Background: It is important to assess contribution of calpain activation and identify substrates affected during neurodegeneration.
Results: Gel-based protease proteomics identified novel substrates that were cleaved in neurotoxin-treated culture and rat brain disease models.
Conclusion: These novel calpain substrates may confer protection against neurodegeneration.
Significance: Our findings contribute to better deciphering the molecular mechanism underlying the progression of protease-mediated neurodegeneration.
Keywords: Calpain, Neurodegeneration, Parkinson Disease, Protein Degradation, Proteomics, Substrates
Abstract
Calpains are a family of calcium-dependent cysteine proteases that are ubiquitously expressed in mammals and play critical roles in neuronal death by catalyzing substrate proteolysis. Here, we developed two-dimensional gel electrophoresis-based protease proteomics to identify putative calpain substrates. To accomplish this, cellular lysates from neuronal cells were first separated by pI, and the immobilized sample on a gel strip was incubated with a recombinant calpain and separated by molecular weight. Among 25 altered protein spots that were differentially expressed by at least 2-fold, we confirmed that arsenical pump-driving ATPase, optineurin, and peripherin were cleaved by calpain using in vitro and in vivo cleavage assays. Furthermore, we found that all of these substrates were cleaved in MN9D cells treated with either ionomycin or 1-methyl-4-phenylpyridinium, both of which cause a calcium-mediated calpain activation. Their cleavage was blocked by calcium chelator or calpain inhibitors. In addition, calpain-mediated cleavage of these substrates and its inhibition by calpeptin were confirmed in a middle cerebral artery occlusion model of cerebral ischemia, as well as a stereotaxic brain injection model of Parkinson disease. Transient overexpression of each protein was shown to attenuate 1-methyl-4-phenylpyridinium-induced cell death, indicating that these substrates may confer protection of varying magnitudes against dopaminergic injury. Taken together, the data indicate that our protease proteomic method has the potential to be applicable for identifying proteolytic substrates affected by diverse proteases. Moreover, the results described here will help us decipher the molecular mechanisms underlying the progression of neurodegenerative disorders where protease activation is critically involved.
Introduction
Parkinson disease (PD)4 is a common neurodegenerative disorder characterized by the progressive loss of dopaminergic neurons in the substantia nigra pars compacta, and these neurons develop proteinaceous inclusions called Lewy bodies (1). Mutations in α-synuclein, parkin, DJ-1, PINK1, and LRRK2 genes appear to be associated with familial forms of PD, but the majority of cases are sporadic. Oxidative stress, mitochondrial dysfunction, and accumulation of abnormal protein aggregates are all thought to contribute to PD pathogenesis (2). Gene- and neurotoxin-based models of PD have been widely used to elucidate the molecular mechanisms associated with neuronal cell death in PD. For example, both apoptotic and necrotic mechanisms have been implicated in neurotoxin-based models established with 6-hydroxydopamine, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP; its active metabolite, MPP+), rotenone, and paraquat. Activation of various proteases, including caspase and calpain, has been shown to play a critical role in neuronal death in these model systems. Consequently, inhibition of protease activation within neurons has been developed as a neuroprotective strategy (1). Calpains belong to a family of intracellular Ca2+-dependent, nonlysosomal cysteine proteases (reviewed in Ref. 3). They are highly conserved, structurally related, and ubiquitously expressed in mammals, as well as many other organisms. Among 16 known genes, calpain 1 (μ-calpain) and calpain 2 (m-calpain) represent two isoforms that are the best characterized members of the calpain family. Structurally, these two heterodimeric isoforms share an identical small regulatory subunit (28 kDa) but have distinct large catalytic subunits (80 kDa) (3). Both isoforms are highly expressed in neurons and glia in the central nervous system (4). Among several proposed functional implications, Ca2+-triggered activation of calpain has been demonstrated to play an important role in the initiation, regulation, and execution of different forms of neuronal death, including apoptosis, necrosis, and autophagy (5). Considering that calpains exert their regulatory action by proteolytic processing of endogenous substrates, it is important to assess the contribution of calpain activation and identify substrates affected during neurodegeneration. Previously, several independent approaches, including proteomic analyses (6–9), were performed to identify endogenous calpain substrates. However, it is not clearly understood whether the putative substrates are directly cleaved by calpain or other proteolytic enzymes. Here, we described a novel protease proteomic analysis that employs conventional gel-based two-dimensional gel electrophoresis (2DE). We used the MN9D dopaminergic neuronal cell line that is a fusion product of embryonic mesencephalic dopaminergic neurons and N18TG neuroblastoma cells (10). First, MN9D cellular lysates were extracted without any protease inhibitor treatment and subjected to isoelectric focusing (IEF). The proteins were immobilized on a strip and incubated with or without active recombinant m-calpain to ensure that only the direct substrates would be cleaved. Following separation by SDS-PAGE, several protein spots that were either up- or down-regulated were subjected to mass spectral analysis using MALDI-TOF. Among these altered protein spots, we selected arsenical pump-driving ATPase (ASNA1), optineurin, and peripherin for further validation. We subsequently confirmed that these proteins are cleaved by activated calpain both in cultured cells and in rat models of neurodegenerative diseases. Our protease proteomic analysis seems to be useful and broadly applicable to identifying novel protease substrates that play critical regulatory roles in neuronal cell death.
EXPERIMENTAL PROCEDURES
Cell Culture, Drug Treatment, and Cell Viability
Cells were plated at a density of either 1.0 × 106, 1.0 × 105, or 2.5 × 104 cells on 25-μg/ml poly-d-lysine (Sigma)-coated P-100 dishes (Corning Glass Works, Corning, NY), 4-well culture dishes (Nunc, Roskilde, Denmark) or 24-well culture plates (Corning Glass Works), respectively. Cells were maintained for 3 days in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) in an incubator with an atmosphere of 10% CO2 at 37 °C. Media were changed to serum-free N2 medium before drug treatment. Cells were treated with 50 μm MPP+ (Research Biochemicals International, Natick, MA) or 5 μg/ml ionomycin (Sigma) in the presence or absence of 40 μm BAPTA/AM (Molecular Probes, Eugene, OR), 50 μm PD150606 (Calbiochem, La Jolla, CA), or 50 μm calpeptin (Calbiochem). Following MPP+ treatment, the rate of cell survival was determined by colorimetric measurement using MTT reduction assays (11). In brief, cells were incubated for 1 h at 37 °C with a final concentration of 1 mg/ml MTT solution and lysed in 20% SDS in 50% aqueous dimethylformamide for 24 h. The optical density of the dissolved formazan grains was measured at 540 nm using a microplate reader (Molecular Devices, Palo Alto, CA). Viabilities were represented as percentages of values from MPP+-treated cells over the untreated control cells (100%).
Middle Cerebral Artery Occlusion (MCAO) and Stereotaxic Surgery
All experimental procedures applied to animals were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Committee of Korea University and Kyung Hee University. To establish a focal cerebral ischemic model, Sprague-Dawley male rats (260–270 g) were anesthetized with a mixture of 3% isoflurane in 70% N2O and 30% O2 (v/v) via facemask and maintained with 2% isoflurane. A rectal temperature probe was introduced, and body temperature was maintained at 37 °C during the entire surgery period. Focal cerebral ischemia was achieved by right-sided endovascular MCAO, as described previously (12). The occlusion was released after 1.5 h, and the animals were allowed to recover. For actual experiments, rats randomly divided into two groups received intracerebroventricle infusion of either 50 μg of calpeptin dissolved in 5 μl of Me2SO (Sigma) or 5 μl of Me2SO alone 30 min before MCAO as basically described (13). The rats were reanesthetized with isoflurane in N2O and O2 24 h after ischemia onset, and their brains were removed for immunoblot analysis. For stereotaxic surgery, Sprague-Dawley female rats were anesthetized with an intraperitoneal injection of chloral hydrate (360 mg/kg) and positioned in a stereotaxic apparatus (Kopf Instruments, Tujunga, CA). Each rat received a unilateral injection of MPP+ (7.4 μg in 2 μl of PBS; Sigma) for 3 days into the right medial forebrain bundle (anteroposterior, 3.6 mm; mediolateral, 2.0 mm; dorsoventral, 7.5 mm from bregma). All injections were performed with a Hamilton syringe equipped with a 26 S gauge beveled needle attached to a syringe pump (KD Scientific, Holliston, MA) at a rate of 0.2 μl/min. One minute after injection, the needle was slowly retracted. The bilateral substantia nigral tissues were dissected out for immunoblot analysis.
2DE
All chemicals used for 2DE were purchased from Sigma unless otherwise stated. Gel-based proteomic analysis was performed as we described previously with some modifications (14). Briefly, MN9D cells were solubilized in sample buffer containing 5 m urea, 2 m thiourea, 2% CHAPS, 0.25% Tween 20, 100 mm DTT, 10% isopropyl alcohol, 12.5% water-saturated butanol, 5% glycerol, and 1% immobilized pH gradient buffer for pH 4–7 linear Immobiline DryStrip (24 cm; GE Healthcare). Protein content was determined with two-dimensional Quantikits (GE Healthcare). DryStrips were rehydrated in sample buffer containing 1.5 mg of cellular lysate. Gels were run for a total of 100 kV-h using progressively increasing voltage on an Ettan IPGphor (GE Healthcare). After IEF, the strips were washed with distilled water and incubated with or without 83.25 units of recombinant m-calpain (Calbiochem: catalog no. 208718) or m-calpain from porcine kidney (Calbiochem; catalog no. 208715) in calpain activation buffer (25 mm Tris, 10 mm KCl, 10 mm CaCl2, and 0.1% Triton X-100) in a final volume of 500 μl for 2 h at 37 °C. Prior to SDS-PAGE, the strips were washed three times with distilled water, and an equilibration step was carried out. SDS-PAGE was performed on 8–18% gradient gels in an Ettan Dalt II System (GE Healthcare). Coomassie Brilliant Blue G-250-stained gels were scanned using a densitometer (Powerlook 2100XL; UMAX, Dallas, TX), and the gel images were analyzed with ProteomeWeaver software version 2.1 (DEFINIENS, München, Germany). To minimize variation of background staining intensity, we simultaneously stained and destained gels in a multiple chamber. Fold changes of the protein spots were normalized by a background subtraction. For calpain activation measurement, a mini-2DE method was conducted. Briefly, DryStrips (7 cm; GE Healthcare) were rehydrated in sample buffer containing 100 μg of cellular lysate. Gels were run for a total of 35 kV-h using progressively increasing voltage on an Ettan IPGphor. After IEF, the strips were washed with distilled water and incubated with or without 5.55 units of porcine kidney m-calpain in calpain activation buffer in a final volume of 125 μl for 2 h at 37 °C.
Protein Identification
In-gel digestion and mass spectrometric analysis were performed as we described previously (14). Briefly, the gel containing protein spots of interest was manually excised and destained with 50% acetonitrile in 25 mm ammonium bicarbonate buffer, pH 8.0. The destained gel slices were completely dried in a SpeedVac evaporator dryer (BioTron Inc., Gyeonggi-do, Korea). Gel slices were rehydrated and digested for 18 h at 37 °C with 10 μg/ml porcine trypsin (842.51 and 2,211.10 m/z; Promega, Madison, WI) in 25 mm ammonium bicarbonate buffer, pH 8.0. Tryptic peptides were purified with a POROS R2 column (Applied Biosystems, Foster City, CA) and spotted onto stainless steel MALDI plates (Applied Biosystems). The peptide mixtures were acquired in reflectron mode (mass range from 800 to 4,000 Da, 20-keV accelerating voltage, 500-ns delayed extraction, averaging 1,000 laser shots/spectrum) using a 4700 Proteomic Analyzer equipped with a 355-nm nitrogen laser (Applied Biosystems). The spectra were analyzed with GPS ExplorerTM software ver. 3.5 (Applied Biosystems) using ProteinProspector 4.0.7 (MS-Fit; University of California, San Francisco) and the following parameters: signal to noise threshold, 10; mass exclusion tolerance, 0.2 m/z; isotope cluster (maximum charge, +1; isotope spacing tolerance, 0.1); Monoisotopic peaks (Adduct: H, Genetic formula: C6H5NO-peptides). The trypsin autodigestion ion picks were used as internal standards. Matrix and/or autoproteolytic trypsin fragments or known contaminant ions were excluded. The resulting peptide mass lists were used to query the SwissProt database (2012.12.3, 453,850 sequences; taxonomy, Mus musculus, 16,581 sequences). The following criteria were used for search parameters: significant protein MOWSE score at p < 0.05, 1 missed cleavage site allowed, 1+ peptide charges allowed, trypsin as enzyme, 50–100 ppm as precursor tolerance, peptide N-terminal Gln to pyroGlu, oxidation of methionine, protein N terminus acetylated, and acrylamide-modified Cys as fixed and variable modifications. Further classification based on indicated criteria of the identified proteins was performed using the program known as the PANTHER (Protein ANalysis THrough Evolutionary Relationships).
Constructs and RNA Interference
Vectors containing mouse ASNA1 or mouse peripherin cDNA sequences were kindly provided by Dr. P. Naredi at Umea University Hospital and Dr. J. Robertson at Toronto University, respectively. The following sequences were amplified by PCR using specific sets of forward and reverse primers: T7-ASNA1-FLAG forward, 5′-CCCAAGCTTATGGCTAGCATGACTGGTGGACAGCAAATGGGTATGGCGGCGGGGGTGGCCGG-3′; T7-ASNA1-FLAG reverse, 5′-AAGGAAAAAAGCGGCCGCTTACTTGTCATCATCGTCCTTATAGTCCTGGGTGCTGGGGGGCTTGTA-3′; T7-peripherin-FLAG forward, 5′-CCCAAGCTTATGGCTAGCATGACTGGTGGACAGCAAATGGGTATGCCATCTTCCGCCAGCATGA-3′; T7-peripherin-FLAG reverse, 5′-AAGGAAAAAAGCGGCCGCTTACTTGTCATCATCGTCCTTATAGTCGTAGCTGTGGATAGAAGACTTGT-3′; peripherin forward, 5′-GCGAATTCATGCCATCTTCCGCCAGCATG-3′; peripherin reverse, 5′-CGTCTAGAGTAGCTGTGGATAGAAGACTTG-3′; optineurin forward, 5′-CGGGGTACCATGTCCCATCAACCTCTGAGCT-3′; and optineurin reverse, 5′-CCGGAATTCAATGATGCAGTCCATCACATGGA-3′. N-terminal T7-tagged and C-terminal FLAG-tagged ASNA1 were subcloned into pcDNA3.1(+), whereas peripherin was subcloned into pcDNA6/Myc-His. Mouse optineurin sequences were subcloned into pcDNA3.1(+)/GST and pcDNA3.1(+)/V5-His. For gene silencing studies, MN9D cells were transfected with the indicated shRNA construct that contains firefly luciferase or optineurin in pRNAT-U6.1/Neo. The sequences were designed through Genescript. The targeting sequences for firefly luciferase and optineurin were CTTACGCTGAGTACTTCGA and GCTCGTTTACAGTAACAAGAA, respectively.
In Vitro and Cell-based Calpain Cleavage Assay
For in vitro calpain cleavage assays, the vectors encoding ASNA1, optineurin, and peripherin were transcribed and translated in vitro in the presence of [35S]methionine (PerkinElmer Life Sciences) by using transcription and translation-coupled reticulocyte lysate system (Promega). For cell-based calpain cleavage assays, MN9D cells were transiently transfected with the above-stated eukaryotic expression vectors using Lipofectamine 2000 (Invitrogen). The cells were lysed in radioimmunoprecipitation assay buffer containing 50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, and 0.1% SDS without protease inhibitors. Cells were homogenized in a Dounce homogenizer on ice and centrifuged at 13,000 × g for 15 min at 4 °C. Protein contents were measured using a Bio-Rad protein assay kit. Predetermined amounts (10 μl for transcription and translation reactants and 100 μg for cellular lysates) were incubated for 2 h at 37 °C with up to 5.55 units of m-calpain or 3.95 units of μ-calpain from porcine erythrocytes (Calbiochem; catalog no. 208712) in calpain activation buffer at a final volume of 20 μl. When necessary, a calpain inhibitor (500 μm PD150606) was added to the reaction mixture. The reactions were stopped by adding 5 μl of 5× sample buffer (250 mm Tris-HCl, 500 mm DTT, 10% SDS, 0.5% bromphenol blue, and 50% glycerol, pH 6.8) followed by boiling for 5 min. The resulting products were separated by 12% SDS-PAGE and processed for autoradiography or immunoblot analysis.
Immunoblot Analysis
MN9D cells were washed with chilled PBS containing 2 mm EDTA and subjected to lysis on ice in radioimmunoprecipitation assay buffer containing complete protease inhibitor mixture (Roche Applied Science). For MN9D cells treated with ionomycin, cells were lysed on ice in a 1% Triton X-100 buffer containing 1% SDS and complete protease inhibitor mixture. The bilateral cortex were dissected and homogenized in radioimmunoprecipitation assay buffer containing complete protease inhibitor mixture. Protein (50–80 μg) was separated on 8–12% SDS-PAGE gels, blotted onto prewetted PVDF membranes (Pall Corp., Ann Arbor, MI), and processed for immunoblot analyses as described previously (15). The primary antibodies used were rabbit anti-Bax antibody (1:3,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), goat anti-ASNA1 antibody (1:1,000; Everest Biotech, Oxfordshire, UK), rabbit anti-optineurin antibody (1:1,000; Cayman Chemical, Ann Arbor, MI), rabbit anti-peripherin antibody (1:1,000; Millipore Corp., Billerica, MA), mouse anti-fodrin antibody (1:4,000; Enzo Life Sciences Inc., Farmingdale, NY), mouse anti-T7 antibody (1:4,000; Novagen, Madison, WI), rabbit anti-GST antibody (1:1,000; Santa Cruz Biotechnology, Inc.), mouse anti-tyrosine hydroxylase antibody (TH; 1:7,500; Pel-Freez Biologicals, Rogers, AR), HRP-conjugated anti-FLAG antibody (Sigma), and HRP-conjugated anti-V5 antibody (Invitrogen). Both mouse anti-GAPDH (1:3,000; Millipore Corp.) and rabbit anti-actin (1:3,000; Sigma) were used as loading controls. For measurement of calpain activation via mini-2DE, DryStrips were subjected to immunoblot analysis using rabbit anti-Bax (1:3,000; Santa Cruz Biotechnology, Inc.) and rabbit anti-actin (1:3,000; Sigma) antibodies as we described previously (15). Subsequently, the membranes were incubated with appropriate HRP-conjugated secondary antibodies (1:3,000; Santa Cruz Biotechnology, Inc.). Specific bands were detected with ECL (PerkinElmer Life Sciences). Relative band intensities were measured using ImageJ software (National Institutes of Health).
Immunofluorescence
To label cytosolic Ca2+, MN9D cells treated with 5 μg/ml ionomycin for 30 min or 50 μm MPP+ for 36 h were stained with 3 μm Fluo-3/AM dye (Molecular Probes) diluted in N2 medium for 30 min at 37 °C in an incubator with a 10% CO2 atmosphere and washed twice with N2 medium as suggested by the manufacturer's protocol. Subsequently, the stained cells were examined under a fluorescence microscope equipped with epifluorescence filters and a digital image analyzer (Axio Observer A1 Microscope; Carl Zeiss). For immunocytochemical localization of ASNA1, optineurin, and peripherin, MN9D cells were cultured at a density of 6.0 × 104 cells on 25 μg/ml poly-d-lysine-coated aclar film (Electron Microscopy Science, Fort Washington, PA). Thirty-six hours after 50 μm MPP+ treatment in the presence or absence of 50 μm calpeptin, cells were washed with PBS (Lonza, Verviers, Belgium) followed by fixation with 4% paraformaldehyde (Electron Microscopy Science) for 15 min at room temperature. The cells were blocked for 1 h at room temperature in PBS containing 5% normal goat serum (Invitrogen) and 0.2% Triton X-100. The cells were incubated with a goat anti-ASNA1 antibody (1:200; Everest Biotech), rabbit anti-optineurin antibody (1:200; Cayman Chemical), or rabbit anti-peripherin antibody (1:200; Millipore Corp.) in PBS containing 1% normal goat serum and 0.2% Triton X-100 overnight at 4 °C followed by incubation with Alexa Flour 488-conjugated rabbit anti-goat IgG (1:200) or Alexa Flour 488-conjugated goat anti-rabbit IgG (1:200; both from Molecular Probes) for 1 h at room temperature. For nuclear counterstaining, the cells were further incubated with 1 μg/ml Hoechst 33258 (Molecular Probes) for 10 min at room temperature. Slides were mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA) and observed under an LSM 510 META confocal laser scanning microscope equipped with epifluorescence filters and a digital image analyzer (Carl Zeiss).
Semi-quantitative RT-PCR
Purified RNAs (5 μg) were mixed with Moloney murine leukemia virus reverse transcriptase and 0.5 μg of random primer (all from Promega). For the PCR, 5 μl of cDNA was mixed with the following specific primers: ASNA1 forward, 5′-CTCAGGCTCCTGAACTTCCC-3′; ASNA1 reverse, 5′-ACAGATCGGATGACAGGCAA-3′; optineurin forward, 5′-GAGCCTTGGAGGAGGAAGTG-3′; optineurin reverse, 5′-CGGTCATCCTGATCTCAACG-3′; peripherin forward, 5′-TGCAGCAGGTGGAGGTAGAG-3′; and peripherin reverse, 5′-TCGTTCATCTCTTGCTTGGC-3′. Universal 18 S rRNA primers (Ambion Inc., Austin, TX) were used in the reaction mixtures as a control.
Statistics
All data are expressed as the means ± S.E. of the indicated number of experiments. Significant differences among groups were determined by one-way analysis of variance and Tukey's post hoc test using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). A value of p < 0.05 was considered statistically significant.
RESULTS
Gel-based Protease Proteomics to Identify Putative Calpain Substrates
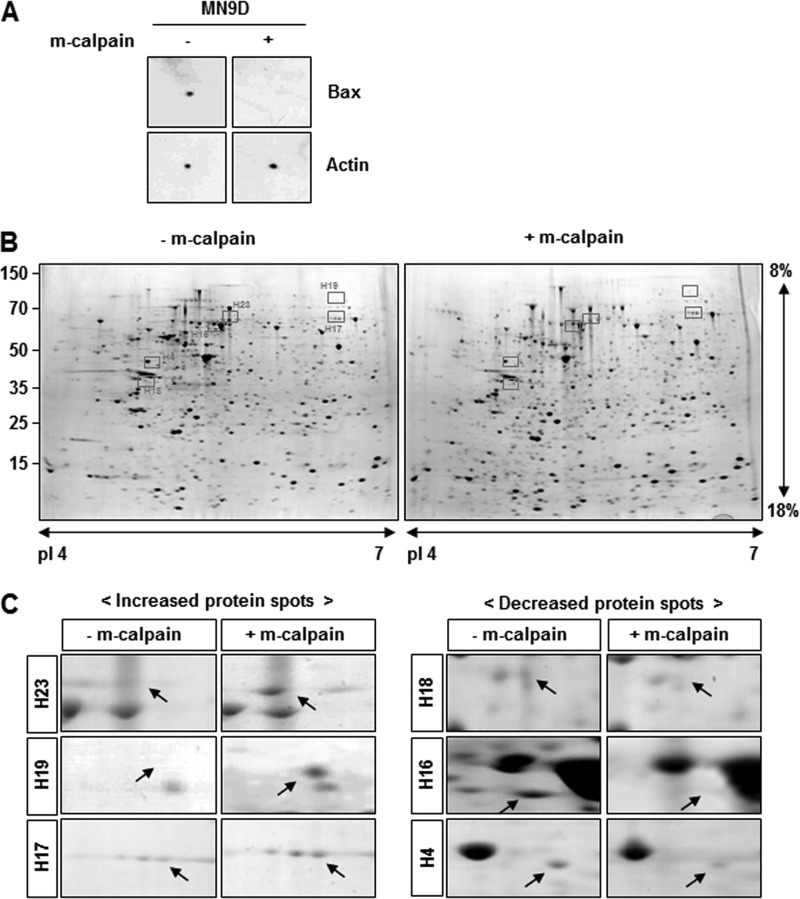
We attempted to develop a simple 2DE-based procedure that can identify substrates that are directly cleaved by an applied protease. To accomplish this, cellular lysates obtained from MN9D dopaminergic neuronal cells were subjected to IEF, and the resulting strips were subsequently incubated with or without recombinant m-calpain (Fig. 1). Thereafter, the strips were further processed for conventional 2DE followed by a mass spectral analysis of the altered protein spots. This modified approach ensured that only calpain-specific substrates can be cleaved among immobilized proteins on a strip after IEF. First, we attempted to empirically set up conditions in which the applied amount of calpain can successfully cleave the known calpain substrate on a strip. Based on previous reports, including one from our laboratory demonstrating that Bax is cleaved by Ca2+-mediated calpain activation (15, 16), isoelectrically separated proteins on a strip were incubated with m-calpain and processed for mini-2DE as described previously (14). Immunoblot analyses using anti-Bax antibody showed that incubation with a total of 5.55 units of recombinant m-calpain for 2 h was sufficient to cleave Bax and led to a disappearance of the Bax protein spot (Fig. 2A). Using this condition, proteome analyses of calpain-treated samples and matching controls were performed. Representative Coomassie Brilliant Blue-stained protein profiles following IEF separation of 1.5-mg protein samples and incubation with or without a total of 83.25 units of recombinant m-calpain are shown in Fig. 2B. Approximately 1,500 distinct protein spots ranging from 10 to 100 kDa were typically resolved in pI 4–7 linear gels. By using densitometric analysis of gel images, we initially identified more than 100 altered protein spots in calpain-treated samples as compared with expression levels in untreated samples. Among these altered protein spots, 25 protein spots with differential expression levels by at least 2-fold were subjected to mass spectral analysis. Information on these proteins is described in Table 1.
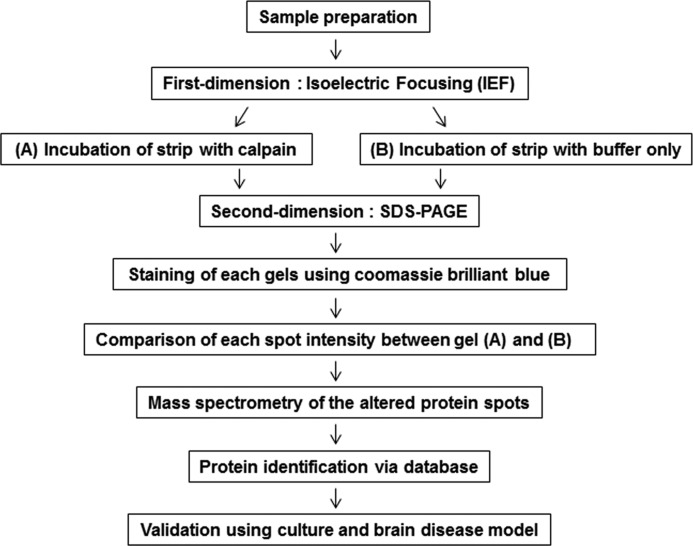
FIGURE 1.
Schematic flow of a gel-based protease proteomic study to identify putative calpain substrates. We developed a gel-based protease proteomic strategy for identifying putative calpain substrates. After IEF separation, immobilized proteins on DryStrips were incubated with (A) or without (B) purified recombinant m-calpain followed by separation by SDS-PAGE. Altered protein spots in Coomassie Brilliant Blue G-250-stained gels were subjected to in-gel digestion followed by MALDI-TOF mass spectrometry. Lists of putative calpain substrates were made after searching through databases as described in the text. Using both culture and rat brain disease models, the identified candidates were subjected to further validation as calpain substrates.
FIGURE 2.
Representative 2DE gel images of putative calpain substrates. A, mini-2DE analysis was carried out to confirm that calpain effectively degraded Bax, one of its known substrates (15, 16). Lysates (100 μg) obtained from MN9D cells were loaded on 7-cm Drystrips and separated by IEF ranging from pI 4 to 7. Next, samples in the ministrips were incubated with 5.55 units of m-calpain for 2 h at 37 °C, subjected to 12% SDS-PAGE, and probed with anti-Bax antibody. Actin was used as a loading and a negative control. B, representative gel images are shown for samples treated with or without m-calpain and further processed for 2DE (24 cm, pI 4–7, and 8–18%). Briefly, total protein lysates (1.5 mg) obtained from MN9D cells were separated by IEF and incubated with or without 83.25 units of m-calpain for 2 h at 37 °C. After separation by SDS-PAGE, the gels were stained with 0.1% Coomassie Brilliant Blue G-250. Approximately 1,500 protein spots ranging from 10 to 100 kDa were routinely detected. C, comparative close-up views of the typically altered protein spots boxed in Fig. 2B, before and after calpain treatment. Arrows indicate the protein spots that were either increased (left) or decreased (right) following calpain treatment. Detailed molecular information for these protein spots is shown in Table 2.
TABLE 1.
Lists of proteins differentially expressed by at least 2-fold after incubation with calpain
| Spot | Protein name | Gene name | Accession number | Function |
|---|---|---|---|---|
| 1 | SAP domain-containing ribonucleoprotein | Sarnp Hcc1 | Q9D1J3 | Nucleic acid binding |
| 2 | Tubby protein, FGFR-3 | Fgfr3 Mfr3 Sam3 | Q61851 | Not classified |
| 3 | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (mSTI1) | Stip1 | Q60864 | Chaperone |
| 4 | A-kinase anchor protein 4 precursor (major fibrous sheath protein) (FSC1) (AKAP82) (p82) | Akap4 Akap82 Fsc1 | Q60662 | Enzyme modulator |
| 5 | 14-3-3 protein ζ/δ (protein kinase C inhibitor protein 1) (K CIP-1) (SE Z-2) | Ywhaz | P63101 | Chaperone |
| 6 | Phosphatase and actin regulator 3 (Scapinin) (scaffold-associated PP1-inhibiting protein) | Phactr3 Scapin1 | Q8BYK5 | Enzyme modulator |
| 7 | ATP synthase D chain, mitochondrial (EC 3.6.3.14) | Atp5b | P56480 | Hydrolase |
| 8 | 6-Phosphogluconolactonase (EC 3.1.1.31) (6PGL) | Pgls | Q9CQ60 | Hydrolase |
| 9 | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) | Casp8 | O89110 | Enzyme modulator |
| 10 | Ras-GTPase-activating protein-binding protein1 (EC 3.6.1.-) (ATP-dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP-1) (HDH-VIII) | G3bp1 G3bp | P97855 | Nucleic acid binding |
| 11 | Sterile alpha and TIR motif-containing protein1 (Tir-1 homolog) | Sarm1 Kiaa0524 | Q6PDS3 | Enzyme modulator |
| 12 | Frizzled-9 precursor (Fz-9) (mF z9) (mF z3) | Fzd9 Fzd3 | Q9R216 | Receptor |
| 13 | GMP synthase (glutamine-hydrolyzing) | Gmps | Q3THK7 | Transferase |
| 14 | ATPase ASNA1 | Asna1 Arsa | O54984 | Hydrolase |
| 15 | Protein MICAL-3 | Mical3 Kiaa0819 Kiaa1364 | Q8CJ19 | Not classified |
| 16 | Optineurin | Optn | Q8K3K8 | Not classified |
| 17 | CDK5 regulatory subunit-associated protein1 (CDK5 activator-binding protein C42) | Cdk5rap1 | Q8BTW8 | Not classified |
| 18 | Zinc finger protein 271 | Znf271 Zfp-35 Zfp35 | P15620 | Transcription factor |
| 19 | Phosphoinositol 3-phosphate-binding protein-2 | Plekha5 | E9Q6H8 | Not classified |
| 20 | Mitogen-activated protein kinase kinase kinase kinase 1 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 1) (MEK kinase kinase 1) (MEKKK 1) (hematopoietic progenitor kinase) (HPK) | Map4k1 Hpk1 | P70218 | Kinase |
| 21 | Glucose-6-phosphate 1-dehydrogenaseX (EC 1.1.1.49) (G6PD) | G6pdx G6pd G6pd-1 | Q00612 | Oxidoreductase |
| 22 | Piwi-like protein 1 | Piwill Miwi | Q9JMB7 | Not classified |
| 23 | Dynamin-1-like protein (EC 3.6.5.5) (Dynamin-related protein 1) (Dynamin family member proline-rich carboxyl-terminal domain less) (Dymple) | Dnml1 Drp1 | Q8K1M6 | Cytoskeletal protein |
| 24 | Peripherin | Prph Prph1 | P15331 | Not classified |
| 25 | Splicing factor, arginine/serine-rich 1 | Srsf1 Sfrs1 | Q6PDM2 | Nucleic acid binding |
Validation of the Identified Calpain Substrates in Dopaminergic Neuronal Cells
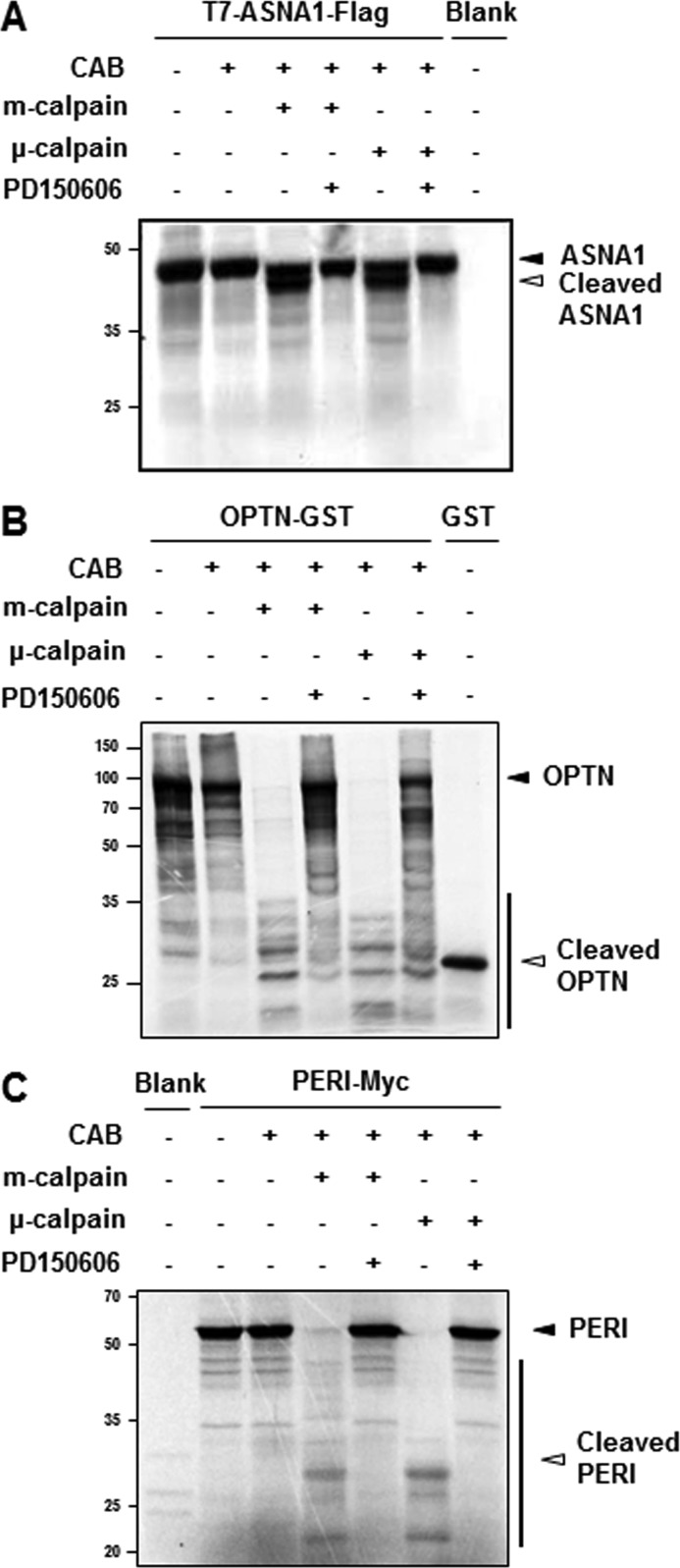
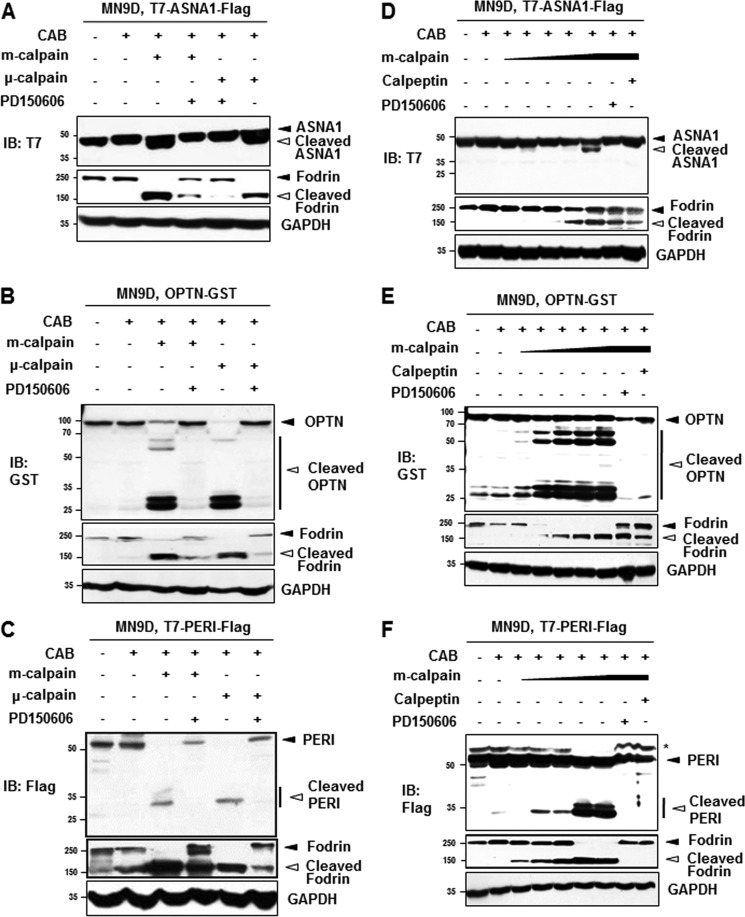
Among these 25 altered proteins, we focused on protein spots that were significantly decreased or increased at least by 3-fold with p < 0.05 (n > 3). Fig. 2C showed representative close-up images of these protein spots boxed in Fig. 2B (also refer to Table 2). We then attempted to further verify each protein spot by performing repeated peptide mass fingerprint analysis, searching against different databases, considering the protein score, and referring to their actual location on the 2DE gel. To unambiguously identify putative calpain substrates, we used parameters for MALDI-TOF mass spectral analysis and mass spectrometry profiling as described under “Experimental Procedures.” Table 2 summarizes the identified protein spots boxed in Fig. 2B and highlighted in Fig. 2C with accession number, molecular mass with pI, fold change in intensity over the untreated control spot, MOWSE score, number of the matched peptides, and sequence coverage. We then searched the available database that predicts putative protease substrates. None of these six proteins were listed as putative calpain substrates from database searches through Peptidecutter and PESTfind (data not shown). In contrast, CaMPDB (17) predicts that ASNA1, optineurin, and peripherin each contain a few sites that could possibly be cleaved by calpain. Therefore, we examined whether these three proteins may be endogenous calpain substrates by performing in vitro calpain cleavage assays using metabolically [35S]methionine-radiolabeled proteins. As shown in Fig. 3A, ASNA1 was directly cleaved by both m- and μ-calpain, generating prominent fragments that ran just below the full-length form. This calpain-mediated cleavage was blocked in the presence of PD150606, a cell-permeable, selective, non-peptide Ca2+-dependent calpain inhibitor (18). For optineurin, treatment with m- or μ-calpain generated several smaller fragments. Again, this phenomenon was largely blocked by PD150606 (Fig. 3B). As shown in Fig. 3C, external addition of m- or μ-calpain also caused generation of several smaller peripherin bands along with two prominent fragments that were blocked in the presence of PD150606. To further confirm these findings, MN9D dopaminergic neuronal cells were transiently transfected with eukaryotic expression vectors containing T7-ASNA1-FLAG tagged, optineurin-GST tagged, or T7-peripherin-FLAG tagged sequences. Lysates obtained from transfected cells were treated with m- or μ-calpain in the presence or absence of PD150606 and subjected to immunoblot analyses using anti-T7, -GST, and -FLAG antibodies (Fig. 4). Immunoblot analyses for fodrin were run in parallel to confirm the state of calpain activation in each sample (see the middle panels of Fig. 4, A–C). Indeed, both m- and μ-calpain cleaved the transiently expressed proteins, and this cleavage was significantly blocked by PD150606. The calpain-mediated cleavage pattern of transiently expressed ASNA1 was found to be quite similar to that observed with the in vitro calpain cleavage assay (compare Fig. 4A with Fig. 3A). However, the cleavage patterns for optineurin and peripherin were somewhat different (compare Fig. 4, B and C, with Fig. 3, B and C, respectively). In any case, calpain-mediated cleavage of the transiently expressed proteins was significantly decreased in the presence of PD150606. To further examine the dose dependence of calpain-mediated cleavage of the identified substrates, we performed cell-based cleavage assays with increasing doses of m-calpain (1.11 to 5.55 units; Fig. 4, D–F). Dose-dependent cleavage of all three proteins was significantly blocked by co-treatment with PD150606 or calpeptin, another membrane-permeable calpain inhibitor. Taken together, these data indicate that all three identified proteins are most likely novel substrates that are directly cleaved by both m- and μ-calpain.
TABLE 2.
Summary of proteins further identified after incubation with recombinant calpain
| Spot | Protein namea | Accession numberb | Molecular mass/pIc | Foldd | MOWSE scoree,f | Masses matchedf,g | Coveragef |
|---|---|---|---|---|---|---|---|
| Da | % | ||||||
| H23 | Ras-GTPase-activating protein-binding protein 1 | P97855 | 51,829/5.4 | 5.55 | 1.84E + 15 | 22 (11) | 48.8 |
| H19 | GMP synthase (glutamine-hydrolyzing) | Q3THK7 | 76,724/6.3 | 8.58 | 5.75E + 07 | 13 (7) | 23.4 |
| H17 | Stress-induced phosphoprotein 1 | Q60864 | 62,583/6.4 | 4.09 | 9.56E + 06 | 18 (9) | 36 |
| H18 | ATPase ASNA1 | O54984 | 38,823/4.8 | 0.38 | 1.00E + 09 | 14 (8) | 37.6 |
| H16 | Optineurin | Q8K3K8 | 67,018/5.2 | 0.17 | 4.19E + 13 | 35 (12) | 39 |
| H4 | Peripherin | P15331 | 54,268/5.4 | 0.23 | 4.78E + 08 | 21 (10) | 44 |
a Each protein spot in Fig. 2 was identified based on mass spectra of tryptic peptides obtained by MALDI-TOF mass spectrometry.
b Accession numbers from SwissProt data base (2012.12.3).
c Theoretical molecular mass and pI.
d Values present change of spots intensity in m-calpain-treated gel in comparison to untreated gel (value = 1).
e MOWSE scores were considered significant for identity (p < 0.05).
f These values were considered from SwissProt.
g The numbers in parentheses indicate percentages.
FIGURE 3.
In vitro calpain cleavage assays. Among the altered protein spots identified as putative calpain substrates, T7-ASNA1-FLAG (A), OPTN-GST (B), and PERI-Myc (C) were radiolabeled with [35S]methionine using an in vitro transcription/translation kit. Reactants were incubated for 2 h at 37 °C with the indicated amount of recombinant calpains (5.55 and 3.95 units for m- and μ-calpain, respectively) in the presence or absence of a calpain inhibitor, PD150606 (500 μm) in a calpain activation buffer (CAB). After incubation, all samples were separated by SDS-PAGE and subjected to autoradiography. Closed arrowheads indicate full-length proteins, and open arrowheads represent cleaved fragments. Autoradiographic images are representative from at least three independent experiments.
FIGURE 4.
Cell-based calpain cleavage assays. Lysates from MN9D cells overexpressing T7-ASNA1-FLAG (A), OPTN-GST (B), or T7-PERI-FLAG (C) were incubated for 2 h at 37 °C with the indicated calpains (5.55 unit m-calpain or 3.95 units μ-calpain) in the presence or absence of 500 μm PD150606. After separation by SDS-PAGE, gels were processed for immunoblot (IB) analysis using anti-T7, -GST, or -FLAG antibody, respectively. Closed and open arrowheads indicate full-length and fragments of each protein, respectively. Fodrin and GAPDH were used as the calpain activation and loading control, respectively. To examine whether cleavage of ASNA1 (D), OPTN (E), and PERI (F) was achieved in a calpain concentration-dependent manner, all protein lysates (100 μg) from MN9D cells transiently overexpressing tagged proteins were incubated with increasing doses of m-calpain (1.11–5.55 units) in the presence or absence of 500 μm PD150606 or 50 μm calpeptin. Immunoblot analyses were performed using anti-T7, -GST, or -FLAG antibody. The asterisk in F indicates a nonspecific band. Representative blots from three independent experiments are shown.
Ionomycin- and MPP+-induced Cleavage of ASNA1, Optineurin, and Peripherin
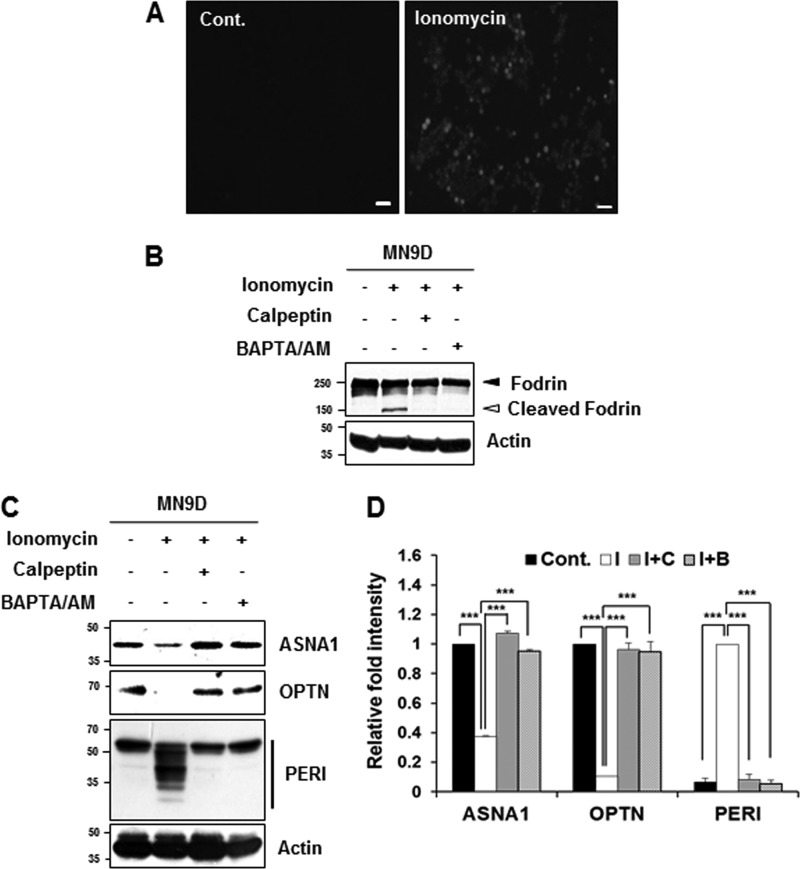
Dysregulation of intracellular free Ca2+ causes perturbed energy metabolism, oxidative stress, and neuronal death (19, 20). Alterations in Ca2+ homeostasis lead to pathologic activation of calpain, and inhibition of calpain is considered a therapeutic target for a number of neurodegenerative disorders, including PD (5, 21, 22). We therefore investigated whether increased intracellular Ca2+ in drug-induced neurodegeneration models was associated with cleavage of the newly identified substrates. To accomplish this, MN9D dopaminergic neuronal cells were first treated with ionomycin, an ionophore produced by Streptomyces conglobatus that is widely used to increase intracellular free Ca2+ and subsequently activates calpain (23). Treatment with 5 μg/ml ionomycin for 30 min apparently increased the number of Fluo-3-positive cells (Fig. 5A) and led to Ca2+-activated, calpain-dependent cleavage of fodrin that was completely inhibited in the presence of 50 μm calpeptin or 40 μm BAPTA/AM, a membrane-permeable Ca2+ chelator (Fig. 5B). As shown in Fig. 5C, levels of full-length forms of ASNA1 and optineurin were decreased following ionomycin treatment. In the case of peripherin, many smaller fragments appeared following ionomycin treatment. Notably, statistical analysis indicated that these ionomycin-induced changes were significantly attenuated in the presence of calpeptin or BAPTA/AM (Fig. 5D).
FIGURE 5.
Cleavage of the identified substrates during ionomycin-induced cell death. A, MN9D cells were incubated with or without ionomycin (5 μg/ml) for 30 min. After incubation, cells were stained with 3 μm Fluo-3/AM dye and examined under a fluorescence microscope equipped with epifluorescence filters and a digital analyzer. Scale bars represent 50 μm. B, MN9D cells were incubated for 30 min with ionomycin (5 μg/ml) in the presence or absence of calpeptin (50 μm) or BAPTA/AM (40 μm). After incubation, cellular lysates (50 μg) from each condition were subjected to SDS-PAGE followed by immunoblot analysis using the indicated antibodies. Ionomycin-induced calpain activation was confirmed with the anti-fodrin antibody. Actin was used as a loading control. C, MN9D cells were incubated for 30 min at 37 °C with ionomycin (5 μg/ml) in the presence or absence of 50 μm calpeptin or 40 μm BAPTA/AM. To measure levels of ASNA1, OPTN, and PERI, cellular lysates (100 μg) were subjected to immunoblot analysis using the indicated antibodies that specifically recognize the endogenous protein. D, after normalization to actin in each condition, the relative change in fold intensity of the full-length (for ASNA1 and optineurin) was calculated over the untreated control (value = 1). Changes in fold intensity of total peripherin fragments were determined over the ionomycin-treated sample (value = 1). Each bar represents the mean ± S.E. from three independent experiments. ***, p < 0.001. Cont., no treatment; I, ionomycin; C, calpeptin; B, BAPTA/AM.
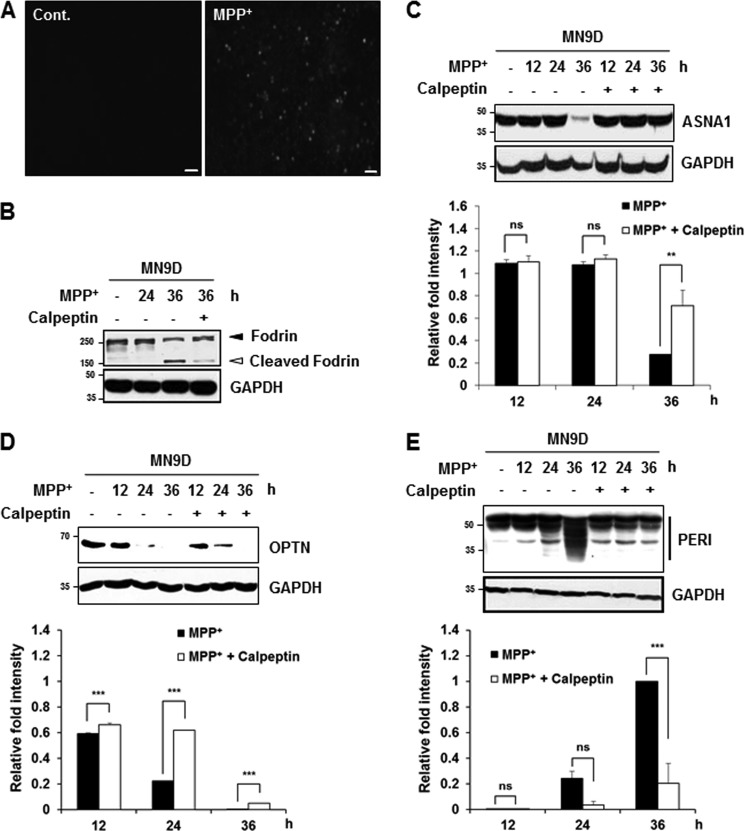
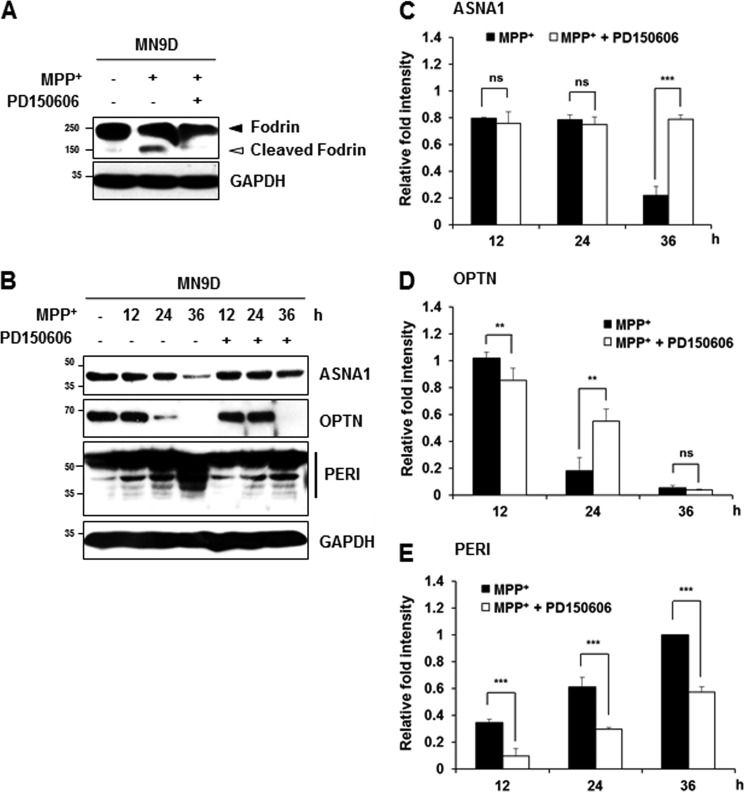
Neurotoxin-based experimental models have been widely used to mimic biochemical changes that occur in PD patients (1). Among these, systemic administration of the mitochondrial complex I inhibitor MPTP to non-human primates and mice and treatment of cells with its active metabolite MPP+ have been used to establish experimental models of PD. Previous reports in our laboratory indicate that MPP+ increases intracellular Ca2+ and causes subsequent calpain activation in cultured cells (15, 24). Furthermore, we reported that MPP+-induced cell death does not activate caspase in MN9D dopaminergic neuronal cells as well as in primary cultures of dopaminergic neurons (25). Therefore, we attempted to investigate whether MPP+ treatment causes calpain-mediated cleavage of the three newly identified substrates. Again, increases of intracellular Ca2+ and calpain-mediated fodrin cleavage and decreased levels of ASNA1 and optineurin full-length forms were detected in MN9D cells after 50 μm MPP+ treatment for 36 h (Fig. 6, A–D). Statistical analyses indicated that MPP+-mediated decrease in ASNA1 and optineurin full-length forms were significantly attenuated in the presence of calpeptin (Fig. 6, C and D). MPP+ did not lead to any detectable change in Apaf-1 expression level, indicating that drug-induced decrease of ASNA1 and optineurin levels was not simply due to cell loss (data not shown). Quite similar to the pattern observed in ionomycin-treated cells, MPP+ treatment for 36 h led to the generation of several peripherin fragments with smaller molecular weights, and this was significantly blocked in the presence of calpeptin (Fig. 6E). Furthermore, MPP+-induced alterations of these three substrates were also significantly blocked in the presence of PD150606 (Fig. 7). In sum, we demonstrated that all three endogenous substrates were processed by drug-induced calpain activation in MN9D cells. We found that the processing patterns generated by ionomycin or MPP+ treatment were quite similar.
FIGURE 6.
MPP+-mediated cleavage of the identified substrates is attenuated by calpeptin. A, MN9D cells were incubated with or without 50 μm MPP+ for 36 h. Cells were stained with 3 μm Fluo-3/AM dye and examined under a fluorescence microscope equipped with epifluorescence filters and a digital analyzer. Scale bars represent 50 μm. B, to measure calpain activity in MN9D cells treated with 50 μm MPP+ in the presence or absence of 50 μm calpeptin for the indicated time periods, cellular lysates (50 μg) were separated and processed for immunoblot analysis using an anti-fodrin antibody. C–E, MN9D cells were incubated in 50 μm MPP+ in the presence or absence of 50 μm calpeptin for the indicated time periods. Cellular lysates (100 μg) were subjected to immunoblot analysis using the indicated antibodies. Changes in fold intensity were calculated as described in the legend Fig. 5. All values represent the means ± S.E. from three independent experiments. ***, p < 0.001; **, p < 0.01. ns, not significant; Cont., control.
FIGURE 7.
MPP+-mediated cleavage of the identified substrates is attenuated by PD150606. A, to measure calpain activity in MN9D cells treated with 50 μm MPP+ for 36 h in the presence or absence of 50 μm PD150606, cellular lysates (50 μg) were separated and processed for immunoblot analysis using an anti-fodrin antibody. B–E, following drug treatment indicated in each lane, cellular lysates (100 μg) were subjected to immunoblot analysis using the indicated antibodies. Relative fold intensity was calculated as described in the legend to Fig. 5. All values represent the means ± S.E. from three independent experiments. ***, p < 0.001; **, p < 0.01. ns, not significant.
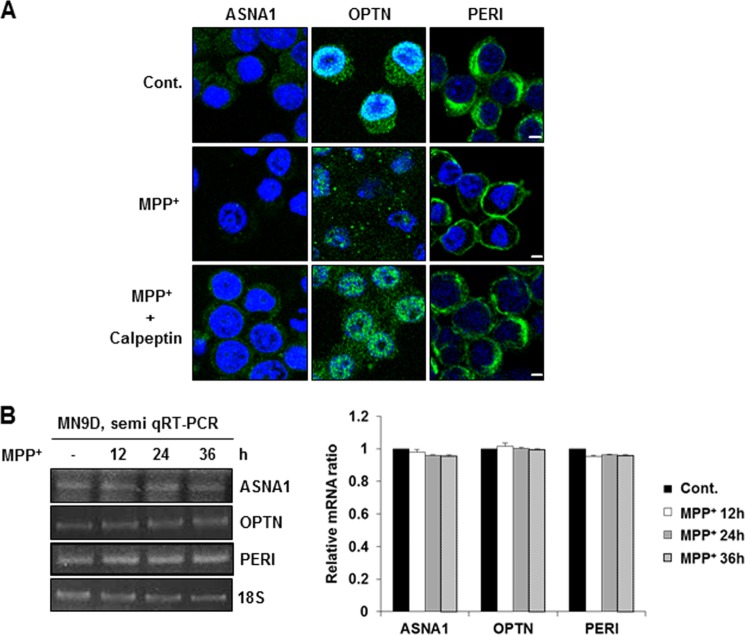
We next used immunofluorescent labeling to measure the cellular distribution patterns of these substrates before and after MPP+ treatment. Overall, the punctate staining intensities of ASNA1 and optineurin were apparently reduced in all areas of the cells upon MPP+ treatment, and these changes were attenuated in the presence of calpeptin (Fig. 8A, left and middle columns). Intriguingly, the diffuse cytosolic staining pattern of peripherin in untreated control cells shifted to the cytoplasmic membrane after MPP+ treatment, and this staining pattern of peripherin was restored in MN9D cells co-treated with calpeptin (Fig. 8A, right column). Based on previous observations demonstrating that expression levels of some proteins in MPP+-treated MN9D cells can be regulated at the transcriptional level (26), we carried out semi-quantitative RT-PCR to compare their mRNA levels in cells treated with or without MPP+. As shown in Fig. 8B, no discernible changes were found, indicating that MPP+-mediated calpain activation is responsible for decreased levels and altered cellular distribution of these endogenous substrates without directly affecting their transcription rates.
FIGURE 8.
Changes in cellular distribution pattern and mRNA levels of the identified substrates in MPP+-induced cell death. A, immunofluorescent localization of each protein was measured in MN9D cells treated with 50 μm MPP+ for 36 h in the presence or absence of 50 μm calpeptin. Scale bars represent 5 μm. B, semi-quantitative RT-PCR was performed using mRNA prepared from MN9D cells treated with 50 μm MPP+ for the indicated time periods. Levels of 18 S rRNA were used as loading controls. Relative ratio of mRNA for ASNA1, OPTN, or PERI was calculated over the untreated controls (value = 1) after normalization to 18 S rRNA. All values represent the means ± S.E. from three independent experiments. Note that no significant changes in mRNA levels were detected. Cont., control.
Cleavage of ASNA1, Optineurin, and Peripherin in Rat Disease Models
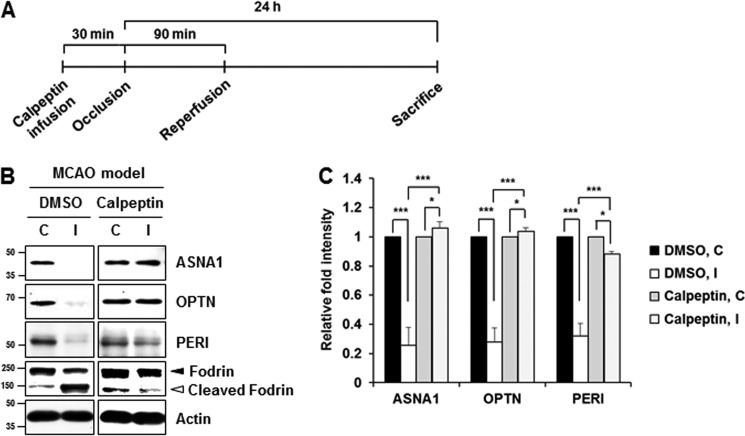
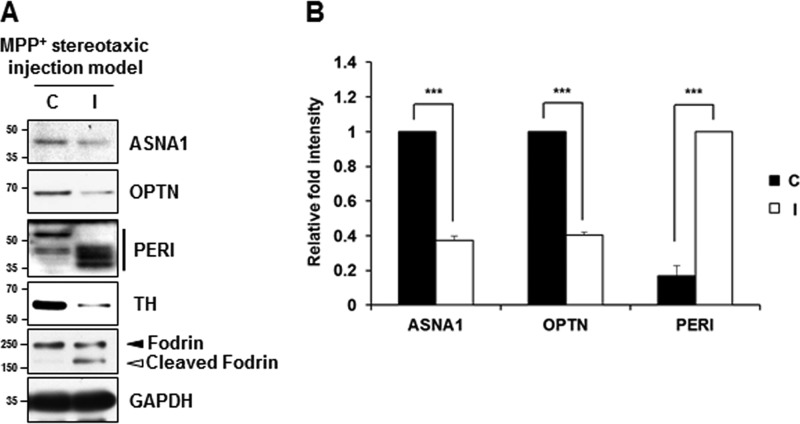
We then tested whether these phenomena could be recapitulated in rat disease models of neurodegeneration. Increased intracellular Ca2+ level caused by disturbance of Ca2+ homeostasis is a major change that occurs following ischemic stroke. This leads to constitutive activation of calpain and accompanying excessive proteolysis that result in neuronal cell death (13, 27). Therefore, we examined the calpain-induced cleavage of ASNA1, optineurin, and peripherin in the cortical tissue in MCAO rats. For actual experiments, rats received intracerebroventricle infusion of either 50 μg of calpeptin dissolved in 5 μl of Me2SO or Me2SO alone 30 min before MCAO (Fig. 9A). Immunoblot analysis using anti-fodrin antibody showed that the ipsilateral hemisphere showed the calpain-cleaved fodrin band, suggesting a certain degree of calpain activation. We also found that full-length optineurin was significantly decreased in the ipsilateral cortex 18 h after ischemic injury, whereas decreases in ASNA and peripherin were not apparent (data not shown). However, all of these proteins were clearly decreased in the ipsilateral cortex 24 h after ischemic injury (Fig. 9B). To unambiguously confirm that this phenomenon is a calpain-dependent event, rats received intracerebroventricle infusion of calpeptin before MCAO. As shown in Fig. 9 (B and C), MCAO-induced change in all three substrates was significantly blocked in the ipsilateral cortex infused with calpeptin. Taken together, these data indicate that ASNA1, optineurin, and peripherin cleavage occurred following brain injury that disturbs Ca2+ homeostasis and increases calpain activation in vivo. As shown in Fig. 10, MPP+ injection into the medial forebrain bundle for 3 days caused appearance of calpain-cleaved fodrin in the substantia nigra pars compacta, indicating calpain activation. In the ipsilateral side of stereotaxic injection model, the full-length forms of ASNA1 and optineurin were reduced, whereas several peripherin fragments with smaller molecular sizes were detected, supporting calpain-mediated alterations of the newly identified substrates. At present, it was not clearly understood how different cleavage products were observed when metabolically labeled substrates or cell lysates were incubated with calpain versus when cultures were treated with drugs or rat brain disease models were established. Search through the Peptidecutter database indicates that all of these identified substrates can be cleaved by several other proteases, explaining a partial blockage of drug- or surgery-mediated alterations of the putative substrates by calpain inhibitors. Similarly, this may explain why the cleaved fragments of the identified substrates are not detected in our culture models of neurodegeneration and rat brain disease models. Although it is highly speculative, therefore, complex regulation of protease activation in neurotoxin-based culture models and in rat neurodegenerative models may be responsible for different patterns of cleavage.
FIGURE 9.
Confirmation of calpain-mediated cleavage of the identified substrates in MCAO model. A, schematic flow diagram for MCAO model infused with or without calpeptin. B, rats randomly divided into two groups received intracerebroventricle infusion of 50 μg of calpeptin in 5 μl of Me2SO or 5 μl of Me2SO alone 30 min before MCAO. The rats were then exposed to MCAO for 1.5 h and subsequent reperfusion for 24 h. Tissue lysates (50 μg) obtained from the contralateral (C) and the ipsilateral (I) side of the cortex to the right-sided endovascular MCAO were subjected to immunoblot analyses using the indicated antibodies. C, relative fold intensity of each protein after normalization to actin in each contralateral (C) and ipsilateral (I) sides of cortex was calculated over the contralateral side of Me2SO group (value = 1). Each bar represents the mean ± S.E. from four independent experiments. ***, p < 0.001; *, p < 0.05.
FIGURE 10.
Cleavage of the identified substrates in MPP+-stereotaxic injection model. A, at 3 days post-injection, the contralateral (C) and ipsilateral (I) side of tissue samples (substantia nigra pars compacta in midbrain) obtained from the MPP+-stereotaxic injection rats were processed for immunoblot analyses using the indicated antibodies that recognize endogenous proteins. TH was used to assess the extent of dopaminergic neurodegeneration, and fodrin and GAPDH were used as a calpain activation and loading control, respectively. B, after normalization to GAPDH, relative change in fold intensity of the indicated protein was determined over the full-length protein in the contralateral side (for ASNA1 and OPTN; value = 1) or the total fragments of the ipsilateral side (for PERI; value = 1). Each bar represents the mean ± S.E. from four independent samples. ***, p < 0.001.
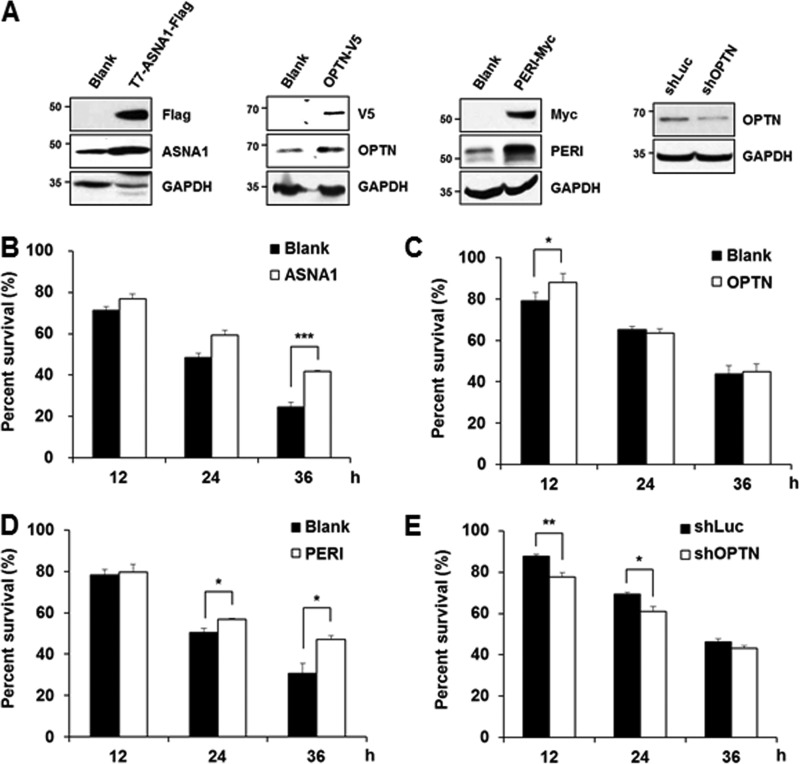
Potentially Protective Effect for ASNA1, Optineurin, and Peripherin in MPP+-induced Cell Death
Previous reports have indicated that calpain activation following neurotoxin challenge is related to various cell death signaling pathways and that many calpain-cleaved substrates play a critical role in maintaining cell survival under physiological conditions (23, 28, 29). We have also demonstrated that MPP+-mediated cell death is blocked in the presence of calpeptin and overexpressed calbindin-D28K (15, 24). We therefore examined whether MPP+-mediated cleavage of these identified substrates causes loss of function or has any pathophysiological consequences. Eukaryotic expression vectors containing ASNA1, optineurin, or peripherin cDNA sequences were transiently expressed in MN9D cells, and its expression level was confirmed by immunoblot analysis (Fig. 11A). Although it varied depending on each transient transfection trial, expression levels of these exogenous ASNA1, optineurin, and peripherin were at least 2-fold higher as compared with levels of the corresponding endogenous proteins. These cells were exposed to 50 μm MPP+ for various time periods and subjected to MTT reduction assay. Data indicated that ASNA1 or peripherin overexpression protected cells from MPP+-induced death, and the protection rate was statistically significant at 36 h post-treatment (Fig. 11, B and D). In contrast, overexpressed optineurin provided slight but meaningful protection at only 12 h after MPP+ treatment (Fig. 11C). To further confirm the protective effect of optineurin, MN9D cells were transiently transfected with the construct containing the shRNA sequence for optineurin. Partial knockdown of optineurin accelerated MPP+-induced cell death at 12 and 24 h post-treatment (Fig. 11, A and E). A similar protective pattern of all substrates was observed in neutral red-based colorimetric assays and morphological examination under a phase contrast microscope (data not shown). Considering the notion that the consequences of protease-mediated cleavage of proteins can be gain of function or loss-of-function or sometimes just bystander events (30), further biochemical studies remain to be performed to clearly argue that calpain-mediated cleavage of ASNA1, peripherin, and perhaps optineurin may be indeed associated with the progression of MPP+-induced neurodegeneration.
FIGURE 11.
Overexpression of ASNA1, optineurin, or peripherin renders cell less vulnerable to MPP+ treatment. A, cellular lysates obtained from MN9D cells transiently transfected with or without the vector containing T7-ASNA1-FLAG, OPTN-V5, PERI-Myc, or shRNA target sequence for optineurin (shOPTN) were subjected to an immunoblot analysis using the indicated antibodies to confirm overexpression of each tagged protein or knockdown of OPTN. B–E, MN9D cells transiently transfected with T7-ASNA1-FLAG (B), OPTN-V5 (C), PERI-Myc (D), or shRNA target sequence for OPTN (shOPTN, E) were treated with 50 μm MPP+ for the indicated time periods. Viability was measured by the MTT reduction assay. Each bar represents the mean ± S.E. from three independent experiments. Blank represents the cells transfected with an empty vector. The shRNA sequences that target firefly luciferase (shLuc) were used for negative control. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
DISCUSSION
Regardless of recent progress in our understanding, attempts to elucidate the molecular mechanisms by which protease activation induces cell death have been greatly hampered by the lack of and/or ineffectiveness of systematic and broadly applicable strategies to identify endogenous substrates that underlie neuronal survival/death mechanisms. To date, several screening methods have been attempted to identify protease substrates, including the expression cloning strategy (31), the yeast two-hybrid screening method (32), and more recently, proteomic identification (for review, see Ref. 33). For proteomic approaches, both gel-based methods (e.g., one- and two-dimensional analyses) and gel-free proteomic techniques (e.g., negative and positive selections of N-terminal identifications, combined fractional diagonal chromatography) have been applied (34, 35). Many of these approaches have proved to be effective in that tens and hundreds of putative substrates have been identified. However, only a fraction has been confirmed as protease substrates, and its valid application remains to be carefully examined in pathophysiologically relevant disease models. A great majority of the proteomic analyses employed samples that were prepared either by direct addition of protease into lysates (36, 37) or using protease knockdown systems (38). However, these approaches may also identify substrates that are cleaved as a consequence of sequential activation of other downstream proteases (39).
In the present study, we described a novel protease proteomic analysis based on conventional, gel-based 2DE, that only identifies putative substrates that are directly cleaved by calpains. This was achieved by incubating immobilized protein spots on IEF strips with calpains under specifically optimized conditions. This protocol ensures that only the direct substrates of calpain are cleaved, eliminating the possibility of identifying substrates that may be cleaved by combinatorial or sequential activation of proteolytic enzymes. Using cellular lysates obtained from the MN9D dopaminergic neuronal cell line that has been used as a culture model of PD, we initially found more than 100 altered protein spots and identified 25 proteins that were differentially expressed by at least 2-fold following calpain treatment. Among these spots, we selected three proteins: ASNA1, optineurin, and peripherin, that were not previously identified as calpain substrates. We performed in vitro and cell-based calpain cleavage assays and confirmed that these three substrates are cleaved by exogenous calpain. Furthermore, calpain-mediated cleavage was also observed in cultured cells and two rat brain models of neurodegeneration. The existing literature indicates that many calpain substrates have a typical PEST sequence (for review, see Ref. 3). However, ASNA1, optineurin, and peripherin do not contain any PEST sequences. Based on previous reports indicating that several calpain substrates do not possess PEST sequences (40), it is plausible to assume that ASNA1, optineurin, and peripherin may have other calpain-specific cleavage sites. Although more detailed study may be needed to verify the cleavage and/or degradation patterns of these identified substrates in vivo as well as in vitro, the present data suggest that our protease proteomic analysis is useful for identifying novel calpain substrates involved in neurodegeneration. In our separate study using the same strategy, we have identified more than 46 novel caspase-3 substrates,5 indicating potentially broad applicability of our gel-based protease proteomic approaches.
In addition to caspases, calpains are the most extensively characterized cysteine proteases and are involved in maintaining neuronal physiology and perpetuating pathophysiology (3). They have roles in several fundamental processes, including cell signaling, cytoskeletal remodeling, and cell death/survival. In various brain disorders including PD, Alzheimer disease, Huntington disease, stroke, and spinal cord injury, growing evidence supports the involvement of Ca2+-mediated, nonlysosomal calpain activation (5, 21, 41–43). This notion is strongly supported by data obtained from the brains of patients with neurodegenerative disorders, showing increased calpain activity compared with age-matched control brains. Consequently, dysregulated calpain activation in these pathological circumstances results in the cleavage of a number of crucial substrates that play important roles in maintaining neuronal structure and function (21), which can lead to injury progression. For example, p25, a proteolytic cleavage product of the neuron-specific activator of cyclin-dependent kinase 5 (Cdk5), has been found to accumulate in the brains of Alzheimer disease patients (44). In brain lysates treated with Ca2+ or in primary cultures of cortical neurons challenged with excitotoxins, hypoxic injury, or amyloid-β peptide, Ca2+-activated calpain is responsible for cleavage of p35 to p25 (23, 45). Furthermore, calpain-mediated cleavage of p35 to p25 and subsequent Cdk5 activation has been implicated in cytoskeletal disruption and neuronal death, consequently leading to Alzheimer disease progression (23). Similarly, calpain-mediated conversion of p35 to p25 also plays a central role in MPTP-induced dopaminergic neuronal death via phosphorylation of critical factors and enzymes (46). More recently, it has been demonstrated that calpain-mediated generation of toxic fragments affects neuronal survival during glutamate receptor-mediated excitotoxicity (47). Therefore, understanding of how calpain-mediated cleavage of critical substrates is regulated is considered important for blocking disease progression, and inhibition of calpain activation has consequently emerged as a potential therapeutic target for neurodegenerative disorders (21).
All three identified proteins have been demonstrated to affect fundamental cellular functions such as cell growth, survival, and death. For example, ASNA1 is evolutionary conserved and homologous to Escherichia coli arsA, a well characterized ATPase involved in arsenite and antimonite efflux (48). It is found throughout the cell but is mainly localized in the cytoplasm. ASNA1 is also known as an essential ATPase for the insertion of tail-anchored proteins into endoplasmic reticulum membranes, including Golgi to endoplasmic reticulum traffic, and is a novel positive regulator of insulin secretion (49). Therefore, the depletion or down-regulation of full-length ASNA1 can significantly affect cell growth and survival/death. Optineurin is a ubiquitously expressed multifunctional cytoplasmic protein and is also localized in the Golgi apparatus, where it contributes to post-Golgi and vesicular trafficking and Golgi organization (50). Therefore, overexpression of optineurin and mutant optineurin (E50K) highlighted its potential roles in trafficking, Golgi integrity, and cell death (51). Indeed, it negatively regulates the induction of interferon-β in response to RNA virus infection and tumor necrosis factor-α-induced nuclear factor-κB activation by competing with NF-κB essential modulator (NEMO) for polyubiquitinated receptor-interacting protein (RIP) (52). Recently, it was reported that optineurin enhances cell survival via reactive oxygen species-induced nuclear translocation of optineurin, whereas fragmentation of nigral neuron Golgi apparati typically found in optineurin mutants is increased in PD patients (53, 54). Finally, peripherin is a type of neuronal intermediate filament that is mainly cytoplasmic and is associated with pathological aggregates in the motor neurons in patients with amyotrophic lateral sclerosis (55). However, it remains controversial whether peripherin is involved in regulating cell survival and death in various neural systems (56, 57). Nevertheless, it has not been well understood how their physiological and pathological functions may be regulated. Based on data from both in vitro and in vivo experiments, we hypothesize that cleavage of these substrates may result in a loss of function phenomena that would associated with calpain-induced neuronal death. In this regard, it is necessary to carefully examine how calpain-mediated cleavage or degradation of ASNA1, optineurin, and peripherin affect their proposed cellular functions, especially in neurodegenerative conditions. Further investigation into the exact mechanisms behind the potential pro-survival roles of these identified proteins will help us to expand our understanding of pathophysiology of various neurodegenerative disorders.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by Ministry of Education, Science, and Technology Grant 2012-0000496 and World Class University Grant R33-10014 (to Y. J. O.). This work was also supported by Bio & Medical Technology Development Program Grant 2011-0019440 through the National Research Foundation of Korea funded by the Ministry of Science and Technology (to W. K. K.).
N. Yun, Y. M. Lee, and Y. J. Oh, unpublished data.
- PD
- Parkinson disease
- MPP+
- 1-methyl-4-phenylpyridinium
- MPTP
- 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- BAPTA/AM
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester)
- 2DE
- two-dimensional gel electrophoresis
- IEF
- isoelectric focusing
- MCAO
- middle cerebral artery occlusion
- OPTN
- optineurin
- PERI
- peripherin.
REFERENCES
- 1. Dauer W., Przedborski S. (2003) Parkinson's disease. Mechanisms and models. Neuron 39, 889–909 [DOI] [PubMed] [Google Scholar]
- 2. Moore D. J., West A. B., Dawson V. L., Dawson T. M. (2005) Molecular pathophysiology of Parkinson's disease. Annu. Rev. Neurosci. 28, 57–87 [DOI] [PubMed] [Google Scholar]
- 3. Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) The calpain system. Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 4. Nakanishi H. (2003) Microglial functions and proteases. Mol. Neurobiol. 27, 163–176 [DOI] [PubMed] [Google Scholar]
- 5. Vosler P. S., Brennan C. S., Chen J. (2008) Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol. Neurobiol. 38, 78–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caberoy N. B., Alvarado G., Li W. (2011) Identification of calpain substrates by ORF phage display. Molecules 16, 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brulé C., Dargelos E., Diallo R., Listrat A., Béchet D., Cottin P., Poussard S. (2010) Proteomic study of calpain interacting proteins during skeletal muscle aging. Biochimie 92, 1923–1933 [DOI] [PubMed] [Google Scholar]
- 8. Bozoky Z., Alexa A., Dancsok J., Gogl G., Klement E., Medzihradszky K. F., Friedrich P. (2009) Identifying calpain substrates in intact S2 cells of Drosophila. Arch. Biochem. Biophys. 481, 219–225 [DOI] [PubMed] [Google Scholar]
- 9. Cuerrier D., Moldoveanu T., Davies P. L. (2005) Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach. The importance of promed side interactions. J. Biol. Chem. 280, 40632–40641 [DOI] [PubMed] [Google Scholar]
- 10. Choi H. K., Won L. A., Kontur P. J., Hammond D. N., Fox A. P., Wainer B. H., Hoffmann P. C., Heller A. (1991) Immortalization of embryonic mesencephalic dopaminergic neurons by somatic cell fusion. Brain Res. 552, 67–76 [DOI] [PubMed] [Google Scholar]
- 11. Galluzzi L., Aaronson S. A., Abrams J., Alnemri E. S., Andrews D. W., Baehrecke E. H., Bazan N. G., Blagosklonny M. V., Blomgren K., Borner C., Bredesen D. E., Brenner C., Castedo M., Cidlowski J. A., Ciechanover A., Cohen G. M., De Laurenzi V., De Maria R., Deshmukh M., Dynlacht B. D., El-Deiry W. S., Flavell R. A., Fulda S., Garrido C., Golstein P., Gougeon M. L., Green D. R., Gronemeyer H., Hajnóczky G., Hardwick J. M., Hengartner M. O., Ichijo H., Jäättelä M., Kepp O., Kimchi A., Klionsky D. J., Knight R. A., Kornbluth S., Kumar S., Levine B., Lipton S. A., Lugli E., Madeo F., Malomi W., Marine J. C., Martin S. J., Medema J. P., Mehlen P., Melino G., Moll U. M., Morselli E., Nagata S., Nicholson D. W., Nicotera P., Nunez G., Oren M., Penninger J., Pervaiz S., Peter M. E., Piacentini M., Prehn J. H., Puthalakath H., Rabinovich G. A., Rizzuto R., Rodrigues C. M., Rubinsztein D. C., Rudel T., Scorrano L., Simon H. U., Steller H., Tschopp J., Tsujimoto Y., Vandenabeele P., Vitale I., Vousden K. H., Youle R. J., Yuan J., Zhivotovsky B., Kroemer G. (2009) Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 16, 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi I. Y., Lee J. C., Ju C., Hwang S., Cho G. S., Lee H. W., Choi W. J., Jeong L. S., Kim W. K. (2011) A3 adenosine receptor agonist reduces brain ischemic injury and inhibits inflammatory cell migration in rats. Am. J. Pathol. 179, 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng S., Kuang Z., Zhang Y., Xu H., Cheng Q. (2011) The protective effects and potential mechanism of Calpain inhibitor Calpeptin against focal cerebral ischemia-reperfusion injury in rats. Mol. Biol. Rep. 38, 905–912 [DOI] [PubMed] [Google Scholar]
- 14. Lee Y. M., Park S. H., Shin D. I., Hwang J. Y., Park B., Park Y. J., Lee T. H., Chae H. Z., Jin B. K., Oh T. H., Oh Y. J. (2008) Oxidative modification of peroxiredoxin is associated with drug-induced apoptotic signaling in experimental models of Parkinson disease. J. Biol. Chem. 283, 9986–9998 [DOI] [PubMed] [Google Scholar]
- 15. Choi W. S., Lee E. H., Chung C. W., Jung Y. K., Jin B. K., Kim S. U., Oh T. H., Saido T. C., Oh Y. J. (2001) Cleavage of Bax is mediated by caspase-dependent or -independent calpain activation in dopaminergic neuronal cells. Protective role of Bcl-2. J. Neurochem. 77, 1531–1541 [DOI] [PubMed] [Google Scholar]
- 16. Wood D. E., Thomas A., Devi L. A., Berman Y., Beavis R. C., Reed J. C., Newcomb E. W. (1998) Bax cleavage is mediated by calpain during drug-induced apoptosis. Oncogene 17, 1069–1078 [DOI] [PubMed] [Google Scholar]
- 17. DuVerle D. A., Ono Y., Sorimachi H., Mamitsuka H. (2011) Calpain cleavage prediction using multiple kernel learning. PLoS One 6, e19035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang K. K., Nath R., Posner A., Raser K. J., Buroker-Kilgore M., Hajimohammadreza I., Probert A. W., Jr., Marcoux F. W., Ye Q., Takano E., Hatanaka M., Maki M., Caner H., Collins J. L., Fergus A., Lee K. S., Lunney E. A., Hays S. J., Yuen P. (1996) An α-mercaptoacrylic acid derivative is a selective nonpeptide cell-permeable calpain inhibitor and is neuroprotective. Proc. Natl. Acad. Sci. U.S.A. 93, 6687–6692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zündorf G., Reiser G. (2011) Calcium dysregulation and homeostasis of neural calcium in the molecular mechanisms of neurodegenerative diseases provide multiple targets for neuroprotection. Antioxid. Redox Signal. 14, 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gleichmann M., Mattson M. P. (2011) Neuronal calcium homeostasis and dysregulation. Antioxid. Redox Signal. 14, 1261–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Camins A., Crespo-Biel N., Junyent F., Verdaguer E., Canudas A. M., Pallàs M. (2009) Calpains as a target for therapy of neurodegenerative diseases. Putative role of lithium. Curr. Drug Metab. 10, 433–447 [DOI] [PubMed] [Google Scholar]
- 22. Crocker S. J., Smith P. D., Jackson-Lewis V., Lamba W. R., Hayley S. P., Grimm E., Callaghan S. M., Slack R. S., Melloni E., Przedborski S., Robertson G. S., Anisman H., Merali Z., Park D. S. (2003) Inhibition of calpains prevents neuronal and behavioral deficits in an MPTP mouse model of Parkinson's disease. J. Neurosci. 23, 4081–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee M. S., Kwon Y. T., Li M., Peng J., Friedlander R. M., Tsai L. H. (2000) Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature 405, 360–364 [DOI] [PubMed] [Google Scholar]
- 24. Choi W. S., Lee E., Lim J., Oh Y. J. (2008) Calbindin-D28K prevents drug-induced dopaminergic neuronal death by inhibiting caspase and calpain activity. Biochem. Biophys. Res. Commun. 371, 127–131 [DOI] [PubMed] [Google Scholar]
- 25. Han B. S., Hong H. S., Choi W. S., Markelonis G. J., Oh T. H., Oh Y. J. (2003) Caspase-dependent and -independent cell death pathways in primary cultures of mesencephalic dopaminergic neurons after neurotoxin treatment. J. Neurosci. 23, 5069–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim H. E., Yoon S. Y., Lee J. E., Choi W. S., Jin B. K., Oh T. H., Markelonis G. J., Chun S. Y., Oh Y. J. (2001) MPP(+) downregulates mitochondrially encoded gene transcripts and their activities in dopaminergic neuronal cells. Protective role of Bcl-2. Biochem. Biophys. Res. Commun. 286, 659–665 [DOI] [PubMed] [Google Scholar]
- 27. Kristián T., Siesjö B. K. (1998) Calcium in ischemic cell death. Stroke 29, 705–718 [DOI] [PubMed] [Google Scholar]
- 28. Tamada Y., Nakajima E., Nakajima T., Shearer T. R., Azuma M. (2005) Proteolysis of neuronal cytoskeletal proteins by calpain contributes to rat retinal cell death induced by hypoxia. Brain Res. 1050, 148–155 [DOI] [PubMed] [Google Scholar]
- 29. Bevers M. B., Lawrence E., Maronski M., Starr N., Amesquita M., Neumar R. W. (2009) Knockdown of m-calpain increases survival of primary hippocampal neurons following NMDA excitotoxicity. J. Neurochem. 108, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Demon D., Van Damme P., Vanden Berghe T., Vandekerckhove J., Declercq W., Gevaert K., Vandenabeele P. (2009) Caspase substrates. Easily caught in deep waters? Trends Biotechnol. 27, 680–688 [DOI] [PubMed] [Google Scholar]
- 31. Cryns V. L., Byun Y., Rana A., Mellor H., Lustig K. D., Ghanem L., Parker P. J., Kirschner M. W., Yuan J. (1997) Specific proteolysis of the kinase protein kinase C-related kinase 2 by caspase-3 during apoptosis. Identification by a novel, small pool expression cloning strategy. J. Biol. Chem. 272, 29449–29453 [DOI] [PubMed] [Google Scholar]
- 32. Kamada S., Kusano H., Fujita H., Ohtsu M., Koya R. C., Kuzumaki N., Tsujimoto Y. (1998) A cloning method for caspase substrates that uses the yeast two-hybrid system. Cloning of the antiapoptotic gene gelsolin. Proc. Natl. Acad. Sci. U.S.A. 95, 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Agard N. J., Wells J. A. (2009) Methods for the proteomic identification of protease substrates. Curr. Opin. Chem. Biol. 13, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. (2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771–777 [DOI] [PubMed] [Google Scholar]
- 36. Klaiman G., Petzke T. L., Hammond J., LeBlanc A. C. (2008) Targets of Caspase-6 activity in human neurons and Alzheimer disease. Mol. Cell. Proteomics 7, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., Núñez G. (2008) Targeted Peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu D., Suenaga N., Edelmann M. J., Fridman R., Muschel R. J., Kessler B. M. (2008) Novel MMP-9 substrates in cancer cells revealed by a label-free quantitative proteomics approach. Mol. Cell. Proteomics 7, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wood D. E., Newcomb E. W. (1999) Caspase-dependent activation of calpain during drug-induced apoptosis. J. Biol. Chem. 274, 8309–8315 [DOI] [PubMed] [Google Scholar]
- 40. Molinari M., Anagli J., Carafoli E. (1995) PEST sequences do not influence substrate susceptibility to calpain proteolysis. J. Biol. Chem. 270, 2032–2035 [DOI] [PubMed] [Google Scholar]
- 41. Ray S. K., Samantaray S., Smith J. A., Matzelle D. D., Das A., Banik N. L. (2011) Inhibition of cysteine proteases in acute and chronic spinal cord injury. Neurotherapeutics 8, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Samantaray S., Ray S. K., Banik N. L. (2008) Calpain as a potential therapeutic target in Parkinson's disease. CNS Neurol. Disord. Drug Targets 7, 305–312 [DOI] [PubMed] [Google Scholar]
- 43. Zatz M., Starling A. (2005) Calpains and disease. New Engl. J. Med. 352, 2413–2423 [DOI] [PubMed] [Google Scholar]
- 44. Tseng H. C., Zhou Y., Shen Y., Tsai L. H. (2002) A survey of Cdk5 activator p35 and p25 levels in Alzheimer's disease brains. FEBS Lett. 523, 58–62 [DOI] [PubMed] [Google Scholar]
- 45. Kusakawa G., Saito T., Onuki R., Ishiguro K., Kishimoto T., Hisanaga S. (2000) Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J. Biol. Chem. 275, 17166–17172 [DOI] [PubMed] [Google Scholar]
- 46. Smith P. D., Mount M. P., Shree R., Callaghan S., Slack R. S., Anisman H., Vincent I., Wang X., Mao Z., Park D. S. (2006) Calpain-regulated p35/cdk5 plays a central role in dopaminergic neuron death through modulation of the transcription factor myocyte enhancer factor 2. J. Neurosci. 26, 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hossain M. I., Roulston C. L., Kamaruddin M. A., Chu P. W., Ng D. C., Dusting G. J., Bjorge J. D., Williamson N. A., Fujita D. J., Cheung S. N., Chan T. O., Hill A. F., Cheng H. C. (2013) A truncated fragment of Src protein kinase generated by calpain-mediated cleavage is a mediator of neuronal death in excitotoxicity. J. Biol. Chem. 288, 9696–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tisa L. S., Rosen B. P. (1990) Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. J. Biol. Chem. 265, 190–194 [PubMed] [Google Scholar]
- 49. Kao G., Nordenson C., Still M., Rönnlund A., Tuck S., Naredi P. (2007) ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell 128, 577–587 [DOI] [PubMed] [Google Scholar]
- 50. Sahlender D. A., Roberts R. C., Arden S. D., Spudich G., Taylor M. J., Luzio J. P., Kendrick-Jones J., Buss F. (2005) Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. J. Cell Biol. 169, 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park B. C., Shen X., Samaraweera M., Yue B. Y. (2006) Studies of optineurin, a glaucoma gene. Golgi fragmentation and cell death from overexpression of wild-type and mutant optineurin in two ocular cell types. Am. J. Pathol. 169, 1976–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu G., Wu C. J., Zhao Y., Ashwell J. D. (2007) Optineurin negatively regulates TNF α-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Curr. Biol. 17, 1438–1443 [DOI] [PubMed] [Google Scholar]
- 53. Fujita Y., Ohama E., Takatama M., Al-Sarraj S., Okamoto K. (2006) Fragmentation of Golgi apparatus of nigral neurons with α-synuclein-positive inclusions in patients with Parkinson's disease. Acta Neuropathol. 112, 261–265 [DOI] [PubMed] [Google Scholar]
- 54. De Marco N., Buono M., Troise F., Diez-Roux G. (2006) Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J. Biol. Chem. 281, 16147–16156 [DOI] [PubMed] [Google Scholar]
- 55. Troy C. M., Muma N. A., Greene L. A., Price D. L., Shelanski M. L. (1990) Regulation of peripherin and neurofilament expression in regenerating rat motor neuron. Brain Res. 529, 232–238 [DOI] [PubMed] [Google Scholar]
- 56. Kedzierski W., Nusinowitz S., Birch D., Clarke G., McInnes R. R., Bok D., Travis G. H. (2001) Deficiency of rds/peripherin causes photoreceptor death in mouse models of digenic and dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 98, 7718–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beaulieu J. M., Nguyen M. D., Julien J. P. (1999) Late onset death of motor neurons in mice overexpressing wild-type peripherin. J. Cell Biol. 147, 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]