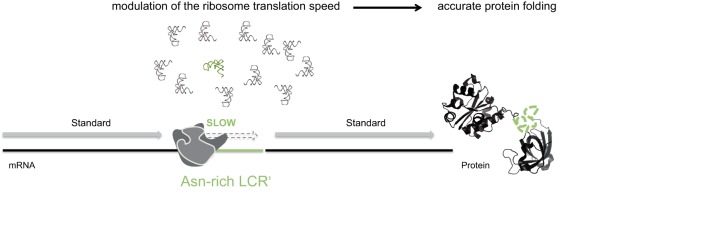
Model for cotranslational folding of proteins in Plasmodium. Asparagine-rich LCRs interspersed in plasmodial proteins likely act as intrinsic chaperones by delaying ribosome translation. Under limiting Asn-tRNAAsn concentrations, translation of asparagine-rich sequences force ribosomes to slow down and allow cotranslational folding of proteins.
Conclusions
Impact on Protein Synthesis versus a Specific Function for Asparagine-rich LCRs
Single amino acid repeats have been found for all 20 amino acids in all three domains of life. Yet, plasmodia are distinct because of the number of amino acid repeats and because they are found in all classes of proteins. The AT content of the P. falciparum, P. yoelii, P. chabaudi, and P. berghei genomes definitely plays a part in the accumulation of asparagine repeats in their proteomes; it does not explain why these codons tend to cluster together in LCRs (6). However, the predilection for alanine repeats in P. knowlesi and P. vivax suggests that the presence of amino acids stretches is important, but the nature of the incorporated amino acid is not. This species-specific usage of amino acid repeats is a further indication that repeats (and LCRs) might not have acquired functions per se but that their presence is important for the proper function of the rest of the protein. We propose that differential accumulation of asparagine repeats could direct the correct folding and/or the stage specific expression of nascent Plasmodium proteins by controlling the rate of the translating ribosome. This could replace what exists in most organisms, where the tightness of secondary and tertiary structures in mRNAs controls ribosome binding and decoding rate (i.e. Refs. 55 and 56). One may indeed wonder whether AU-rich Plasmodium mRNAs can adopt such structures and use the same post-transcriptional regulatory strategies.
Acknowledgments
We thank Tamara Hendrickson for support and comments on the manuscript. We acknowledge Caroline Paulus, Sylvie Perrotey, and Tania Bour for assistance and thank Nizar Saad and Daniel Kern for providing us with asparagine synthetase A.
This work was supported by the European Community's Seventh Framework Programme FP7/2007-2013 under Grant 223024 and the Laboratoire d'Excellence NetRNA Grant ANR-10-LABX-36.
- LCR
- low complexity region
- AsnRS
- asparaginyl-tRNA synthetase
- aaRS
- aminoacyl-tRNA synthetase.
REFERENCES
- 1. Gardner M. J., Shallom S. J., Carlton J. M., Salzberg S. L., Nene V., Shoaibi A., Ciecko A., Lynn J., Rizzo M., Weaver B., Jarrahi B., Brenner M., Parvizi B., Tallon L., Moazzez A., Granger D., Fujii C., Hansen C., Pederson J., Feldblyum T., Peterson J., Suh B., Angiuoli S., Pertea M., Allen J., Selengut J., White O., Cummings L. M., Smith H. O., Adams M. D., Venter J. C., Carucci D. J., Hoffman S. L., Fraser C. M. (2002) Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature 419, 531–534 [DOI] [PubMed] [Google Scholar]
- 2. Dalby A. R. (2009) A comparative proteomic analysis of the simple amino acid repeat distributions in Plasmodia reveals lineage specific amino acid selection. PloS One 4, e6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastien O., Lespinats S., Roy S., Métayer K., Fertil B., Codani J. J., Maréchal E. (2004) Analysis of the compositional biases in Plasmodium falciparum genome and proteome using Arabidopsis thaliana as a reference. Gene 336, 163–173 [DOI] [PubMed] [Google Scholar]
- 4. Aravind L., Iyer L. M., Wellems T. E., Miller L. H. (2003) Plasmodium biology: genomic gleanings. Cell 115, 771–785 [DOI] [PubMed] [Google Scholar]
- 5. DePristo M. A., Zilversmit M. M., Hartl D. L. (2006) On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene 378, 19–30 [DOI] [PubMed] [Google Scholar]
- 6. Pizzi E., Frontali C. (2001) Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 11, 218–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xue H. Y., Forsdyke D. R. (2003) Low-complexity segments in Plasmodium falciparum proteins are primarily nucleic acid level adaptations. Mol. Biochem. Parasitol. 128, 21–32 [DOI] [PubMed] [Google Scholar]
- 8. Karlin S., Brocchieri L., Bergman A., Mrazek J., Gentles A. J. (2002) Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl. Acad. Sci. U.S.A. 99, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferreira M. U., Ribeiro W. L., Tonon A. P., Kawamoto F., Rich S. M. (2003) Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene 304, 65–75 [DOI] [PubMed] [Google Scholar]
- 10. Hughes A. L. (2004) The evolution of amino acid repeat arrays in Plasmodium and other organisms. J. Mol. Evol. 59, 528–535 [DOI] [PubMed] [Google Scholar]
- 11. Cortés A., Mellombo M., Masciantonio R., Murphy V. J., Reeder J. C., Anders R. F. (2005) Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 73, 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verstrepen K. J., Jansen A., Lewitter F., Fink G. R. (2005) Intragenic tandem repeats generate functional variability. Nat. Genet. 37, 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huntley M., Golding G. B. (2000) Evolution of simple sequence in proteins. J. Mol. Evol. 51, 131–140 [DOI] [PubMed] [Google Scholar]
- 14. Frugier M., Bour T., Ayach M., Santos M. A., Rudinger-Thirion J., Théobald-Dietrich A., Pizzi E. (2010) Low Complexity Regions behave as tRNA sponges to help co-translational folding of plasmodial proteins. FEBS Lett. 584, 448–454 [DOI] [PubMed] [Google Scholar]
- 15. Li G. W., Oh E., Weissman J. S. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charneski C. A., Hurst L. D. (2013) Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 11, e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ibba M., Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 [DOI] [PubMed] [Google Scholar]
- 18. López-Barragán M. J., Lemieux J., Quiñones M., Williamson K. C., Molina-Cruz A., Cui K., Barillas-Mury C., Zhao K., Su X. Z. (2011) Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Florens L., Washburn M. P., Raine J. D., Anthony R. M., Grainger M., Haynes J. D., Moch J. K., Muster N., Sacci J. B., Tabb D. L., Witney A. A., Wolters D., Wu Y., Gardner M. J., Holder A. A., Sinden R. E., Yates J. R., Carucci D. J. (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526 [DOI] [PubMed] [Google Scholar]
- 20. Bour T., Akaddar A., Lorber B., Blais S., Balg C., Candolfi E., Frugier M. (2009) Plasmodial aspartyl-tRNA synthetases and peculiarities in Plasmodium falciparum. J. Biol. Chem. 284, 18893–18903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roy H., Becker H. D., Reinbolt J., Kern D. (2003) When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc. Natl. Acad. Sci. U.S.A. 100, 9837–9842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J. P., Florentz C., Giegé R. (1990) Conformation in solution of yeast tRNAAsp transcripts deprived of modified nucleotides. Biochimie 72, 735–743 [DOI] [PubMed] [Google Scholar]
- 23. Fechter P., Rudinger J., Giegé R., Théobald-Dietrich A. (1998) Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 436, 99–103 [DOI] [PubMed] [Google Scholar]
- 24. Trager W., Jensen J. B. (1997) Continuous culture of Plasmodium falciparum: its impact on malaria research. Int. J. Parasitol. 27, 989–1006 [DOI] [PubMed] [Google Scholar]
- 25. Campanacci V., Dubois D. Y., Becker H. D., Kern D., Spinelli S., Valencia C., Pagot F., Salomoni A., Grisel S., Vincentelli R., Bignon C., Lapointe J., Giegé R., Cambillau C. (2004) The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity. J. Mol. Biol. 337, 273–283 [DOI] [PubMed] [Google Scholar]
- 26. Moras D. (1992) Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem. Sci. 17, 159–164 [DOI] [PubMed] [Google Scholar]
- 27. Crepin T., Peterson F., Haertlein M., Jensen D., Wang C., Cusack S., Kron M. (2011) A hybrid structural model of the complete Brugia malayi cytoplasmic asparaginyl-tRNA synthetase. J. Mol. Biol. 405, 1056–1069 [DOI] [PubMed] [Google Scholar]
- 28. Kron M. A., Wang C., Vodanovic-Jankovic S., Howard O. M., Kuhn L. A. (2012) Interleukin-8-like activity in a filarial asparaginyl-tRNA synthetase. Mol. Biochem. Parasitol. 185, 66–69 [DOI] [PubMed] [Google Scholar]
- 29. Beaulande M., Tarbouriech N., Härtlein M. (1998) Human cytosolic asparaginyl-tRNA synthetase: cDNA sequence, functional expression in Escherichia coli and characterization as human autoantigen. Nucleic Acids Res. 26, 521–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frugier M., Moulinier L., Giegé R. (2000) A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J. 19, 2371–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheong H. K., Park J. Y., Kim E. H., Lee C., Kim S., Kim Y., Choi B. S., Cheong C. (2003) Structure of the N-terminal extension of human aspartyl-tRNA synthetase: implications for its biological function. Int. J. Biochem. Cell Biol. 35, 1548–1557 [DOI] [PubMed] [Google Scholar]
- 32. Francin M., Kaminska M., Kerjan P., Mirande M. (2002) The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain. J. Biol. Chem. 277, 1762–1769 [DOI] [PubMed] [Google Scholar]
- 33. Chayen N., Dieckmann M., Dierks K., Fromme P. (2004) Size and shape determination of proteins in solution by a noninvasive depolarized dynamic light scattering instrument. Ann. N.Y. Acad. Sci. 1027, 20–27 [DOI] [PubMed] [Google Scholar]
- 34. Madern D., Anselme J., Härtlein M. (1992) Asparaginyl-tRNA synthetase from the Escherichia coli temperature-sensitive strain HO202. A proline replacement in motif 2 is responsible for a large increase in Km for asparagine and ATP. FEBS Lett. 299, 85–89 [DOI] [PubMed] [Google Scholar]
- 35. Andrulis I. L., Chiang C. S., Arfin S. M., Miner T. A., Hatfield G. W. (1978) Biochemical characterization of a mutant asparaginyl-tRNA synthetase from Chinese hamster ovary cells. J. Biol. Chem. 253, 58–62 [PubMed] [Google Scholar]
- 36. Payne S. H., Loomis W. F. (2006) Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryot. Cell 5, 272–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krugliak M., Zhang J., Ginsburg H. (2002) Intraerythrocytic Plasmodium falciparum utilizes only a fraction of the amino acids derived from the digestion of host cell cytosol for the biosynthesis of its proteins. Mol. Biochem. Parasitol. 119, 249–256 [DOI] [PubMed] [Google Scholar]
- 38. Seignovert L., Härtlein M., Leberman R. (1996) Asparaginyl-tRNA synthetase from Thermus thermophilus HB8. Sequence of the gene and crystallization of the enzyme expressed in Escherichia coli. Eur. J. Biochem. 239, 501–508 [DOI] [PubMed] [Google Scholar]
- 39. Becker H. D., Kern D. (1998) Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. U.S.A. 95, 12832–12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Senger B., Auxilien S., Englisch U., Cramer F., Fasiolo F. (1997) The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochem. 36, 8269–8275 [DOI] [PubMed] [Google Scholar]
- 41. Grosjean H., Sprinzl M., Steinberg S. (1995) Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie 77, 139–141 [DOI] [PubMed] [Google Scholar]
- 42. Marck C., Grosjean H. (2002) tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8, 1189–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Novoa E. M., Ribas de Pouplana L. (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 28, 574–581 [DOI] [PubMed] [Google Scholar]
- 44. Cannarozzi G., Schraudolph N. N., Faty M., von Rohr P., Friberg M. T., Roth A. C., Gonnet P., Gonnet G., Barral Y. (2010) A role for codon order in translation dynamics. Cell 141, 355–367 [DOI] [PubMed] [Google Scholar]
- 45. Urbonavicius J., Qian Q., Durand J. M., Hagervall T. G., Björk G. R. (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marczinke B., Hagervall T., Brierley I. (2000) The Q-base of asparaginyl-tRNA is dispensable for efficient -1 ribosomal frameshifting in eukaryotes. J. Mol. Biol. 295, 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singh G. P., Chandra B. R., Bhattacharya A., Akhouri R. R., Singh S. K., Sharma A. (2004) Hyper-expansion of asparagines correlates with an abundance of proteins with prion-like domains in Plasmodium falciparum. Mol. Biochem. Parasitol. 137, 307–319 [DOI] [PubMed] [Google Scholar]
- 48. Muralidharan V., Oksman A., Pal P., Lindquist S., Goldberg D. E. (2012) Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat. Commun. 3, 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dobson C. M. (2004) Principles of protein folding, misfolding and aggregation. Semin. Cell Dev. Biol. 15, 3–16 [DOI] [PubMed] [Google Scholar]
- 50. Wayne N., Bolon D. N. (2010) Charge-rich regions modulate the anti-aggregation activity of Hsp90. J. Mol. Biol. 401, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zaborske J. M., Narasimhan J., Jiang L., Wek S. A., Dittmar K. A., Freimoser F., Pan T., Wek R. C. (2009) Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 284, 25254–25267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dittmar K. A., Goodenbour J. M., Pan T. (2006) Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryckelynck M., Giegé R., Frugier M. (2005) tRNAs and tRNA mimics as cornerstones of aminoacyl-tRNA synthetase regulations. Biochimie 87, 835–845 [DOI] [PubMed] [Google Scholar]
- 54. Frugier M., Ryckelynck M., Giegé R. (2005) tRNA-balanced expression of a eukaryal aminoacyl-tRNA synthetase by an mRNA-mediated pathway. EMBO Rep. 6, 860–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Geissmann T., Chevalier C., Cros M. J., Boisset S., Fechter P., Noirot C., Schrenzel J., François P., Vandenesch F., Gaspin C., Romby P. (2009) A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 37, 7239–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang L., Wessler S. R. (2001) Role of mRNA secondary structure in translational repression of the maize transcriptional activator Lc(1,2). Plant Physiol. 125, 1380–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aguiló A., Castaño E., Tauler P., Guix M. P., Serra N., Pons A. (2000) Participation of blood cells in the changes of blood amino acid concentrations during maximal exercise. J. Nutr. Biochem. 11, 81–86 [DOI] [PubMed] [Google Scholar]