Background: Many pentatricopeptide repeat (PPR) proteins are RNA site specificity factors and include C-terminal DYW deaminase domains.
Results: ELI1 and DOT4 are required for editing single sites. The DYW deaminase domain binds two zinc atoms.
Conclusion: The C terminus of PLS-type PPR proteins shares molecular characteristics with cytidine deaminase.
Significance: This study provides the first evidence that DYW deaminase domains bind zinc.
Keywords: Chloroplast, Gene Expression, Mass Spectrometry (MS), Metalloenzymes, RNA Modification
Abstract
Many transcripts expressed from plant organelle genomes are modified by C-to-U RNA editing. Nuclear encoded pentatricopeptide repeat (PPR) proteins are required as RNA binding specificity determinants in the RNA editing mechanism. Bioinformatic analysis has shown that most of the Arabidopsis PPR proteins necessary for RNA editing events include a C-terminal portion that shares structural characteristics with a superfamily of deaminases. The DYW deaminase domain includes a highly conserved zinc binding motif that shares characteristics with cytidine deaminases. The Arabidopsis PPR genes, ELI1 and DOT4, both have DYW deaminase domains and are required for single RNA editing events in chloroplasts. The ELI1 DYW deaminase domain was expressed as a recombinant protein in Escherichia coli and was shown to bind two zinc atoms per polypeptide. Thus, the DYW deaminase domain binds a zinc metal ion, as expected for a cytidine deaminase, and is potentially the catalytic component of an editing complex. Genetic complementation experiments demonstrate that large portions of the DYW deaminase domain of ELI1 may be eliminated, but the truncated genes retain the ability to restore editing site conversion in a mutant plant. These results suggest that the catalytic activity can be supplied in trans by uncharacterized protein(s) of the editosome.
Introduction
C-to-U RNA editing takes place in most land plant chloroplasts and mitochondria (1, 2) and requires nuclear proteins (3–5). In flowering plants, the transcripts of chloroplasts and mitochondria are modified post-transcriptionally by C-to-U editing with about 35 C-to-U editing events in chloroplasts and hundreds of editing sites in the mitochondria (6). Editing sites are specifically recognized by a 5′ cis-element of about 20–30 nucleotides (7–10).
Pentatricopeptide repeat (PPR)2 genes have been shown to be required for RNA editing (4, 11–20), and form a large family of protein-coding genes in higher plants with over 400 members in Arabidopsis (21). The PPR genes can be divided into P and PLS subfamilies (22). The P subfamily is composed of tandem arrays of a degenerate 35-amino acid motif (P), whereas the PLS subfamily is characterized by distinct variants of the 35-amino acid repeats, with P type as well as long (L) and short (S) variants of the repeats (21). Specific amino acid residues within the PPRs have been shown to specify the base recognized in the cis-element (23, 24). In addition, the PLS subfamily includes characteristic C-terminal domains known as the E, E+, and DYW. The DYW domain is known to have zinc binding motifs (HXE and CXXC) (25), and analogous zinc-coordinating residues are conserved in nucleotide deaminases (26–29). Phylogenetic analyses have shown that the distribution of PPR proteins with the DYW domain coincides with the distribution of C-to-U editing in land plants (25), in the protist, Naegleria gruberi (30, 31), and possibly also in diverse lower eukaryotes such as Acanthamoeba, Physarum, Nitella, and rotifers (32). A bioinformatics analysis of the deaminase superfamily has shown that the C-terminal region exhibits structural characteristics of a deaminase domain and places the DYW deaminase domain in the deaminases superfamily (27). The deaminase fold is represented by sequences composed of part of the E, the entire E+, and most of the DYW domain, and has been identified as the DYW family of nucleic acid deaminases (Pfam: Pf14432). The entire region is referred to as the “DYW deaminase domain” in this work.
Several PPR genes are known to be responsible for editing various sites, but lack a portion of the DYW deaminase domain (4, 12, 13, 16, 33–36). In addition, portions of the DYW deaminase domain can be eliminated through truncation, but transgenic plants expressing truncated variants are fully capable of editing site conversion (14, 15). These experiments suggest that the PPR proteins act as editing site recognition factors (14, 15) in a genetic complementation assay. Thus, although the entire DYW deaminase domain is under strong selection (37), most of the DYW deaminase domain is dispensable. In at least one case, editing has been shown to require a PPR that provides site specificity, and a separate protein has been shown to supply a portion of the DYW deaminase domain (38). Both PPR proteins CRR4 and DYW1 are required for editing ndhD C2 (38). CRR4 lacks an intact DYW domain and is apparently required as a site recognition factor, whereas DYW1 has an intact DYW domain and may contribute the catalytic activity. In addition, RIP/MORF proteins have been shown to interact with PPR proteins required for editing (3, 5) and may be involved in forming editing complexes with PPR proteins.
In this study, we demonstrate that two PPR genes, ELI1 and DOT4/FLV, are each required for editing single chloroplast editing sites. ELI1 has a full-length DYW domain, and mass spectrometry (MS) combined with inductively coupled optical emission spectroscopy (ICP-OES) demonstrates that the DYW deaminase domain binds two zinc atoms per subunit. A full-length recombinant DYW1 protein has been analyzed and also binds two zinc atoms per polypeptide. This is the first demonstration of zinc binding by a plant editing factor, and it provides additional support for a catalytic function in C deamination. In addition, truncated variants of ELI1 that lack most of the structural features of the deaminase fold are capable of restoring editing in a mutant plant. A critical 15-amino acid region must be retained for editing site conversion. Based on these results, we propose a bipartite model where one PPR protein is required for site recognition, and a second PPR or other protein provides the catalytic activity for editing site conversion.
EXPERIMENTAL PROCEDURES
Plant Materials
Seeds of Brassica napus (PI 470086 named 79-389, PI 601282 named Lindora-00, and PI 547112 named Humus) were obtained from the United States Department of Agriculture (USDA) through the Germplasm Resources Information Network (GRIN). Seeds of Brassica rapa (PI 279857, PI 418984, PI 257239, PI 508422 named Late Spring, PI 508418 named Early Spring, PI 508408 named Green Pakchoi, PI 633176 named Patria, PI 633163 also called NU 51085, PI 649168 named I.B.468, Ames 2795 named Lembkeho Tabor, and PI 649197 named BC29) were also obtained through GRIN. B. rapa seeds Rubicon F1 were obtained from Johnny's Selected Seeds (Winslow, ME). B. rapa Chiifu-401 seeds were received from Kerry-Sue Peplow at the University of Warwick (UK). Arabidopsis T-DNA lines SALK_099381C, SALK_143246C, SALK_058528C, SALK_021211C, SALK_136027C, SALK_128858C, CS877857 (eli1-1), CS100280 (eli1-2), SALK_121602C, SALK_067071C, CS860142, CS860130, SALK_034662, SALK_144651, CS3254 (flv), and SALK_139995 (dot4-2) were obtained from the Arabidopsis Biological Resource Center (ABRC).
PPR Ortholog Identification
A reciprocal BLAST approach was used to identify putative orthologs in a comparative genomics survey (17). A similar approach was used to identify the function of ELI1; the amino acid sequence for each Arabidopsis thaliana PPR gene was queried in the TBLASTN program to the nonredundant database at Phytozome 7.0 or the Brassica database (BRAD) to identify orthologs. To distinguish orthologs from nonorthologous PPR-containing genes, the amino acid sequences of initial hits was queried by TBLASTN back to the genome of A. thaliana using a nonredundant nucleotide database at GenBankTM. Gene sequences were aligned with ClustalW with the MEGA5 software (39). Cladograms were constructed using MEGA5 (39) for a neighbor-joining tree with the default parameters except that gaps and missing data were analyzed with pairwise deletion.
RNA Editing Analysis
Bulk sequencing was performed at the University of California, Berkeley DNA Sequencing Facility. Quantitation of editing site conversion was performed using the raw trace file. The peak height for C and T traces at the edited position was used to measure editing site conversion as the percentage of T. Percentage of editing was reported to the nearest 10%.
Complementation of the Arabidopsis Mutant eli1-1
Arabidopsis T-DNA seed line CS877857 was grown until vigorously flowering. The full-length ELI1 gene was amplified using genomic DNA and primers that included a 5′ BglII and a 3′ XhoI restriction site. The insert was cloned into the BamHI and SalI sites of pCHF1 (40). Likewise, transformation vectors were constructed for expressing truncated gene fragments of ELI1: ELI1-trc1 (encodes amino acid residues Met-1-Thr-599), ELI1-trc2 (encodes Met-1–Glu-566), ELI1-trc3 (Met-1-Ser-533), ELI1-trc4 (encodes Met-1–Phe-512), and ELI1-trc5 (encodes Met-1–Lys-496). The full-length CRR21 gene was amplified from Nasturtium officinale and Barbarea verna genomic DNA and cloned into pCHF1 (40).
The transformation vectors were electroporated into competent Agrobacterium strain ASE (41). The bacteria were grown to an absorbance of 0.8 at 600 nm and resuspended in 5% sucrose with 0.05% Silwet V77. Flowering plants were dipped into the Agrobacterium culture and allowed to recover 1 day. Seeds were sterilized and selected on agar plates containing ½ Murashige and Skoog salts with 100 mg/liter gentamicin.
DYW Deaminase Domain Expression and Zinc Analysis
Sequences encoding the DYW deaminase domain from ELI1 (residues Lys-490–Trp-632) and DYW1 (Ala-36–Trp-239) were cloned into the BamHI and SalI restriction sites of pET28a. Amplicons for DYW1Δzn1 encoded residues Ala-36 to Lys-166 of DYW1 and were also cloned into pET28a. Plasmids were transformed into Escherichia coli strain Rosetta 2 (DE3) pLysS from Novozymes (Bagsvaerd, Denmark). Strains were grown at 37 °C to an absorbance of 0.5 at 600 nm and then cooled to 18 °C, induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside, and maintained at 18 °C with shaking for 4 h. Bacterial pellets were resuspended in 50 mm Tris, pH 8.0, 250 mm NaCl and sonicated in six bursts of sonication for 20 s. Crude lysates were purified using nickel-nitrilotriacetic acid resin from Thermo Fisher Scientific. Purified protein was dialyzed against 50 mm Tris, pH 8.0, 250 mm NaCl. For the analysis of ELI1-DYW, zinc was provided by an additional dialysis against 50 mm Tris, pH 8.0, 250 mm NaCl, 50 μm ZnCl2. The N-terminal His-tagged portion was removed by digestion with thrombin. One unit of thrombin (BD Biosciences) was added per 1 mg of protein and incubated overnight at 4 °C. Protein extracts were treated with 1 mm PMSF and dialyzed against 20 mm ammonium acetate. For ICP analysis, purified protein was submitted to the Center for Applied Isotope Studies at the University of Georgia (Athens, GA). Highly purified bovine carbonic anhydrase (catalogue number C2624) was purchased from Sigma-Aldrich.
The mass spectrometry analysis of the proteins was performed with a Waters QTOF2 instrument. The instrument uses time-of-flight analysis with an electrospray ion source, and ionization conditions were manipulated to occur under native or denatured conditions. Under native ionization conditions, the protein samples were dialyzed against 20 mm ammonium acetate and ionized in 20 mm ammonium acetate with an electrospray ion voltage of 3.6 kV, a cone voltage of 40 V, and a desolvation temperature of 120 °C. For the denatured ionization condition, the protein sample was denatured with 50% acetonitrile in the presence of 0.1% formic acid. The denatured protein samples were ionized under the same conditions as native samples.
RESULTS
ELI1 Is a DYW Class PPR Gene Required for Editing ndhB C830
A comparative genomics approach was used to identify the gene responsible for editing ndhB C830. A. thaliana has 34 known editing sites in the chloroplast, and B. rapa var. Chiifu 401-42 shares 29 of these sites (Table 1). Three Arabidopsis editing sites (psbE C214, ndhB C836, and rps14 C80) are absent in B. rapa as a result of a genomically encoded T at those positions (Table 1). Two Arabidopsis editing sites are represented as a genomic C in B. rapa, but are not converted to a U: ndhB C830 and a site in the intron of rps12 (A. thaliana plastid genome nucleotide position 69553) (Table 1). Thus, a total of five editing sites including ndhB C830 are present in A. thaliana but are absent in B. rapa.
TABLE 1.
Editing site comparison between A. thaliana and B. rapa Chiifu-401
| Editing site | PPR loci | AT % of C>Ua | BR % of C>Ua | Ref. |
|---|---|---|---|---|
| atpF C92 | 90 | 100 | ||
| accD C794 | RARE1; VAC1 | 100 | 90 | 17, 18 |
| accD C1403b | T | 80 | ||
| At 58642c | VAC1 | 80 | 10 | 18 |
| clpP C559 | CLB19 | 100 | 100 | 12 |
| matK C640 | 90 | 70 | ||
| ndhB C149 | 70 | 100 | ||
| ndhB C467 | CRR28 | 90 | 100 | 14 |
| ndhB C586 | 100 | 100 | ||
| ndhB C746 | CRR22 | 100 | 100 | 14 |
| ndhB C830 | ELI1d | 100 | 0 | |
| ndhB C836 | OTP82 | 90 | T | 15 |
| ndhB C872 | 100 | 90 | ||
| ndhB C1255 | 90 | 20 | ||
| ndhB C1481 | OTP84 | 90 | 100 | 13 |
| ndhD C2 | CRR4 | 20 | 50 | 4 |
| ndhD C383 | CRR21 | 100 | 90 | 16 |
| ndhD C674 | OTP85 | 90 | 100 | 13 |
| ndhD C878 | CRR28 | 70 | 100 | 14 |
| ndhD C887 | CRR22 | 90 | 70 | 14 |
| ndhF C290 | OTP84; VAC1 | 100 | 100 | 13, 18 |
| ndhG C50 | OTP82 | 90 | 90 | 15 |
| petL C5 | 90 | 90 | ||
| psbE C214 | 90 | T | ||
| psbF C77 | LPA66 | 100 | 90 | 11 |
| psbZ C50 | OTP84 | 90 | 80 | 13 |
| rpl23 C89 | OTP80 | 80 | 80 | 13 |
| rpoA C200 | CLB19 | 70 | 90 | 12 |
| rpoB C338 | YS1 | 90 | 80 | 20 |
| rpoB C551 | CRR22 | 90 | 80 | 14 |
| rpoB C566b | N/A | 70 | ||
| rpoB C2432 | 90 | 80 | ||
| rpoC1 C488 | DOT4d | 50 | 100 | |
| At 69553c | OTP81 | 30 | 0 | 13 |
| rps14 C80 | OTP86 | 100 | T | 13 |
| rps14 C149 | 90 | 80 |
a For % C>U, 0 indicates a C present in the transcript and a T indicates a genomic T.
b Editing sites found in this study.
c A. thaliana plastid genome position.
d Novel PPR functions described in this study.
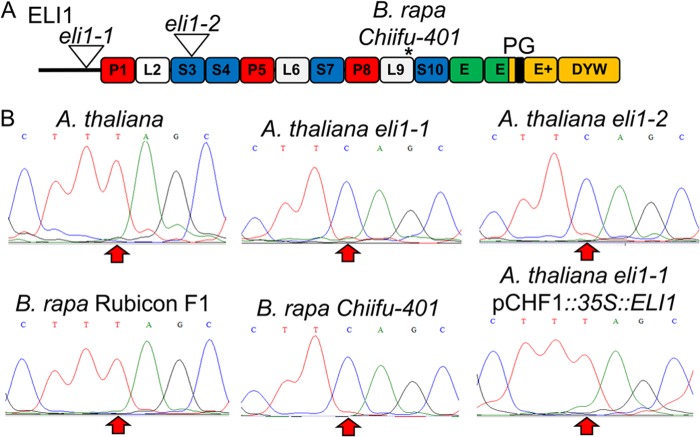
We identified 13 PPR genes that could be responsible for differences in editing between these species based on the presence of a PPR gene in Arabidopsis and the apparent absence of a PPR ortholog in B. rapa (Table 2). Arabidopsis T-DNA insertion lines are available with disruptions in or near 10 Arabidopsis PPR genes that have no apparent ortholog in B. rapa, or the ortholog is predicted to be severely truncated (Table 2). T-DNA insertion lines for 10 candidate PPR genes were screened for editing for the five chloroplast editing sites absent in B. rapa: ndhB C830, ndhB C836, At 69553, rps14 C80, and psbE C214. Two independent T-DNA lines for a DYW type PPR gene (AT4G37380) were identified that were incapable of converting ndhB C830, and thus are called editing lacking insertional mutant 1 (eli1-1 and eli1-2) (Fig. 1). No other defects in chloroplast RNA editing were detected for the 34 known sites in eli1-1 or eli1-2 mutant plants (data not shown). Editing activity was fully restored in eli1-1 mutant plants that express the full-length ELI1 gene (Fig. 1B). The ndhB C830 editing site changes a serine codon to a leucine codon; however, mutant plants do not display a dramatic phenotype under growth chamber conditions (Fig. 1). Chloroplast number and morphology are normal based on confocal imaging.3
TABLE 2.
Candidate genes for novel editing factors
Truncated orthologs are indicated by Trc followed by the location of the truncation.
| Gene | Subclass | B. rapa | T-DNA insertion | ndhB C830 |
|---|---|---|---|---|
| AT1G09410.1 | DYW | Absent | SALK_058528 | U |
| AT1G29710.1 | DYW | Absent | SALK_021211 | U |
| AT1G50270.1 | E+ | Absent | SALK_147964, SALK_019602 | U |
| AT2G25580.1 (MEF8) | DYW | Absent | ||
| T2G27610.1 | DYW | Trc. TP | SALK_121602, SALK_067071 | U |
| AT2G34370.1 | DYW | Absent | SALK_143246a | U |
| AT2G35030.1 | E+ | Trc. E+ | ||
| AT2G36730.1 | E+ | Trc. R4 | ||
| AT2G44880.1 | E+ | Absent | SALK_034662b | U |
| AT3G63370.1 (OTP86) | DYW | Absent | SALK_099381 | U |
| AT4G14820.1 | DYW | Trc. DYW | SALK_136027, SALK_128858b | U |
| AT4G37380.1 (ELI1) | DYW | Trc. R9 | SAIL_899_G08, SET1036–5–3 | C |
| AT5G08490.1 | E+ | Trc. TP | SALK_144651a | U |
a T-DNA insertion in promoter region.
b T-DNA insertion near C terminus.
FIGURE 1.
ELI1 is necessary for editing of ndhB C830. A, PPR gene architecture for ELI1 is shown with the PPR repeat structure. The locations of T-DNA insertions for eli1-1 and eli1-2 are indicated by triangles, and an asterisk indicates the position of an in-frame stop codon in B. rapa Chiifu-401. Red, white, blue, and green boxes represent P, L, S, and E domains, respectively (21). The DYW deaminase domain is colored in gold with the PG box colored in black. B, sequence traces are shown using cDNA templates generated from ndhB transcripts of wild type and transgenic plants. An arrow highlights the position of the editing site (C830).
A truncated ortholog for ELI1 is present in the B. rapa Chiifu-401 genome (Table 2), and these plants are incapable of editing ndhB C830 (Fig. 1). B. rapa Chiifu-401 grows vigorously with no observable growth phenotype under laboratory conditions (data not shown). ELI1 orthologs were sequenced from 16 Brassica species with the same genomic background as the sequenced B. rapa (the A genome). Truncation of the ELI1 gene and loss of editing ndhB C830 are only present in the B. rapa strain Chiifu-401 (data not shown). B. rapa variety Rubicon F1 has an uninterrupted ELI1 ortholog, and ndhB C830 was found to be fully edited (Fig. 1).
DOT4/FLV Is a DYW Class PPR Gene That Is Required for Editing rpoC1 C488
A gene named defectively organized tributaries 4 (DOT4) had been characterized with T-DNA knock-out plants in a DYW class PPR gene, AT4G18750, and has a striking phenotype of serrated white to pale green leaves, and the formation of radially symmetric, needle-shaped leaves (42). There are extensive phenotypic similarities between (dot4-2) and flavodentata (flv) (43) The mutant flv has been described as an editing mutant (44). The similarities between the unusual phenotypes suggest that the same gene is disrupted in both mutants.
Mutant plants dot4-2 (SALK_139995) and flv were screened for editing at all 34 known chloroplast sites. The rpoC1 C488 editing site was found to remain unedited in both mutant lines (Fig. 2), whereas all of the other known editing sites in Arabidopsis chloroplasts were edited (data not shown). In addition, the DOT4 gene was recovered from mutant flv plants, and has a point mutation at Gly-554 (GGA → TGA) that creates a stop codon in repeat 11 (data not shown). Therefore, disruption of the DOT4 gene leads to an inability of plants to edit rpoC1 transcripts and results in the unusual phenotype. Although the mutant has a pronounced leaf phenotype, no gross abnormalities in chloroplast number or morphology are present in green tissues of the mutant compared with wild type plants.3
FIGURE 2.
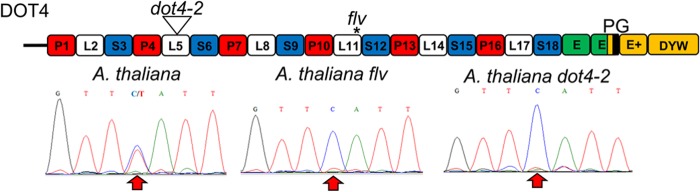
DOT4 is essential for rpoC 488 editing. PPR gene architecture for DOT4 is shown on top. A triangle or asterisk indicates the position of a T-DNA insertion or in-frame stop codon, respectively. Sequence traces are shown using cDNA templates generated from rpoC1 transcripts illustrating the region 485/491 with an arrow indicating the position of the editing site (C488). Red, white, blue, and green boxes represent respective P, L, S, and E domains (21). The DYW deaminase domain is colored in gold with the PG-box colored in black.
The DYW Deaminase Domain Binds Two Zinc Atoms
All of the known PPR genes that are required for RNA editing include an intact or truncated DYW deaminase domain. DYW1 is a small PPR protein with a modified repeat region and an intact C-terminal DYW deaminase domain, and was recently reported to be required for editing ndhD C2 (38). Because the DYW deaminase domains from ELI1 and DYW1 could potentially serve a catalytic function in the editing mechanism, and zinc is a critical cofactor for most nucleotide deaminases, zinc binding by the DYW deaminase domain was investigated.
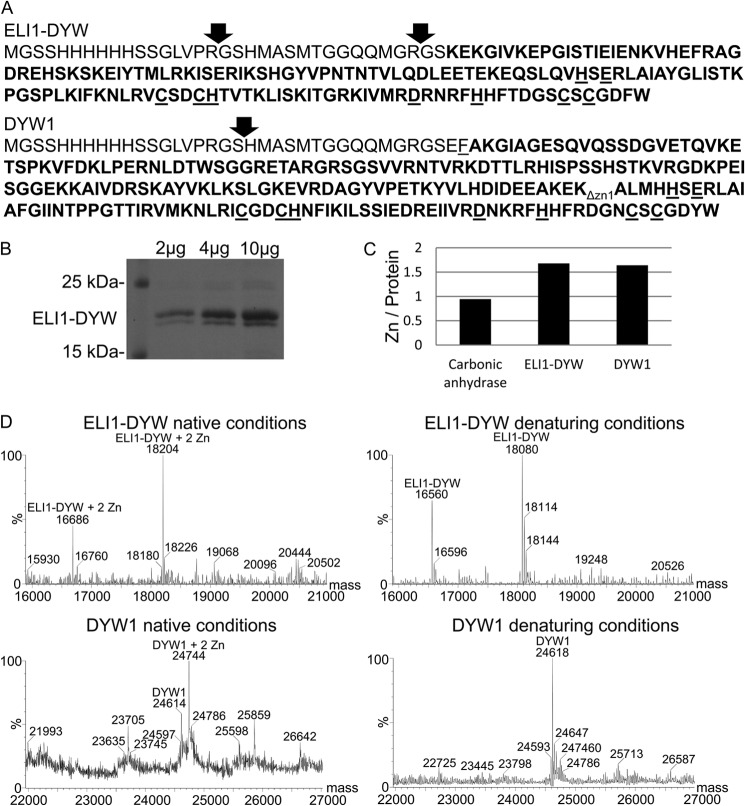
The DYW deaminase domains from ELI1 and DYW1 were expressed as His6-tagged polypeptides, purified by nickel affinity chromatography, treated with thrombin, and dialyzed to remove the N terminus containing the His6 tag (Fig. 3A). For ELI1-DYW, the purified protein preparation ran as a doublet of two polypeptides of about 17 and 18 kDa on SDS-PAGE (Fig. 3B), whereas DYW1 ran as a single band (data not shown). To identify the metal ions present in the recombinant DYW deaminase domain, the polypeptides were subjected to metal analysis by ICP-OES. Molar zinc ratios are 1.7 and 1.6 zinc atoms per recombinant DYW deaminase polypeptide for ELI1-DYW and DYW1, respectively (Fig. 3C). No additional metals were detected above trace amounts (data not shown). A highly purified commercial preparation of bovine erythrocyte carbonic anhydrase was included as a control and was shown to contain 0.9 zinc atoms per monomer. Each molecule of carbonic anhydrase is expected to bind 1 zinc atom.
FIGURE 3.
The recombinant DYW deaminase domain binds two zinc atoms per polypeptide. A, bolded sequences represent the recombinant ELI1-DYW and DYW1 deaminase domain. Thrombin cleavages confirmed by MS are indicated by arrows. Conserved amino acids capable of coordinating zinc are underlined. The C terminus of DYW1ΔZn1 precedes the subscript label ΔZn1. B, a Coomassie Blue-stained SDS-PAGE gel showing the protein used for zinc analysis of ELI1 DYW deaminase domain. C, a bar graph indicates the results of metal analysis by ICP-OES. D, at left, MS spectra indicate the projected mass for recombinant ELI1-DYW and DYW1 under native conditions. At right, spectra are for denaturing conditions.
Native MS can determine the molecular mass of polypeptides under conditions that retain prosthetic groups (45, 46), and covalently bound ligands can be identified based on the difference between native and denaturing MS spectra. Recombinant ELI1-DYW protein was analyzed under native MS conditions, and two polypeptides with average molecular masses of 18,204 and 16,686 Da were identified after application of the deconvolution algorithm (Fig. 3D). Denaturing MS revealed polypeptide masses for ELI1-DYW of 18,080 and 16,560 Da; the mass differences between the native and denaturing analyses are 124 and 126 Da (Fig. 3D). Zinc has an atomic mass of 65.4; however, cysteines coordinate zinc directly to the sulfur atom with loss of a proton (47). The expected mass difference for a polypeptide with two zinc atoms coordinated by four cysteine residues would be about 127 Da. Thus, the observed mass difference is consistent with 2 bound zinc atoms. The native MS spectra of ELI1-DYW did not detect the presence of polypeptides with no zinc or a single zinc atom (Fig. 3D).
Recombinant DYW1 was also analyzed under native conditions. The most prevalent peak corresponds to a polypeptide with a mass of 24,744 Da, and a smaller peak at 24,614 Da was also observed. Under denaturing conditions, a single major form of the polypeptide was detected with a mass of 24,618 Da. The mass difference for the major form observed in the native MS is 126 Da, and is consistent with two zinc atoms per DYW1 polypeptide. In addition, the native MS shows a polypeptide with a mass of 24,614, and this form appears to be DYW1 polypeptides with no bound zinc atoms. Polypeptides with a mass corresponding to a single zinc atom are not observed in the MS spectra.
The region of DYW1 that contains the putative zinc-coordinating residues is in the C terminus (Fig. 3A). A truncated DYW1 protein, DYW1Δzn1, was prepared that places a stop codon just upstream of the first zinc binding motif (HSE) and eliminates the entire zinc binding region. Native and denaturing MS spectra of DYW1ΔZn1 polypeptide showed no significant mass difference (data not shown), and the zinc binding by DYW1 requires the C-terminal region of the DYW deaminase domain.
Truncation of the ELI1 Gene Identifies the 15-amino Acid PG Box That Is Critical for Editing
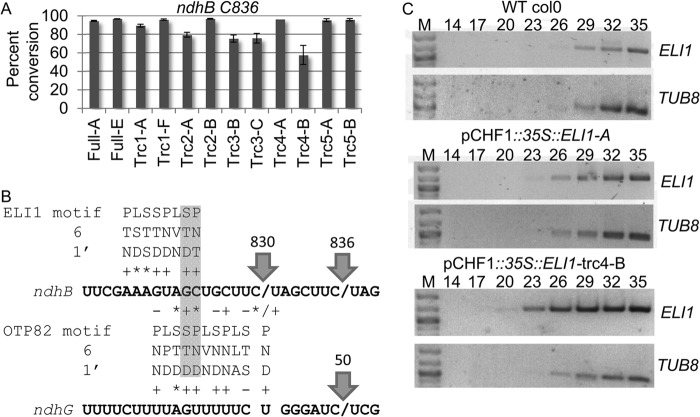
Truncated forms of CRR22, CRR28, and OTP82 that eliminate portions of the DYW deaminase domain are capable of restoring RNA editing in respective PPR knock-out mutants (14, 15). To further examine the role of the ELI1 DYW deaminase domain, C-terminal truncation variants of ELI1 were expressed in eli1-1 plants (Fig. 4A). Five ELI1 variants were generated with truncations in the DYW deaminase domain (Fig. 4A). ELI1 transgenes with truncations C-terminal to the PG box (trc 1–4) were capable of fully restoring editing at C830; however, truncation immediately before the PG box (trc5) resulted in a dramatic decrease in editing site conversion (Fig. 4B). None of these truncations affected editing at ndhB C746 (Fig. 4B) or distal sites in ndhB transcripts (data not shown). Thus, the function of the ELI1 gene in a phenotype complementation assay requires that the gene encode the PG box, but sequences encoding amino acids beyond the PG box are not required for conversion of ndhB C830.
FIGURE 4.
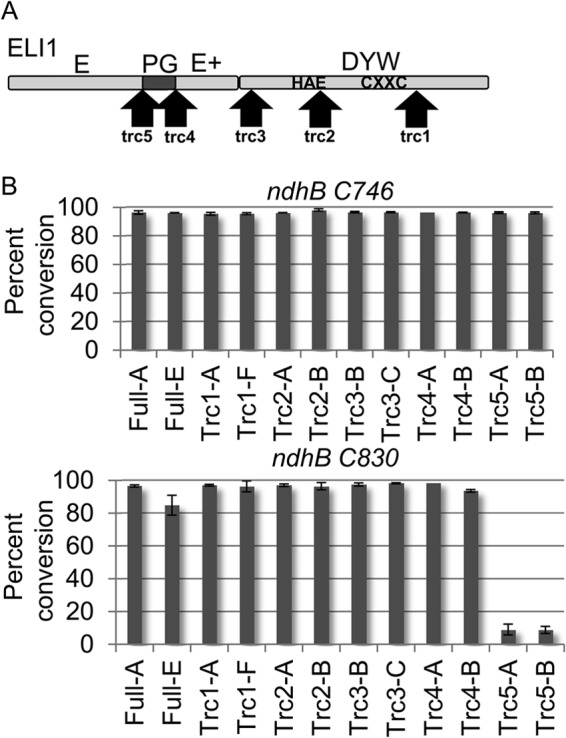
Truncated forms of rELI1 that have the PG box complement eli1-1 mutants. A, the C-terminal portion of ELI1 is shown with arrows indicating the position of five truncations. B, histograms indicate the average percentage of conversion for editing sites in ndhB at C746 and C830. All transformants are in the eli1-1 T-DNA background. For x axis labels, letters after a dash indicate a plant from an independent transformation event.
Overexpression of ELI1 Reduces Editing at ndhB C836 through Competitive Binding with OTP8
Several transgenic plants expressing ELI1 variants convert the adjacent editing site, ndhB C836, to a lower extent than wild type plants (Fig. 5A). The ndhB C836 editing site is only 5 nucleotides away from the ELI1 target and requires a PPR protein, OTP82, for conversion (15). The ELI1-trc4-B plant expresses a truncated variant of ELI1 with the PG box and exhibits the most dramatic decrease in conversion of ndhB C836 (Fig. 5A). The amino acid residues at positions 6 and 1′ in a repeat can be used to infer the nucleotides recognized by each individual PPR (23), and application of the PPR code indicates that ELI1 and OTP82 share overlapping cis-elements (Fig. 5B). Semiquantitative RT-PCR demonstrates that the expression of the ELI1 transgene in the plant with the greatest suppression in ndhB C836 editing (trc4-B) is substantially greater than other transgenics or the native ELI1 gene (Fig. 5C). OTP82 has two editing site targets, and editing of the second target, ndhG C50, was not significantly different between plant trc4-B and wild type plants (data not shown). These results suggest that overexpression of ELI1 suppressed editing of a nearby editing site by binding to overlapping nucleotides in the cis-elements.
FIGURE 5.
An overexpressed PPR protein can compete with a second PPR for an overlapping binding site. A, a histogram displays the average percentage of conversion of ndhB C836 in transgenic plants expressing truncated ELI1. B, putative cis-elements for ELI1 and OTP82 are represented based on the PPR code (23). +, *, and − indicate absolute correlation with the combinatorial code, correlation of position 6 with purine/pyrimidine, and anti-correlation, respectively. Arrows point to known RNA editing sites. C, semiquantitative RT-PCR was used to determine transgene expression levels among transformants relative to a tubulin control (TUB8). The number of PCR cycles is shown at the top of each lane. M indicates molecular mass markers.
The PG Box of CRR21 Is Essential for Editing ndhD C383
To determine whether retention PG box is a general feature of other chloroplast PPR genes that function in editing, we identified T-DNA insertions or natural mutations that truncate PPR genes and examined editing function. The T-DNA insertion in crr21-3 is immediately upstream of the PG box and truncates CRR21 with loss of the PG box (Fig. 6A). The only known editing site target of CRR21 is ndhD C383, and crr21-3 plants fail to edit C383 (Fig. 6B). The CRR21 ortholog in B. verna, BvCRR21, contains a truncation in the 17th PPR repeat (Fig. 6A), and the ndhD C383 editing site is lost in that species through substitution to a genomic T (37). N. officinale is a close relative of B. verna, but edits ndhD C383 and contains a CRR21 ortholog with an intact coding sequence through the end of the E+ domain (37). Transgenic Arabidopsis crr21-3 plants expressing NoCRR21 are capable of editing ndhB C383; however, plants expressing BvCRR21 fail to edit ndhD C383 (Fig. 6C). These results provide an additional example of gene truncation 5′ to the PG box, resulting in loss of editing capability.
FIGURE 6.
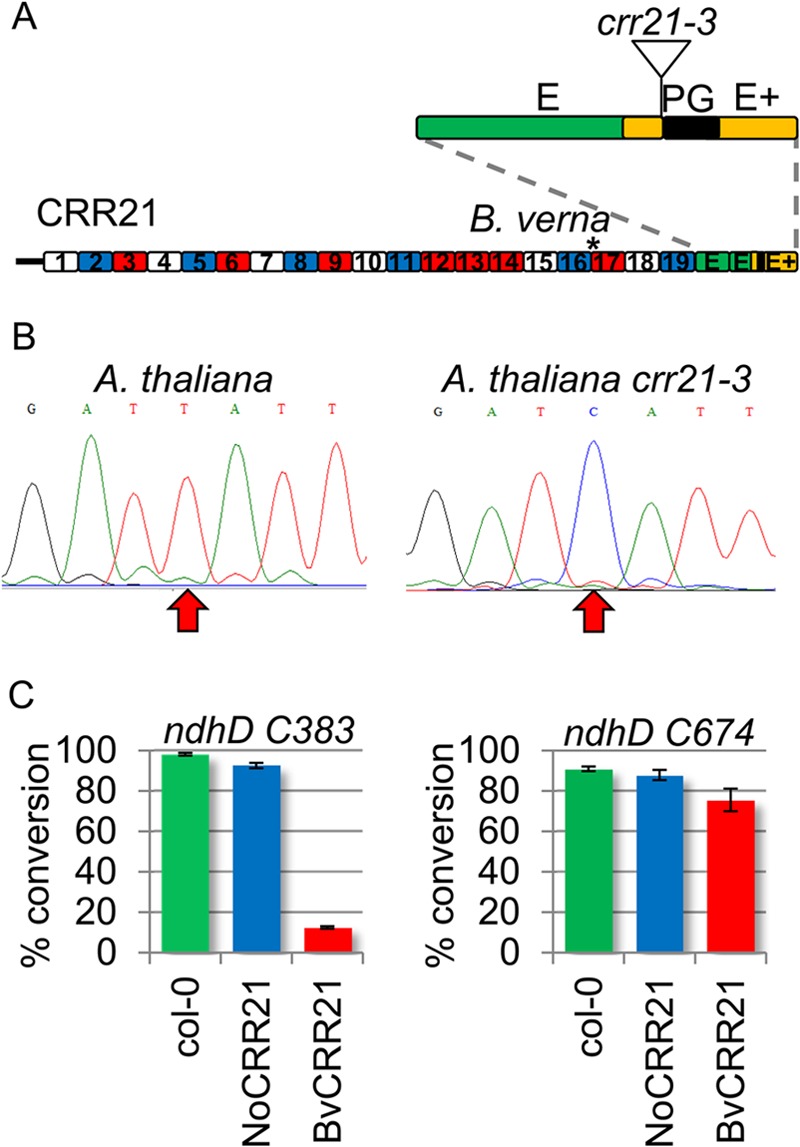
The PG box is essential for CRR21 editing function. A, red, white, blue, and green boxes represent P, L, S, and E designations. The DYW deaminase is colored gold with the PG-box colored black. The C-terminal region is represented as an inset and shows the proximal location of the crr21-3 T-DNA insertion. The location of a truncation in B. verna is indicated by an asterisk. B, sequence traces demonstrate the presence or absence of editing in wild type Arabidopsis and crr21-3. An arrow highlights ndhD C383, the target of CRR21. C, a bar graph shows the percentage of conversion of ndhD editing sites in wild type (green) and transformants in the crr21-3 background expressing CRR21 genes from N. officinale (blue) and B. verna (red).
The PG Box Is Highly Conserved in PPR Proteins Required for Chloroplast RNA Editing
With the addition of ELI1 and DOT4, 17 PPR proteins have been identified to be necessary for editing at least one chloroplast RNA editing site. The amino acid sequence of the PG box exhibits a high degree of sequence identity and conservation in chloroplast PPR proteins that are required for editing (Fig. 7A). The motif is not as highly conserved in mitochondrial proteins required for editing (Fig. 7B). In addition to the high coincidence among Arabidopsis PPR proteins, the PG box is highly conserved in ELI1 orthologs (Fig. 7C). The PG box corresponds to the first β-strand (S1) in the deaminase-fold (27) and is predicted to be in close association with the HAE and CXXC motifs required for zinc binding at the catalytic site (Fig. 7D).
FIGURE 7.
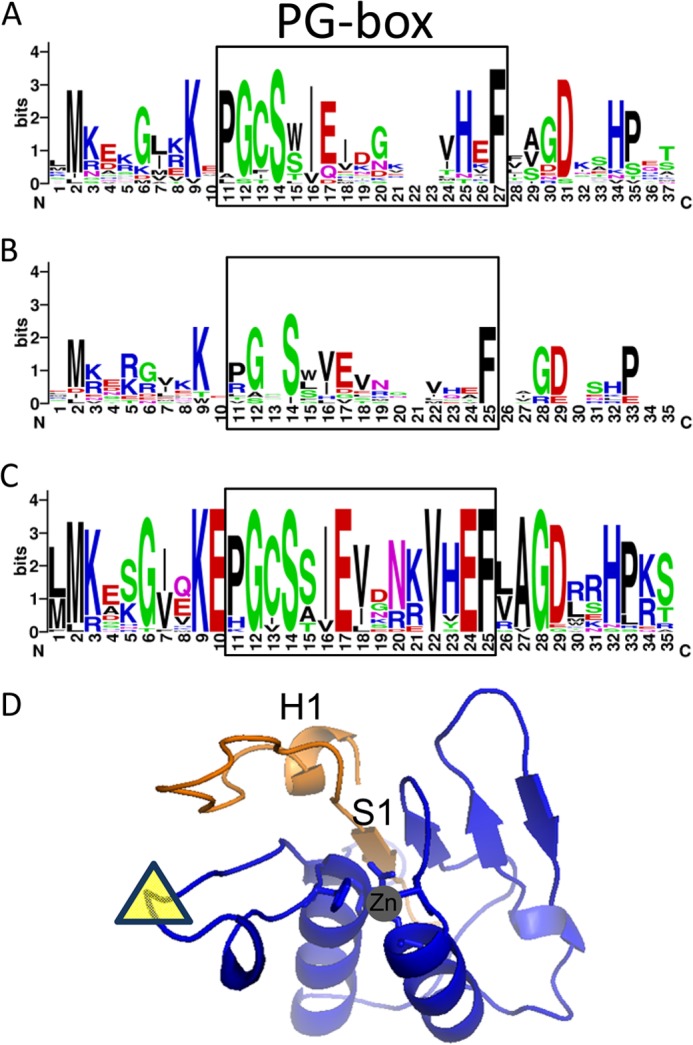
Amino acid residues in the PG Box are highly conserved in chloroplast PPR proteins required for editing. A, the coincidence of amino acid residues in the PG box of 17 PPR proteins that are required for editing chloroplast transcripts is represented by a WebLogo plot. B, a WebLogo plot shows the coincidence of amino acid residues in the PG box of 12 PPR proteins that are required for editing mitochondrial transcripts: REME1, SLO1, MEF1, MEF8, MEF9, MEF18, MEF14, MEF19, MEF20, MEF21, MEF22, and OTP87. C, conservation of amino acid residues is displayed by a WebLogo plot for 25 ELI1 orthologs in plants: A. thaliana, Arabidopsis lyrata, Capsella rubella, B. rapa, Citrus sinensis, Citrus clementine, Eucalyptus grandis, Vitis vinifera, Mimulus guttatus, Aquilegia coerulea, Sorghum bicolor, Zea mays, Setaria italica, Oryza sativa, Brachypodium distachyon, Manihot esculenta, Ricinus communis, Linum usitatissimum, Populus trichocarpa, Phaseolus vulgaris, Glycine max, Cucumis sativus, Prunus persica, and Malus domestica. D, a model of the ELI1 DYW structure based on a DYW deaminase model (27). The location of a large amino acid extension specific to DYW deaminase and thus absent in the model is indicated by a yellow triangle. The minimal portion of the DYW deaminase capable of complementing the eli1-1 mutant is represented by the orange polypeptide chain. The location of the zinc ion based on the deaminase fold is shown by the gray circle.
The DYW Domain Experiences Truncation in Evolution, and OTP80 in the Brassicaceae Is Derived from a Gene with a Full-length DYW
PLS-type PPR proteins have been divided into subtypes based on the presence of the E, E+, or DYW domain at the C terminus of the polypeptide (21). Four PPR proteins (CRR4, CRR21, CLB19, and OTP80) with truncated C-terminal domains are currently known to be required for RNA editing in chloroplasts (4, 12, 13, 16), and numerous mitochondrial editing factors (MEF3, MEF18 MEF19, MEF20, MEF21, OTP87, OTP71, and OTP72) are members of the E-class (33–36). To examine whether DYW class PPR genes have experienced truncation in evolution, the architecture of OTP80 genes was examined in plants with sequenced genomes (Fig. 8A). Among these taxa, truncated orthologs for OTP80 are only observed in the order Brassicales, and they include all sequenced examples from the Brassicaceae (Fig. 8A). Additional C-terminal sequences for OTP80 genes were obtained by 3′ rapid amplification of cDNA ends and demonstrated that Reseda lutea (Resedaceae), a member of the order Brassicales, has a full-length DYW (Fig. 8B). Other members of the Brassicales such as Capparis (Capparaceae) as well as Cleome, Brassica, and Arabidopsis (Brassicaceae) contain truncated orthologs. In all cases, truncation occurred within the E+ domain, and the truncated orthologs retain the PG box. These data suggest that a single truncation event occurred in a branch of the Brassicales that lead to the genera Cleome, Capparis, Brassica, and Arabidopsis (Fig. 8B). The editing target of OTP80, rpl23 C89, is present in these species and was edited in plants with full-length or truncated OTP80 genes (Fig. 8B). Thus, the DYW deaminase domain is susceptible to truncation in evolution, and the truncated orthologs retain the capacity to edit a target site.
FIGURE 8.
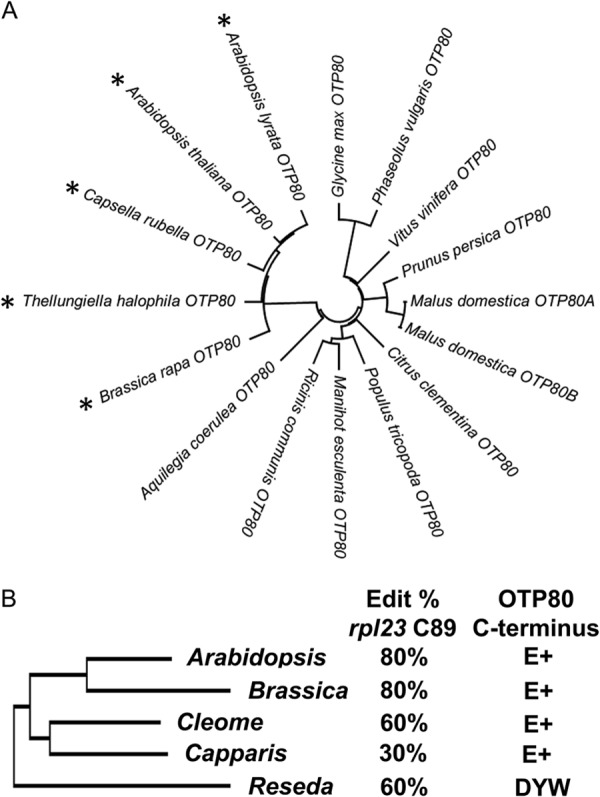
A truncation in OTP80 has occurred in one branch of Brassicales. A, an unrooted neighbor-joining phylogenetic tree indicates the relationship between OTP80 orthologs found in several sequenced dicots. The phylogenetic tree is based on an alignment of codons from Arabidopsis codon Ser-80 to Lys-580. An asterisk indicates species that have predicted stop codons that truncate the OTP80 protein. B, at left is a neighbor-joining phylogenetic tree that was constructed with rbcL sequences from A. thaliana, B. rapa, Cleome hassleriana, Capparis spinosa, and R. lutea and that has the same topology as a phylogenetic survey using rbcL and 18 S sequences (56). The editing status of rpl23 C89 is shown for A. thaliana, B. rapa, Cleome gynandra, C. spinosa, and R. lutea. The C-terminal domains of these OTP80 orthologs are shown on the right.
DISCUSSION
The identity of the catalytic component of the C-to-U editing apparatus has been elusive. The higher plant DYW domain was recognized to bear similarities to deaminases based on the presence and proximity of canonical zinc binding motifs (25), and a recent informatics analysis based on structural motifs places the DYW deaminase domain into a deaminase superfamily (27). Most members of the deaminase superfamily have a single zinc atom at the active site that is coordinated in a tetrahedral configuration by a histidine or cysteine residue ((H/C)XE) and two cysteine residues (CXXC). The fourth ligand of the zinc atom is the substrate water molecule, which is deprotonated by the glutamate residue to facilitate nucleophilic attack in the deamination reaction (26). The detection of zinc in the DYW deaminase domain supports a catalytic function in C-to-U editing.
The presence of a second zinc atom associated with the DYW deaminase is an intriguing result. Most of the deaminase superfamily members bind a single zinc atom (48–50); however, the DYW deaminase domain was predicted to have a second zinc binding site at the C terminus (27). The observation of two zinc atoms confirms the prediction of a second zinc binding site; however, the role of the second zinc atom is unknown. The detection of only DYW1 polypeptides with either two or zero zinc atoms suggests that both zinc atoms are required for protein stability and may bind cooperatively. The region of the DYW domain (HHFTDGSCSCGDFW) is highly conserved in PPR proteins, and the first H residue and the CSC are invariant in Arabidopsis and Naegleria DYW domains. Many zinc binding motifs include cysteine and histidine residues separated by 2 or more residues (51, 52); however, other configurations are well known. The SWIM domain is a zinc finger-like domain that is commonly found in plants in MuDR transposases and in FAR1, a transcription factor (53). The zinc finger-like domain in these proteins employ zinc binding residues separated by a single amino acid (CXC and CXH), and CSC is frequently observed. Thus, the zinc ring finger-like structures in the SWIM domain have a similar signature as the putative second site in the DYW deaminase domain. Because transposases and transcription factors are both likely to directly interact with nucleic acids through these ring finger-like SWIM domains, the highly conserved HX6CSC motif in the DYW deaminase domain might provide an additional site for substrate RNA interactions.
The DYW deaminase domain has been shown to be dispensable for PPR gene function as assayed in genetic complementation assays (Figs. 4 and 6) (14, 15). Many chloroplast editing factors have been reported that lack an intact DYW deaminase domain (4, 12, 13, 16); however, all of these PPR proteins (CRR4, CRR21, CLB19, and OTP80) include the entire PG box plus an additional 11–41 amino acid residues. These results have led investigators to refer to PPRs as site recognition factors (14, 54) In this work, we pinpoint a highly conserved 15-amino acid region, the PG box, as a critical feature that is required for editing site conversion. Although the function of the PG box is unknown, it could be required to form a complex with other editing components. The amino acid sequence of the PG box exhibits heterogeneity between editing factors, especially between PPR proteins that function in chloroplasts and in mitochondria (Fig. 7). The distinguishing characteristics of the PG box may explain in part why the E domains from different editing PPR proteins are not functionally equivalent (33). PPR proteins with different PG boxes may interact with different binding partners in the formation of editing complexes.
Considerable heterogeneity exists in the C-terminal region of the PLS-type PPR genes (21). The evolutionary relationship of variants in the DYW deaminase has been an enigma given the potential enzymatic role of the C terminus. The molecular evolution of four PPRs required for chloroplast editing indicated that all regions within the DYW deaminase domain exhibit strong negative selection (37). In this work, we demonstrate that the DYW deaminase domain of OTP80 is unstable in evolution and has undergone truncation during the evolution of the Brassicales (Fig. 8). These results suggest that the PLS-type PPRs that are required for RNA editing may have dual functions that are revealed both by molecular manipulations and in evolution. The PLS repeat portion of the PPR is required for editing site recognition, and this portion of the gene is retained when the editing site is present; however, when editing sites are lost, the PPR gene may be lost or converted to a pseudogene (37). Thus, portions of the DYW domain are dispensable in recombinant molecular analyses, and the dispensable nature of DYW deaminase domain is also detected in evolution. Apparently the RNA specificity function of the PPR protein provided by the PLS repeat domain is critical, but the DYW domain has a distinct and separable function that may be lost, and apparently provided in trans by another protein.
Thus, there appears to be a conundrum with respect to the structure of the DYW domain and its potential catalytic involvement in the editing reaction. The DYW deaminase domain has key features that suggest that it has a catalytic role in cytidine deamination; however, large portions of the DYW deaminase domain are dispensable. These observations support a model that a PPR protein serves a RNA recognition function and that the editing deaminase activity can be provided in trans (14, 16, 55). A recent study demonstrated that two genes are responsible for editing ndhD C2 (38). CRR4 is a PLS-type PPR protein with a truncated DYW deaminase domain and provides editing site recognition, and DYW1 contains an intact DYW deaminase domain and could serve a catalytic function (38). These results provide an example of two genes acting in trans to perform editing site recognition and editing site conversion. This work, taken together with results presented here, suggests that PPR proteins may be capable of working in pairs with one PPR protein providing the site specificity and a second PPR protein providing the catalytic activity for the deamination reaction. Although there are relatively few examples of multiple PPR genes necessary for the same editing site (13, 17, 19), it is possible that there is functional redundancy of the PPR protein that provides the deaminase activity so that any one of several PPRs could serve a catalytic function. The RIP/MORF protein family members each have broad effects on various subgroups of editing sites (3, 5), and multiple PPR proteins could potentially bind to each RIP/MORF to provide close proximity of PPR proteins for site specificity and catalysis.
Acknowledgments
We thank Chelsea G. Pham for helping clone ELI1-DYW into the pET28a vector. We thank Amir Sattarzadeh, Ph.D. and Prof. Maureen Hanson for examining plastids from eli1 and dot4 mutants using confocal microscopy. We thank Prof. Thomas Poulos for guidance with the expression and analysis of the recombinant ELI1 DYW domain polypeptide.
This work was supported by National Science Foundation Grant MCB-0929423 (to R. M. M.).
M. R. Hanson, personal communication.
- PPR
- pentatricopeptide repeat
- RIP
- RNA editing factor-interacting protein
- MORF
- multiple organellar RNA editing factor
- PI
- Plant Introduction number from the National Plant Germplasm System
- ICP-OES
- inductively coupled optical emission spectroscopy
- trc
- truncation.
REFERENCES
- 1. Covello P. S., Gray M. W. (1989) RNA editing in plant mitochondria. Nature 341, 662–666 [DOI] [PubMed] [Google Scholar]
- 2. Gualberto J. M., Lamattina L., Bonnard G., Weil J. H., Grienenberger J. M. (1989) RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341, 660–662 [DOI] [PubMed] [Google Scholar]
- 3. Bentolila S., Heller W. P., Sun T., Babina A. M., Friso G., van Wijk K. J., Hanson M. R. (2012) RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc. Natl. Acad. Sci. U.S.A. 109, E1453–E1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kotera E., Tasaka M., Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330 [DOI] [PubMed] [Google Scholar]
- 5. Takenaka M., Zehrmann A., Verbitskiy D., Kugelmann M., Härtel B., Brennicke A. (2012) Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bentolila S., Elliott L. E., Hanson M. R. (2008) Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics 178, 1693–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes M. L., Hanson M. R. (2007) Identification of a sequence motif critical for editing of a tobacco chloroplast transcript. RNA 13, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miyamoto T., Obokata J., Sugiura M. (2004) A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts. Proc. Natl. Acad. Sci. U.S.A. 101, 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chaudhuri S., Maliga P. (1996) Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J. 15, 5958–5964 [PMC free article] [PubMed] [Google Scholar]
- 10. Takenaka M., Neuwirt J., Brennicke A. (2004) Complex cis-elements determine an RNA editing site in pea mitochondria. Nucleic Acids Res. 32, 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai W., Ji D., Peng L., Guo J., Ma J., Zou M., Lu C., Zhang L. (2009) LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150, 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chateigner-Boutin A. L., Ramos-Vega M., Guevara-García A., Andrés C., de la Luz Gutiérrez-Nava M., Cantero A., Delannoy E., Jiménez L. F., Lurin C., Small I., León P. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602 [DOI] [PubMed] [Google Scholar]
- 13. Hammani K., Okuda K., Tanz S. K., Chateigner-Boutin A. L., Shikanai T., Small I. (2009) A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21, 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okuda K., Chateigner-Boutin A. L., Nakamura T., Delannoy E., Sugita M., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21, 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okuda K., Hammani K., Tanz S. K., Peng L., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2010) The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 61, 339–349 [DOI] [PubMed] [Google Scholar]
- 16. Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. U.S.A. 104, 8178–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robbins J. C., Heller W. P., Hanson M. R. (2009) A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15, 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tseng C. C., Sung T. Y., Li Y. C., Hsu S. J., Lin C. L., Hsieh M. H. (2010) Editing of accD and ndhF chloroplast transcripts is partially affected in the Arabidopsis vanilla cream1 mutant. Plant Mol. Biol. 73, 309–323 [DOI] [PubMed] [Google Scholar]
- 19. Yu Q. B., Jiang Y., Chong K., Yang Z. N. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 59, 1011–1023 [DOI] [PubMed] [Google Scholar]
- 20. Zhou W., Cheng Y., Yap A., Chateigner-Boutin A. L., Delannoy E., Hammani K., Small I., Huang J. (2009) The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96 [DOI] [PubMed] [Google Scholar]
- 21. Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Le Ret M., Martin-Magniette M. L., Mireau H., Peeters N., Renou J. P., Szurek B., Taconnat L., Small I. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andres C., Lurin C., Small I. D. (2007) The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plantarum 129, 14–22 [Google Scholar]
- 23. Barkan A., Rojas M., Fujii S., Yap A., Chong Y. S., Bond C. S., Small I. (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. (2013) Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8, e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salone V., Rüdinger M., Polsakiewicz M., Hoffmann B., Groth-Malonek M., Szurek B., Small I., Knoop V., Lurin C. (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581, 4132–4138 [DOI] [PubMed] [Google Scholar]
- 26. Betts L., Xiang S., Short S. A., Wolfenden R., Carter C. W., Jr. (1994) The 2.3 Ä crystal structure of an enzyme: Transition-state analog complex. J. Mol. Biol. 235, 635–656 [DOI] [PubMed] [Google Scholar]
- 27. Iyer L. M., Zhang D., Rogozin I. B., Aravind L. (2011) Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 39, 9473–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teng B., Burant C. F., Davidson N. O. (1993) Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260, 1816–1819 [DOI] [PubMed] [Google Scholar]
- 29. Xie K., Sowden M. P., Dance G. S. C., Torelli A. T., Smith H. C., Wedekind J. E. (2004) The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1. Proc. Natl. Acad. Sci. U.S.A. 101, 8114–8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knoop V., Rüdinger M. (2010) DYW-type PPR proteins in a heterolobosean protist: plant RNA editing factors involved in an ancient horizontal gene transfer? FEBS Lett. 584, 4287–4291 [DOI] [PubMed] [Google Scholar]
- 31. Rüdinger M., Fritz-Laylin L., Polsakiewicz M., Knoop V. (2011) Plant-type mitochondrial RNA editing in the protist Naegleria gruberi. RNA 17, 2058–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schallenberg-Rüdinger M., Lenz H., Polsakiewicz M., Gott J. M., Knoop V. (2013) A survey of PPR proteins identifies DYW domains like those of land plant RNA editing factors in diverse eukaryotes. RNA Biol. 10, 1343–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chateigner-Boutin A. L., Colas des Francs-Small C., Fujii S., Okuda K., Tanz S. K., Small I. (2013) The E domains of pentatricopeptide repeat proteins from different organelles are not functionally equivalent for RNA editing. Plant J. 74, 935–945 [DOI] [PubMed] [Google Scholar]
- 34. Hammani K., des Francs-Small C. C., Takenaka M., Tanz S. K., Okuda K., Shikanai T., Brennicke A., Small I. (2011) The pentatricopeptide repeat protein OTP87 is essential for RNA editing of nad7 and atp1 transcripts in Arabidopsis mitochondria. J. Biol. Chem. 286, 21361–21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takenaka M., Verbitskiy D., Zehrmann A., Brennicke A. (2010) Reverse genetic screening identifies five E-class PPR proteins involved in RNA editing in mitochondria of Arabidopsis thaliana. J. Biol. Chem. 285, 27122–27129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Verbitskiy D., van der Merwe J. A., Zehrmann A., Härtel B., Takenaka M. (2012) The E-class PPR protein MEF3 of Arabidopsis thaliana can also function in mitochondrial RNA editing with an additional DYW domain. Plant Cell Physiol. 53, 358–367 [DOI] [PubMed] [Google Scholar]
- 37. Hayes M. L., Giang K., Mulligan R. M. (2012) Molecular evolution of pentatricopeptide repeat genes reveals truncation in species lacking an editing target and structural domains under distinct selective pressures. BMC Evol. Biol. 12, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boussardon C., Salone V., Avon A., Berthomé R., Hammani K., Okuda K., Shikanai T., Small I., Lurin C. (2012) Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24, 3684–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fankhauser C., Yeh K. C., Lagarias J. C., Zhang H., Elich T. D., Chory J. (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541 [DOI] [PubMed] [Google Scholar]
- 41. Fraley R. T., Rogers S. G., Horsch R. B., Eichholtz D. A., Flick J. S., Fink C. L., Hoffmann N. L., Sanders P. R. (1985) The SEV system – a new disarmed Ti plasmid vector system for plant transformation. Biotechnology 3, 629–635 [Google Scholar]
- 42. Petricka J. J., Clay N. K., Nelson T. M. (2008) Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 56, 251–263 [DOI] [PubMed] [Google Scholar]
- 43. Serrano-Cartagena J., Robles P., Ponce M. R., Micol J. L. (1999) Genetic analysis of leaf form mutants from the Arabidopsis Information Service collection. Mol. Gen. Genet. 261, 725–739 [DOI] [PubMed] [Google Scholar]
- 44. Bersoult A., Chateigner- A.-L., Boutin, Hammani K., Yap A., Delannoy E., Kahlau S., Small I. (2010) Roles for RNA editing factors in leaf development. in 20th International Conference on Arabidopsis Research, Abstract C03, p. 54, Edinburgh, United Kingdom [Google Scholar]
- 45. Loo J. A. (1997) Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass. Spectrom. Rev. 16, 1–23 [DOI] [PubMed] [Google Scholar]
- 46. van den Heuvel R. H., Heck A. J. R. (2004) Native protein mass spectrometry: from intact oligomers to functional machineries. Curr. Opin. Chem. Biol. 8, 519–526 [DOI] [PubMed] [Google Scholar]
- 47. Dudev T., Lim C. (2002) Factors governing the protonation state of cysteines in proteins: An ab initio/CDM study. J. Am. Chem. Soc. 124, 6759–6766 [DOI] [PubMed] [Google Scholar]
- 48. Johansson E., Mejlhede N., Neuhard J., Larsen S. (2002) Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 Ä resolution. Biochemistry 41, 2563–2570 [DOI] [PubMed] [Google Scholar]
- 49. Losey H. C., Ruthenburg A. J., Verdine G. L. (2006) Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159 [DOI] [PubMed] [Google Scholar]
- 50. Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S. (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 [DOI] [PubMed] [Google Scholar]
- 51. Maret W. (2012) New perspectives of zinc coordination environments in proteins. J. Inorg. Biochem. 111, 110–116 [DOI] [PubMed] [Google Scholar]
- 52. Namuswe F., Berg J. M. (2012) Secondary interactions involving zinc-bound ligands: Roles in structural stabilization and macromolecular interactions. J. Inorg. Biochem. 111, 146–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Makarova K. S., Aravind L., Koonin E. V. (2002) SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 27, 384–386 [DOI] [PubMed] [Google Scholar]
- 54. Okuda K., Shikanai T. (2012) A pentatricopeptide repeat protein acts as a site-specificity factor at multiple RNA editing sites with unrelated cis-acting elements in plastids. Nucleic Acids Res. 40, 5052–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okuda K., Nakamura T., Sugita M., Shimizu T., Shikanai T. (2006) A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J. Biol. Chem. 281, 37661–37667 [DOI] [PubMed] [Google Scholar]
- 56. Hall J. C., Sytsma K. J., Iltis H. H. (2002) Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Am. J. Bot. 89, 1826–1842 [DOI] [PubMed] [Google Scholar]