Background: Aldo-keto reductase 1B10 (AKR1B10) protein is a new tumor biomarker in humans.
Results: Heat shock protein 90α (HSP90α) is a chaperone molecule that mediates transportation to lysosomes and secretion of AKR1B10. Helix 10 of AKR1B10 protein mediates its interaction with HSP90α.
Conclusion: HSP90α mediates AKR1B10 secretion through binding to its helix 10 domain.
Significance: This finding is significant in exploiting the use of AKR1B10 in cancer clinics.
Keywords: Breast Cancer, HSP90, Lysosomes, Protein Secretion, Tumor Marker, Chaperone
Abstract
Aldo-keto reductase 1B10 (AKR1B10) protein is a new tumor biomarker in humans. Our previous studies have shown that AKR1B10 is secreted through a lysosome-mediated nonclassical pathway, leading to an increase in the serum of breast cancer patients. This study illuminates the regulatory mechanism of AKR1B10 secretion. The cytosolic AKR1B10 associates with and is translocated to lysosomes by heat shock protein 90α (HSP90α), a chaperone molecule. Ectopic expression of HSP90α significantly increased the secretion of endogenous AKR1B10 and exogenous GFP-AKR1B10 fusion protein when cotransfected. Geldanamycin, a HSP90α inhibitor, dissociated AKR1B10-HSP90α complexes and significantly reduced AKR1B10 secretion in a dose-dependent manner. We characterized the functional domain in AKR1B10 and found that helix 10 (amino acids 233–240), located at the C terminus, regulates AKR1B10 secretion. Targeted point mutations recognized that amino acids Lys-233, Glu-236, and Lys-240 in helix 10 mediate the interaction of AKR1B10 with HSP90α. Together, our data suggest that HSP90α mediates AKR1B10 secretion through binding to its helix 10 domain. This finding is significant in exploiting the use of AKR1B10 in cancer clinics.
Introduction
Aldo-keto reductase 1B10 (AKR1B10), also named aldose reductase-like 1 (ARL-1), is a new serum marker and potential therapeutic target of breast cancer (1). This protein is primarily expressed in the human colon and small intestine but overexpressed in breast cancer, hepatocellular carcinoma, non-small cell lung carcinoma, and cervical and endometrial cancers (1–4). AKR1B10 may be implicated in cancer development and progression by activating procarcinogen polycyclic aromatic hydrocarbon in cigarette smoke and the environment (5), regulating cellular retinoic acid levels (6–8), mediating fatty acid synthesis (9, 10), and detoxifying cytotoxic carbonyl compounds (11–17) and therapeutic drugs (18, 19). Recent studies have demonstrated that AKR1B10 expressed in the intestinal epithelium and cultured cancer cells is secreted through a lysosome-mediated nonclassical protein secretion pathway (20), indicating its detoxicating and/or paracrine role locally in the intestine and in distant organs. This study unraveled the regulatory mechanism of AKR1B10 secretion and characterized the functional domain of AKR1B10 that mediates its secretion.
Nonclassical protein export is characterized by the lack of a conventional signal peptide at the N terminus, but a functional domain is often recognized. This functional domain is involved in the interaction with membranes or chaperone proteins for transportation and secretion. For instance, antennapedia is a small peptide secreted by a nonclassical protein export in which a 16-amino acid peptide (the third helix) translocates it through the biological membrane (21), and the phosphatidylserine-binding domain of FGF-1 mediates the protein-phospholipid interaction for transport of FGF-1 through the plasma membrane (22, 23). Similarly, a functional domain of AKR1B10 is also characterized in this study, mediating its secretion.
Proteins transport into organelles, such as mitochondria, Golgi bodies, and, lysosomes, by a leading signal peptide during and after translation. However, cytosolic proteins without a signaling peptide need chaperone molecules, such as heat shock protein 90α (HSP90α), heat shock protein70 (HSP70), and cyclophilin A, for transportation (24–26). HSP90α is a molecular chaperone that aids in the proper folding, maturation, and intracellular trafficking of proteins (25, 27). Proteins that are regulated by HSP90α include important mediators of signal transduction, cell cycle controllers, and pathogenic factors involved in the development and progression of cancer. Inside the cell, HSP90α forms complexes with client proteins, transports them across cellular compartments, and binds them to the plasma membranes of organelles, such as lysosomes (24, 28). In addition to being a cytosolic chaperone, HSP90α is also secretory (29). To date, HSP90α is found to be secreted by keratinocytes, non-small cell lung cancer cells (CL1–5), breast cancer cells (MCF-7), and colon cancer cells (HCT-8) (30, 31). In this study, we found that, as a molecular chaperone, HSP90α associates with AKR1B10, transports it to lysosomes, and is secreted jointly with it. The α-helix 10 of AKR1B10 acts as a functional domain to mediate these events. This study provided critical information for understanding the secretory mechanisms of AKR1B10 and for developing targeted strategies to modulate its association and secretion.
MATERIALS AND METHODS
Cell Cultures
HCT-8 and 293T cells (ATCC) were maintained in RPMI 1640 medium or DMEM supplemented with 10% FBS, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Fisher, FL) at 37 °C and 5% CO2.
Expression Vectors and Transfection
pcDNA3.1/FLAG/HSP90α was provided by Dr. Johannes Buchner (Technische Universität München, Department Chemie, LS Biotechnologie, München, Germany) (32). The EGFP-AKR1B10 vector has been produced previously (14). AKR1B10 peptide expression vectors were produced as follows. pQE80L-AKR1B10 (2) was digested by BamHI plus HindIII, EcoRV, BsaI, or BglI to release AKR1B10 cDNA encoding full-length or aa4 1–39, aa 1–83, aa 1–142, and aa 1–231. These fragments were subcloned into a CMV/GST-tagged vector at the BamHI and NotI sites after the NotI site was filled in by Klenow DNA polymerase. The C-terminal encoding region of AKR1B10 was subcloned by PCR, and EGFP-tagged AKR1B10 mutants at K233A, E236A, or K240A (single or combinations) were generated by PCR-based target mutagenesis. All constructs were confirmed by DNA sequencing. The primers for PCR are summarized in supplemental Table 1. Transfections were carried out as described previously (14).
Sandwich ELISA
A sandwich ELISA was used to measure AKR1B10 in medium, as described previously (20).
Lysosome Isolation and Proteinase K Protection
Lysosome isolation and proteinase protection were carried out as described previously, and cathepsin D was assessed as an indicator of lysosomes (20).
Western Blot Analysis
Proteins were separated on 8–12% SDS-PAGE and blotted onto nitrocellulose membranes at 260 mA for 150 min using a Mini-Protean II transfer apparatus (Bio-Rad). Anti-AKR1B10 (generated in our laboratory), anti-cathepsin D, and anti-HSP90α (Cell Signaling Technology), or anti-β-actin (Sigma-Aldrich Inc.) antibodies were probed and exposed as described previously (10).
Coimmunoprecipitation
Cells were lysed in IP buffer (150 mm NaCl, 50 mm Tris-HCl (pH 7.2), 2 mm EDTA, 0.2% Nonidet P-40, 10% glycerol). Soluble proteins (200 μg) were incubated at 4 °C overnight with 5 μg of the indicated antibodies, followed by incubation with 20 μl of slurry-Sepharose protein G beads at 4 °C for 1 h with gentle shaking. Beads were collected by brief centrifugation and washed five times with IP buffer. Beads were resuspended in 1× SDS loading buffer and heated at 95 °C for 5 min. Supernatants were subjected to Western blot analysis.
Fluorescent Colocalization
HCT-8 cells on coverslips were fixed for 15 min in methanol at −20 °C and incubated with a mouse monoclonal antibody against HSP90α (Abcam) and a rabbit antibody against AKR1B10, followed by a FITC-conjugated goat anti-mouse and a Rhodamine-conjugated donkey anti-rabbit antibody (Sigma-Aldrich), respectively. Images were taken with an Olympus confocal microscope (Olympus, Japan).
HSP90α Protein Preparation and Pull-down Assays
N-terminal His-tagged human HSP90α was prepared using a prokaryotic system, a gift from Dr. David Agard (Howard Hughes Medical Institute and Department of Biochemistry and Biophysics, University of California) (33). For pull-down assays, cells were lysed in IP buffer, and HSP90α protein (5 μg) was added into 500 μg of soluble proteins and incubated with gentle shaking on ice for 1 h. nickel-nitrilotriacetic acid-agarose beads (20 μl) were equilibrated with IP buffer and added to the HSP90α-cell lysate mixture on ice for 1 h with gentle shaking. Nickel-nitrilotriacetic acid-agarose beads were collected by brief centrifugation and washed five times with IP buffer. Proteins were released by heating at 95 °C in 1× SDS-PAGE loading buffer for 5 min and subjected to Western blot analysis.
Statistical Analysis
Statistical analysis was performed using Student's t test or Chi square tests, as appropriate, with the INSTAT statistical analysis package (GraphPad Software). Data were considered statistically significant at p < 0.05.
RESULTS
AKR1B10 Associates with HSP90α
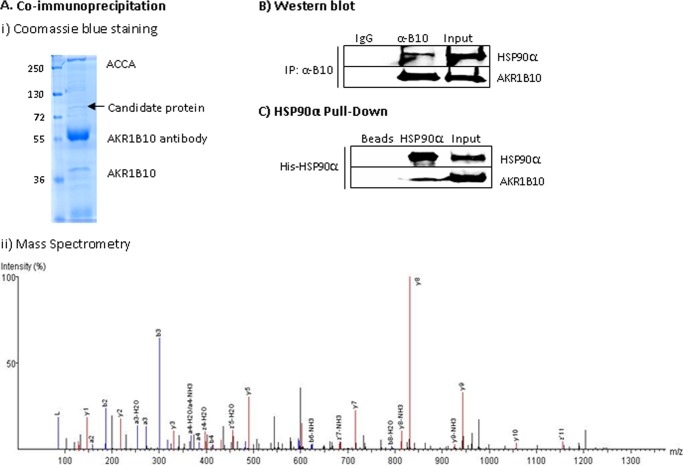
Chaperone molecules such as HSP90α are often involved in the transportation of proteins from the cytosol to lysosomes (24). AKR1B10 is a cytosolic protein but is secreted through lysosomes (20). To understand the mechanism of AKR1B10 translocation to lysosomes, we carried out coimmunoprecipitation with a specific anti-AKR1B10 antibody and protein mass spectrometry analyses of the coprecipitates. As shown in Fig. 1A, i, acetyl-CoA carboxylase α was coimmunoprecipitated with AKR1B10, as shown previously (10). Additionally, an ∼90-kDa and an ∼130-kDa protein band appeared in the immunoprecipitates. Protein mass spectrometry recognized that the 90-kDa protein was a chaperone molecule, HSP90α (Fig. 1A, ii), which was further confirmed by Western blot analysis (Fig. 1B). With the addition of 6× histidine-tagged HSP90α protein into the cell lysate from HCT8 cells, the soluble AKR1B10 was successfully pulled down (Fig. 1C). Furthermore, an intracellular fluorescent study showed that cytosolic AKR1B10 was colocalized with HSP90α in the cytosol (Fig. 2). These data suggest that AKR1B10 associates with HSP90α inside the cells.
FIGURE 1.
AKR1B10 association with HSP90 α. A, coimmunoprecipitation. AKR1B10 protein complexes in HCT-8 cells were immunoprecipitated with a specific anti-AKR1B10 antibody and separated in 12% SDS-PAGE. i, Coomassie Blue staining showing a candidate protein at ∼90 kDa (arrow). The numbers on the left show the molecular weight (in kilodalton) of protein markers. ACCA, acetyl-CoA carboxylase-α. ii, matrix-assisted laser desorption ionization mass spectrometry of the candidate protein. The image shows the identified peptide that is identical to residues 101–112 (ADLINNLGTIAK) of HSP90α. B, Western blot analysis. Immunoprecipitated AKR1B10 complexes were subjected to Western blot analysis as described under “Materials and Methods” to confirm the presence of HSP90α. Lane 1, rabbit IgG negative control. Lane 2, immunoprecipitates with anti-AKR1B10 antibody (α-B10). Lane 3, 20% input. C, HSP90α pull-down carried out with purified HSP90α protein as described under “Materials and Methods.”
FIGURE 2.
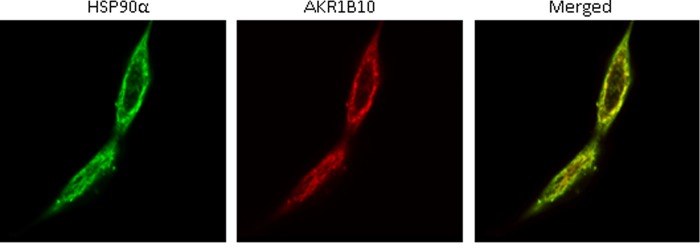
Fluorescent colocalization. Fluorescent immunocytochemistry and laser confocal imaging were carried out as described under “Materials and Methods.” The secondary antibodies against the primary antibodies to HSP90α and AKR1B10 were labeled with green FITC and red Rhodamine, respectively, and the merged yellow image indicates the colocalization of AKR1B10 and HSP90α in cytosol.
HSP90α Modulates AKR1B10 Secretion
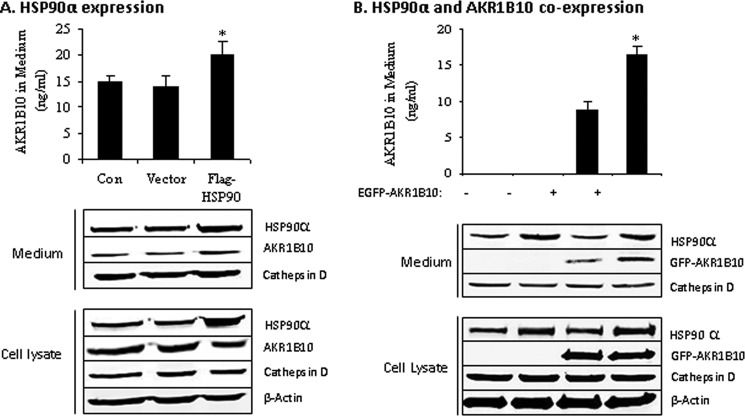
To understand the function of AKR1B10-HSP90α association, we assessed the effect of HSP90α expression on the secretion of AKR1B10. As shown in Fig. 3A, ectopic expression of HSP90α in HCT-8 cells significantly stimulated endogenous AKR1B10 secretion. We further confirmed this finding by coexpression of EGFP-AKR1B10 and HSP90α in 293T cells that do not express AKR1B10 (10). Results showed that HSP90α also stimulated the secretion of the EGFP-AKR1B10 fusion protein ectopically expressed in 293T cells when cotransfected (Fig. 3B). In contrast, geldanamycin (GA), an inhibitor of HSP90α (34), inhibited AKR1B10 secretion in a dose-dependent manner (Fig. 4A). Consistently, coimmunoprecipitation with an AKR1B10- or HSP90α-specific antibody showed that the HSP90α inhibitor GA dissociated AKR1B10, leading to a significant reduction of AKRB10 in coimmunoprecipitates (Fig. 4B). These data suggest that the chaperone protein HSP90α mediates AKR1B10 secretion. It was noted that HSP90α overexpression in HCT-8 cells had only a modest effect on AKR1B10 secretion, which may be due to the high expression of endogenous HSP90α.
FIGURE 3.
Stimulation of AKR1B10 secretion by HSP90α. HSP90α alone or in combination with EGFP-AKR1B10 was delivered into cells, and AKR1B10 in medium was measured using ELISA as described under “Materials and Methods.” A, endogenous AKR1B10 secretion in HCT8 cells with ectopic expression of HSP90α. Con, control. B, secretion of exogenous EGFP-AKR1B10 fusion protein in 293T cells, alone or coexpressed with HSP90α. Data represent the mean ± S.D. from three independent assays. *, p < 0.01 compared with EFGP-AKR1B10 alone.
FIGURE 4.
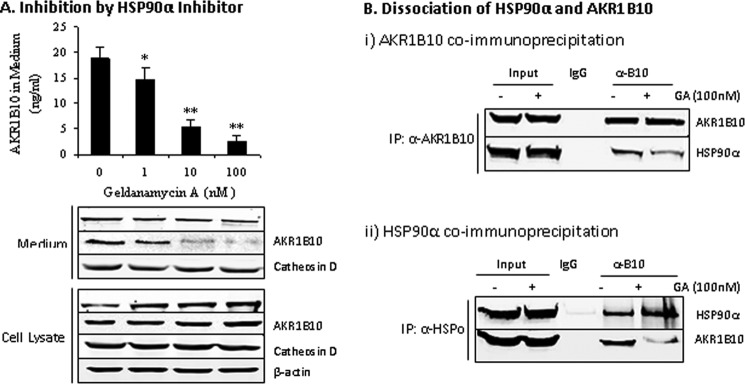
Inhibition of AKR1B10 secretion by an HSP90α inhibitor. HCT-8 cells (2.5 × 105) were incubated for 12 h with the HSP90α inhibitor GA at the indicated concentrations and then fed with fresh serum-free medium for 30 min. The medium and cells were collected for sandwich ELISA, Western blot analysis, or immunoprecipitation. A, inhibition of HSP90α secretion. *, p < 0.05; **, p < 0.01 compared with vehicle control. B, dissociation of the HSP90 and AKR1B10 complexes by GA. α-B10, anti-AKR1B10 antibody.
HSP90α Translocates AKR1B10 to Secretory Lysosomes
AKR1B10 is secreted by lysosomal exocytosis (20). HSP90α associates with AKR1B10 and modulates its secretion. Therefore, HSP90α may participate in the translocation of AKR1B10 to lysosomes. We isolated lysosomes, and conducted a protease-protection assay. As shown in Fig. 5A, together with AKR1B10, HSP90α was present in lysosomes and partially protected by lysosomes from proteinase degradation. HSP90α inhibitor GA did not affect the translocation of HSP90α to lysosomes, but AKR1B10 in the lysosomes was significantly reduced (Fig. 5B), indicating that GA dissociates HSP90-AKR1B10 complexes, leading to a decrease of AKR1B10 in lysosomes. It is noteworthy that, when lysosomes were exposed to proteinase K (0.0125 mg/ml) to remove outside proteins, HSP90α leftover was decreased markedly, whereas AKR1B10 was not altered significantly. This result suggests that some HSP90α releases the associated AKR1B10 and stays outside of the lysosomes. This finding is consistent with previous reports that a chaperone protein may relocate and release the client protein to membrane transporters and that the chaperone is then recycled (35).
FIGURE 5.
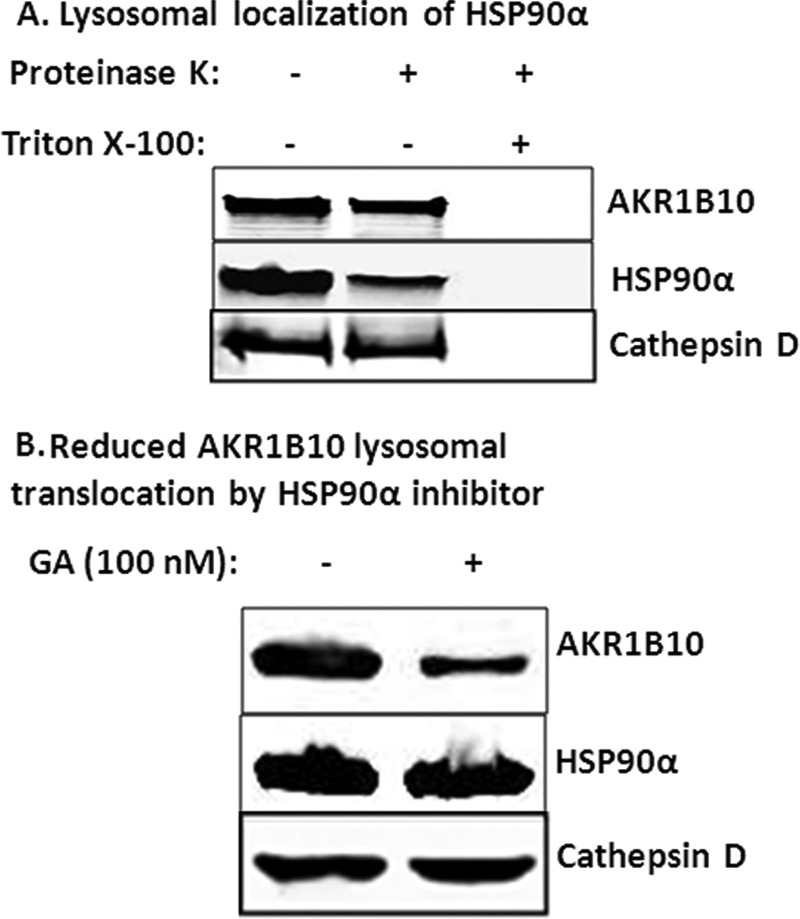
Lysosomal localization of HSP90α and AKR1B10. A, proteinase K protection. HCT-8 cells (5 × 107) were broken down using a Dounce homogenizer, and lysosomes were isolated for proteinase K protection assays as described under “Materials and Methods.” Triton X-100 (0.5%) was added in a group to destroy biomembranes. B, HSP90α inhibition. HCT-8 cells (2.5 × 107) were incubated for 12 h with 100 nm HSP90α inhibitor GA. Lysosomes were isolated, and AKR1B10 and HSP90α in the lysosomes were detected by Western blot analysis.
Helix 10 (aa 233–240) of AKR1B10 Mediates Its Association with HSP90α and Secretion
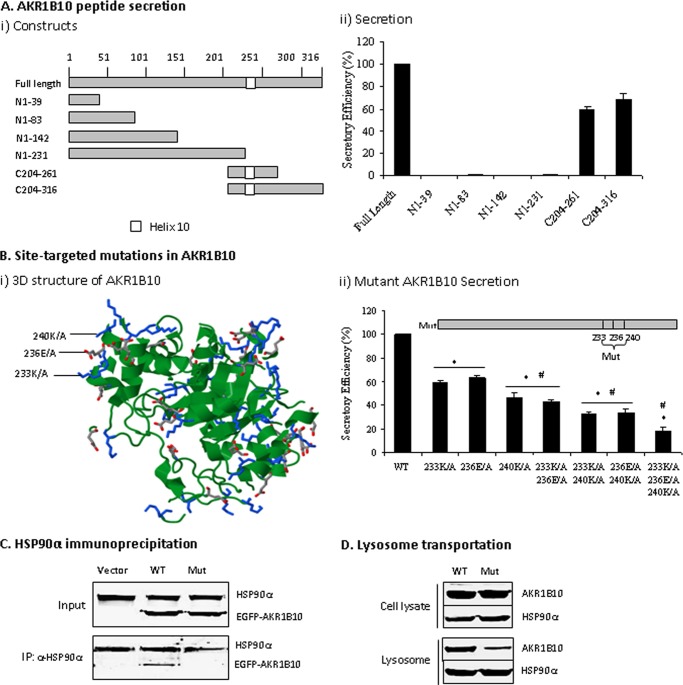
An N-terminal signal peptide is lacking in proteins secreted through a nonclassical secretion pathway, but a functional domain is often identified (22, 23). To identify the domain that mediates AKR1B10 secretion, we first tested the secretory activity of different N- and C-terminal peptides, with full-length AKR1B10 (316 aa) as a control. As shown in Fig. 6A, GST-tagged expression vectors of N/C-terminal peptides were constructed, delivered into 293T cells, and tested for secretory activity. Results showed that N-terminal peptides (N1–39 to N1–231) had no secretory activity, whereas the C-terminal peptides (C204–261 and C204–316) were secreted with an efficiency of 74.2 ± 3.0% and 81.5 ± 5.5% of full-length AKR1B10. These data suggest that the functional domain of AKR1B10 may be located in the C-terminal peptide C204–263.
FIGURE 6.
Functional domain mediating AKR1B10 secretion. A, AKR1B10 peptide secretion activity. AKR1B10 full-length (316 aa) and peptide segments, as indicated, were subcloned into a GST-tagged vector (i) and transfected into 293T cells for a secretory activity assay (ii) as described under “Materials and Methods.” Peptide secretory activity was expressed as a percentage of full-length AKR1B10. Data represent the mean ± S.D. from three independent assays. B, site-targeted mutations. i, stereo view of targeted mutation sites in AKR1B10. The image was produced using the Protein Data Bank (see “Materials and Methods”) and shows the position of three targeted mutation sites (Lys-233, Glu-236, and Lys-240) in the AKR1B10 crystal structure. ii, mutational AKR1B10 secretion. Single point mutations (Mut) or combinations, as indicated, were cloned into the CMV/GST-tag vector and transfected into 293T cells for a secretory activity assay as described under “Materials and Methods.” The data indicate the percentage of wild-type AKR1B10. *, p < 0.01 compared with wild-type AKR1B10; #, p < 0.05 compared with the adjacent group on the left. C, HSP90α immunoprecipitation. Wild-type and mutant AKR1B10 (fused with EGFP) with three point mutations were transfected into 293T cells. After 36 h, immunoprecipitation with HSP90α antibody was conducted, and precipitates were subjected to Western blot analysis as described under “Materials and Methods.” D, mutant AKR1B10 in lysosomes. The 293T cells transfected with wild-type or mutant AKR1B10 at three amino acid points were broken by a Dounce homogenizer; lysosomes were isolated from the cells, and HSP90α and AKR1B10 proteins were detected by Western blot analysis.
The predicted crystal structure of AKR1B10 in the Protein Data Bank shows that helix 10 (aa 233–240) is located in the secretory peptide C204–261 and stretches out of the α-helix/β-sheet catalytic pocket. This structural feature suggests that helix 10 is most likely the domain involved in the association with HSP90α and secretion of AKR1B10. In helix 10, the amino acids Lys-233, Glu-236, and Lys-240 are reaching out (Fig. 6B, i) and may be the key residues binding to HSP90α. Therefore, a panel of targeted mutants (single or different combinations) were constructed and tested for secretory activity (Fig. 6B, ii). The results showed that the single mutation of K233A, E236A, or K240A decreased the secretory efficiency of AKR1B10 protein to 59.2 ± 2.1%, 63.3 ± 1.1%, and 46.9 ± 3.1%, respectively. Combinations of any two-site mutations further reduced the secretory activity to 43.15 ± 1.5% for K233A plus E236A, 33.30 ± 1% for K233A plus K240A, and 33.52 ± 3.1% for E236A plus K240A. All three-site mutations further lowered AKR1B10 secretory efficiency to 18.2 ± 3.2%. Furthermore, the AKR1B10 mutant with targeted mutations at all three amino acids lost its capability of binding to HSP90α (Fig. 6C), and, thus, the translocation to lysosomes was decreased (D). These data suggest that helix 10 (aa 233–240) acts as a functional domain for the association with HSP90α and secretion of AKR1B10 and that the amino acids Lys-233, Glu-236, and Lys-240 in this helix are the key residues for binding to HSP90α.
DISCUSSION
With more than 100 members, AKRs represent a super protein family. AKR1B10 is one of the most important proteins in the AKR family, profoundly involved in human tumors. Very recently, we reported that AKR1B10 is a secretory protein and potential serum marker of breast cancer (1, 20). AKR1B10 is also considered as oncogenic, promoting tumor growth and metastasis, and, thus, may be a new potential therapeutic target (1). This study recognized HSP90α as a chaperone protein that transports AKR1B10 to lysosomes and characterized helix 10 (aa 233–240) of AKR1B10 as the functional domain that mediates its association with HSP90α and secretion. These results are valuable for the development of AKR1B10-targeted cancer management.
In the lysosome-mediated protein secretion pathway, a target protein needs to be translocated to lysosomal compartments. HSPs are multifunctional molecular chaperones that are implicated in protein folding, assembly, activation, and transportation (24, 27, 28, 36). This study stamped HSP90α as a chaperone protein of AKR1B10 for transportation to lysosomes. It is of interest to test whether HSP90β also interacts with AKR1B10.
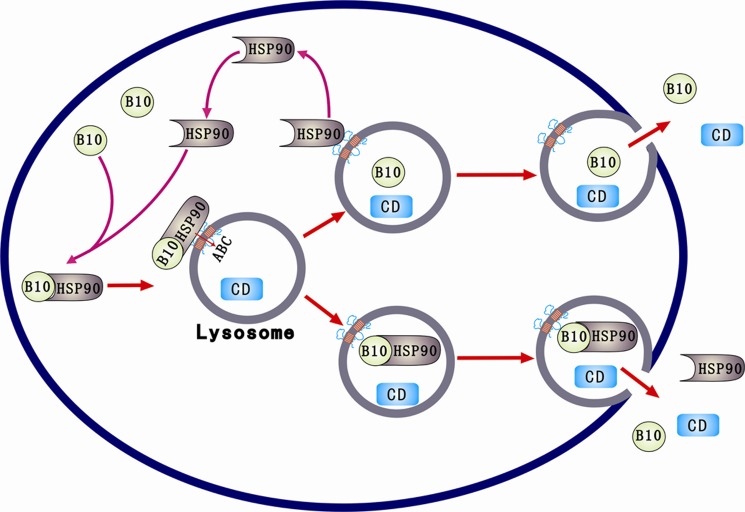
There are two types of lysosomes, conventional and secretory lysosomes (37, 38). HSP90α often transports misfolded or denatured proteins into conventional lysosomes for clearance. AKR1B10 secreted into medium has the same molecular weight as in cytosol and retains enzymatic activity (20), indicating that AKR1B10 is transported into secretory lysosomes by HSP90α for secretion. Furthermore, the proteinase protection and semiquantitative Western blot analysis showed that lysosomal HSP90α protein was largely reduced by proteinase treatment, whereas AKR1B10 was altered slightly (Fig. 5A). This suggests that some HSP90α protein releases AKR1B10, stays outside of the lysosomes, and, thus, is removed by proteinase. Taken together, we propose a functioning model of HSP90α (Fig. 7). HSP90α associates with AKR1B10 through its helix 10 and transports AKR1B10 to secretory lysosomes, where AKR1B10 is released from HSP90α and imported into the lumen of lysosomes by ATP-binding cassette (ABC) transporter transporters (20). HSP90α is recycled for new protein transportation. HSP90α may also import into lysosomes and is secreted together with AKR1B10.
FIGURE 7.
Hypothetical model of AKR1B10 secretion. The entire secretory process of AKR1B10 includes trafficking of AKR1B10 to the lysosomal membrane, import into the lysosomal lumen, and lysosomal exocytosis. A portion of HSP90 enters into lysosomes and is secreted with AKR1B10, but the other remains outside of lysosomes and is recycled. B10, AKR1B10; CD, cathepsin D.
Nonclassical secretary proteins often contain a functional domain to mediate their secretion. This study identified and characterized helix 10 (aa 233–240) of AKR1B10 as the functional domain that mediates its interaction with HSP90α and secretion (Fig. 6). Using site-targeted mutagenesis, we further defined the amino acids Lys-233, Glu-236, and Lys-240 as the key residues for AKR1B10 association with HSP90α. It is noteworthy that the AKR1B10 mutant with all three amino acid mutations retained 18.2 ± 3.2% secretory activity. This may be due to the contribution of the hydrophobic helix itself to the interaction with HSP90α. The characterization of this interaction domain is of significance in the development of small chemical inhibitors.
In summary, this study characterized HSP90α as a chaperone molecule that associates with AKR1B10 and mediates its translocation to secretory lysosomes and secretions. This study also characterized the functional domain and key amino acid residues that mediate AKR1B10 interaction with HSP90α protein. AKR1B10 is expressed in several human tumors and may be a new serum marker (1–3). In breast cancer, AKR1B10 promotes tumor growth and lymph node metastasis, leading to worse disease-related survival (1). Therefore, AKR1B10 may be a potential prognostic marker and therapeutic target of breast cancer. Characterization of its functional domain and definition of its chaperone protein (HSP90α) would lay a critical basis for validating the potential use of AKR1B10 in cancer clinical settings.
This work was supported by Department of Defense Breast Cancer Research Program Grant BC083555 (to D. C.), by National Natural Science Foundation of China Grants 81372825 (to D. L.) and 81272918 (to D. C.), by China Hunan Provincial Science and Technology Department Grant 2012SK3065 (to D. L.), by Health Department of China Hunan Province Grant B2012-157 (to D. L.), and by China Hunan Provincial Education Department Grant 13C882 (to D. L.).

This article contains supplemental Table 1.
- aa
- amino acids
- IP
- immunoprecipitation
- GA
- geldanamycin
- AKR
- aldo-keto reductase
- HSP
- heat shock protein.
REFERENCES
- 1. Ma J., Luo D. X., Huang C., Shen Y., Bu Y., Markwell S., Gao J., Liu J., Zu X., Cao Z., Gao Z., Lu F., Liao D. F., Cao D. (2012) AKR1B10 overexpression in breast cancer. Association with tumor size, lymph node metastasis and patient survival and its potential as a novel serum marker. Int. J. Cancer 131, E862–871 [DOI] [PubMed] [Google Scholar]
- 2. Cao D., Fan S. T., Chung S. S. (1998) Identification and characterization of a novel human aldose reductase-like gene. J. Biol. Chem. 273, 11429–11435 [DOI] [PubMed] [Google Scholar]
- 3. Fukumoto S., Yamauchi N., Moriguchi H., Hippo Y., Watanabe A., Shibahara J., Taniguchi H., Ishikawa S., Ito H., Yamamoto S., Iwanari H., Hironaka M., Ishikawa Y., Niki T., Sohara Y., Kodama T., Nishimura M., Fukayama M., Dosaka-Akita H., Aburatani H. (2005) Overexpression of the aldo-keto reductase family protein AKR1B10 is highly correlated with smokers' non-small cell lung carcinomas. Clin. Cancer Res. 11, 1776–1785 [DOI] [PubMed] [Google Scholar]
- 4. Yoshitake H., Takahashi M., Ishikawa H., Nojima M., Iwanari H., Watanabe A., Aburatani H., Yoshida K., Ishi K., Takamori K., Ogawa H., Hamakubo T., Kodama T., Araki Y. (2007) Aldo-keto reductase family 1, member B10 in uterine carcinomas. A potential risk factor of recurrence after surgical therapy in cervical cancer. Int. J. Gynecol. Cancer 17, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 5. Quinn A. M., Harvey R. G., Penning T. M. (2008) Oxidation of PAH trans-dihydrodiols by human aldo-keto reductase AKR1B10. Chem. Res. Toxicol. 21, 2207–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruiz F. X., Gallego O., Ardèvol A., Moro A., Domínguez M., Alvarez S., Alvarez R., de Lera A. R., Rovira C., Fita I., Parés X., Farrés J. (2009) Aldo-keto reductases from the AKR1B subfamily. Retinoid specificity and control of cellular retinoic acid levels. Chem. Biol. Interact. 178, 171–177 [DOI] [PubMed] [Google Scholar]
- 7. Gallego O., Ruiz F. X., Ardèvol A., Domínguez M., Alvarez R., de Lera A. R., Rovira C., Farrés J., Fita I., Parés X. (2007) Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc. Natl. Acad. Sci. U.S.A. 104, 20764–20769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crosas B., Hyndman D. J., Gallego O., Martras S., Parés X., Flynn T. G., Farrés J. (2003) Human aldose reductase and human small intestine aldose reductase are efficient retinal reductases. Consequences for retinoid metabolism. Biochem. J. 373, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C., Yan R., Luo D., Watabe K., Liao D. F., Cao D. (2009) Aldo-keto reductase family 1 member B10 promotes cell survival by regulating lipid synthesis and eliminating carbonyls. J. Biol. Chem. 284, 26742–26748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma J., Yan R., Zu X., Cheng J. M., Rao K., Liao D. F., Cao D. (2008) Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-α in breast cancer cells. J. Biol. Chem. 283, 3418–3423 [DOI] [PubMed] [Google Scholar]
- 11. Yan R., Zu X., Ma J., Liu Z., Adeyanju M., Cao D. (2007) Aldo-keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells. Implication for cancer intervention. Int. J. Cancer 121, 2301–2306 [DOI] [PubMed] [Google Scholar]
- 12. Zhong L., Liu Z., Yan R., Johnson S., Zhao Y., Fang X., Cao D. (2009) Aldo-keto reductase family 1 B10 protein detoxifies dietary and lipid-derived α, β-unsaturated carbonyls at physiological levels. Biochem. Biophys. Res. Commun. 387, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balendiran G. K., Martin H. J., El-Hawari Y., Maser E. (2009) Cancer biomarker AKR1B10 and carbonyl metabolism. Chem. Biol. Interact. 178, 134–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zu X., Yan R., Robbins S., Krishack P. A., Liao D. F., Cao D. (2007) Reduced 293T cell susceptibility to acrolein due to aldose reductase-like-1 protein expression. Toxicol. Sci. 97, 562–568 [DOI] [PubMed] [Google Scholar]
- 15. Shen Y., Zhong L., Markwell S., Cao D. (2010) Thiol-disulfide exchanges modulate aldo-keto reductase family 1 member B10 activity and sensitivity to inhibitors. Biochimie 92, 530–537 [DOI] [PubMed] [Google Scholar]
- 16. Shen Y., Zhong L., Johnson S., Cao D. (2011) Human aldo-keto reductases 1B1 and 1B10. A comparative study on their enzyme activity toward electrophilic carbonyl compounds. Chem. Biol. Interact. 191, 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin H. J., Maser E. (2009) Role of human aldo-keto-reductase AKR1B10 in the protection against toxic aldehydes. Chem. Biol. Interact. 178, 145–150 [DOI] [PubMed] [Google Scholar]
- 18. Martin H. J., Breyer-Pfaff U., Wsol V., Venz S., Block S., Maser E. (2006) Purification and characterization of akr1b10 from human liver. Role in carbonyl reduction of xenobiotics. Drug Metab. Dispos. 34, 464–470 [DOI] [PubMed] [Google Scholar]
- 19. Zhong L., Shen H., Huang C., Jing H., Cao D. (2011) AKR1B10 induces cell resistance to daunorubicin and idarubicin by reducing C13 ketonic group. Toxicol. Appl. Pharmacol. 255, 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo D. X., Huang M. C., Ma J., Gao Z., Liao D. F., Cao D. (2011) Aldo-keto reductase family 1, member B10 is secreted through a lysosome-mediated non-classical pathway. Biochem. J. 438, 71–80 [DOI] [PubMed] [Google Scholar]
- 21. Derossi D., Joliot A. H., Chassaing G., Prochiantz A. (1994) The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 269, 10444–10450 [PubMed] [Google Scholar]
- 22. Tarantini F., Gamble S., Jackson A., Maciag T. (1995) The cysteine residue responsible for the release of fibroblast growth factor-1 residues in a domain independent of the domain for phosphatidylserine binding. J. Biol. Chem. 270, 29039–29042 [DOI] [PubMed] [Google Scholar]
- 23. Jackson A., Tarantini F., Gamble S., Friedman S., Maciag T. (1995) The release of fibroblast growth factor-1 from NIH 3T3 cells in response to temperature involves the function of cysteine residues. J. Biol. Chem. 270, 33–36 [DOI] [PubMed] [Google Scholar]
- 24. Lotz G. P., Brychzy A., Heinz S., Obermann W. M. (2008) A novel HSP90 chaperone complex regulates intracellular vesicle transport. J. Cell Sci. 121, 717–723 [DOI] [PubMed] [Google Scholar]
- 25. Agarraberes F. A., Dice J. F. (2001) A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114, 2491–2499 [DOI] [PubMed] [Google Scholar]
- 26. Zimmermann R. (1998) The role of molecular chaperones in protein transport into the mammalian endoplasmic reticulum. Biol. Chem. 379, 275–282 [PubMed] [Google Scholar]
- 27. Gething M. J., Sambrook J. (1992) Protein folding in the cell. Nature 355, 33–45 [DOI] [PubMed] [Google Scholar]
- 28. Csermely P., Schnaider T., Soti C., Prohászka Z., Nardai G. (1998) The 90-kDa molecular chaperone family. Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 79, 129–168 [DOI] [PubMed] [Google Scholar]
- 29. Tsutsumi S., Neckers L. (2007) Extracellular heat shock protein 90. A role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 98, 1536–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen J. S., Hsu Y. M., Chen C. C., Chen L. L., Lee C. C., Huang T. S. (2010) Secreted heat shock protein 90α induces colorectal cancer cell invasion through CD91/LRP-1 and NF-κB-mediated integrin αV expression. J. Biol. Chem. 285, 25458–25466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W., Li Y., Guan S., Fan J., Cheng C. F., Bright A. M., Chinn C., Chen M., Woodley D. T. (2007) Extracellular heat shock protein-90α. Linking hypoxia to skin cell motility and wound healing. EMBO J. 26, 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Retzlaff M., Stahl M., Eberl H. C., Lagleder S., Beck J., Kessler H., Buchner J. (2009) Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Rep. 10, 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krukenberg K. A., Böttcher U. M., Southworth D. R., Agard D. A. (2009) Grp94, the endoplasmic reticulum Hsp90, has a similar solution conformation to cytosolic Hsp90 in the absence of nucleotide. Protein Sci. 18, 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ochel H. J., Eichhorn K., Gademann G. (2001) Geldanamycin. The prototype of a class of antitumor drugs targeting the heat shock protein 90 family of molecular chaperones. Cell Stress Chaperones 6, 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gross M., Hessefort S. (1996) Purification and characterization of a 66-kDa protein from rabbit reticulocyte lysate which promotes the recycling of hsp 70. J. Biol. Chem. 271, 16833–16841 [DOI] [PubMed] [Google Scholar]
- 36. Young J. C., Hoogenraad N. J., Hartl F. U. (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 [DOI] [PubMed] [Google Scholar]
- 37. Blott E. J., Griffiths G. M. (2002) Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3, 122–131 [DOI] [PubMed] [Google Scholar]
- 38. Holt O. J., Gallo F., Griffiths G. M. (2006) Regulating secretory lysosomes. J Biochem. 140, 7–12 [DOI] [PubMed] [Google Scholar]