Background: The in vivo importance of PCSK7 in the vertebrates is currently poorly understood.
Results: Inhibiting PCSK7 in zebrafish results in various developmental defects and dysregulation of gene expressions.
Conclusion: PCSK7 is essential for zebrafish development and regulates the expression and proteolytic cleavage of TGFβ1a.
Significance: PCSK inhibitors are considered future therapeutics for human diseases; understanding the biological role of PCSK7 is therefore critical.
Keywords: Development, Gene Expression, Homology Modeling, Protease, Transforming Growth Factor β (TGFβ), PC7, Otolith, PCSK7, Proprotein Convertase, Zebrafish
Abstract
Proprotein convertase subtilisin/kexin (PCSK) enzymes convert proproteins into bioactive end products. Although other PCSK enzymes are known to be essential for biological processes ranging from cholesterol metabolism to host defense, the in vivo importance of the evolutionarily ancient PCSK7 has remained enigmatic. Here, we quantified the expressions of all pcsk genes during the 1st week of fish development and in several tissues. pcsk7 expression was ubiquitous and evident already during the early development. To compare mammalian and zebrafish PCSK7, we prepared homology models, which demonstrated remarkable structural conservation. When the PCSK7 function in developing larvae was inhibited, we found that PCSK7-deficient fish have defects in various organs, including the brain, eye, and otic vesicle, and these result in mortality within 7 days postfertilization. A genome-wide analysis of PCSK7-dependent gene expression showed that, in addition to developmental processes, several immune system-related pathways are also regulated by PCSK7. Specifically, the PCSK7 contributed to the mRNA expression and proteolytic cleavage of the cytokine TGFβ1a. Consequently, tgfβ1a morphant fish displayed phenotypical similarities with pcsk7 morphants, underscoring the importance of this cytokine in the zebrafish development. Targeting PCSK activity has emerged as a strategy for treating human diseases. Our results suggest that inhibiting PCSK7 might interfere with normal vertebrate development.
Introduction
Seven proprotein convertase subtilisin/kexin (PCSK1,3 PCSK2, FURIN, and PCSK4–PCSK7) enzymes modulate the biological activity of immature proproteins by catalyzing limited proteolysis at sites containing a stretch of basic amino acid residues. Consequently, PCSK enzymes are important regulators of a multitude of biological events, including development, host defense, and hormone function (1). Although the archetype PCSKs possess closely related, even redundant, biochemical properties in vitro and often share substrate molecules, studies with genetically targeted animals and humans carrying inactive PCSK alleles argue for substrate specificity. FURIN (2), PCSK5 (3), and PCSK6 (4) are essential for normal mammalian development, whereas the inactivation of PCSK1 (5), PCSK2 (6), and PCSK4 (7) result in tissue-restricted phenotypes that range from infertility to defects in the neuroendocrine system. Genetic inactivation has also demonstrated a specific function for the more recently identified and biochemically unique PCSK family members MBTPS1 (8) and PCSK9 (9) in cholesterol and lipid metabolism. In contrast, the biological function of the evolutionarily most ancient enzyme, PCSK7, has remained enigmatic. Scattered references in the literature postulate that PCSK7-deficient mice have little, if any, phenotypic abnormalities, but a comprehensive analysis of these animals has not yet been published (10–12).
It has been demonstrated that similarly to FURIN, PCSK5, and PCSK6 the expression of PCSK7 is ubiquitous and that it exerts its function mainly in the trans-Golgi network and on the cell surface (13, 14). Importantly, previous biochemical studies indicate that PCSK7 operates often redundantly with other PCSKs, especially FURIN. For example, the activity of PCSK7 can be replaced in the site-specific cleavage of pro-BMP4 (15), pro-PDGF (16), pro-NGF (17, 18), and pro-VEGF-C (19). However, PCSK7 seems to be solely responsible for rescuing an unstable MHC I in post-endoplasmic reticulum compartments (20) and for the proteolysis of pro-EGF (21). In addition, a genome-wide association study recently revealed that an SNP (rs236918) in the PCSK7 gene region is associated with an increase in the serum level of soluble transferrin receptor, thus implying that PCSK7 could have a role in iron metabolism in humans (22). This observation was later confirmed by showing that PCSK7 is the only convertase that sheds transferrin receptor into the medium in several cell lines (23).
Most mammalian proteins and genes have orthologs in the zebrafish, which makes them a feasible model for studying the role of PCSK enzymes in vertebrate biology in vivo. Previously, the functions of FURIN (24), PCSK5 (25), MBTPS1 (26), and PCSK9 (27) have been assessed in developing zebrafish. Inactivation of FURIN and MBTPS1 in zebrafish results in defective skeletal and cartilage formation, respectively, whereas PCSK5 and PCSK9 are important for neurological development. To decode the poorly defined PCSK7 function in vertebrate biology in vivo, we inhibited PCSK7 in zebrafish using morpholinos (MOs). The phenotypes of the pcsk7 morphants clearly demonstrate that PCSK7 has a critical and non-redundant role in early zebrafish development.
EXPERIMENTAL PROCEDURES
Genes Studied
Blast searches were used to look for zebrafish orthologies for human PCSK genes. The following zebrafish pcsk genes were found: pcsk1, ENSDARG00000002600; pcsk2, ENSDARG00000019451; furinA, ENSDARG00000062909; furinB, ENSDARG00000070971; pcsk5a, ENSDARG00000067537; pcsk5b, ENSDARG00000060518; pcsk7, ENSDARG00000069968; mbtps1, ENSDARG00000014634; and pcsk9, ENSDARG00000074185.
Zebrafish Lines
Embryos used in all experiments were obtained through natural crosses of wild-type AB strain individuals. Adult fish used in QRT-PCR expression analyses as well as in MO experiments were also of the wild-type AB strain. All fish used in the experiments were maintained under standard conditions at 28.5 °C. The care and analyses of the animals were in accordance with the Finnish Laboratory Animal Welfare Act 62/2006, the Laboratory Animal Welfare Ordinance 36/2006, and Authorization LSLH-2007-7254/Ym-23 by the national Animal Experiment Board.
QRT-PCR Expression Analyses
Expression of pcsk genes was measured both in various adult zebrafish tissues (female/male gonads, liver, kidney, intestine, eye, gill, brain, skin, and tail) as well as in whole developing embryos of different ages (1–7 days postfertilization (dpf)). Conventional quantitative real time PCR with a reverse transcribed cDNA template from total RNA was used. ef1a gene (ENSDARG00000020850) was utilized as a housekeeping gene (28). Primers used in the QRT-PCR analyses are presented in Table 1.
TABLE 1.
Primers used in QRT-PCR analyses
F, forward; R, reverse.
| Gene | Sequence 5′–3′ |
|---|---|
| pcsk1 | F CGGGAAAAGGAGTGGTCAT |
| R GGTGGAGTCGTATCTGGG | |
| pcsk2 | F CGGATCTGTATGGAAACTGC |
| R GCCGGACTGTATTTTATGAATG | |
| furinA | F AGCATGTTCAAGCGCAG |
| R CCAGTCATTGAAGCCCTCA | |
| furinB | F CCAAGGCATCTACATCAACAC |
| R ACACCTCTGTGCTGGAAA | |
| pcsk5a | F AAGCCATGGTACCTGGAAGA |
| R GGTCAGAGCTGGATTTGCTT | |
| pcsk5b | F TGTTCCTCGACCCTTACCAC |
| R ATCTCGCCATGTCAGGAAAG | |
| pcsk7 | F AGAGTGTTGGACGGGC |
| R TGCCTAATGGATGCGGT | |
| mbtps1 | F GATGTTATAGGTGTTGGAGGG |
| R TCACGATGTCAGGCTTCA | |
| pcsk9 | F AGGGCAAGGGTACTGTG |
| R TGTTTAGGGTGCGACTGA | |
| ef1a | F CTGGAGGCCAGCTCAAACAT |
| R ATCAAGAAGAGTAGTACCGCTAGCATTAC | |
| tgfβ1A | F CAGGATGAGGATGAGGACTA |
| R CAGCCGGTAGTCTGGAATA |
Morpholinos
Prior to morpholino design 8–10 individual fish of the AB strain were sequenced for each gene to verify the sequence identity with the published sequence and to find genomic regions homomorphic enough for MO design. The furinA and furinB MOs were designed to hit ATG sites, whereas the pcsk7 MOs target exon-intron boundaries in exon 3 (pcsk7 e3 MO) and exon 8 (pcsk7 e8 MO). These exons contain codons for amino acids of the catalytic triad of the PCSK7 enzyme. The morpholino for p53 was commercially predesigned, and the random control (RC) morpholino is a random base mixture at every position intended for use as a negative control.
The sequences for the gene-specific MOs used were as follows: furinA, TCAATGAGGCAAGCCTGAGATCCAT; furinB, ACAGCAGGATCAAGCGGCCCTCCAT; pcsk7 e3, AGGACTCTGGAAAACACACAGGTTT; pcsk7 e8, CTTTATGGTTTGTGGATGTACCTGT; p53, GCGCCATTGCTTTGCAAGAATTG; and tgfβ1a, TCAGCACCAAGCAAACCAACCTCAT. Morpholinos were designed and synthesized by GeneTools (Philomath, OR) and stored dissolved in distilled water (−20 °C) at a 1 mm concentration.
Morpholino and RNA Injections
For injections, 0.25–1.0 pmol of each MO was used. A rhodamine dextran tracer was used to control the injections, and unsuccessfully injected embryos were removed at 1 dpf. All injections (1–2 nl) were administered into the yolk sac of a one- to four-cell stage zebrafish embryo. 0.2 m KCl was used as buffer in the injection solutions. The RNA used in the RNA rescue experiments was transcribed from pcsk7 cDNA with the SP6 mMESSAGE mMACHINE kit (Ambion, Austin, TX) according to the manufacturer's instructions (imaGenes, Berlin, Germany). All MO injection experiments were controlled with embryos injected with RC MO (GeneTools) as well as with non-injected embryos. Injections were carried out using a PV830 Pneumatic PicoPump (World Precision Instruments) and a Nikon SM7645 microscope. For visual analysis and live fish pictures, a SteREO Lumar V12 microscope with the AxioVision Rel. 4.8 program (Carl Zeiss) was used.
pcsk7 Histological Staining
The larvae were fixed in 4% paraformaldehyde, PBS solution and embedded in 2% agarose. Next, the samples were dehydrated in an alcohol series (70%, 96%, absolute ethanol, and xylene; 1 h in each). The dehydrated samples were embedded in paraffin, and 5-μm transverse sections were cut. These were then fixed on glass slides at 58 °C, deparaffinized in xylene (2 × 4 min) and in absolute, 96, and 70% ethanol (each 1–2 × 3 min), and rinsed with distilled H2O. The hematoxylin-eosin staining was performed as follows: Mayer's hematoxylin, 2 min; running tap water, 2 min; water, 1.5 min; 70% ethanol, 15 s; eosin Y, 15 s; 96% ethanol, 30 s twice; absolute ethanol, 1 min twice; and xylene, 1–4 min. Thereafter, the slides were mounted, and pictures were taken using an Olympus SZX16 microscope and a Color View Soft Imaging System camera.
pcsk7 in Situ Hybridization
Whole mount in situ hybridization was performed for pcsk7 as described in detail in Thisse and Thisse (29) to analyze the detailed expression pattern of the gene. 2-dpf embryos were used. An antisense probe was used for specific detection of the pcsk7 mRNA, and a sense probe was used to control for unspecific background staining.
RNA Microarray
100 ng of total RNA (triplicate samples pooled from 18–35 embryos/group; two time points of 6 and 24 hpf) isolated from fish injected with RC (0.5 pmol) or pcsk7 e3 (0.5 pmol) + p53 (0.75 pmol) MO was amplified and Cy3-labeled. Altogether, 1.65 μg of sample was then hybridized on the arrays (Agilent 4x44K Zebrafish GE v3; 65 °C, overnight). Chips were scanned using an Agilent Technologies G2565CA Scanner, and numeric data were obtained from Agilent Feature Extraction software v10.7.1 (The Finnish Microarray and Sequencing Centre, Turku, Finland). Only features having corresponding Entrez ID available were retained. The expression value for each gene is calculated in three phases. 1) If the same probe appears in the raw data file more than once, the row from the data file that has the highest average expression across all samples is used to represent that probe. 2) If the same gene appears in the raw data file more than once, the probe that has the highest average expression across all samples is used to represent that gene. 3) The resulting gene expression values are quantile-normalized across samples. Genes with a median expression >10 on a linear scale in either the pcsk7 morphant or control samples were included in further analyses. A two-sample t test was used to test the differential expression of each gene between the morphant and control samples using the log2 gene expression values. To control for the false discovery rate, the resulting p values from the t test were used to calculate q values for each gene as described (30). Genes with a q value <0.05 were retained. A log2 ratio between the median of pcsk7 morphant samples and controls was calculated for each gene, and genes with an absolute log2 ratio ≥2 were considered differentially expressed. A gene ontology enrichment analysis was conducted using hypergeometric distribution testing. Here, we tested whether the number of differentially expressed genes annotated to a gene ontology (GO) term was larger than could be expected by chance. The resulting p values serve as an indication of the possible enrichment of each GO term.
To summarize the GO categories, each enriched term was annotated to a high level GO term. A high level GO term refers to terms that are directly linked to the root term biological process, molecular function, or cellular component. Finally, the number of terms under each high level GO term was calculated. Here, we included the 15 most enriched terms into the analysis. The original raw data of the microarray are available in the Gene Expression Omnibus database.
Cloning, cDNA Constructs, and Western Blotting
FURIN-deficient RPE.40 cells (a kind gift from Prof. J. Creemers, KU Leuven, Belgium) were grown in Ham's F-12 medium supplemented with 10% fetal bovine serum and antibiotics. The cDNA sequence encoding the zebrafish tgfβ1a gene was amplified from the wild-type AB zebrafish cDNA (7 dpf) by PCR (forward oligo, 5′-GGAGAATTCGCCATGAGGTTGGTTTGCTTGGTGCTG; and reverse oligo, 5′-CATGGTGGTGAGGAACTGCAAGTGCAGTGGTACCGGA). The insert was subcloned into pcDNA3.1-myc-His plasmid and validated by sequencing. Human TGFβ1 (ATCC) subcloned into pcDNA3.1-myc-His, pSVL-huFURIN (ATCC), pcDNA3-huPCSK7-FLAG (a gift from Prof. J. Creemers), pME18S-FL3-zffurinA, pME18S-FL3-zffurinB, or pCMV-SPORT6.1-zfpcsk7 (imaGenes) was transfected in RPE.40 cells using FuGENE® 6 transfection reagent (Promega). 48 h after transfections, supernatants were collected, and cells were lysed in Triton X-100 lysis buffer (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20% glycerol, 0.1% Triton X-100, 1 mm EDTA, 50 mm NaF, 1 mm tris(2-carboxyethyl)phosphine hydrochloride) supplemented with protease inhibitors (Complete Mini, Roche Applied Science). Equal amounts of proteins were separated by SDS-PAGE, and immunodetection was performed using anti-myc (M5546 Sigma) primary antibody and anti-mouse HRP secondary antibody (HAF007, R&D Systems). Visualization was done using the ECLTM Western blotting detection kit (GE Healthcare) and AGFA CP1000 imaging system. Signal intensities were analyzed using NIH ImageJ software.
RESULTS
Quantification of pcsk Gene Expression in Zebrafish
The zebrafish genome contains orthologs for most mammalian genes. We blasted the human PCSK protein sequences against zebrafish genome databases and found unambiguous orthologs for all but two members (PCSK4 and PCSK6) of the mammalian PCSK family. Two isoforms for both human FURIN (furinA and furinB) and PCSK5 (pcsk5a and pcsk5b),which probably arose from a genome duplication that occurred early in the evolution of teleost fish, were present in the zebrafish genome (31).
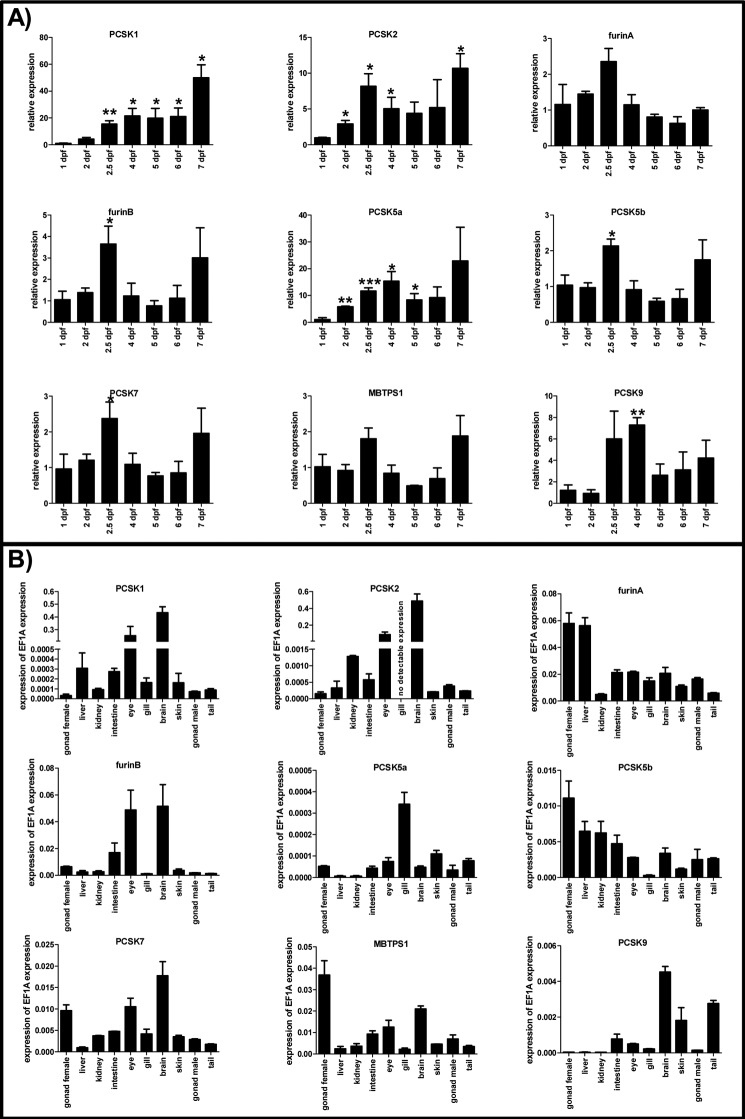
We first analyzed the expression of pcsk genes in the whole developing zebrafish using a QRT-PCR approach (1–7 dpf). In these experiments, different pcsk genes showed variable expression patterns. For example, the pcsk family members with reported implications in neuronal development (pcsk1, pcsk2, pcsk5a, and pcsk9) were significantly up-regulated during the early development (Fig. 1A). In contrast, the expression of other pcsk genes remained relatively constant during the first 7 days of life. However, a subtle but significant up-regulation of the furinB, pcsk5b, and pcsk7 mRNAs could be observed at 2.5 dpf. In accordance with previous studies in mammals, our experiments assessing tissue-specific pcsk expression in adult fish showed particularly high expression of pcsk1 and pcsk2 in eye and brain (32, 33) (Fig. 1B). Apart from the lack of pcsk2 in gill tissue, variable degrees of expression of all proprotein convertase enzymes could be detected with QRT-PCR in the tested tissues. Interestingly, two orthologs of FURIN and PCSK5 showed (to some extent) complementary tissue expression profiles. The pcsk7 expression was highest in female gonads, brain, and eye in adult zebrafish. The magnitude of pcsk5a/b and pcsk7 expression levels was generally lower than that of the furin genes. These data corroborate that pcsk7 is present in both developing larvae and multiple adult zebrafish tissues together with other convertase enzymes. This allows the reliable assessment of the specific role of the pcsk7 in vertebrate biology.
FIGURE 1.
Expression of pcsk genes in zebrafish. QRT-PCR analysis was used to analyze the relative expression of the pcsk genes in developing zebrafish larvae (1–7 dpf) (A) and various adult zebrafish tissues (B). A, pcsk gene expression levels were normalized to the housekeeping gene ef1a, and the normalized gene expression on 1 dpf was given a value of 1. Other time points are shown as relative to this. Asterisks denote statistical significance for differences in comparisons between 1 dpf and other time points: *, p < 0.05; **, p < 0.01; ***, p < 0.001 in Welch-corrected two-tailed Student's t tests. Experiments in A were performed with three biological replicates (each sample consisted of 15–30 individual larvae depending on the age of larvae). B, in adult tissue analyses, gene expression levels related to that of ef1a are shown. Experiments in B were performed twice in technical replicates with essentially similar results. Error bars represent S.D. Note the diverse scales on the y axis.
Homology Modeling of PCSK7
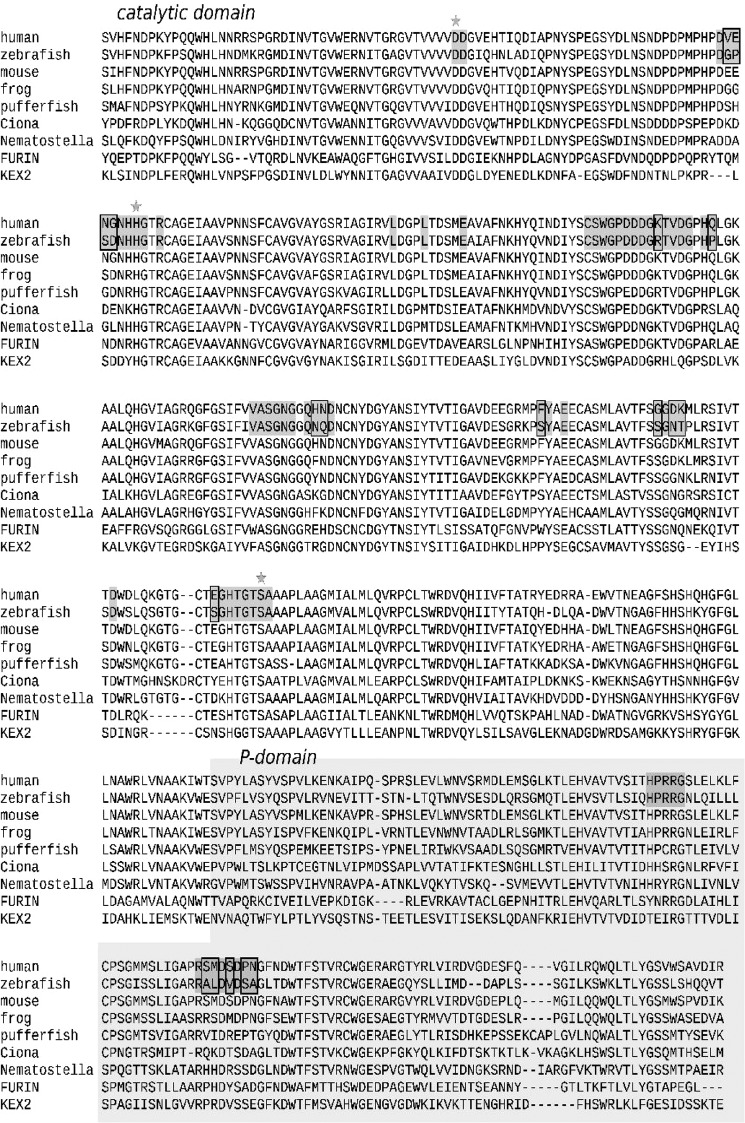
To assess putative structural and electrostatic differences between mammalian and zebrafish PCSK7, we prepared a homology model of the protein with Modeler 9v10 (34) using published crystallographic structures of mouse FURIN and yeast Kexin as templates. Our modeling efforts showed evident structural conservation in catalytic and P domains of PCSK7 between the human and zebrafish (Fig. 2). We then analyzed the binding of a substrate to PCSK7 by superimposing the crystallographic structure of subtilisin (Protein Data Bank code 1CSE (35)) with the PCSK7 models. These data demonstrated that most of the potential substrate-binding residues are also conserved between human and zebrafish. The alignment of the PCSK7 catalytic and P domains of several species further demonstrated that the active sites are close to identical except for a non-conserved loop preceding the catalytic histidine (Fig. 3). The human sequence VENG in this loop is replaced by GPSD in zebrafish, which potentially affects substrate specificity at the P2 or P1′ site (Figs. 2 and 3). Notably also, a little further away from the active site, differences in the charge distribution can be observed (Figs. 2, lower part, and 3). In conclusion, PCSK7s in less developed organisms display remarkable structural similarities but subtle electrostatic differences with mammalian counterparts. This indicates that studying the zebrafish PCSK7 function can also give insights into other vertebrate homologs.
FIGURE 2.
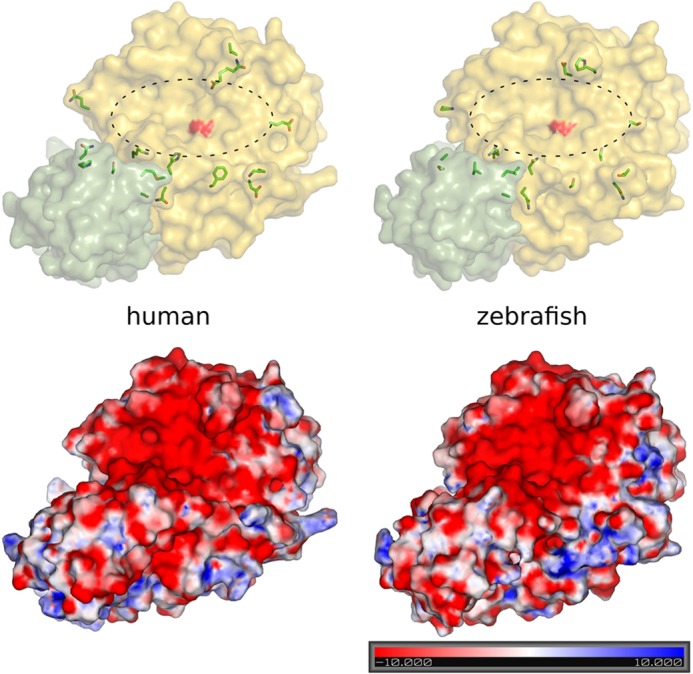
Homology models of human and zebrafish PCSK7. Upper part, yellow, catalytic domain; green, P domain. The catalytic site is marked with an oval, and the surface of the catalytic serine is highlighted in red. Amino acid differences close to the conserved substrate binding site are shown in stick presentation. Lower part, electrostatic potentials (kT/e) for the models were calculated using APBS 1.3 (57) and visualized in PyMOL 2.7. Red-white-blue color indicates the ±10 kT/e electrostatic potential plotted on the protein surface.
FIGURE 3.
ClustalW alignment of PCSK7 homologs. Human (Homo sapiens, UniProt Q16549), zebrafish (Danio rerio, RefSeq NP_001076494.1), mouse (Mus musculus, UniProt Q61139), frog (Xenopus laevis, RefSeq NP_001090019.1), pufferfish (Tetraodon nigroviridis, Ensembl ENSTNIP00000017367), vase tunicate (Ciona intestinalis, RefSeq XP_002125956.1), and starlet sea anemone (Nematostella vectensis, RefSeq XP_001638665.1) PCSK7 catalytic and P domains were aligned with those of mouse FURIN (Protein Data Bank code 1P8J (58)) and yeast Kexin (Saccharomyces cerevisiae, Protein Data Bank code 2ID4 (59)). The catalytic triad is marked with stars, and potential substrate-binding residues are in dark gray. Residues showing differences in Fig. 2 are highlighted with rectangles.
Inactivation of PCSK7 in Zebrafish Larvae Results in Developmental Lethality
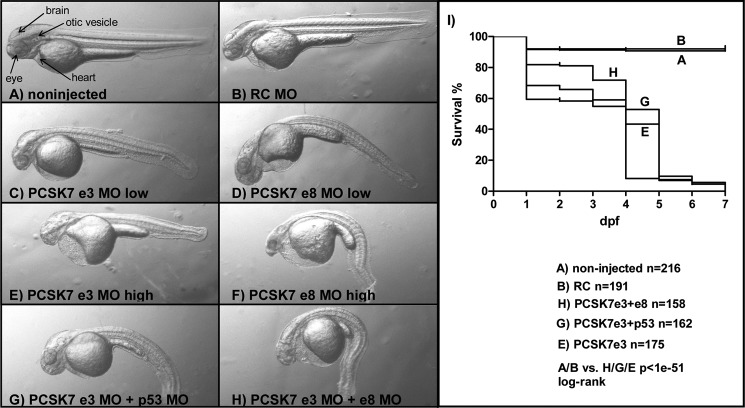
Previous in vitro experimental data show that FURIN can often replace PCSK7 (19, 36, 37). To compare and contrast how these enzymes regulate zebrafish development, we first interfered with furinA and furinB translation using morpholino technology. In accordance with a previous report (24), the simultaneous blocking of furinA and furinB with MOs resulted in a reduction in ventral jaw structures and consequently an open mouth phenotype (data not shown). Disruption of furinA/B translation also reduced the survival of fish larvae; only 30.6% (22 of 72 fish) of MO-injected fish were alive on 7 dpf. We next designed two distinct morpholinos that block the pre-messenger RNA splicing at exon-intron boundaries around the catalytic site containing exons 3 and 8 in the zebrafish pcsk7 gene (e3 and e8 MOs). Injecting the pcsk7-targeting MOs alone or in combination resulted in numerous defects that reduced the survival of fish larvae dramatically and resulted in 100% mortality by 7 dpf (Fig. 4). The survival rate and gross phenotypes observed were MO dose-dependent, and the development of several organs, including the brain, eye, heart, otolith, and tail, was severely affected.
FIGURE 4.
Phenotypes and survival of pcsk7 morphant fish. Zebrafish larvae (2 dpf) were non-injected (A) or injected with RC MO (0.5 pmol) (B), pcsk7 e3 MO (0.25 pmol) (C), pcsk7 e8 MO (0.25 pmol) (D), pcsk7 e3 MO (0.5 pmol) (E), pcsk7 e8 MO (0.5 pmol) (F), pcsk7 e3 + p53 MO (0.5 + 0.75 pmol) (G), or pcsk7 e3 + e8 MO (0.25 + 0.25 pmol) (H). A–H, 35× magnification. I, survival of fish injected with pcsk7 e3 MO (0.5 pmol), pcsk7 e3 + e8 MOs (0.25 + 0.25 pmol), and pcsk7 e3 + p53 MOs (0.5 + 0.75 pmol) was significantly lower than that of RC morphant (0.5 pmol) or uninjected fish (p < 1e−51 for all comparisons by log rank test). pcsk7 e8 MO reduced the survival of larvae similarly to e3 MO: all larvae died by 6 dpf (n = 98 larvae; e8 MO, 0.5 pmol; data not shown).
Because p53-dependent off-target neural toxicity has been estimated to affect 15–20% of all morpholino injections (38, 39), we co-injected a p53 MO together with the pcsk7-silencing MOs. The phenotypes and survival of morphant fish remained similar compared with fish not injected with the p53 MO (Fig. 4). This indicates that the observed lethality of the PCSK7 morphant was not due to p53-mediated toxicity. To further validate the specificity of the pcsk7 MO phenotypes, we set up an RNA rescue experiment. When in vitro transcribed pcsk7 RNA was co-injected together with e3 MO into the developing larvae, the severity of the MO phenotypes was reduced, and the survival improved (p < 0.0001, log rank χ2 = 38.94, df = 1) (Fig. 5, A and B). To verify that the pcsk7 MOs specifically disrupt the splicing of the pcsk7 pre-mRNA molecules, we amplified and sequenced the affected exon regions from the morphant fish (Fig. 6, A and B, and data not shown). Our results demonstrated that injecting the e3 MO results in three mRNA products with different lengths, whereas the e8 MO deletes a 58-bp fragment at the end of exon 8. These data show that pcsk7 morpholinos target pcsk7 pre-mRNA and that pcsk7 has a critical and non-redundant role in zebrafish development.
FIGURE 5.
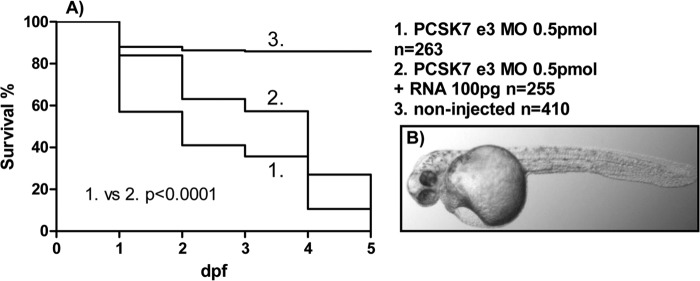
pcsk7 mRNA improves the survival and reduces the severity of the pcsk7 morphant phenotype. A, survival of zebrafish uninjected or injected with pcsk7 e3 MO alone or together with in vitro transcribed pcsk7 mRNA was monitored daily. Data are pooled from two independent experiments. B, 2-dpf zebrafish larvae co-injected with pcsk7 e3 MO (0.5 pmol) and pcsk7 mRNA (100 pg).
FIGURE 6.
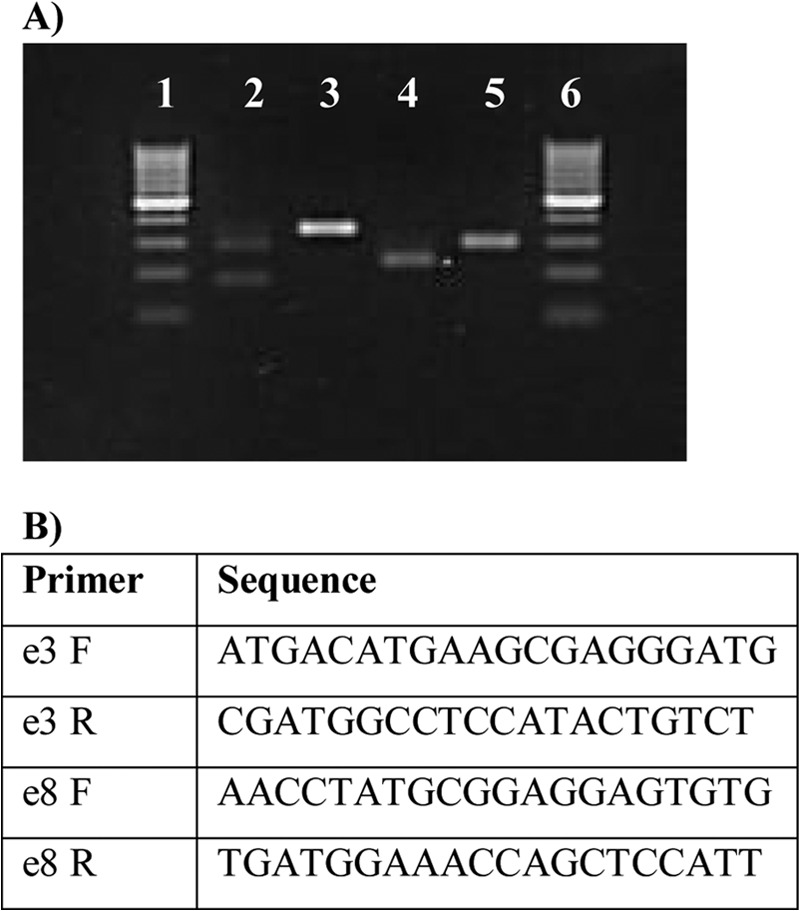
pcsk7 MO injections result in erratic splicing of the pre-mRNA. A, total RNA was isolated from the control (RC MO) and different pcsk7 morphant fish and reverse transcribed into cDNA, which was amplified by PCR and run on an agarose gel. From the left, lane 1, 100-bp molecular weight marker; lane 2, pcsk7 e3 MO; lane 3, control (=RC MO with pcsk7 e3 primers); lane 4, pcsk7 e8 MO; lane 5, control (=RC MO with pcsk7 e8 primers); lane 6, 100-bp molecular weight marker. Sequencing showed that 345- and 288-bp DNA fragments represent intact, wild-type pcsk7 mRNA that can be detected in RC MO samples with the e3 (lane 3) and e8 primers (lane 5), respectively. The pcsk7 e3 MO injection resulted in three differentially sized fragments: (i) a fragment corresponding to the completely deleted exon 3 (the shortest band in lane 2), (ii) a fragment with a 58-bp deletion from the start of exon 3 followed by a 49-bp polymorphic region either from exon 3 or from exon 4 and further supplemented with the untouched end of exon 3 (the middle band in lane 2), and (iii) a faint band of wild-type exon 3 (the longest band in lane 2). The pcsk7 e8 MO injection deleted a 58-bp fragment from the end of exon 8 (lane 4) resulting in a truncated pcsk7 mRNA molecule. B, sequences for the primers used for the sequencing described in A. F, forward; R, reverse.
PCSK7 Regulates Brain, Otolith, and Eye Development
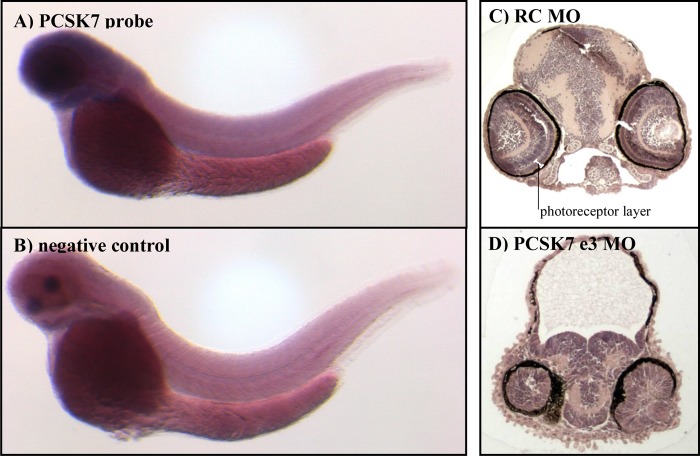
Our expression analyses implicated pcsk7 as prominently present in adult zebrafish eye and brain tissues. To investigate the pcsk7 expression pattern in larvae, we used RNA in situ hybridization on wild-type fish at 2 days postfertilization. These experiments demonstrated that pcsk7 is most abundant in the entire cranial area and the eye already in developing wild-type zebrafish (Fig. 7, A and B).
FIGURE 7.
pcsk7 is expressed in the head region, and pcsk7 morphant fish have cranial developmental defects. pcsk7 expression (dark blue) was analyzed with RNA in situ hybridization in wild-type zebrafish larvae at 2 dpf. A, pcsk7 antisense probe. B, negative control (pcsk7 sense probe). Right-hand panels show hematoxylin-eosin-stained transverse sections of zebrafish cranial region of RC (C) and pcsk7 e3 morphant (D) (3 dpf).
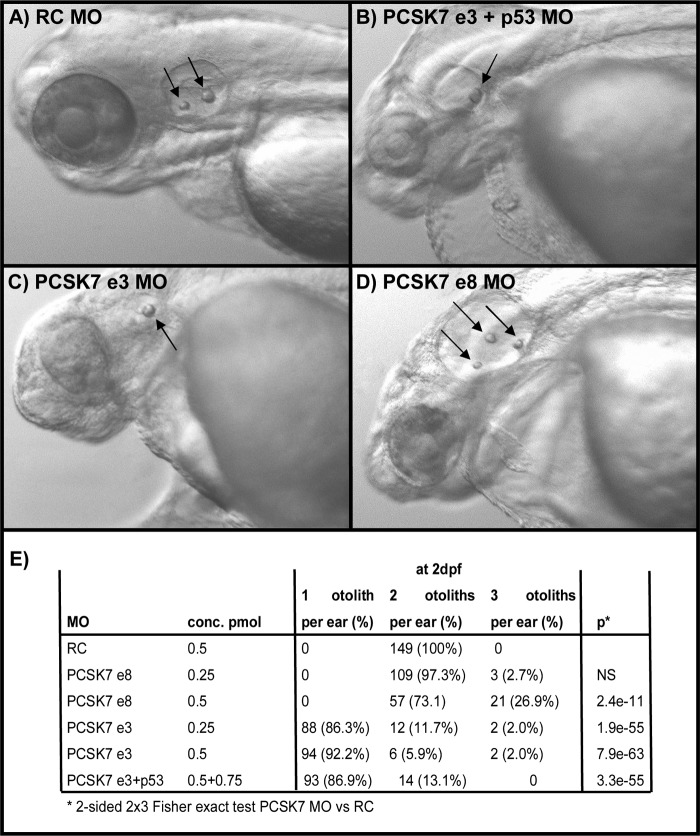
The observed gross anatomical differences and location of early pcsk7 expression prompted us to analyze in more detail the head region phenotypes seen in the developing zebrafish. A dramatic underdevelopment of the entire brain and eyes was observed in histological sections (Fig. 7, C and D). The cranium of pcsk7 morphants contained mostly a non-cellular, meshlike substance, whereas the brain remained underdeveloped and was located abnormally downward between the eyes. In addition, the cellular structure in the eyes of MO-injected fish was unorganized, and photoreceptor, outer and inner plexiform, and inner nuclear and ganglion cell layers were undefined. However, the retinal pigment epithelium and lens structures were also evident in morphant fish. The zebrafish with a non-functional PCSK7 also had an abnormal number of otoliths. Injection of the e3 MO resulted in a reduction in otolith number (one otolith per ear in >92% of fish, p = 7.9e−63), whereas the fish injected with the e8 MO in contrast had an increased amount of otoliths per ear (three otoliths per ear in >26% of fish, p = 2.4e−11). All evaluated 149 control fish had the normal two otoliths per ear (Fig. 8, A–E). These data show that in accordance with its spatiotemporal expression PCSK7 is important for several developmental processes of the vertebrate cranial organs.
FIGURE 8.
pcsk7 morphant fish have an abnormal number of otoliths. Zebrafish were injected with RC MO (0.5 pmol) (A), pcsk7 e3 + p53 MO (0.5 + 0.75 pmol) (B), pcsk7 e3 MO (0.5 pmol) (C), or pcsk7 e8 MO (0.5 pmol) (D), and otoliths (arrows) were visualized on 3 dpf. Quantification of otoliths is presented in E. NS, not significant.
Analysis of PCSK7-dependent Gene Networks
Genome-wide expression analyses can be used to clarify the biological relevance of gene-specific studies. Therefore, to comprehend the gene networks that are affected in the pcsk7 morphant fish, we performed a genome-wide mRNA expression study in whole fish larvae after PCSK7 inhibition. We chose to evaluate gene expression at two different time points, 6 and 24 hpf. At the gastrula stage (6 hpf), most of the zygotic genes have commenced transcript accumulation and often peak in their expression. In contrast, after 24 hpf, the genome-wide gene expression profile remains relatively steady as was demonstrated in a previous analysis with wild-type zebrafish (40).
Conventional PCSK enzymes are highly redundant in substrate processing. Therefore, it is important to evaluate whether disrupting PCSK7 function causes any significant changes in the expression of other proprotein convertases (41), which might contribute to or even compensate for the observed phenotype. At both 6 and 24 hpf, the other conventional pcsk genes were not found to be significantly up-regulated in pcsk7 morphant fish. pcsk7 expression, however, was significantly reduced in the morphant zebrafish (log ratio of −1.95 and p = 5.77e−6 at 24 hpf). The decrease in the pcsk7 mRNA level is presumably due to the cellular deletion of the defectively spliced pcsk7 pre-mRNA molecule.
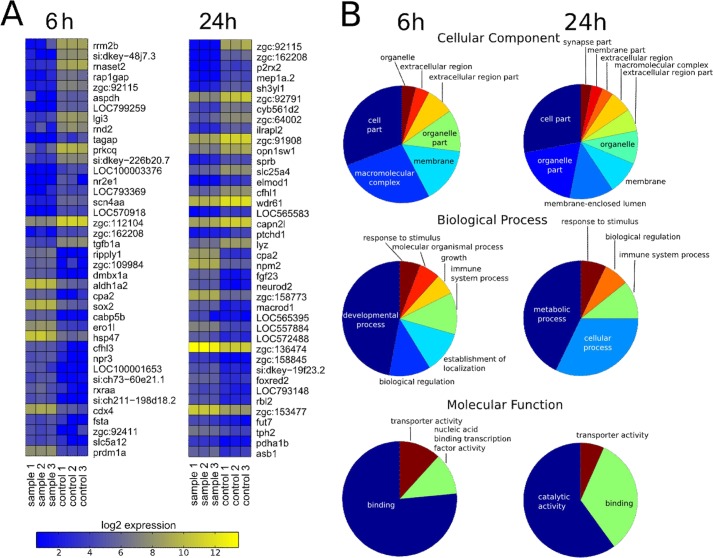
To understand which major biological, molecular, and cellular processes are dependent on the intact PCSK7 enzyme, we first performed a GO enrichment analysis using hypergeometric distribution testing. At 6 hpf, as expected from the observed morphant phenotypes, terms related to development (for example otic placode formation) and transcription regulation were abundantly enriched. Consequently, at 24 hpf, genes related to metabolism were also commonly up-regulated. Strikingly, at both 6 and 24 hpf, genome-wide mRNA expression samples showed a clear enrichment in immune system-related terms with immune system being the most enriched (p = 0.0004) term at 24 hpf. Several cytokine signaling-affiliated terms could also be found among the most significantly enriched terms at both time points (supplemental Table S1 and Fig. 9).
FIGURE 9.
Heat map of the most differentially expressed genes and high level summary of gene ontology enrichments between control and pcsk7 morphant fish. Samples were prepared, and data were analyzed as described under “Experimental Procedures.” A, zebrafish genes that have an identifiable human homolog are shown in the heat maps. B, summaries from the enriched gene ontology terms. Pie charts show enriched high level categories for cellular component, biological process, and molecular function at 6 and 24 hpf. In each chart, the size of the wedge corresponds to the number of terms enriched under the given high level category.
A more detailed analysis of the PCSK7-dependent genes in the microarray revealed the dysregulation of several immune system-, neurological system-, and otolith/otic vesicle-related genes (supplemental Table S2 and Fig. 9). For example, the expression levels of important regulators of both the adaptive and innate immunity, stat4, tgfβ1a, and csfra, were greatly reduced at 6 hpf in the pcsk7 morphants, whereas otolith/ear-related foxi1 (down-regulated at 6 hpf) and pax2a, msxc, and fsta (up-regulated at 6 hpf) were also dysregulated. In addition, a multitude of differentially expressed neurological genes, including several protocadherin and fibroblast growth factor genes, were found to be dependent on a functional PCSK7.
We have demonstrated previously that another PCSK family member, FURIN, is a key regulator of both T helper 1 type immune responses and T regulatory cell-mediated peripheral immune tolerance (42, 43). These crucial events of adaptive immunity are dependent on transcription factor STAT4 function and cytokine TGFβ1 signaling, respectively. We noticed that in our microarray experiment both of the aforementioned genes were significantly repressed when pcsk7 expression was blocked. To confirm this finding, we performed a QRT-PCR analysis that revealed a down-regulation of both tgfβ1a and stat4 by 11.7- and 5.1-fold, respectively, at 6 h postfertilization in an RC versus pcsk7 + p53 comparison (three biological replicates, one-tailed p = 0.037 for both genes in Student's t test with Welch correction; data not shown). These findings imply that, in addition to FURIN, PCSK7 may also have an important regulatory role in developing adaptive immune responses.
PCSK7 Regulates the Expression of tgfβ1a in Zebrafish
In mammals, FURIN is the major proprotein convertase enzyme that regulates the bioavailability of the anti-inflammatory TGFβ1 cytokine (43, 44). From previous experiments, it is also noteworthy that TGFβ1 can directly up-regulate the expression of furin (45). However, PCSK7 can also proteolytically process the TGFβ superfamily cytokines. Mammalian PCSK7 is suggested to cleave pro-BMP4 in a developmentally regulated fashion, and it possesses up to one-third of the capacity of FURIN in pro-TGFβ1 processing in vitro (15, 44). Furthermore, the mRNA expression correlation of human PCSK7 with TGFβ1 is very strong, which may indicate a physiological role for PCSK7 in pro-TGFβ1 processing (46). As described above, we observed that tgfβ1a (ENSDARG00000041502), the zebrafish counterpart for the mammalian TGFβ1, was markedly down-regulated in the microarray analysis at 6 hpf in pcsk7 morphant fish (supplemental Table S2 and Fig. 9). This coordinated expression of pcsk7 and tgfβ1a prompted us to address the importance of PCSK7 for tgfβ1a expression and activation and to assess whether the lack of TGFβ1a could contribute to the PCSK7-dependent phenotype.
We first wanted to investigate whether zfPCSK7 can directly proteolytically process zfpro-TGFβ1a. To this end, the cDNA encoding TGFβ1a was amplified from wild-type zebrafish and subcloned into a myc-His expression plasmid. After verification by sequencing (data not shown), we then co-expressed tgfβ1a together with zebrafish furinA, furinB, or pcsk7 in FURIN-deficient RPE.40 cells. For comparison, we also co-expressed human TGFβ1 together with human FURIN and PCSK7 cDNAs. Our results clearly demonstrate that in addition to FURIN both zebrafish and human PCSK7s are able to process and promote the release of bioactive TGFβ1 (zebrafish, 16 kDa; human, 14 kDa) into cell culture supernatants (Fig. 10A). In line with previous reports, the PCSK7s possessed approximately one-third of the activity of FURIN in the TGFβ1 maturation (44).
FIGURE 10.
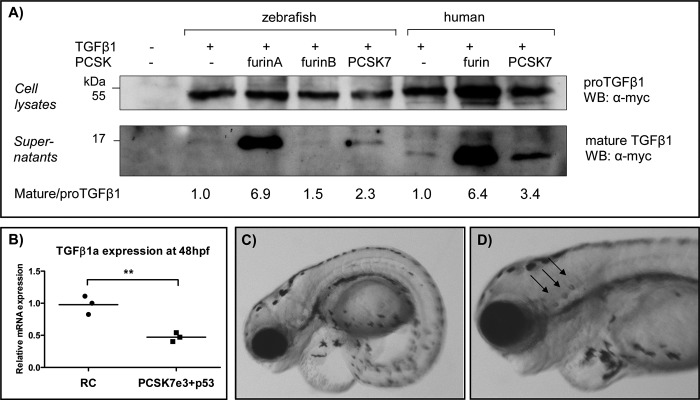
PCSK7 regulates TGFβ1a, which affects the zebrafish larva development and otolith formation. A, FURIN-deficient RPE.40 cells were transiently transfected with zebrafish tgfβ1a-myc or human TGFβ1-myc together with zffurinA, zffurinB, zfpcsk7, huFURIN, or huPCSK7. Pro-TGFβ1 (45 kDa) and mature TGFβ1 (16 kDa in zebrafish and 14 kDa in human) expressions were detected with Western blotting (WB). Mature/pro-TGFβ1 ratios were quantified using NIH ImageJ software, and ratios in cells transfected only with tgfβ1 cDNAs were given an arbitrary value of 1. Equal loading of cell lysates and supernatants was verified by Ponceau S staining (data not shown). The experiment was repeated twice with similar results. B, tgfβ1a mRNA expression was measured by QRT-PCR from pcsk7 e3 + p53 and RC morphant embryos at 48 hpf (**, p = 0.0052, two-tailed Student's t test). C and D depict the phenotypes (4 dpf) of zebrafish injected with tgfβ1a + p53 MOs (tgfβ1a MO, 1.0 pmol; p53 MO, 1.5 pmol). tgfβ1a morphants had incorrect otolith numbers: tgfβ1a MO, 65 fish with two otoliths per ear, 21 fish with three otoliths per ear; RC MO, 120 fish with two otoliths per ear, 0 fish with three otoliths per ear (p = 2.16e−9 in Fisher 2 × 2 test).
We then assessed whether the tgfβ1a mRNA expression also remains down-regulated in pcsk7 morphant fish in a later developmental stage and found that tgfβ1a gene expression is significantly repressed also at 48 hpf (p = 0.0052; Fig. 10B). To investigate how the lack of TGFβ1a contributes to the pcsk7 morphant phenotype, we injected zebrafish with a TGFβ1a-blocking MO. In these experiments, we observed clear similarities between the tgfβ1a and pcsk7 morphants; both showed tail abnormalities, pericardial swelling, and an abnormal number of otoliths (24.4% of tgfβ1a morphants (21 of 86) had three otoliths per ear (p = 2.16e−9 in Fisher 2 × 2 test; Fig. 10, C and D)). Taking these observations together, our data suggest that in zebrafish the intact PCSK7 enzyme is important for the expression and bioavailability of mature TGFβ1a and that a defect in this contributes to the observed phenotype of the pcsk7 morphant fish.
DISCUSSION
Regardless of extensive studies on proprotein convertase enzymes in vertebrate biology, the function and significance of the evolutionarily ancient PCSK7 has remained largely unclear. In an effort to fill this gap, we studied the function of PCSK7 in zebrafish and observed that it is critical for the development of zebrafish larvae. The pcsk7 morphant fish display severe developmental defects that lead to 100% mortality within the first 7 days of life. The lack of functional PCSK7 enzyme interferes with the organogenesis of several key elements, including the brain, eyes, and otic vesicles. In addition, our genome-wide gene expression and biochemical analyses demonstrate that PCSK7 regulates genes important for organogenesis and immunology and that it is specifically capable of contributing to the function of cytokine TGFβ1a in developing fish larvae.
To first validate the feasibility of zebrafish as a model for the analysis of the PCSK7 function in vertebrate biology, we surveyed the expression of all identifiable proprotein convertases. In accordance with previous reports in mammals, the zebrafish PCSK enzymes also show variation in their developmental and tissue-specific expression. Zebrafish homologs for the neuroendocrine system-specific PCSK1 and PCSK2 were particularly highly expressed in the fish neural tissues, and the homologs of previously reported ubiquitous enzymes, such as FURIN, PCSK5, and PCSK7, were found to be relatively widely expressed similarly to earlier reports using mammals (17, 18, 47). PCSK7 shares several common substrate molecules with PCSK5 and FURIN at least in in vitro analyses (48). We found that pcsk7 was co-expressed in different tissues and developing fish together with other PCSK enzymes that have been reported to compensate its biological function. Taken together, our quantitative pcsk expression data demonstrated obvious similarities between the mammalian and fish pcsk expression. Consequently, deleting the PCSK7 function in developing fish can also give important insights into the specific biological function of this poorly defined proprotein convertase in other vertebrates.
We and others have previously reported notable evolutional conservation of the catalytic and P domains in the PCSK enzymes (46, 49). To specifically address structural and electrostatic properties in the PCSK7 enzymes, we generated homology models of mammalian and fish PCSK7s and investigated the PCSK7 sequences in several species. In conclusion, our modeling data support the idea that the catalytic and P domains in zebrafish PCSK7 share most structural features with the human counterpart. Therefore again, investigating the fish PCSK7 function is also likely to generate novel insights into the PCSK7 function in other vertebrates.
The crucial role of many of the PCSK enzymes in vertebrate development is indisputable. For example, FURIN-deficient mouse embryos show defective ventral closure and axial rotation and die during the 2nd week of embryonic development (2). In addition, a mutation in furinA causes significant embryonic lethality in zebrafish despite the duplication of the furin gene (24). These fundamental phenotypes can be explained by a lack of processing of PCSK substrate molecules; in early mouse development, the significance of the proper activation of the TGFβ family cytokines, such as BMPs and NODAL (50), is particularly emphasized. In contrast, a thorough functional analysis of a mammalian model for PCSK7 is not available in the literature. A few scattered references to Pcsk7 knock-out mice, however, suggest either complete redundancy or at least non-critical functions in mammalian development (10–12). Our analysis of the pcsk7 morphant fish and a recent publication by Senturker et al. (51) using a Xenopus model system demonstrate that PCSK7 is indispensable at least in lower vertebrates. Both studies come to the conclusion that a lack of PCSK7 function leads to severe defects especially in the neural system and eye. In zebrafish, these defects lead to 100% mortality within the 1st week postfertilization.
A complete knock-out or an inactive mutation of the gene would give the definitive answer for the biological relevance of PCSK7 in zebrafish. However, to our knowledge, PCSK7-null zebrafish have not been produced. To overcome this limitation, we analyzed how the lack of an active PCSK7 protein affects the larva development. Using morpholinos to assess the biological function of a protein has provoked some controversy. In some cases, morpholinos can cause unspecific phenotypes due to p53-dependent off-target neural toxicity. However, in our experiments, silencing the p53 pathway did not alter the conclusive role of PCSK7. In addition, sequencing the MO-truncated pcsk7 mRNA showed that the e8 morpholino efficiently removes the end of exon 8 (58 bp) and shifts the reading frame in the catalytic domain. This replaces roughly 50 C-terminal residues of the catalytic domain by 23 residues of non-native sequence followed by a stop codon. In contrast, the e3 morpholino results in the complete deletion of exon 3 of PCSK7. This removes a codon encoding one of the amino acids of the catalytic triad. In addition, the correct reading frame is lost for the remaining protein. As a consequence, PCSK7 proteins translated in both morphants lack the entire P domain, which is needed for correct folding of the enzyme (52). A PCSK7 without the P domain is unlikely to traverse to the secretory pathway, and it might eventually be degraded in the cell.
Co-injecting pcsk7 mRNA with morpholino significantly improved the survival of morphant larvae and partially restored the defected phenotypes, but a complete rescue could not be achieved. The partial improvement of phenotype by RNA rescue has also been reported by others (53). Therefore, it is probable that a more natural spatiotemporal RNA expression of pcsk7 than what could be achieved using RNA injections into the yolk sac would be needed to cancel out all the effects of pcsk7 MO.
Little information has been available on PCSK7-dependent biological processes at the genomic level. Our GO enrichment analysis revealed several biological, molecular, and cellular functions that were significantly altered if PCSK7 function was inhibited. These findings corroborated the observed gross phenotype of the pcsk7 morphants by highlighting the enrichment of several development-related GO terms. Noteworthy, the enriched GO terms also included the otic placode formation, which is a novel finding and suggests, together with the abnormal amount of otoliths in the pcsk7 morphants, a specific and non-redundant role for PCSK7 in ear development. In the future, it will be interesting to assess whether genetic alterations in human PCSK7 (22) play a role in hearing- or balance-related phenotypes.
One of the few specific PCSK7 functions in vitro is the rescue of an unstable MHC I-peptide complex (20). Analyses of microarray data on our pcsk7 morphant fish revealed that PCSK7 in addition to having a role in several developmental pathways is indeed linked strongly to immunological processes. Differentially expressed genes included several genes that are key regulators of both the adaptive and innate immunity, such as tgfβ1a, stat4, csf1a, ccr7, and various MHC genes. The host defense-linked GO terms containing these genes were among the most enriched categories already at 6 hpf and remained significantly overrepresented until 24 hpf. It has been shown earlier that TGFβ1 up-regulates the convertase FURIN, which is the major proteolytic activator of this central anti-inflammatory cytokine (44). Interestingly, our genome-wide expression analysis demonstrated that when functional PCSK7 is not available during early development tgfβ1a was among the most down-regulated genes at 6 hpf. The strength of down-regulation was weaker at later phases of development, but tgfβ1a expression remained significantly down-regulated up to 2 days postfertilization.
Importantly, tgfβ1a and pcsk7 morphant fish shared several phenotypic similarities, further suggesting that PCSK7 has a role in the regulation of the TGFβ1a cytokine in developing zebrafish. The detailed mechanisms behind the PCSK7-dependent regulation of the TGFβ1a bioavailability are not fully understood. Mature TGFβ1 cytokine is known to promote its own function by up-regulating the expressions of its own mRNA and the converting enzyme FURIN. Accordingly, because we and others have shown that PCSK7 can also process and activate pro-TGFβ1 at one-third of FURIN proteolytic capacity, the reduced PCSK7 expression can interfere with this feed-forward loop and result in reduced tgfβ1a expression (Fig. 10) (44). However, it is equally probable that in vivo PCSK7 promotes the TGFβ1a function in an indirect manner that does not involve direct proteolysis (21).
In addition to its fundamental inhibitory role in immunity (54, 55), TGFβ1 directly controls cell differentiation and proliferation. It is noteworthy that TGFβ1 deficiency in mice causes significant intrauterine lethality (56). Our experiments concur with this multifunctionality of TGFβ1 and suggest that in zebrafish TGFβ1a plays a role in various developmental processes, including otolith formation.
Proprotein convertases have a fundamental role in both health and disease. Interfering with PCSK activity holds promise for future treatment of a plethora of diseases ranging from infections to atherosclerosis. These efforts are often compromised by the lack of specificity of inhibitors of PCSK family members. If the inhibitors are considered for clinical use, it is of utmost importance to also understand the biological significance of PCSK7. Our data presented here underscore the importance of PCSK7 in zebrafish neural development and more specifically genomic processes associated with immunity. These can also be important factors to take into account when extrapolating the unwanted effects of general PCSK inhibitors in disease settings.
Acknowledgments
CSC (IT Center for Science Ltd.) is acknowledged for the computational resources, and the Finnish Microarray and Sequencing Centre, Turku, Finland is acknowledged for the analysis service. The zebrafish work was carried out at the Tampere Zebrafish Core Facility funded by the Biocenter Finland, Tampere Tuberculosis Foundation, and Emil Aaltonen Foundation. Leena Mäkinen, Matilda Martikainen, and Hannaleena Piippo are acknowledged for technical assistance and advice in zebrafish work. Timo Kauppila is acknowledged for the technical analyses of the pcsk genes and fruitful discussions.
This work was supported by Academy of Finland Projects 128623, 135980 (to M. Pesu), 121003 (to M. Parikka), 140978 (to V. H.), and 132877 (to M. N.); a Marie Curie International Reintegration Grant within the 7th European Community Framework Programme (to M. Pesu); the Emil Aaltonen Foundation (to M. Pesu); the Sigrid Jusélius Foundation (to M. Pesu); the Tampere Tuberculosis Foundation (to M. Pesu); the Tampere Graduate Program in Biomedicine and Biotechnology (to A. O. and S. K.); the Finnish Cultural Foundation (to A. O. and V. K.); the Finnish Foundation for Technology Promotion (to V. K.); Laboratoriolääketieteen edistämissäätiö (to A. O.); and Competitive Research Funding of the Tampere University Hospital Grants 9M080 and 9N056 (to M. Pesu).

This article contains supplemental Tables S1 and S2.
- PCSK
- proprotein convertase subtilisin/kexin
- MO
- morpholino
- RC
- random control
- dpf
- days postfertilization
- hpf
- hours postfertilization
- GO
- gene ontology
- hu
- human
- zf
- zebrafish
- QRT-PCR
- quantitative RT-PCR
- e
- exon
- BMP
- bone morphogenetic protein.
REFERENCES
- 1. Seidah N. G., Prat A. (2012) The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 11, 367–383 [DOI] [PubMed] [Google Scholar]
- 2. Roebroek A. J., Umans L., Pauli I. G., Robertson E. J., van Leuven F., Van de Ven W. J., Constam D. B. (1998) Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development 125, 4863–4876 [DOI] [PubMed] [Google Scholar]
- 3. Essalmani R., Hamelin J., Marcinkiewicz J., Chamberland A., Mbikay M., Chrétien M., Seidah N. G., Prat A. (2006) Deletion of the gene encoding proprotein convertase 5/6 causes early embryonic lethality in the mouse. Mol. Cell. Biol. 26, 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Constam D. B., Robertson E. J. (2000) SPC4/PACE4 regulates a TGFβ signaling network during axis formation. Genes Dev. 14, 1146–1155 [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu X., Zhou A., Dey A., Norrbom C., Carroll R., Zhang C., Laurent V., Lindberg I., Ugleholdt R., Holst J. J., Steiner D. F. (2002) Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc. Natl. Acad. Sci. U.S.A. 99, 10293–10298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gagnon J., Mayne J., Chen A., Raymond A., Woulfe J., Mbikay M., Chrétien M. (2011) PCSK2-null mice exhibit delayed intestinal motility, reduced refeeding response and altered plasma levels of several regulatory peptides. Life Sci. 88, 212–217 [DOI] [PubMed] [Google Scholar]
- 7. Mbikay M., Tadros H., Ishida N., Lerner C. P., De Lamirande E., Chen A., El-Alfy M., Clermont Y., Seidah N. G., Chrétien M., Gagnon C., Simpson E. M. (1997) Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc. Natl. Acad. Sci. U.S.A. 94, 6842–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patra D., Xing X., Davies S., Bryan J., Franz C., Hunziker E. B., Sandell L. J. (2007) Site-1 protease is essential for endochondral bone formation in mice. J. Cell Biol. 179, 687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mbikay M., Sirois F., Mayne J., Wang G. S., Chen A., Dewpura T., Prat A., Seidah N. G., Chretien M., Scott F. W. (2010) PCSK9-deficient mice exhibit impaired glucose tolerance and pancreatic islet abnormalities. FEBS Lett. 584, 701–706 [DOI] [PubMed] [Google Scholar]
- 10. Constam D. B., Calfon M., Robertson E. J. (1996) SPC4, SPC6, and the novel protease SPC7 are coexpressed with bone morphogenetic proteins at distinct sites during embryogenesis. J. Cell Biol. 134, 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villeneuve P., Feliciangeli S., Croissandeau G., Seidah N. G., Mbikay M., Kitabgi P., Beaudet A. (2002) Altered processing of the neurotensin/neuromedin N precursor in PC2 knock down mice: a biochemical and immunohistochemical study. J. Neurochem. 82, 783–793 [DOI] [PubMed] [Google Scholar]
- 12. Besnard J., Ruda G. F., Setola V., Abecassis K., Rodriguiz R. M., Huang X. P., Norval S., Sassano M. F., Shin A. I., Webster L. A., Simeons F. R., Stojanovski L., Prat A., Seidah N. G., Constam D. B., Bickerton G. R., Read K. D., Wetsel W. C., Gilbert I. H., Roth B. L., Hopkins A. L. (2012) Automated design of ligands to polypharmacological profiles. Nature. 492, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Declercq J., Meulemans S., Plets E., Creemers J. W. (2012) Internalization of proprotein convertase PC7 from plasma membrane is mediated by a novel motif. J. Biol. Chem. 287, 9052–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas G. (2002) Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nelsen S. M., Christian J. L. (2009) Site-specific cleavage of BMP4 by furin, PC6, and PC7. J. Biol. Chem. 284, 27157–27166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siegfried G., Basak A., Prichett-Pejic W., Scamuffa N., Ma L., Benjannet S., Veinot J. P., Calvo F., Seidah N., Khatib A. M. (2005) Regulation of the stepwise proteolytic cleavage and secretion of PDGF-B by the proprotein convertases. Oncogene 24, 6925–6935 [DOI] [PubMed] [Google Scholar]
- 17. Stawowy P., Marcinkiewicz J., Graf K., Seidah N., Chrétien M., Fleck E., Marcinkiewicz M. (2001) Selective expression of the proprotein convertases furin, pc5, and pc7 in proliferating vascular smooth muscle cells of the rat aorta in vitro. J. Histochem. Cytochem. 49, 323–332 [DOI] [PubMed] [Google Scholar]
- 18. Marcinkiewicz M., Marcinkiewicz J., Chen A., Leclaire F., Chrétien M., Richardson P. (1999) Nerve growth factor and proprotein convertases furin and PC7 in transected sciatic nerves and in nerve segments cultured in conditioned media: their presence in Schwann cells, macrophages, and smooth muscle cells. J. Comp. Neurol. 403, 471–485 [DOI] [PubMed] [Google Scholar]
- 19. Siegfried G., Basak A., Cromlish J. A., Benjannet S., Marcinkiewicz J., Chrétien M., Seidah N. G., Khatib A. M. (2003) The secretory proprotein convertases furin, PC5, and PC7 activate VEGF-C to induce tumorigenesis. J. Clin. Investig. 111, 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leonhardt R. M., Fiegl D., Rufer E., Karger A., Bettin B., Knittler M. R. (2010) Post-endoplasmic reticulum rescue of unstable MHC class I requires proprotein convertase PC7. J. Immunol. 184, 2985–2998 [DOI] [PubMed] [Google Scholar]
- 21. Rousselet E., Benjannet S., Marcinkiewicz E., Asselin M. C., Lazure C., Seidah N. G. (2011) Proprotein convertase PC7 enhances the activation of the EGF receptor pathway through processing of the EGF precursor. J. Biol. Chem. 286, 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oexle K., Ried J. S., Hicks A. A., Tanaka T., Hayward C., Bruegel M., Gögele M., Lichtner P., Müller-Myhsok B., Döring A., Illig T., Schwienbacher C., Minelli C., Pichler I., Fiedler G. M., Thiery J., Rudan I., Wright A. F., Campbell H., Ferrucci L., Bandinelli S., Pramstaller P. P., Wichmann H. E., Gieger C., Winkelmann J., Meitinger T. (2011) Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum. Mol. Genet. 20, 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guillemot J., Canuel M., Essalmani R., Prat A., Seidah N. G. (2013) Implication of the proprotein convertases in iron homeostasis: proprotein convertase 7 sheds human transferrin receptor 1 and furin activates hepcidin. Hepatology 57, 2514–2524 [DOI] [PubMed] [Google Scholar]
- 24. Walker M. B., Miller C. T., Coffin Talbot J., Stock D. W., Kimmel C. B. (2006) Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev. Biol. 295, 194–205 [DOI] [PubMed] [Google Scholar]
- 25. Chitramuthu B. P., Bennett H. P. (2011) Use of zebrafish and knockdown technology to define proprotein convertase activity. Methods Mol. Biol. 768, 273–296 [DOI] [PubMed] [Google Scholar]
- 26. Schlombs K., Wagner T., Scheel J. (2003) Site-1 protease is required for cartilage development in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 100, 14024–14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poirier S., Prat A., Marcinkiewicz E., Paquin J., Chitramuthu B. P., Baranowski D., Cadieux B., Bennett H. P., Seidah N. G. (2006) Implication of the proprotein convertase NARC-1/PCSK9 in the development of the nervous system. J. Neurochem. 98, 838–850 [DOI] [PubMed] [Google Scholar]
- 28. Tang R., Dodd A., Lai D., McNabb W. C., Love D. R. (2007) Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. 39, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thisse C., Thisse B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 30. Storey J. D. (2002) A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 64, 479 [Google Scholar]
- 31. Freeman J. L., Adeniyi A., Banerjee R., Dallaire S., Maguire S. F., Chi J., Ng B. L., Zepeda C., Scott C. E., Humphray S., Rogers J., Zhou Y., Zon L. I., Carter N. P., Yang F., Lee C. (2007) Definition of the zebrafish genome using flow cytometry and cytogenetic mapping. BMC Genomics 8, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Day R., Schafer M. K., Watson S. J., Chrétien M., Seidah N. G. (1992) Distribution and regulation of the prohormone convertases PC1 and PC2 in the rat pituitary. Mol. Endocrinol. 6, 485–497 [DOI] [PubMed] [Google Scholar]
- 33. Marcinkiewicz M., Day R., Seidah N. G., Chrétien M. (1993) Ontogeny of the prohormone convertases PC1 and PC2 in the mouse hypophysis and their colocalization with corticotropin and α-melanotropin. Proc. Natl. Acad. Sci. U.S.A. 90, 4922–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 35. Bode W., Papamokos E., Musil D. (1987) The high-resolution x-ray crystal structure of the complex formed between subtilisin Carlsberg and eglin c, an elastase inhibitor from the leech Hirudo medicinalis. Structural analysis, subtilisin structure and interface geometry. Eur. J. Biochem. 166, 673–692 [DOI] [PubMed] [Google Scholar]
- 36. McColl B. K., Paavonen K., Karnezis T., Harris N. C., Davydova N., Rothacker J., Nice E. C., Harder K. W., Roufail S., Hibbs M. L., Rogers P. A., Alitalo K., Stacker S. A., Achen M. G. (2007) Proprotein convertases promote processing of VEGF-D, a critical step for binding the angiogenic receptor VEGFR-2. FASEB J. 21, 1088–1098 [DOI] [PubMed] [Google Scholar]
- 37. Basak A., Zhong M., Munzer J. S., Chrétien M., Seidah N. G. (2001) Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bedell V. M., Westcot S. E., Ekker S. C. (2011) Lessons from morpholino-based screening in zebrafish. Brief. Funct. Genomics 10, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eisen J. S., Smith J. C. (2008) Controlling morpholino experiments: don't stop making antisense. Development 135, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 40. Mathavan S., Lee S. G., Mak A., Miller L. D., Murthy K. R., Govindarajan K. R., Tong Y., Wu Y. L., Lam S. H., Yang H., Ruan Y., Korzh V., Gong Z., Liu E. T., Lufkin T. (2005) Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 1, 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim W., Essalmani R., Szumska D., Creemers J. W., Roebroek A. J., D'Orleans-Juste P., Bhattacharya S., Seidah N. G., Prat A. (2012) Loss of endothelial furin leads to cardiac malformation and early postnatal death. Mol. Cell. Biol. 32, 3382–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pesu M., Muul L., Kanno Y., O'Shea J. J. (2006) Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon γ. Blood 108, 983–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pesu M., Watford W. T., Wei L., Xu L., Fuss I., Strober W., Andersson J., Shevach E. M., Quezado M., Bouladoux N., Roebroek A., Belkaid Y., Creemers J., O'Shea J. J. (2008) T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature 455, 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dubois C. M., Blanchette F., Laprise M. H., Leduc R., Grondin F., Seidah N. G. (2001) Evidence that furin is an authentic transforming growth factor-β1-converting enzyme. Am. J. Pathol. 158, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Blanchette F., Day R., Dong W., Laprise M. H., Dubois C. M. (1997) TGFβ1 regulates gene expression of its own converting enzyme furin. J. Clin. Investig. 99, 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turpeinen H., Kukkurainen S., Pulkkinen K., Kauppila T., Ojala K., Hytönen V. P., Pesu M. (2011) Identification of proprotein convertase substrates using genome-wide expression correlation analysis. BMC Genomics. 12, 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pearton D. J., Nirunsuksiri W., Rehemtulla A., Lewis S. P., Presland R. B., Dale B. A. (2001) Proprotein convertase expression and localization in epidermis: evidence for multiple roles and substrates. Exp. Dermatol. 10, 193–203 [DOI] [PubMed] [Google Scholar]
- 48. Remacle A. G., Shiryaev S. A., Oh E. S., Cieplak P., Srinivasan A., Wei G., Liddington R. C., Ratnikov B. I., Parent A., Desjardins R., Day R., Smith J. W., Lebl M., Strongin A. Y. (2008) Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J. Biol. Chem. 283, 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henrich S., Lindberg I., Bode W., Than M. E. (2005) Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J. Mol. Biol. 345, 211–227 [DOI] [PubMed] [Google Scholar]
- 50. Mesnard D., Donnison M., Fuerer C., Pfeffer P. L., Constam D. B. (2011) The microenvironment patterns the pluripotent mouse epiblast through paracrine Furin and Pace4 proteolytic activities. Genes Dev. 25, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Senturker S., Thomas J. T., Mateshaytis J., Moos M., Jr. (2012) A homolog of subtilisin-like proprotein convertase 7 is essential to anterior neural development in Xenopus. PLoS One 7, e39380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu X., Muller L., Mains R. E., Lindberg I. (1998) Structural elements of PC2 required for interaction with its helper protein 7B2. J. Biol. Chem. 273, 1158–1164 [DOI] [PubMed] [Google Scholar]
- 53. Shu X., Zeng Z., Gautier P., Lennon A., Gakovic M., Cheetham M. E., Patton E. E., Wright A. F. (2011) Knockdown of the zebrafish ortholog of the retinitis pigmentosa 2 (RP2) gene results in retinal degeneration. Invest. Ophthalmol. Vis. Sci. 52, 2960–2966 [DOI] [PubMed] [Google Scholar]
- 54. Shull M. M., Ormsby I., Kier A. B., Pawlowski S., Diebold R. J., Yin M., Allen R., Sidman C., Proetzel G., Calvin D. (1992) Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 359, 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prud'homme G. J., Piccirillo C. A. (2000) The inhibitory effects of transforming growth factor-β-1 (TGF-β1) in autoimmune diseases. J. Autoimmun. 14, 23–42 [DOI] [PubMed] [Google Scholar]
- 56. Kulkarni A. B., Huh C. G., Becker D., Geiser A., Lyght M., Flanders K. C., Roberts A. B., Sporn M. B., Ward J. M., Karlsson S. (1993) Transforming growth factor β1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. U.S.A. 90, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henrich S., Cameron A., Bourenkov G. P., Kiefersauer R., Huber R., Lindberg I., Bode W., Than M. E. (2003) The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat. Struct. Biol. 10, 520–526 [DOI] [PubMed] [Google Scholar]
- 59. Wheatley J. L., Holyoak T. (2007) Differential P1 arginine and lysine recognition in the prototypical proprotein convertase Kex2. Proc. Natl. Acad. Sci. U.S.A. 104, 6626–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]