Background: Exosomes, secreted from cells, have immunomodulatory capacities.
Results: NFκB- and STAT3-mediated cytokine release is triggered by various types of ex vivo exosomes in a TLR-dependent fashion.
Conclusion: Exosomes have inherent signaling capacities important for global inflammatory responses.
Significance: Detailed knowledge about intercellular communication in cancer and inflammatory diseases is crucial for development of new therapeutic approaches.
Keywords: Cytokine Induction, Exosomes, NFκB Transcription Factor, STAT3, Toll-like receptor (TLR), Amniotic Fluid, Malignant Ascites
Abstract
Tumor-derived exosomes have been shown to induce various immunomodulatory effects. However, the underlying signaling pathways are poorly understood. Here, we analyzed the effects of ex vivo-derived exosomes on monocytic cell differentiation/activation using THP-1 cells as model. We isolated exosomes from various body fluids such as amniotic fluid, liver cirrhosis ascites, and malignant ascites of ovarian cancer patients. We observed that exosomes were internalized by THP-1 cells and induced the production of IL-1β, TNF-α, and IL-6. Analysis of the signaling pathways revealed a fast triggering of NFκB and a delayed activation of STAT3. Pharmacologic and antibody-blocking experiments showed that the initial production of IL-6 was instrumental for subsequent activation of STAT3. Importantly, triggering of cell signaling was not a unique property of tumor exosomes but was also observed with exosomes of noncancerous origin. Exosomal signaling was TLR-dependent as the knockdown of Toll-like receptor 2 (TLR2) and TLR4 blocked NFκB and STAT3 activation. Similar results were obtained with TLR-neutralizing antibodies. Exosomes also triggered the release of cytokines from mouse bone marrow-derived dendritic cells or macrophages. This process was MyD88-dependent, further supporting a role of TLR signaling. Our results suggest that exosomes trigger TLR-dependent signaling pathways in monocytic precursor cells but possibly also in other immune cells. This process could be important for the induction of immunosuppressive mechanisms during cancer progression and inflammatory diseases.
Introduction
Exosomes are small membrane vesicles released from many different cell types in the body such as erythrocytes, platelets, lymphocytes, dendritic cells (DCs)3 and tumor cells (1–3). Exosomes are formed by invagination and budding from the limiting membrane of late endosomes (4, 5). It is believed that they accumulate in cytosolic multivesicular bodies from where they are released by fusion with the plasma membrane in a Rab-dependent manner (4–6). This process is very active in proliferating cells, such as cancer cells (7). Depending on the cellular origin, the protein composition of exosomes can differ from that of the plasma membrane. Exosomal proteins include MHC molecules, tetraspanins, adhesion molecules, and metalloproteinases (1, 2, 8). In addition to functional proteins, exosomes carry mRNAs as well as miRNAs (9–12). Exosomes can be engulfed by other cells, and it was proposed that this transfer of protein and RNAs from one to another cell might represent a novel mechanism of intercellular communication (4, 9, 13).
Exosomes play an important role in modulating immune responses (14, 15). When derived from dendritic cells, they bear MHC proteins and co-stimulatory molecules (2, 16). These vesicles are effective in stimulating antigen-specific T cell responses in vitro and in vivo. In contrast, exosomes from tumor cells or those released from the fetus during pregnancy can suppress immune responses (15, 17–22). It was found that tumor-derived exosomes can directly inhibit natural killer cell or T cell function (19, 23), block dendritic cell maturation (24), induce MDSCs (15, 25, 26), or augment the activity of regulatory T cells (17, 27). Cancer-derived microvesicles can modify the surrounding stroma and promote the process of fibroblast differentiation into myofibroblasts (28). They foster the interplay of stroma cells with breast tumor cells (29). These effects support tumor growth and suppress innate and adaptive immune responses.
It is presently unknown which signal pathways exosomes trigger in recipient cells to induce an immunosuppressive phenotype. Recent work has shown that the induction of MDSCs from precursors involved TLR signaling and STAT3 activation (25, 30). Another study has shown that miRNAs in cancer-released exosomes can reach and bind TLRs and induce prometastatic inflammatory responses (31). However, it is unclear whether these properties are unique to cancer-derived exosomes or are an intrinsic feature of all exosomes.
In this paper we have addressed the questions of whether exosomes can modulate the biological functions of monocytic cells and which signaling pathways are activated by exosomes. We show that microvesicles are taken up by human monocytic THP-1 cells and stimulate cytokine release and NFκB/STAT3 activation. We also show that the initiation of this signaling is TLR-dependent and can be elicited not only by tumor-derived exosomes.
EXPERIMENTAL PROCEDURES
Chemicals and Antibodies
Antibodies to human CD24 (SWA11), ADAM10 (11G2), and CD9 (TS9) have been described (32–34). The mAbs to HSP70, Annexin-1, Tsg101, and CD9 were from BD Transduction. The antibodies to GAPDH or p65 were from Santa Cruz Biotechnologies. The antibodies against STAT3, p-STAT3 (Y705) and phospho-p65 (S536) were obtained from Cell Signaling. Neutralizing antibodies to IL-6 and IL-6 receptor were a kind gift from Prof. Stefan Rose-John, University of Kiel, Germany. TLR2 and TLR4 neutralizing antibodies and TLR agonists (LPS, PAM3CSK4, and R848) were obtained from Invivogen. Fluorochrome-conjugated mAbs for FACS analysis of TLR2 (Biolegend 121805) or TLR4 (E Bioscience 17-9917) were used at a dilution of 1:200. Lentiviral shRNA particles (each five shRNA clones) for stable knockdown of TLR2 (NM_003264.2) or TLR4 (NM_003266.2) as well as control particles (shEGFP) were obtained from Sigma-Aldrich. Phorbol 12-myristate 13-acetate (PMA) was from Sigma-Aldrich. Parthenolide (IκB kinase inhibitor), P6 (pyridone 6/JAK2 inhibitor), and AG490 (JAK1 inhibitor) were from Calbiochem.
Isolation of Exosomes
Analysis of patient-derived material was under the approval of the ethics commission of the University of Heidelberg. Human amniotic fluid was collected for routine amniocentesis and analyzed after removal of cells. Exosomes were isolated by differential centrifugation as described (32). The purification of exosomes from the ascites of liver cirrhosis patients was described before (12, 32). All purified exosomes were dissolved in PBS, and the protein concentration was determined using a Bradford protein quantification kit (Bio-Rad). BSA was used as standard. NanoSight analysis was carried out by courtesy of the company (NanoSight, Amesbury, Wiltshire, UK).
Treatment of THP-1 Cells with Exosomes
THP-1 cells were seeded into 15-ml conical tubes (THP-1, 2 × 106 cells/tube). Depending on the experiment, cells were treated with body fluid exosomes (40–200 ng/μl) in a total volume of 500 μl of serum-free RPMI 1640 medium for different periods of time (30 min to 48 h). After treatment, lysates were analyzed by Western blotting or RT-PCR. In case of co-incubation of cells with exosomes and antibodies, antibodies were used at a concentration of 1–3 μg/ml unless otherwise indicated. Treatment of cells with heat-inactivated or enzymatically digested exosomes were performed accordingly (40 ng/μl exosomes/2 × 106 cells in a total volume of 500 μl of serum-free medium). Co-treatment of cells with exosomes and inhibitors was performed with 20 μg (40 ng/μl) of exosomes in the presence of either 10 μm parthenolide, 2 μm pyridone 6 (P6), or 10 μm AG490, respectively.
Heat Inactivation and Enzymatic Digestion of Exosomes
Purified exosomes were heat-inactivated by incubation at 95 °C for 5–10 min on a heating block. For enzymatic digestion, 100–170 μg of exosomes was digested in the presence of 10 μg/ml DNase I, 10 μg/ml RNase A, or 10 μg/ml protease K, respectively, for 1.5 h at 37 °C. To inhibit protease K after digestion, the digestion mix was supplied with 100 μm PMSF.
Lentiviral Transduction of THP-1 Cells
For the transduction of THP-1 monocytes with lentiviral particles, 2 × 105 cells were pelleted in 15-ml conical tubes (1000 × g, 5 min). Five different shTLR2- and shTLR4-viral particle clones as well as one shEGFP control clone were used to transduce cells. Cells were spinoculated (1000 × g, 1 h at 22 °C) in the presence of particles at a respective multiplicity of infection of 1. Cells undergoing spinoculation in the absence of virus further served as cytotoxicity control under selection conditions. After centrifugation and aspiration of virus-containing supernatant, cells were resuspended in growth medium (DMEM, 10% FCS) and transferred to 12-well plates. Transduced cells were grown under standard cell culture conditions for another 72 h before being placed in selection medium (0.6 μg/ml puromycin in growth medium). Expansion under selective conditions was continued for 14 days.
FACS Analysis
FACS analysis of isolated vesicles was done after adsorbing of isolated vesicles to 4 nm (surfactant-free) aldehyde-sulfate latex beads (Interfacial Dynamics, Portland, OR) as described (34). The staining of beads or cultivated cells with mAbs and phycoerythrin-conjugated secondary antibodies has been described (32, 34). Stained beads or cells were analyzed with a FACScan using Cellquest software (BD Biosciences).
Cell Adhesion Assay
THP-1 cells (1.0 × 105) were incubated for 24 h in the presence of malignant ascites (AS) exosomes (50 μg/ml) in a 96-well format. Plastic-adherent cells were washed with PBS and incubated for 90 min at 37 °C with 10 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/ml) in 100 μl of RPMI 1640 medium. After incubation, the plate was centrifuged, and supernatant was discarded. Per well, 100 μl of dimethyl sulfoxide was added and incubated for 10 min at room temperature. The amount of released formazan was measured at 570 nm.
Cell Migration Assay
THP-1 cells (2.5 × 105) were seeded in 100 μl of RPMI 1640 medium + 50 μg/ml exosomes in the upper chamber of a Boyden chamber (pore size 5 μm). The lower chamber was filled with 400 μl of RPMI 1640 medium. After a 24-h incubation, migrated cells were detached from the bottom side of the membrane by incubation with trypsin/EDTA and combined with the cell containing medium of the lower chamber. Migrated cells were quantified with CASY® Cell Analyzer systems.
Mouse Cells and Treatment with Exosomes
Bone marrow cells from 8–12-week-old C57BL/6 WT or C57BL/6-MyD88−/− mice were collected from femur bone marrow elutions. DCs were matured by incubation with supernatant from the X63 plasmocytoma cell line producing murine GM-CSF in RPMI 1640 medium, 10% FCS, 1% penicillin/streptomycin, 0.05 mm β-mercaptoethanol for 8 days. Macrophages were differentiated in DMEM supplemented with 30% L929 supernatant, 10% FCS, and 1% penicillin/streptomycin for 7 days. DCs and macrophages (each 2 × 105/well) were stimulated with 8 μg of amniotic fluid (AF) exosomes or various TLR agonists (1 μg/ml) in a 96-well (DCs) or 48-well (macrophages) format for 16 h. Cell-free supernatants were used for detection of indicated cytokines using specific ELISAs (BD Biosciences).
Quantitative Real-time PCR
RT-PCR was performed as described before (35). Briefly, total RNA from cells was isolated and transcribed using the ReverseAid H Minus First Strand cDNA Synthesis kit (Fermentas, St. Leon-Rot, Germany), purified on Microspin G-50 columns (GE Healthcare) and quantified by a NanoDrop Spectrophotometer (ND-2000; Thermo Scientific). Primers were designed using the IDT primer quest program and were produced by MWG (Ebersberg, Germany). β-Actin was used as an internal standard. The sequences of primers are available on request.
Cell Lysis and Western Blot Analysis
Cell pellets were solubilized in lysis buffer (250 mm NaCl, 50 mm HEPES, 0.5% Nonidet P-40, 10% glycerol, 2 mm EDTA, 10 mm NaF, 1 mm sodium orthovanadate, 1 mm PMSF, 10 mg/ml each leupeptin and aprotinin) for 30 min on ice. Lysates were cleared by centrifugation and boiled with reducing or nonreducing SDS loading buffer. Samples were separated on SDS-polyacrylamide gels and transferred to Immobilon membranes using semidry blotting. After blocking with 4% BSA in Tween 20/TBS, membranes were probed with primary antibodies followed by horseradish peroxidase-conjugated secondary antibody and ECL detection (Amersham Biosciences).
Bio-Plex Assay
Cytokines in cell culture supernatants were analyzed using a Bio-Plex Pro Human Cytokine 27-plex assay (Bio-Rad).
Statistical Analysis
Data are presented as the mean ± S.E. Student's unpaired t test was used to evaluate the difference between groups. p < 0.05 was considered statistically significant.
RESULTS
Exosomes from Body Fluids Are Internalized and Affect Cellular Properties of THP-1 Cells
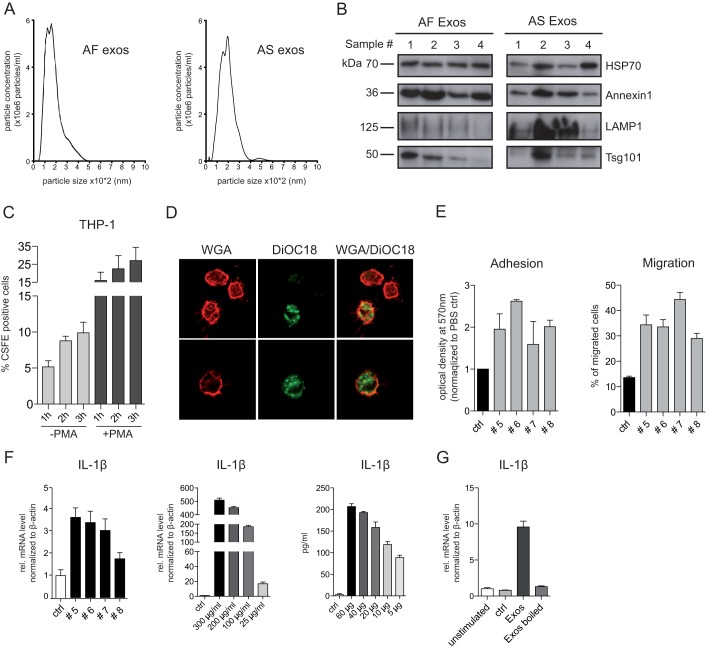
We previously isolated exosomes by differential centrifugation from human AS, serum, and urine (11, 12). A similar protocol was used to collect exosomes from AF. An average of 7.21 μg/ml (range 1.6–13 μg/ml, n = 64) of protein was recovered per AF sample, which is comparable with the yield reported for AS. Density gradient centrifugation and transmission EM analysis for AS and liver cirrhosis (LC) exosomes were performed previously (12). Nanosight analysis showed a size distribution of 100–300 nm for both types of exosomes (Fig. 1A). Biochemical analysis revealed that AS and AF exosomes were positive for the established exosomal marker proteins HSP70, Annexin-1, and LAMP1 as well as the ESCRT component Tsg101 (Fig. 1B).
FIGURE 1.
Characterization of AF and AS exosomes. A, nanosight analysis of isolated AF and AS exosomes is shown. Exosomes were isolated by differential centrifugation from AF or AS of ovarian carcinoma patients. B, representative Western blot analysis of exosomes isolated from four different AF or AS donors is shown. C, AS exosomes were labeled with CFSE and 1 mg of exosomes and exposed to THP-1 cells treated or nontreated before with PMA at 37 °C. After the indicated time point, aliquots of cells were washed and analyzed by cytofluorographic analysis. Mean fluorescence of labeled cells after uptake from three exosomal donors is plotted. D, THP-1 cells that had internalized CFSE exosomes for 1 h were analyzed by fluorescent microscopy. To visualize the plasma membrane the cells were counterstained with phycoerythrin-coupled wheat germ agglutinin (WGA). Note the intracellular localization of labeled exosomes. E, AS exosomes from different donors (#5–8) trigger plastic adhesiveness and cell migration of THP-1 cells. Motility of cells was determined after 24 h by Boyden chamber analysis. F, exosomes induced IL-1β expression. THP-1 cells were incubated with the AS exosomes (10 μg/ml) (donors #5–8) for 24 h at 37 °C. IL-1β mRNA expression was measured by RT-PCR. THP-1 cells were incubated with the indicated amounts of AS exosomes (donor #5) for 24 h at 37 °C. Levels of IL-1β mRNA were measured by RT-PCR, and the cell culture supernatant was analyzed by IL-1β-specific ELISA. G, boiling destroys the stimulating activity. THP-1 cells incubated with 50 μg/ml AS exosomes either boiled (5 min, 95 °C) or nonboiled were analyzed for IL-1β by RT-PCR after 24 h at 37 °C. Error bars, S.E.
Exosomes are released from cells by fusion of multivesicular bodies with the plasma membrane and have an orientation of antigens facing outside (1, 5). We confirmed the orientation of antigens by FACS analysis after immobilization of the vesicles on latex beads. We observed stronger staining with antibodies to CD24 and ADAM10 and weaker staining for CD9 and β1-integrin. These proteins were previously described to occur in exosomes (1, 12). The uptake of CFSE-labeled exosomes was investigated in the human monocytic precursor cell line THP-1. As detected by FACS analysis, the uptake proceeded in a time-dependent fashion and was augmented when THP-1 cells were pretreated with PMA before the assay (Fig. 1C). The degree of uptake was variable and patient-dependent. Fluorescent microscopy showed that labeled exosomes localized inside of the cells (Fig. 1D).
Upon differentiation into macrophages THP-1 cells become plastic-adherent. We examined whether the incubation with exosomes affected the adhesion and motility of THP-1 cells. When tested 24 h after exosome exposure, THP-1 cells showed enhanced adhesiveness to plastic and augmented migration in Boyden chamber assays (Fig. 1E). Although to a different extent, this was observed with exosomes from several patients.
Exosomes Induce Production of Cytokines in THP-1 Cells
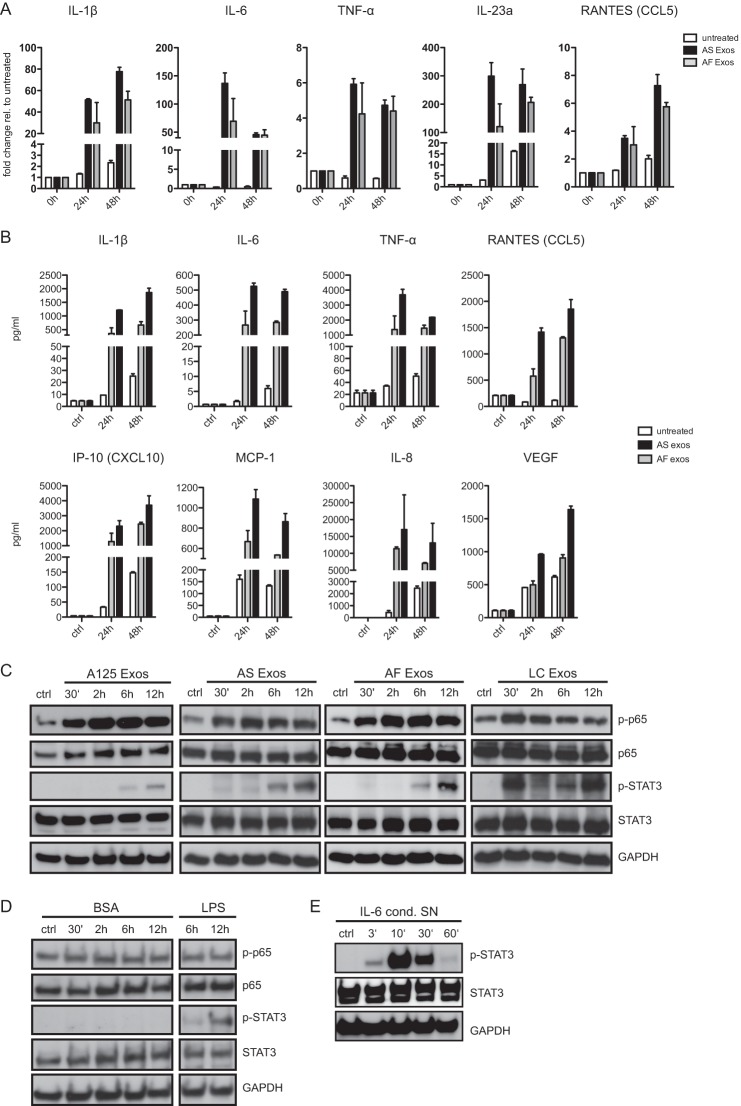
We next analyzed THP-1 cells for changes in gene expression after exosome exposure. We found that exosomes induced a dose-dependent up-regulation of IL-1β as measured by RT-PCR (Fig. 1F). Enhanced release of IL-1β was detected by ELISA (Fig. 1F). The induction was again dose-dependent and abolished by boiling exosomes before addition to the cells (Fig. 1G). In human blood monocytes tumor-derived exosomes trigger the release of cytokines, including IL-6, TNF-α, and TGF-β (26). We therefore extended the analysis of cytokines released by THP-1 cells. To find out whether also non-tumor-derived exosomes could trigger cytokine release, we compared AS-derived with AF-derived exosomes. By RT-PCR (Fig. 2A) and Bio-Plex analysis (Fig. 2B) we found strong induction of cytokines such as IL-6, IL-1β, TNF-α, IL-23a, and IL-8. Importantly, at the dose tested (40 μg/ml) both types of exosomes enhanced production and secretion of cytokines in a comparable fashion.
FIGURE 2.
Exosomes trigger cytokine production and cell signaling in THP-1 cells. A, cells were incubated with 40 μg/ml AF or AS exosomes for the indicated times at 37 °C and analyzed by RT-PCR for the expression of cytokines. Pooled results of n = 3 experiments are shown. RANTES, regulated on activation normal T cell expressed and secreted. Error bars, S.E. B, exposure of THP-1 cells to exosomes induces secretion of cytokines. Cells were incubated with 40 μg/ml exosomes for the indicated length of time at 37 °C, and the culture supernatant was analyzed by multiplex ELISA. A representative experiment of n = 3 experiments is shown. C, cells were incubated with 40 μg/ml A125, AS, AF, or LC exosomes for the indicated length of time at 37 °C. Cell lysates were analyzed by Western blotting. A representative of n > 3 experiments is shown. D, THP-1 cells were incubated with BSA (40 μg/ml, as negative control) or LPS (1 μg/ml, as positive control) for the indicated length of time. Note that BSA does not activate STAT3 phosphorylation whereas LPS does. E, THP-1 cells were incubated with IL-6 containing cell culture supernatant (SN) for the indicated length of time followed by Western blot analysis.
Exosomes Exposure Triggers NFκB and STAT3 Activation
Previous work had shown that ex vitro-derived exosomes from tumor cell lines could trigger NFκB signaling (31, 36) and activate STAT3 (25, 30). In THP-1 cells we investigated the signaling events triggered by exosomes. We observed a rapid activation of NFκB as detected by phosphorylation of the p65 subunit that peaked at 30–120 min after exosome exposure (Fig. 2C). STAT3 activation was delayed and became only obvious after 6–12 h (Fig. 2C). The kinetic of activation was similar for AF-, AS-, and LC-derived exosomes. Additionally, exosomes from in vitro cultured A125 lung cancer cells showed comparable activation of NFκB and STAT3. Phosphorylation of STAT3 after 12 h was also seen when cells were stimulated for control with LPS but not with BSA (Fig. 2D). Rapid activation of STAT3 (3–10 min) was shown when THP-1 cells were treated with IL-6-enriched supernatant (Fig. 2E). This clearly demonstrated that exosomal activation of STAT3 was temporally different from classical activation by IL-6.
Interestingly, for LC exosomes an early STAT3 phosphorylation after 30 min was also noticed (Fig. 2C). Of note, when body fluid-derived exosomes were examined for the content of cytokines, LC exosomes carried IL-6. Most likely this reflected their origin from an inflammatory background. We concluded that the property to stimulate NFκB and late STAT3 activation in THP-1 cells is not a unique property of tumor-derived exosomes but is shared by exosomes from other sources.
STAT3 Activation Is Mediated by Autocrine/Paracrine Function of IL-6
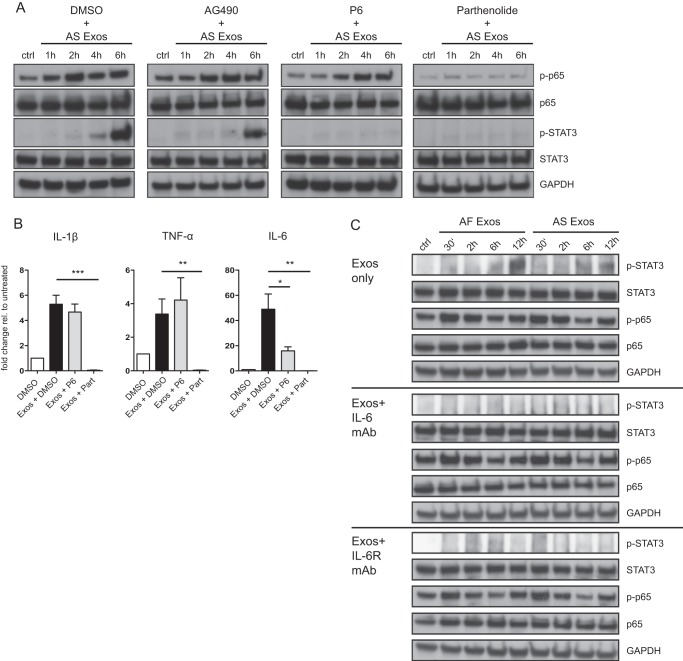
STAT3 phosphorylation is classically induced by the activation of the IL-6/JAK2/STAT3 pathway (37). We argued that IL-6 produced in the first wave of NFκB activation would subsequently trigger STAT3 phosphorylation in an autrocrine/paracrine fashion. In support of this, we found that STAT3 activation was blocked in the presence of the specific NFκB inhibitor parthenolide and the JAK2-specific inhibitor P6 but not by the JAK1 inhibitor AG490 (Fig. 3A).
FIGURE 3.
Role of NFκB and STAT3 in THP-1 cell signaling. A, THP-1 cells were incubated with 40 μg/ml AS exosomes for the indicated length of time at 37 °C in the absence or presence of the JAK2 inhibitor P6 (2 μm), the JAK1 inhibitor AG490 (20 μm), or the specific NFκB inhibitor parthenolide (10 μm). Dimethyl sulfoxide (DMSO) served as solvent control. Cell lysates were prepared and analyzed by Western blotting with the indicated primary antibodies followed by peroxidase-conjugated secondary antibodies and ECL detection. B, RT-PCR analysis was performed on mRNAs isolated from cells treated in A. Error bars, S.E. C, THP-1 cells were incubated with exosomes as described above in the presence or absence of neutralizing antibodies to IL-6 and IL-6 receptor (each 1 μg/ml). Cell lysates were prepared and analyzed by Western blotting as described. ***, p < 0.001; **, p <0.01; *, p <0.05.
Strikingly, the treatment with parthenolide abolished the induction of cytokine genes by exosomes as determined for IL-6, TNF-α, and IL-1β by RT-PCR (Fig. 3B). Treatment with P6 also reduced the level of IL-6 in agreement with the notion that IL-6 is also a STAT3-dependent gene (38). These findings suggested that a cytokine was the likely mediator between the initial NFκB and delayed STAT3 activation and that the observed signaling events were linked to the induction of cytokine genes.
To further solidify the important role of IL-6, we employed neutralizing mAbs to IL-6 and IL-6 receptor. When THP-1 cells were stimulated with exosomes in the presence of anti-IL-6 or anti-IL-6 receptor antibodies no phosphorylation of STAT3 was observed (Fig. 3C).
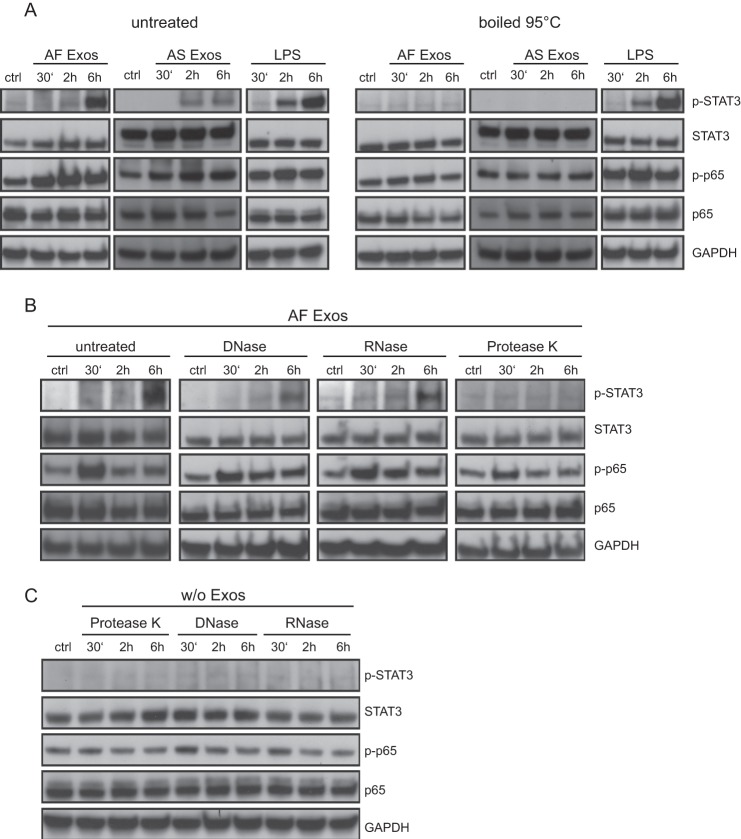
NFκB and STAT3 Activation by Exosomes Is Proteinase-sensitive
To find out which determinant on exosomes was responsible for signal initiation, we treated exosomes with DNase, RNase, or proteinase K before exposure to THP-1 cells. Only boiling at 95 °C (Fig. 4A) or proteinase K treatment (Fig. 4B) completely abolished NFκB and STAT3 activation. In contrast, boiling of LPS did not destroy the ability to trigger NFκB and STAT3 activation (Fig. 4A). Importantly, the enzymes alone did not trigger activation of THP-1 cells (Fig. 4C). Collectively, these findings together with those presented in Fig. 1G emphasized that an LPS contamination was unlikely to be the source of triggering. Our data rather suggested that a protein determinant associated with exosomes is responsible for the induction of p65 phosphorylation.
FIGURE 4.
Proteinase-sensitive determinant(s) on exosomes trigger THP-1 cells. A, effect of heat on stimulation efficacy of exosomes and LPS. Exosomes (40 μg/ml) or LPS (1 μg/ml) was boiled at 95 °C for 5 min, added to THP-1 cells, and incubated for the indicated length of time. Cell lysates were prepared and analyzed by Western blotting. B, effect of enzymatic treatment on stimulation efficacy. AF exosomes in a volume of 100 μl were preincubated with DNase (10 mg/ml), RNAs (10 mg/ml), or proteinase K (10 mg/ml) and then added to THP-1 cells. Cells were incubated for the indicated length of time. Cell lysates were prepared and analyzed by Western blotting. C, the same amount of enzymes as used in B added to THP-1 cells to exclude unspecific effects.
TLRs Are Involved in Exosome-mediated Signaling
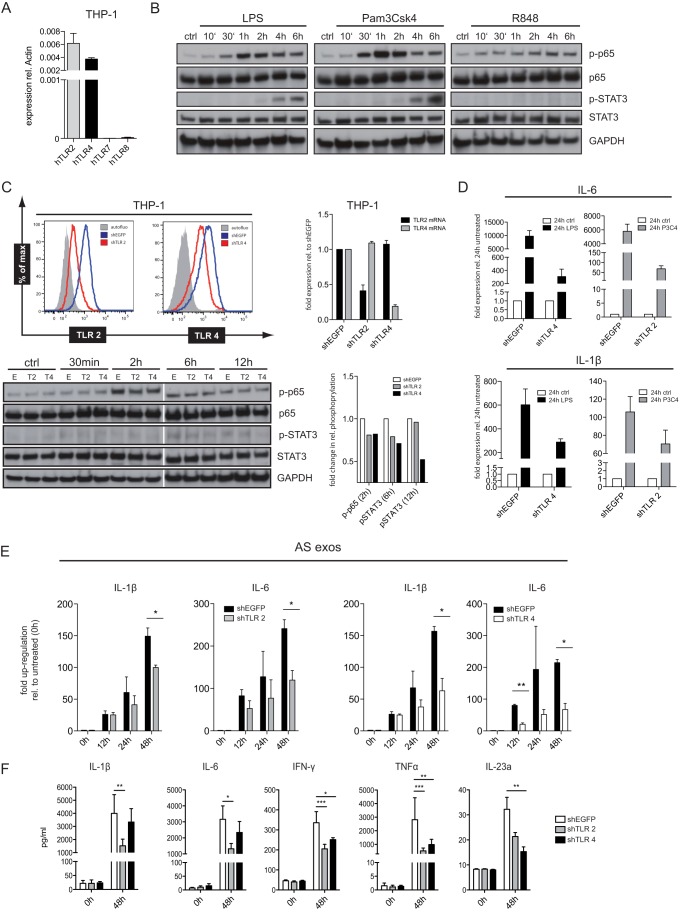
It is known that TLRs can activate the NFκB pathway. Indeed, we noticed that the effects of exosomes on cell signaling were similar to TLR agonists. This prompted us to study more closely the role of TLRs in exosome-mediated signaling. THP-1 cells expressed TLR2 and TLR4 but were negative for TLR7 and TLR8 as detected by RT-PCR (Fig. 5A). In agreement with this expression profile, strong stimulation was observed with the TLR1/2 agonist PAM3CSK4 (P3C4) and LPS, a known ligand to TLR4 (Fig. 5B). In contrast, no stimulation was seen with the TLR7/8 ligand R848 (Fig. 5B).
FIGURE 5.
Role of TLR2 and TLR4 in exosomal signaling. A, characterization of TLR expression in THP-1 cells was done by RT-PCR analysis. B, THP-1 cells were stimulated with TLR agonists (1 μg/ml) for the indicated length of time, and cell lysates were analyzed by Western blotting. C, THP-1 cells were analyzed after lentiviral shRNA-mediated TLR2 and TLR4 knockdown. Cells transduced with shEGFP served as control. FACS analysis was performed with specific dye-coupled mAb to TLR2 and TLR4. Isotype control Ab served as background control (gray curve). Expression levels of TLR2 and TLR4 mRNA in THP-1/shTLR cells were determined by RT-PCR. p65 and STAT3 activation by exosomes (40 μg/ml) was checked by Western blotting following quantification of p-p65 and p-STAT3 bands. D, THP-1/shTLR2 and THP-1/shTLR4 cells were stimulated with TLR agonists LPS and P3C4 for 24 h. Cells were lysed, and mRNA was isolated and subjected to RT-PCR analysis using primers for IL-6 and IL-1β. E, THP-1/shTLR2, THP-1/shTLR4, or shEGFP control cells were stimulated with exosomes as described above for the indicated length of time. The level of IL-1β or IL-6 mRNA was determined by RT-PCR. F, the level of cytokines released into the medium was analyzed via multiplex ELISA. Pooled data from n = 3 experiments are shown. ***, p < 0.001; **, p <0.01; *, p <0.05. Error bars, S.E.
To find out which TLRs were involved in exosome signaling, we stably silenced TLR2 and TLR4 expression by lentiviral-transduced shRNA. As determined by FACS analysis, the THP-1/shTLR2-transduced cells showed significantly diminished expression of TLR2 (67% decrease) compared with the control cells (shEGFP) (Fig. 5C). A similar reduction of TLR4 was seen in THP-1/shTLR4 (52% decrease) (Fig. 5C). The knockdown was specific for the respective TLRs as confirmed by RT-PCR analysis (Fig. 5C). Activation of p65 and STAT3 was also reduced in THP-1/shTLR2 and THP-1/shTLR4 cells (Fig. 5C) The stimulation of knockdown cells with the TLR agonists P3C4 and LPS, respectively, was reduced by >95% for IL-6 and 30–50% for IL-1β (Fig. 5D).
Importantly, the exosome signaling was also impaired. In TLR4 knock-out cells the production of IL-6 and IL-1β was strongly decreased compared with the control cells (Fig. 5E). In THP-1/shTLR2 cells the exosomal stimulation was slightly less affected but also reached significance at the 48-h time point (Fig. 5E). In addition, both types of knockdown cells secreted less IL-1β, IL-6, INF-γ, TNF-α, and IL-23a which was mostly apparent after 48 h (Fig. 5F).
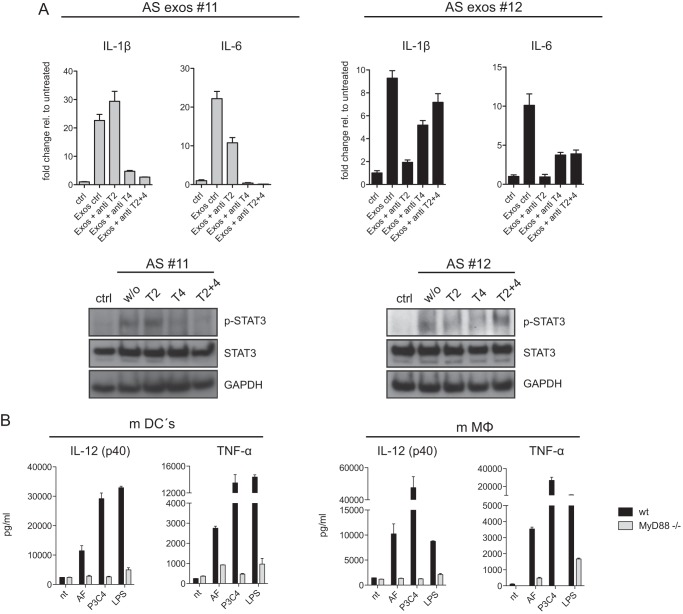
Finally, we investigated whether functional blocking antibodies to TLR2 or TLR4 could inhibit exosome-mediated signaling. Indeed, we observed that antibodies to TLR2 and TLR4 could partially inhibit the exosome-mediated phosphorylation of STAT3 and induction of IL-1β and IL-6 mRNA and that this effect was additive when both antibodies were combined (Fig. 6A). Collectively, these data implicate that TLR2 and TLR4 are relevant for exosome-mediated signaling.
FIGURE 6.
TLR-specific antibody blocking and exosome signaling in mouse dendritic cells. A, antibodies to TLR2 (T2), TLR4 (T4), or in combination (each 3 μg/ml) were added to THP-1 cells followed by stimulation with AS exosomes. After 48 h mRNA was prepared, and the level of IL-1β and IL-6 was determined by RT-PCR. Data from two representative experiments of n = 4 are shown. STAT3 phosphorylation was examined by Western blotting. B, dendritic cells (mDCs) or macrophages (mMΦ) from B6 WT or MyD88−/− mice were matured from bone marrow cells and stimulated with AF exosomes or TLR agonists LPS and P3C4. After a 48-h culture supernatants were collected, and the levels of TNF-α and IL-12 were determined. Error bars, S.E.
Exosome-mediated Triggering of Mouse Bone Marrow-derived Cells Is TLR-dependent
To further demonstrate the importance of TLRs we made use of knock-out mice deficient for the TLR downstream signaling-component MyD88. We stimulated mouse DCs and macrophages with human exosomes. Cytokine release revealed induction of TNF-α and IL-12 in DCs and macrophages of wild-type mice. Importantly, in mice deficient for MyD88 no activation was observed (Fig. 6B). These findings further supported a role for TLRs in exosome-mediated signaling.
DISCUSSION
Membrane vesicles in the form of exosomes are released from normal and cancer cells. However, significantly enhanced levels of exosomes are found in the sera or AS of cancer patients, and the amount is often correlated with the stage of the disease (12, 39, 40). There is evidence that exosomes released by the tumor can suppress the immune system of the patient and prepare niches for metastatic spread (41). In the present report we have asked how exosomes can interact and stimulate immune cells using monocytic THP-1 cells as a model. We find that (i) exosomes are internalized by THP-1 cells and stimulate the expression and secretion of cytokines; (ii) this feature is not restricted to cancer-derived exosomes but is also observed with exosomes from nontumorous sources; (iii) for cytokine release the signaling via the NFκB and STAT3 pathways is instrumental; (iv) TLR2 and TLR4 on the cell surface and a proteinase-sensitive determinant on exosomes are responsible for signal initiation. Our results suggest that exosomes have inherent signaling capacity that could be important for the induction of immunosuppression.
Previous studies showed immunosuppressive effects of tumor-derived exosomes from cell culture supernatants. Such exosomes are derived from cells that have undergone multiple passages ex vivo in the absence of any selective pressure from the immune system. Contrary to exosomes from cell culture, we assumed that exosomes from body fluid are more likely to reflect the in vivo situation. Thus, in the current study, a main goal was to deepen the understanding of immune cell stimulation by ex vivo-derived exosomes. Indeed, microvesicles isolated from various body fluids including those from AS, LC ascites, urine, and amniotic fluid have been previously characterized in our laboratory and identified as exosomes (12, 32, 33, 42). Surprisingly, we found in the present study that exposure of such exosomes to THP-1 cells could trigger NFκB and STAT3 activation and cytokine release. The secretion of proinflammatory mediators could be induced with AS exosomes coming from a tumor background. Intriguingly, however, exosomes from non-cancer-associated body fluids such as AF or LC exosomes were at least equally capable of triggering gene expression and secretion of proinflammatory mediators. In the course of this study, we purified and tested exosomes from many different ascites and amniotic fluid samples and found that the potency to activate THP-1 cells could differ between exosomal batches. This could be because exosome number was estimated by means of protein concentrations in exosome preparations. Protein concentration, in turn, might be a poor indicator of exosome number. However, this could also reflect differences in the composition of exosomes. Nevertheless, all tested sources of exosomes were generally potent and reliable activators of THP-1 cells. Thus, we hypothesized that exosomes from different body fluids, malignant or nonmalignant, carry common determinant(s) that enable them to activate THP-1 cells.
We found that STAT3 was not directly activated by exosomes. Pharmacological and antibody-mediated inhibition experiments indicated that the initial activation of NFκB leading to the production of IL-6 was required for subsequent STAT3 activation in a paracrine/autocrine fashion. Similar conclusions were reported previously in the study of Chalmin et al. (25). We also provide evidence that activation of the NFκB and STAT3 pathways were necessary for the induction of cytokine genes. Collectively, these data provide novel insights into the signaling potential of exosomes.
We also show that in THP-1 cells the TLRs are key receptors for exosome-mediated signaling. This is based on the following findings: (i) the stable knockdown of TLR2 or TLR4 led to a partial reduction of cytokine gene induction and release; (ii) antibodies to TLR2 and TLR4 alone could block in part the phosphorylation of STAT3 and subsequent induction of IL-1β and IL-6 transcription, but the effect was strongest when both antibodies were used in combination; (iii) human exosomes could trigger secretion of cytokines in mouse DCs and macrophages, but this was abolished in cells deficient for MyD88, an adaptor protein required for TLR signaling. Our results confirm and extend previous work demonstrating a functional role of TLR2 (25, 30). For the first time we also show an involvement of TLR4. Previous studies have focused mostly on mouse systems and tissue culture-derived exosomes, and a role of TLR4 was not investigated. Meanwhile Fabbri et al. reported that miRNAs in exosomes can trigger the endosomal TLR7/8 leading to cytokine secretion (31). Due to the absence of these receptors in THP-1 cells we were unable to investigate this. Our data do not exclude the possibility that in addition to TLRs other molecules can serve as exosomal receptors on monocytic or other immune cells.
An important question is which determinants on exosomes trigger TLRs and cytokine production. Previous studies have reported conflicting results. Xiang et al. proposed that exosomes isolated from in vivo grown breast adenocarcinomas were able to induce expansion of MDSCs via a mechanism dependent on the exosomal presence of prostaglandin E2 (43). Chalmin et al. used cell-culture derived exosomes and found that activation of MDSCs was dependent on the presence of HSP72 on exosomes whereas no exosomal prostaglandin E2 was found in their study (25). Using body fluid-derived exosomes we observed that the stimulating potential was destroyed by proteinase K but not with DNase or RNase treatment, supporting the notion that signals come from proteins. These determinant(s) need to be further characterized. It should be borne in mind that beside HSPs other alarmins including HMGB1 or S100 proteins were shown to be potentially secreted on exosomes (44–46).
A limitation of our current study is due to the THP-1 cells used as model system being a transformed cell line. Additional studies with primary monocytes are clearly needed. In part such studies have been already performed. Work from the Rivoltini laboratory has demonstrated that CD14+ monocytes isolated from healthy donors and differentiated with IL-4 and GM-CSF in the presence of tumor-derived microvesicles turned into HLA-DR−/low cells, retaining CD14 expression and failed to up-regulate co-stimulatory molecules, such as CD80 and CD86. The phenotypic changes were paralleled by a significant release of different cytokines, including IL-6, TNF-α, and TGF-β (47). Thus, cytokine production by exosomes has also been seen in nontransformed cells, but the exact signaling pathways were not elucidated. Although our data lay the ground for a better understanding of signaling pathways engaged by exosomes in monocytic cells, we can presently not explain how tumor-derived exosomes induce immunosuppression under physiological conditions. Perhaps a critical factor in the process is the amount of exosomes and the presence of other immunosuppressive factors in the tumor microenvironment. Increased amounts of exosomes might surpass a certain threshold of activation. As illustrated in Fig. 7, it is our hypothesis that cancer-released exosomes initiate a signaling cascade involving cytokines such as IL-6 and activation of STAT3 in tumor cells and immune cells. This milieu could alter tumor-infiltrating immune cells in a way favoring immunosuppression and the escape of the tumor from immunosurveillance (14).
FIGURE 7.
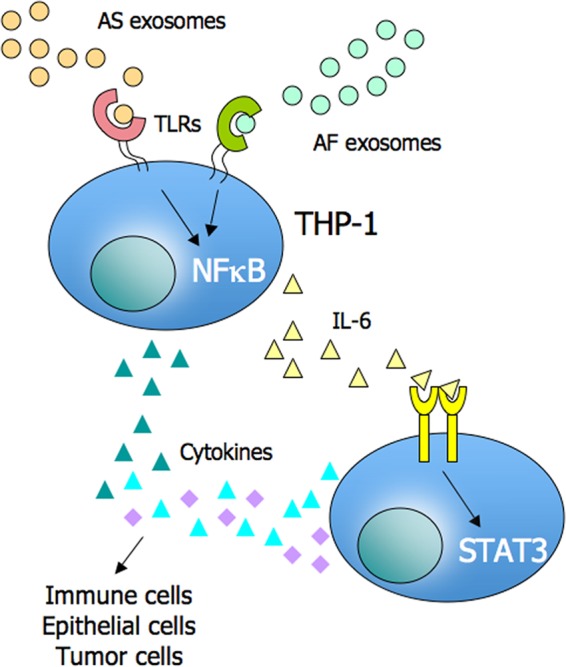
A proposed model for exosomal effects on the tumor microenvironment. Exosomes released from tumor cells into the microenvironment can stimulate via TLRs the cytokine production including IL-6 in monocytic cells. IL-6 can activate STAT3 in an autocrine/paracrine fashion on immune cells, stromal cells and tumor cells. This leads to a cytokine environment favoring immune escape of tumor cells.
Acknowledgments
We thank Natalie Erbe-Hoffmann for excellent technical assistance and Prof. Alexander Dalpke, University of Heidelberg, and Dr. Kai Doberstein for continuous support and discussions.
This work was supported by Deutsche Krebshilfe Project 109745 and by the Deutsches Krebsforschungszentrum/Tumorzentrum Collaborative Program (to P. A. and F. M.).
- DC
- dendritic cell
- AF
- amniotic fluid
- AS
- malignant ascites
- LC
- liver cirrhosis
- MDSC
- myeloid-derived suppressor cell
- miRNA
- micro RNA
- PMA
- phorbol 12-myristate 13-acetate
- TLR
- toll-like receptor
- CFSE
- carboxyfluorescein succinimidyl ester.
REFERENCES
- 1. Théry C., Zitvogel L., Amigorena S. (2002) Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 2. Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., Tursz T., Amigorena S., Raposo G., Angevin E., Zitvogel L. (2002) Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360, 295–305 [DOI] [PubMed] [Google Scholar]
- 3. Hendrix A., Westbroek W., Bracke M., De Wever O. (2010) An ex(o) citing machinery for invasive tumor growth. Cancer Res. 70, 9533–9537 [DOI] [PubMed] [Google Scholar]
- 4. Raiborg C., Rusten T. E., Stenmark H. (2003) Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446–455 [DOI] [PubMed] [Google Scholar]
- 5. Stoorvogel W., Kleijmeer M. J., Geuze H. J., Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 6. Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C. F., Schauer K., Hume A. N., Freitas R. P., Goud B., Benaroch P., Hacohen N., Fukuda M., Desnos C., Seabra M. C., Darchen F., Amigorena S., Moita L. F., Thery C. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 [DOI] [PubMed] [Google Scholar]
- 7. Ginestra A., La Placa M. D., Saladino F., Cassarà D., Nagase H., Vittorelli M. L. (1998) The amount and proteolytic content of vesicles shed by human cancer cell lines correlate with their in vitro invasiveness. Anticancer Res. 18, 3433–3437 [PubMed] [Google Scholar]
- 8. Keller S., Sanderson M. P., Stoeck A., Altevogt P. (2006) Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107, 102–108 [DOI] [PubMed] [Google Scholar]
- 9. Lotvall J., Valadi H. (2007) Cell to cell signalling via exosomes through esRNA. Cell Adh. Migr. 1, 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keller S., Ridinger J., Rupp A. K., Janssen J. W., Altevogt P. (2011) Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 9, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rupp A. K., Rupp C., Keller S., Brase J. C., Ehehalt R., Fogel M., Moldenhauer G., Marmé F., Sültmann H., Altevogt P. (2011) Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol. 122, 437–446 [DOI] [PubMed] [Google Scholar]
- 13. Camussi G., Deregibus M. C., Bruno S., Cantaluppi V., Biancone L. (2010) Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 78, 838–848 [DOI] [PubMed] [Google Scholar]
- 14. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 15. Valenti R., Huber V., Iero M., Filipazzi P., Parmiani G., Rivoltini L. (2007) Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 67, 2912–2915 [DOI] [PubMed] [Google Scholar]
- 16. Viaud S., Théry C., Ploix S., Tursz T., Lapierre V., Lantz O., Zitvogel L., Chaput N. (2010) Dendritic cell-derived exosomes for cancer immunotherapy: what's next? Cancer Res. 70, 1281–1285 [DOI] [PubMed] [Google Scholar]
- 17. Whiteside T. L. (2005) Tumour-derived exosomes or microvesicles: another mechanism of tumour escape from the host immune system? Br. J. Cancer 92, 209–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clayton A., Mitchell J. P., Court J., Linnane S., Mason M. D., Tabi Z. (2008) Human tumor-derived exosomes down-modulate NKG2D expression. J. Immunol. 180, 7249–7258 [DOI] [PubMed] [Google Scholar]
- 19. Clayton A., Mitchell J. P., Court J., Mason M. D., Tabi Z. (2007) Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 67, 7458–7466 [DOI] [PubMed] [Google Scholar]
- 20. Taylor D. D., Akyol S., Gercel-Taylor C. (2006) Pregnancy-associated exosomes and their modulation of T cell signaling. J. Immunol. 176, 1534–1542 [DOI] [PubMed] [Google Scholar]
- 21. Taylor D. D., Gercel-Taylor C. (2011) Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 33, 441–454 [DOI] [PubMed] [Google Scholar]
- 22. Clayton A. (2012) Cancer cells use exosomes as tools to manipulate immunity and the microenvironment. Oncoimmunology 1, 78–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C., Yu S., Zinn K., Wang J., Zhang L., Jia Y., Kappes J. C., Barnes S., Kimberly R. P., Grizzle W. E., Zhang H. G. (2006) Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J. Immunol. 176, 1375–1385 [DOI] [PubMed] [Google Scholar]
- 24. Yu S., Liu C., Su K., Wang J., Liu Y., Zhang L., Li C., Cong Y., Kimberly R., Grizzle W. E., Falkson C., Zhang H. G. (2007) Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J. Immunol. 178, 6867–6875 [DOI] [PubMed] [Google Scholar]
- 25. Chalmin F., Ladoire S., Mignot G., Vincent J., Bruchard M., Remy-Martin J. P., Boireau W., Rouleau A., Simon B., Lanneau D., De Thonel A., Multhoff G., Hamman A., Martin F., Chauffert B., Solary E., Zitvogel L., Garrido C., Ryffel B., Borg C., Apetoh L., Rébé C., Ghiringhelli F. (2010) Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Invest. 120, 457–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valenti R., Huber V., Filipazzi P., Pilla L., Sovena G., Villa A., Corbelli A., Fais S., Parmiani G., Rivoltini L. (2006) Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-β-mediated suppressive activity on T lymphocytes. Cancer Res. 66, 9290–9298 [DOI] [PubMed] [Google Scholar]
- 27. Szajnik M., Czystowska M., Szczepanski M. J., Mandapathil M., Whiteside T. L. (2010) Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PloS One 5, e11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Webber J., Steadman R., Mason M. D., Tabi Z., Clayton A. (2010) Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 70, 9621–9630 [DOI] [PubMed] [Google Scholar]
- 29. Luga V., Zhang L., Viloria-Petit A. M., Ogunjimi A. A., Inanlou M. R., Chiu E., Buchanan M., Hosein A. N., Basik M., Wrana J. L. (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 [DOI] [PubMed] [Google Scholar]
- 30. Xiang X., Liu Y., Zhuang X., Zhang S., Michalek S., Taylor D. D., Grizzle W., Zhang H. G. (2010) TLR2-mediated expansion of MDSCs is dependent on the source of tumor exosomes. Am. J. Pathol. 177, 1606–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G. J., Zanesi N., Crawford M., Ozer G. H., Wernicke D., Alder H., Caligiuri M. A., Nana-Sinkam P., Perrotti D., Croce C. M. (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keller S., Rupp C., Stoeck A., Runz S., Fogel M., Lugert S., Hager H. D., Abdel-Bakky M. S., Gutwein P., Altevogt P. (2007) CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 72, 1095–1102 [DOI] [PubMed] [Google Scholar]
- 33. Keller S., König A. K., Marmé F., Runz S., Wolterink S., Koensgen D., Mustea A., Sehouli J., Altevogt P. (2009) Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 278, 73–81 [DOI] [PubMed] [Google Scholar]
- 34. Stoeck A., Keller S., Riedle S., Sanderson M. P., Runz S., Le Naour F., Gutwein P., Ludwig A., Rubinstein E., Altevogt P. (2006) A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 393, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Riedle S., Kiefel H., Gast D., Bondong S., Wolterink S., Gutwein P., Altevogt P. (2009) Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/γ-secretase activity. Biochem. J. 420, 391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marton A., Vizler C., Kusz E., Temesfoi V., Szathmary Z., Nagy K., Szegletes Z., Varo G., Siklos L., Katona R. L., Tubak V., Howard O. M., Duda E., Minarovits J., Nagy K., Buzas K. (2012) Melanoma cell-derived exosomes alter macrophage and dendritic cell functions in vitro. Immunol. Lett. 148, 34–38 [DOI] [PubMed] [Google Scholar]
- 37. Yu H., Pardoll D., Jove R. (2009) STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dauer D. J., Ferraro B., Song L., Yu B., Mora L., Buettner R., Enkemann S., Jove R., Haura E. B. (2005) STAT3 regulates genes common to both wound healing and cancer. Oncogene 24, 3397–3408 [DOI] [PubMed] [Google Scholar]
- 39. Taylor D. D., Gercel-Taylor C. (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 [DOI] [PubMed] [Google Scholar]
- 40. Peinado H., Alečkovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B. A., Callahan M. K., Yuan J., Martins V. R., Skog J., Kaplan R. N., Brady M. S., Wolchok J. D., Chapman P. B., Kang Y., Bromberg J., Lyden D. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Filipazzi P., Bürdek M., Villa A., Rivoltini L., Huber V. (2012) Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin. Cancer Biol. 22, 342–349 [DOI] [PubMed] [Google Scholar]
- 42. Runz S., Keller S., Rupp C., Stoeck A., Issa Y., Koensgen D., Mustea A., Sehouli J., Kristiansen G., Altevogt P. (2007) Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 107, 563–571 [DOI] [PubMed] [Google Scholar]
- 43. Xiang X., Poliakov A., Liu C., Liu Y., Deng Z. B., Wang J., Cheng Z., Shah S. V., Wang G. J., Zhang L., Grizzle W. E., Mobley J., Zhang H. G. (2009) Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 124, 2621–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gardella S., Andrei C., Ferrera D., Lotti L. V., Torrisi M. R., Bianchi M. E., Rubartelli A. (2002) The nuclear protein HMGB1 is secreted by monocytes via a nonclassical, vesicle-mediated secretory pathway. EMBO Rep. 3, 995–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krysko D. V., Garg A. D., Kaczmarek A., Krysko O., Agostinis P., Vandenabeele P. (2012) Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer 12, 860–875 [DOI] [PubMed] [Google Scholar]
- 46. Ji H., Greening D. W., Barnes T. W., Lim J. W., Tauro B. J., Rai A., Xu R., Adda C., Mathivanan S., Zhao W., Xue Y., Xu T., Zhu H. J., Simpson R. J. (2013) Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 13, 1672–1686 [DOI] [PubMed] [Google Scholar]
- 47. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]