Abstract
Patients with obstructive lung diseases display abnormal circadian rhythms in lung function. We determined the mechanism whereby environmental tobacco/cigarette smoke (CS) modulates expression of the core clock gene BMAL1, through Sirtuin1 (SIRT1) deacetylase during lung inflammatory and injurious responses. Adult C57BL6/J and various mice mutant for SIRT1 and BMAL1 were exposed to both chronic (6 mo) and acute (3 and 10 d) CS, and we measured the rhythmic expression of clock genes, circadian rhythms of locomotor activity, lung function, and inflammatory and emphysematous responses in the lungs. CS exposure (100–300 mg/m3 particulates) altered clock gene expression and reduced locomotor activity by disrupting the central and peripheral clocks and increased lung inflammation, causing emphysema in mice. BMAL1 was acetylated and degraded in the lungs of mice exposed to CS and in patients with chronic obstructive pulmonary disease (COPD), compared with lungs of the nonsmoking controls, linking it mechanistically to CS-induced reduction of SIRT1. Targeted deletion of Bmal1 in lung epithelium augmented inflammation in response to CS, which was not attenuated by the selective SIRT1 activator SRT1720 (EC50=0.16 μM) in these mice. Thus, the circadian clock, specifically the enhancer BMAL1 in epithelium, plays a pivotal role, mediated by SIRT1-dependent BMAL1, in the regulation of CS-induced lung inflammatory and injurious responses.— Hwang, J.-W., Sundar, I. K., Yao, H., Sellix, M. T., Rahman, I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway.
Keywords: oxidative stress, cytokines, chronic obstructive pulmonary disease, locomotor activity, emphysema, mouse
Circadian rhythms are biological oscillations that occur with a near-24-h period and are synchronized or entrained to environmental cues, such as the day–night transition (1). In mammals, the central circadian pacemaker is localized in the suprachiasmatic nucleus (SCN) of the hypothalamus (1). In addition to the central pacemaker, peripheral tissues, including liver, heart, and lung, are autonomous circadian oscillators (2). The clock in each of these tissues plays a critical role in optimizing cellular function and responses to environmental stimuli (1, 2). The coordinated activity of these oscillators can be referred to as the circadian timing system (2). Disruption of the circadian timing system has been shown to cause cellular dysfunction and chronic diseases (3). Circadian rhythms of respiratory function have been described in both experimental animals (4, 5) and healthy human subjects (6, 7). However, the effect of environmental stress on clock function in the lung and its role in lung pathophysiology are unknown.
The molecular clock drives intrinsic daily rhythms of physiology and behavior. At the cellular level, the clock is defined as a transcriptional and translational feedback loop (TTFL) oscillator (1). The core loop consists of the CLOCK:BMAL1 heterodimer, which activates the transcription of Period (PER) and Cryptochrome (CRY). PER and CRY proteins form heterodimeric complexes that inhibit their own transcription by suppressing the activity of CLOCK:BMAL1 (1). Emerging evidence suggests that the molecular clock is intimately associated with the response to environmental stress in pathophysiology (8–11). However, the role of the circadian clock gene BMAL1 in oxidative stress–induced inflammation remains elusive.
It is well known that tobacco and cigarette smoke (CS) induces oxidative stress and consequently leads to excessive pulmonary inflammation in the pathogenesis of chronic obstructive pulmonary disease (COPD) and emphysema (12). Daily rhythms of lung function, bronchodilator responses, surfactant protein levels, steroid efficacy, mucus secretion, and chronic cough are hallmarks of obstructive lung diseases known to be clock dependent (5, 7, 12–17). Patients with COPD display rhythmic variation in respiratory symptoms, including nocturnal breathlessness, insomnia, and an early-morning decline in lung function associated with increased cough and mucus hypersecretion (18–20). These symptoms may stem from altered clock function in lung tissue, and it is reasonable to suspect that disrupted clock function can have a profound influence on lung function and lung pathology. However, it is not known whether CS directly alters clock function during the pathogenesis of COPD.
Sirtuin1 (SIRT1), an NAD+-dependent deacetylase, affects clock function by binding with CLOCK:BMAL1 complexes and deacetylating BMAL1 and PER2 proteins (21–25). CS reduces the levels and activity of SIRT1, and patients with COPD have reduced lung levels of SIRT1 (26–28). Hence, it is likely that CS-mediated reduction of SIRT1 leads to increased acetylation of circadian clock proteins, culminating in abnormal clock gene expression and proinflammatory responses in the lung. We hypothesize that environmental CS affects molecular clock function in lung tissue via the reduction of SIRT1, which in turn leads to inflammatory and injurious responses in the pathogenesis of COPD and emphysema. To address this hypothesis, we determined whether BMAL1, as a substrate of SIRT1, has a role in the regulation of CS-induced lung inflammatory and injurious responses.
MATERIALS AND METHODS
Animals
Male C57BL/6J (C57) and Bmal1-floxed mutant mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). To generate conditional deletion of Bmal1 in lung epithelial cells (CC10-Cre/Bmal1floxed/floxed, hereafter referred as BMAL1-CC10 cre), Bmal1-floxed mutant mice were crossed with mice expressing a Cre transgene driven by the CC10 promoter (obtained from Dr. Thomas Mariani, University of Rochester, Rochester, NY, USA; refs. 29, 30). Transgenic (Tg) mice overexpressing Sirt1 (SIRT1 Tg mice) were obtained from Dr. Leonard Guarente (Massachusetts Institutes of Technology, Cambridge, MA, USA) and Dr.Wei Gu (Columbia University, New York, NY, USA), and SIRT1 knockout mice (SIRT1-deficient mice) were from Dr. Michael McBurney (University of Ottawa, Ottawa, ON, Canada; refs. 30, 31). SIRT1 heterozygous knockout mice were used, because SIRT1 homozygous knockout mice have a low perinatal survival rate (30). The mice were housed under a 12:12 light:dark (L:D) cycle with lights on at 6 AM, and were fed a regular diet and water ad libitum, unless otherwise indicated. For acute (10 d) CS exposure, the mice were entrained for a minimum of 2 wk to an inverted L:D cycle (lights on from 6 PM to 6 AM), except for wheel-running experiments, before CS exposure during their active phase. For 3 d and chronic (6 mo) CS exposure, the mice were kept in a standard 12:12 L:D cycle with lights on from 6 AM to 6 PM throughout the experiment. All of the procedures described in this study were approved by the University Committee on Animal Research at the University of Rochester.
Tobacco/CS exposure
Eight-week-old mice were used for tobacco/CS exposure (27, 32, 33). We used both acute (3 and 10 d exposure, which induces lung inflammatory responses) and chronic (6 mo exposure, which causes pulmonary emphysema) mouse models of CS exposure to determine the effect and mechanism of both acute and chronic CS exposure on circadian clock function and lung inflammation. The mice were exposed to CS from a mainstream-delivering Baumgartner-JaegerCSM2082i cigarette-smoking machine (CH Technologies, Westwood, NJ, USA) for 3 d or an environmental sidestream-delivering Teague TE-10 smoking machine (Teague Enterprises, Davis, CA, USA) for 10 d and 6 mo of CS exposure in the Inhalation Facility at the University of Rochester Medical Center. For 3 d CS exposure, the mice were placed in individual compartments of a wire cage, which was then placed inside a closed plastic box connected to the smoke source. The smoke was generated from 3R4F research cigarettes containing 11.0 mg total particulate matter (TPM), 9.4 mg tar, and 0.73 mg nicotine/cigarette (University of Kentucky, Lexington, KY, USA). The TPM per cubic meter of air in the exposure chamber was monitored in real time with a MicroDust Proaerosol monitor (Casella CEL, Bedford, UK) and verified daily by gravimetric sampling (33). The smoke concentration was set at ∼300 mg/m3 TPM by adjusting the flow rate of the diluted medical air, and the level of carbon monoxide in the chamber was 350 ppm (33). The mice received two 1-h exposures per day (1 h apart), daily for 3 consecutive days according to the Federal Trade Commission protocol (1 puff/min of 2 s duration and 35 ml volume) and were euthanized 24 h after the last exposure (33, 34). The control mice were exposed to filtered air in a chamber and protocol identical to those used for CS exposure. For 10 d and chronic 6 mo CS exposure, 3R4F cigarettes were used to generate a mixture of sidestream (89%) and mainstream (11%) smoke at a concentration of ∼100 mg/m3 TPM, so as to avoid possible toxicity to the mice at a high concentration of long-term CS exposure (32). Each smoldering cigarette was puffed for 2 s, once every minute for a total of 5 puffs, at a flow rate of 1.05 L/min, to provide a standard puff of 35 cm3. Mice received 5 h exposure per day, 5 d/wk for the duration of exposure and were euthanized at 6-h intervals 24 h after the last CS exposure. The 6 h sampling interval was based on prior studies on circadian gene expression in rodents (35).
Locomotor activity recording
The mice were individually housed and allowed free access to food and water. Locomotor activity was measured with the Photobeam Activity System (San Diego Instruments, San Diego, CA, USA), a computerized system that measures the frequency of photobeam breaks along the side of the cage. For wheel-running behavior, the mice were housed singly in standard mouse cages fitted with a stainless-steel running wheel (Coulbourn Instruments, Flushing, NY, USA) placed within a light-tight chamber (Phenome Technologies, Lincolnshire, IL, USA). Total cage activity (photobeam break) and wheel-running activity were recorded in 1 min intervals and analyzed with ClockLab software (Actimetrics, Evanston, IL, USA). For wheel-running analyses, the mice were exposed to CS during their inactive phase for 10 d and then transferred to recording chambers. They remained under a 12:12 L:D cycle for 2 d followed by release into constant darkness. The free-running period of wheel-running activity was determined during 2 wk of constant darkness with a χ2 periodogram (P<0.001), and data were analyzed with an unpaired t test (GraphPad Prism, San Diego, CA, USA).
Real-time luminescence recording
Adult male Period2::luciferase knock-in (PER2::LUC) mice (36) were euthanized by excess CO2 exposure 3 h before lights off [zeitgeber time (ZT) 9–12; lights off=ZT12]. Portions of the lung and the brain were removed and collected in cold, sterile Hanks' balanced salt solution (HBSS). After the tissue was blocked, the brain was sliced in the coronal plane with a vibrating microtome at a thickness of 300 μm. A section containing the bilateral SCN near the midpoint of the rostrocaudal extent was recovered, and a minimum of tissue, including the paired SCNs was removed with fine scalpels. The paired SCNs were then separated by a precise midline cut, producing 2 hemi-SCN tissue explants. Small (5 mm3) fragments of lung tissue were isolated. The tissue specimens were placed in 35-mm culture dishes with 1.2 ml of culture medium [DMEM supplemented with B27 (Life Technologies, Inc., Grand Island, NY, USA), 10 mM HEPES, 352.5 μg/ml NaHCO3, 3.5 mg/ml d-glucose, 25 U/ml penicillin, 25 μg/ml streptomycin, and 0.1 mM luciferin (Promega, Madison, WI, USA)]. Cultures were prepared with clean medium as described above (control) or with the same medium containing cigarette smoke extract (CSE) and sealed with sterile vacuum grease and a glass coverslip. Sealed cultures were maintained at 35°C in a light-tight incubator, and luminescence was continuously recorded (counts/second) with an automated luminometer (LumiCycle; Actimetrics). Raw luminescence data were detrended (24 h moving average) and smoothed (2 h moving average; Origin Pro 8.5, OriginLabs, Northampton, MA, USA).
Lung morphometry
Mouse lungs (which had not been lavaged) were inflated by 1% low-melting-point agarose at a pressure of 25 cmH2O, and then fixed with 4% neutral buffered formalin (27, 32). Fixed lung tissues were dehydrated, embedded in paraffin, and sectioned (4 μm) with a rotary microtome (Microm International GmbH, Walldorf, Germany). The lung sections were deparaffinized, rehydrated by passing them through a series of xylene and graded alcohol, and stained with hematoxylin and eosin (H&E). Alveolar size was estimated from the mean linear intercept (Lm) of the airspace, which is a measure of airspace enlargement or emphysema performed with MetaMorph software (Molecular Devices, Sunnyvale, CA, USA; refs. 27, 32). Lm was calculated for each sample on the basis of 10 random fields/slide, observed at ×200. The airway and vascular structures were eliminated from the analysis.
Measurements of lung mechanical properties
The mechanical properties of the mouse lungs were determined with the FlexiVent apparatus (Scireq, Montreal, QC, Canada; refs. 27, 32). Quasi-static compliance (QsC), lung resistance (R), and tissue elastance (E) were measured in the mice, anesthetized by sodium pentobarbital [50 mg/kg body weight (BW), intraperitoneally]]. A tracheotomy was performed, and an 18-gauge cannula was inserted 3 mm into an anterior nick in the exposed trachea and connected to a computer-controlled rodent ventilator (FlexiVent; Scireq). Initially, the mice were ventilated with room air (150 breaths/min) at a volume of 10 ml/kg body mass. After 3 min of ventilation, measurement of lung mechanical properties was initiated by a computer-generated program to measure QsC, R, and E at 3 cmH2O positive end expiratory pressure obtained by fitting a model to each impedance spectrum. The calibration procedure removed the impedance of the equipment and tracheal tube within this system (37, 38). These measurements were repeated 3 times for each animal.
Bronchoalveolar lavage (BAL)
The mice were anesthetized at 24 h after the last exposure by an intraperitoneal injection of 100 mg/kg BW pentobarbital sodium (Abbott Laboratories, Abbott Park, IL, USA) and killed by exsanguination. The heart and lungs were removed en bloc, and the lungs were lavaged 3 times with 0.6 ml of saline (0.9% sodium chloride) via a cannula inserted into the trachea (4). The lavaged fluid was centrifuged, and the cell-free supernatants were frozen at −80°C for later analysis. The BAL inflammatory cell pellet was resuspended in 1 ml saline, and the total number of cells was counted with a hemocytometer. Cytospin slides (Thermo Shandon, Pittsburgh, PA, USA) were prepared at 50,000 cells/slide, and differential cell counts (∼500 cells/slide) were performed on cytospin-prepared slides stained with Diff-Quik (Dade Behring, Newark, DE, USA).
RNA isolation and quantitative PCR (qPCR)
Total RNA was isolated from brain and nonlavaged lung tissue specimens (stored in RNAlater; Ambion, Austin, TX, USA) with an RNeasy kit (Qiagen, Valencia, CA, USA). RNA yields were determined by UV absorbance with a NanoDrop instrument (ND-1000 Spectrophotometer; NanoDrop Technologies, Wilmington, DE, USA). cDNA was synthesized from 0.5 μg of total RNA by using the RT2 First Strand Kit (SABioscience, Frederick, MD, USA). To validate the expression of diverse genes in brain and lung tissues, qPCR was performed with a Bio-Rad iCycler real-time system and SYBR Green qPCR Master mix from SABioscience. In chronic CS exposure, qPCR data were gathered from circadian gene and proinflammatory cytokine gene expression at the ZT24 time point (n=2/air group) in all datasets. All the specific primers were purchased from SABioscience. Gene expression was normalized to 18s rRNA levels. Relative RNA abundance was quantified by the comparative 2−ΔΔCt method. Significant rhythms of gene expression were verified with CircWave software (version 1.4). In addition, the center of gravity (COG), or peak phase, was determined for each rhythm by using CircWave (39).
Proinflammatory mediator analysis
The levels of proinflammatory mediators, such as CCL2/monocyte chemotactic protein-1 (MCP-1), CXCL1/keratinocyte derived chemokine (KC), and CXCL10/interferon-γ inducible protein 10 (IP-10) in lung homogenates were measured by enzyme-linked immunosorbent assay (ELISA) performed with the respective duo-antibody kits (R&D Systems, Minneapolis, MN, USA), according to the manufacturer's instructions. The results were expressed in the samples as picograms per milligram protein.
Immunoblot and immunoprecipitation
Protein samples from lung homogenates were separated by 6–10% SDS-PAGE. The separated proteins were electroblotted onto nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ, USA). The membranes were blocked for 1 h at room temperature with 5% BSA and then probed with anti-SIRT1, anti-Ac Lys (1:1000; Cell Signaling Technology, Danvers, MA, USA), anti-BMAL1 and anti-CLOCK, (1:1000-1:2000; Abcam, Cambridge, MA, USA), and anti-acetyl BMAL1 (1:1000; Millipore, Billerica, MA, USA) antibodies, to determine the respective proteins. After 3 washing steps (10 min each), the levels of protein were detected with secondary antibody (1:5000 dilution) in 2.5% BSA in PBS containing 0.1% Tween 20 (v/v) for 1 h linked to horseradish peroxidase (Dako, Carpinteria, CA, USA), and bound complexes were detected by ECL (Perkin Elmer, Wellesley, MA, USA). Equal loading of the samples was determined by quantitation of proteins and by stripping and reprobing the membranes for β-actin (Oncogene, Cambridge, MA, USA). Immunoprecipitation was performed to determine the acetylation of BMAL1 in human whole-lung homogenates from nonsmokers, smokers, and patients with COPD, as described previously (26, 27, 34). ImageJ 1.41densitometry software (U.S. National Institutes of Health, Bethesda, MD, USA) was used for gel band quantitative densitometry.
Administration of SRT1720
SRT1720 (100 mg/kg, >95% pure by C-13 NMR and LCMS, synthesized by Life Chemicals, Niagara-on-the-Lake, ON, Canada) was administered to the acute air- and CS-exposed mice daily for 3 d through oral gavage 1 h before CS exposure (40).
Human samples
Lung tissue specimens from nonsmokers, smokers, and patients with COPD were obtained as described previously (26, 27, 34).
Statistical analysis
The period of PER2::LUC expression in each tissue explant was determined with a χ2 periodogram analysis (LumiCycle Analysis Software; Actimetrics). A minimum of 3 d of data were used to calculate the period of PER2::LUC expression in each explant of lung and SCN tissue. Period data from the lung tissue were analyzed with 1-factor analysis of variance (ANOVA) and the Newman-Keuls post hoc test. Period data from the SCN tissue (2 groups) were analyzed with an unpaired t test. Data are presented as the mean ± se. For statistical analysis of qPCR data, CircWave 1.4 software was used. In addition to CircWave analysis, statistical significance between the air- and CS-exposed groups was calculated by Fisher's multiple comparison, with 2-way ANOVA. To emphasize the waveform of the data, representative protein expression was subjected to nonlinear regression analysis with a sixth-order polynomial (Prism; Graphpad). Statistical analysis of significance was calculated by StatView software in a 1-way ANOVA followed by Tukey's post hoc test for multigroup comparisons. Values of P < 0.05 represented a significant difference.
RESULTS
CS exposure differentially affected the amplitude and phase of circadian clock gene expression in the lungs and brain
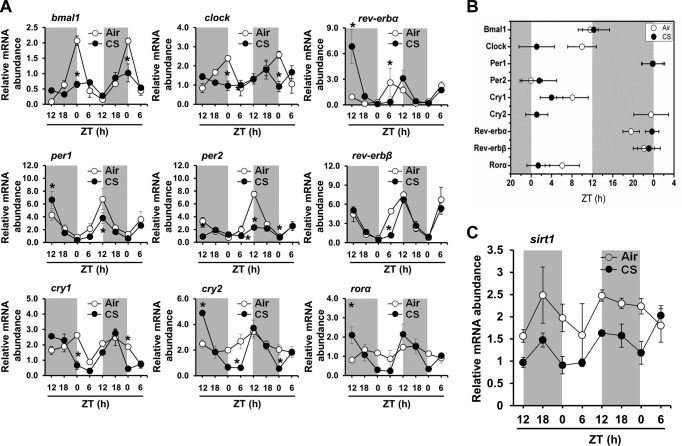
We used qPCR to investigate the effect of environmental CS exposure on rhythmic expression of core clock genes in mouse lungs. Both chronic (6 mo exposure, which causes pulmonary emphysema) and acute (3 and 10 d exposure, which induces a lung inflammatory response) environmental CS exposure models were used to determine the effects of CS on rhythmic expression of core clock genes in lung tissue and the functional relationship between altered clock function and lung inflammation. Most of the core clock genes (bmal1, clock, per1, per2, cry1, cry2, rev-erbα, and rev-erbβ) were rhythmically expressed in lung tissue from mice exposed to air or CS, either acutely (10 d; Fig. 1A, B) or chronically (6 mo; Supplemental Fig. S1A, B). CircWave analysis confirmed statistically significant (P<0.001) rhythms of bmal1, clock, per1, per2, rev-erbα, and rev-erbβ gene expression (Fig. 1A, B, Table 1, Supplemental Fig. S1A, and Table 2). Of the known core clock genes, only rorα mRNA was not rhythmic in the lungs of the mice exposed to air or CS for 6 mo (Supplemental Fig. S1A). In both the air- and CS-exposed mice, the expression of bmal1, clock, and cry1 displayed nocturnal acrophases, peaking in the mid to late portions of the dark phase (ZT18–24; ZT0=lights on; ZT12=lights off; Fig. 1A, B and Supplemental Fig. S1A, B). As expected, in both groups, the peaks of per1, per2, cry2, rev-erbα, and rev-erbβ were nearly antiphase to bmal1, which peaked between ZT6 and ZT12 (Fig. 1A, B and Supplemental Fig. S1A, B). Chronic CS exposure caused a modest reduction in the amplitude of bmal1 and rev-erbα expression and substantially reduced the amplitude of per1 expression in the lungs (Supplemental Fig. S1A, B). Further, CircWave analyses indicated that chronic CS exposure altered the phase of per1 and per2 expression in the lungs (Supplemental Fig. S1B).
Figure 1.
Acute CS exposure differentially affected clock and sirt1 gene expression in mouse lungs. Acute (10 d) air or CS exposure was performed as described in Materials and Methods. On the day after the last exposure, lungs were harvested every 6 h for 42 h beginning at ZT12. Total RNA was extracted from lung tissues, and cDNA was prepared for gene expression analysis by qPCR. A) Expression of core clock genes (bmal1, clock, per1, per2, cry1, cry2, rev-erbα, rev-erbβ, and rorα) was examined by qPCR. CircWave analysis confirmed statistically significant rhythms of each clock gene (P<0.05 for bmal1; P<0.001 for per1, cry1, cry2, rev-erbα, rev-erbβ, and rorα) in CS-exposed mice. B) COG or peak phase values for each gene expression rhythm were obtained by using CircWave and plotted on a horizontal phase map. Gray shading indicates the relative dark phase (ZT12–24). C) Circadian rhythm of sirt1 gene expression in lung tissue was analyzed by qPCR in lung tissue from mice after acute air or CS exposure. Data from air-exposed (open circle) and CS-exposed (solid circle) mice are shown as the mean ± se (n=3–4 mice/group) for each time point. *P < 0.05 vs. corresponding air-exposed mice.
Table 1.
COG and significance for each clock gene expression rhythm, as determined by CircWave analysis in acute air- or CS-exposed mouse lung
| Clock gene | Air |
CS |
||
|---|---|---|---|---|
| COG | P | COG | P | |
| Bmal1 | 11.6 ± 1.5 | 0.001*** | 12.3 ± 3.1 | 0.05* |
| Clock | 10.0 ± 2.8 | 0.001*** | 1.1 ± 3.5 | NS |
| Per1 | 23.9 ± 2.3 | 0.001*** | 23.8 ± 2.0 | 0.001*** |
| Per2 | 0.0 ± 2.18 | 0.001*** | 1.6 ± 3.4 | NS |
| Cry1 | 8.1 ± 3.1 | 0.001*** | 4.0 ± 2.2 | 0.001*** |
| Cry2 | 23.5 ± 3.4 | NS | 1.1 ± 2.2 | 0.001*** |
| Rev-erb α | 19.6 ± 1.6 | 0.001*** | 23.7 ± 1.3 | 0.001*** |
| Rev-erb β | 22.1 ± 2.1 | 0.001*** | 23.1 ± 2.2 | 0.001*** |
| Ror α | 6.1 ± 3.34 | NS | 1.4 ± 2.2 | 0.001*** |
Data are shown as mean NS ± se (n=3–4/group). NS, not significant.
P < 0.05,
P < 0.001.
Table 2.
COG values and significance for each clock gene expression rhythm, as determined by CircWave analysis in chronic air- or CS-exposed mouse lung
| Clock gene | Air |
CS |
||
|---|---|---|---|---|
| COG | P | COG | P | |
| Bmal1 | 21.7 ± 1.4 | 0.001*** | 22.6 ± 1.9 | 0.001*** |
| Clock | 21.4 ± 2.58 | 0.001*** | 21.8 ± 2.6 | NS |
| Per1 | 14.2 ± 3.1 | 0.001*** | 3.6 ± 3.3 | NS |
| Per2 | 14.6 ± 2.2 | 0.001*** | 11.0 ± 2.3 | 0.001*** |
| Cry1 | 19.9 ± 2.5 | NS | 19.0 ± 2.5 | 0.001*** |
| Cry2 | 16.0 ± 3.4 | NS | 0.5 ± 3.0 | NS |
| Rev-erb α | 6.2 ± 2.1 | 0.001*** | 3.3 ± 1.7 | 0.001*** |
| Rev-erb β | 11.6 ± 3.1 | 0.05* | 10.8 ± 3.1 | NS |
| Ror α | 19.1 ± 3.3 | NS | 18.9 ± 3.4 | NS |
Data are shown as means ± se (n=3–4/group). NS, not significant.
P < 0.05,
P < 0.001.
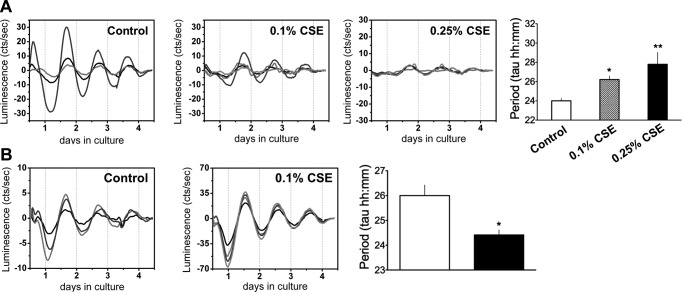
As with the chronic-exposure model, acute CS exposure also reduced the amplitude of bmal1, clock, and per2 mRNA expression, but only modestly affected the amplitude of per1 expression in the lungs (Fig. 1A). Surprisingly, acute CS exposure appeared to have time-dependent suppressive effects on cry1, cry2, and rev-erbβ expression (Fig. 1A). Finally, CircWave analyses indicated that acute CS slightly delayed the phase of rev-erbα expression in the lungs (Fig. 1B). Together, data from both the acute and chronic models reveal that the amplitude and phase of clock gene expression in the lungs were differentially affected by CS exposure, although the effects of acute exposure were more robust. Parallel effects of CS exposure on the phase and amplitude of clock gene expression were observed in brain tissue of the acutely CS-exposed mice (Supplemental Fig. S3), suggesting that the effects of CS on clock function are not limited to lung tissue. The effect of acute CS exposure on clock gene expression in the lung (Fig. 2A) and SCN (Fig. 2B) was confirmed by real-time monitoring of PER2::LUC expression in lung and SCN tissue explants. Medium containing CSE (0.1-0.25%) dramatically and dose dependently reduced the amplitude of PER2::LUC expression in the lung tissue explants (Fig. 2A). CSE treatment dose dependently increased the period of PER2::LUC expression in the lung tissue explants (Fig. 2A). Of note, treatment with 0.1% CSE did not affect the amplitude of PER2::LUC expression in the SCN explants, compared to that in the lung tissue (Fig. 2B). However, the same treatment significantly shortened the period of PER2::LUC expression in the CS-exposed SCN explants when compared with that in the untreated control (Fig. 2B). Overall, CSE treatment reduced the amplitude and differentially affected the period of PER2::LUC expression in brain and lung tissue explants ex vivo.
Figure 2.
Effects of CSE on the amplitude and period of PER2::LUC expression in lung and SCN tissue explants. A) PER2::LUC expression in representative lung tissue explants treated with medium alone (controls). Treatment at the time of culture with CSE at 0.1 and 0.25% significantly and dose dependently dampened the rhythm of PER2::LUC expression in lung tissue. Treatment with CSE dose dependently increased the period of PER2::LUC expression in lung explants (control, 24±0.25; 0.1% CSE, 26.21±0.37; 0.25% CSE, 27.79±1.24). B) PER2::LUC expression rhythms in representative hemi-SCN tissue explants treated with medium alone (controls). Treatment with 0.1% CSE did not reduce the amplitude of PER2::LUC expression in SCN explants, but 0.1% CSE significantly shortened the period of PER2::LUC expression in the SCN when compared with controls (controls, 26±0.43 vs. 0.1% CSE, 24.42±0.20). Each line represents tissue from different animals. Data from control and CSE treatment represent means ± se (n=4–6/group). *P < 0.05, **P < 0.01 vs. corresponding untreated control (medium alone).
CS exposure reduced total locomotor activity and shortened the free-running period of wheel-running behavior in mice
Seeing that CS exposure altered the amplitude and period of clock gene expression in lung and brain tissue, we investigated its effect on the circadian rhythms of behavior in mice. The total cage activity of individual C57BL/6J mice was recorded by infrared-beam breaks during the 6 mo of CS exposure (17 d before starting and continuing until the end of chronic CS exposure). Mice exposed to chronic CS showed a significant reduction in total cage locomotor activity (Supplemental Fig. S2A). Analysis of total ambulatory counts across the 24 h day revealed that chronic CS exposure significantly reduced activity in the WT mice within 5 d of the first exposure, and the decrease persisted until the beginning of the 6th mo of CS exposure (Supplemental Fig. S2B). When daytime and nighttime activity were analyzed separately, the amplitude of the response increased, indicating that the primary effect of CS exposure was a reduction in nighttime activity (Supplemental Fig. S2B).
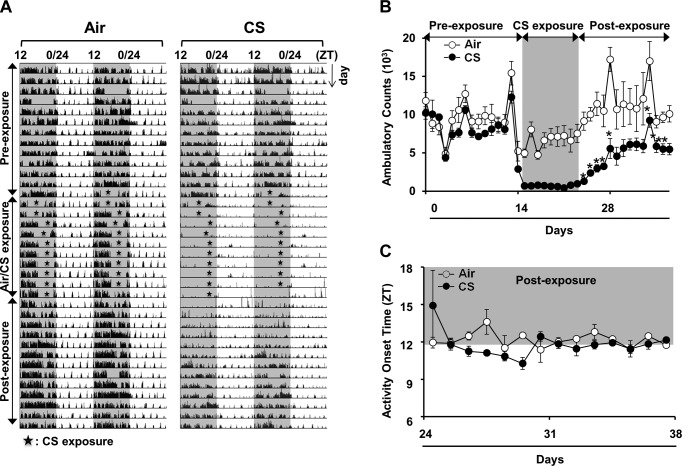
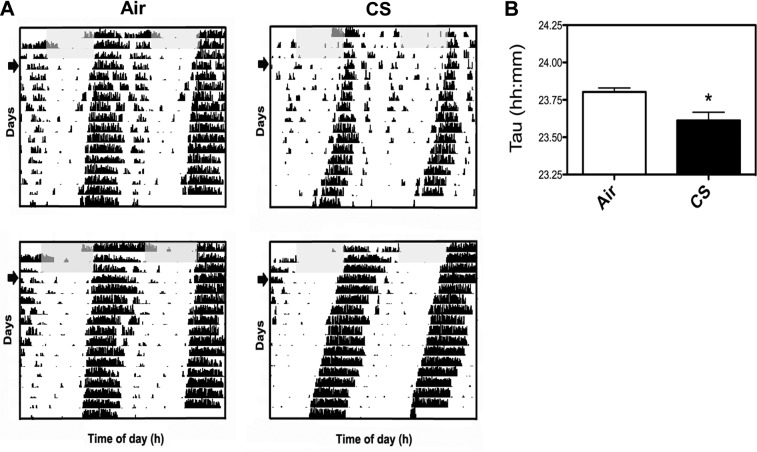
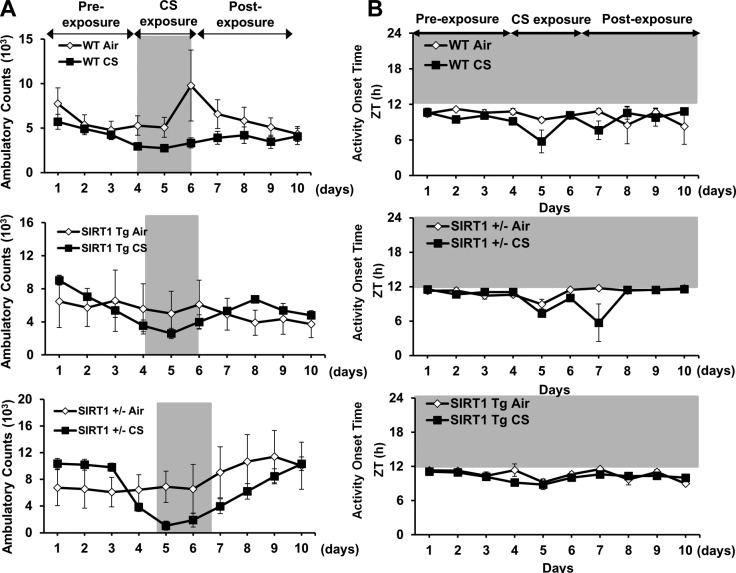
The effect of acute CS exposure on total cage locomotor activity and wheel-running behavior were measured to determine whether the reduction in activity that we observed is initiated at an early stage of smoking. We used our beam break monitor to record total cage activity for 2 wk before CS exposure, during the active phase for 10 d of CS exposure and during a 2 wk period after CS exposure (clean air). The mice were exposed to CS during their active phase for 5 h/d over 10 consecutive days. Locomotor activity was almost immediately attenuated in the CS-exposed mice, and was persistently lower after CS exposure ended (Fig. 3A). Although activity increased during recovery, the mice managed to approach only ∼70% of their original activity level after 2 wk of breathing clean air (Fig. 3B). Activity onset time or phase angle of entrainment was estimated with Clocklab software (Actimetrics) and did not appear to be affected by CS (Fig. 3C). Analysis of wheel-running behavior revealed a small, but significant, reduction in the free-running period of activity in mice after acute exposure to CS (τ=23.61±0.06) during their inactive phase when compared with the air-exposed mice (τ=23.80±0.03; P<0.05) (Fig. 4). Shortening of the free-running period of wheel-running activity suggests that CS alters clock gene expression in the central clocks that regulate behavioral rhythms, a finding supported by our analysis of PER2::LUC expression in SCN tissue explants (Fig. 2B) and clock gene expression in whole brain (Supplemental Fig. S3).
Figure 3.
Acute CS exposure reduced activity, but did not affect the phase angle of entrainment or activity consolidation. Acute (10 d) air or CS exposure was performed as described in Materials and Methods. Locomotor activity was recorded from individual mice for a period of 38 d. A) Representative double-plotted actograms showing considerable reduction in activity of CS-exposed mice (right) relative to air-exposed mice (left). Gray shading indicates the relative dark phase (ZT12-24). During the periods of CS exposure (ZT3-9), activity was not recorded (stars). B) Nocturnal activity was plotted in ambulatory counts. CS reduced locomotor activity, which recovered up to 70% of the activity level in air-exposed mice during the postexposure phase. C) Activity onset relative to lights off (ZT12; phase angle entrainment) during the postexposure. Shading indicates the dark phase (ZT12-24). Data from air- and CS-exposed groups represent the mean ± se (n=3–4 mice/group) for each time point. *P < 0.05 vs. corresponding air-exposed mice.
Figure 4.
Effect of acute CS exposure on circadian rhythms of wheel-running activity. A) Representative actograms of mice that were exposed to 10 d of CS (see Materials and Methods) and then placed in cages with attached running wheels. Mice were entrained to a 12:12 L:D cycle for 2 d and then released into constant darkness (D:D) for 14 d. Gray shaded region indicates the light phase. Arrow: first full day in D:D. B) Free-running period of running activity in D:D was calculated across the 2 wk period with a χ2 periodogram. Period was significantly shorter in the CS group (τ=23.61±0.06) compared with that in the air-exposed group (τ=23.80±0.03). *P < 0.05 vs. corresponding air-exposed mice.
Disruption of circadian clock function in the lungs resulted in increased inflammatory responses
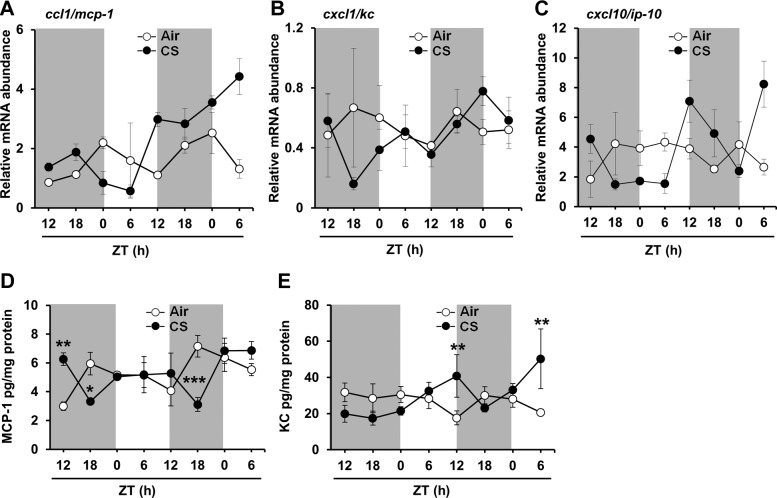
To investigate whether CS alters circadian rhythmicity of proinflammatory cytokine responses, we determined the expression of proinflammatory cytokine genes after chronic and acute CS exposure. In air-exposed mice from the acute exposure experiments, lung mRNA expression of both monocyte chemotactic protein–1 (ccl1/mcp-1) and keratinocyte chemoattractant (cxcl1/kc) peaked during the dark phase (Fig. 5A–C and Supplemental Fig. S4A–C). CircWave analysis confirmed significant circadian rhythms of both genes in air-exposed mice (P<0.001). In contrast, the expression of interferon-γ-induced protein 10 (cxcl10/IP10) peaked during the light phase, but did not have a significant circadian rhythm of expression in air-exposed mice (Fig. 5C and Supplemental Fig. S4C). Acute and chronic CS exposure disrupted rhythms of ccl1/mcp-1 and cxcl1/kc expression in the lungs (Fig. 5A, B and Supplemental Fig. S4A, B). These results suggest that proinflammatory cytokine gene expression is rhythmic in the lungs, and is disrupted by both acute and chronic CS exposure.
Figure 5.
Acute CS exposure affected circadian rhythms of proinflammatory cytokine gene expression and abundance in mouse lungs. Acute (10 d) air or CS exposure was performed as described in Materials and Methods. At the end of CS or air exposure, lungs were harvested every 6 h for 42 h beginning at ZT12 on the day following the last CS exposure. Total RNA was extracted from lung tissue, and cDNA was prepared for gene expression analysis by qPCR. Expression of proinflammatory cytokine genes including ccl1/mcp-1 (A), cxcl1/kc (B), and cxcl10/ip-10 (C) was determined by qPCR. Levels of the proinflammatory mediators MCP-1 (D) and KC (E) were also measured in lung homogenates obtained from acute air- or CS-exposed mice. Significant circadian rhythms of cytokine gene expression were detected by CircWave analysis. Data from air-exposed (open circle) and CS-exposed (solid circle) mice represent means ± se (n=3 mice/group) for each time point. *P < 0.05, **P < 0.01, ***P < 0.001 vs. corresponding air-exposed mice.
To understand the functional link between CS-mediated alterations in circadian rhythmicity and proinflammatory cytokine responses, we determined the abundance of proinflammatory cytokines (ccl1/mcp-1, cxcl1/kc, and MIP-2) in mouse lungs after CS exposure. Acute CS exposure appeared to cause a phase shift in the rhythms of MCP-1 and KC release, such that MCP-1 peaked in the middle of the day (ZT6 vs. ZT18 in air-exposed group), and KC levels peaked closer to lights off (ZT12; Fig. 5D, E). Acute CS also appeared to increase the amplitude of KC release in the mouse lung tissue (Fig. 5E). Chronic CS exposure had differential and cytokine-specific effects on MCP-1, KC, and MIP-2 levels in the tissue (Supplemental Fig. S4D–F). The levels of MCP-1 and KC were increased in the latter portion of the dark phase (ZT18-24) after chronic CS exposure.
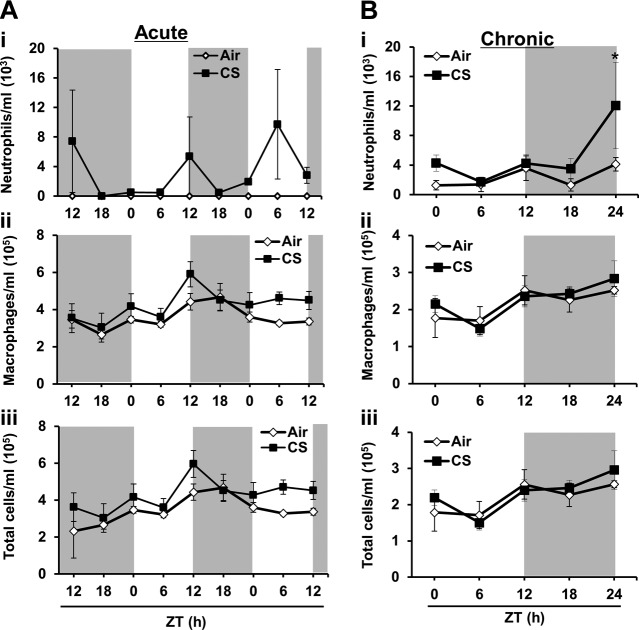
The increased level of cytokines was associated with a rhythmic influx of inflammatory cells in the lung in response to acute (Fig. 6A) and chronic (Fig. 6B) CS exposure. Neutrophil influx into BAL fluid increased significantly during the late night (ZT24), whereas there was no significant difference in the number of macrophages and total cells in BAL fluid (Fig. 6B). Thus, our data show that the disruption of circadian clock function in the lung was associated with augmented proinflammatory responses during both acute and chronic CS exposure.
Figure 6.
A) Acute CS had time-dependent effects on rhythms of inflammatory cell influx in mouse lungs. Acute (10 d) air or CS exposure was performed as described in Materials and Methods. At the end of CS or air exposure, lungs were harvested every 6 h for 42 h, beginning at ZT12 on the day following the last CS exposure. The number of total cells in BAL fluid from acute air- or CS-exposed mice was determined. At least 500 cells in the BAL fluid were counted with a hemocytometer to determine the number of neutrophils (i), macrophages (ii), and total cells (iii) on cytospin slides stained with Diff-Quik. Data from air- and CS-exposed lungs are shown as means ± se (n=3–4/group) for each time point. B) Chronic CS had time-dependent effects on rhythms of inflammatory cell influx in CS-exposed mouse lung. Environmental tobacco/chronic CS exposure (6 mo air or CS) was performed as described in Materials and Methods. On the day after the last exposure, lungs were harvested every 6 h for 24 h beginning at ZT0, and then at ZT6, ZT12, ZT18, and ZT24. The number of total cells in BAL fluid from chronic air- or CS-exposed mice was determined. At least 500 cells in the BAL fluid were counted with a hemocytometer, to determine the number of neutrophils (i), macrophages (ii), and total cells (iii) on cytospin slides stained with Diff-Quik. Data from air- and CS-exposed are shown as means ± se (n=3–4/group) for each time point. *P < 0.05 vs. corresponding air-exposed mice.
Disruption of circadian clock function was associated with airspace enlargement and emphysema along with alteration in lung mechanical properties and function
To investigate the potential effect of circadian clock dysfunction due to CS-induced airspace enlargement and emphysema, we subjected mice to chronic CS exposure and determined their susceptibility to CS-induced airspace enlargement and emphysema by lung histopathologic and functional measurements. There was a significant difference in lung histopathology (H&E) between air- and CS-exposed mice after 6 mo, which was assessed by determining the Lm (air 49.56±3.25 vs. CS 61.40±1.60; P<0.01, n=4–6 mice/group). Similarly, the overall lung E was significantly decreased and R was decreased, but not significantly, in the mice exposed to chronic CS. However, lung QsC was increased significantly in chronic CS-exposed mice as compared to that in air-exposed mice (Table 3). We also found a significant decrease in lung E and R in the chronic CS-exposed mice at ZT18, but not at other times of the day (ZT0, ZT6, ZT12, and ZT24), when compared to those parameters in air-exposed mice (Tables 3 and 4). However, when we combined the data from day (ZT0 and ZT6) and night (ZT12 and ZT18), E was significantly reduced, whereas lung compliance increased in the chronic CS-exposed mice (Table 3). There was no statistically significant difference in BW or mortality between the chronic CS- and air-exposed mice.
Table 3.
Lung mechanical properties measured at different day and night ZTs in WT mice exposed to chronic CS
| Day/night (ZT) | QsC (ml/cmH2O) |
R (cmH2O/s/ml) |
E (cmH2O/ml) |
|||
|---|---|---|---|---|---|---|
| Air | CS | Air | CS | Air | CS | |
| ZT0 + ZT6 | 0.051 ± 0.002 | 0.051 ± 0.002 | 0.747 ± 0.045 | 0.726 ± 0.036 | 19.680 ± 0.792 | 19.654 ± 0.633 |
| ZT12 + ZT18 | 0.046 ± 0.001 | 0.052 ± 0.002* | 0.747 ± 0.045 | 0.708 ± 0.038 | 21.905 ± 0.359 | 19.502 ± 0.565** |
Data are shown as as means ± se (n=4/group).
P < 0.05,
P < 0.01 vs. air-exposed mice.
Table 4.
Lung mechanical properties measured at different ZTs in WT mice exposed to chronic CS
| ZT | Compliance (ml/cmH2O) |
R (cmH2O/s/ml) |
E (cmH2O/ml) |
|||
|---|---|---|---|---|---|---|
| Air | CS | Air | CS | Air | CS | |
| ZT0 | 0.048 ± 0.001 | 0.051 ± 0.002 | 0.766 ± 0.105 | 0.741 ± 0.052 | 20.968 ± 0.647 | 19.759 ± 0.718 |
| ZT6 | 0.054 ± 0.001 | 0.052 ± 0.003 | 0.729 ± 0.015 | 0.711 ± 0.061 | 18.392 ± 0.169 | 19.548 ± 1.217 |
| ZT12 | 0.045 ± 0.002 | 0.050 ± 0.002 | 0.730 ± 0.043 | 0.725 ± 0.008 | 22.027 ± 0.840 | 19.963 ± 0.917 |
| ZT18 | 0.046 ± 0.0003 | 0.053 ± 0.002 | 0.861 ± 0.038 | 0.691 ± 0.083* | 21.783 ± 0.189 | 19.041 ± 0.736* |
| ZT24 | 0.048 ± 0.001 | 0.053 ± 0.002 | 0.736 ± 0.016 | 0.682 ± 0.021 | 20.776 ± 0.525 | 18.853 ± 0.751 |
Data are shown as means ± se (n=4/group).
P < 0.05 vs. air-exposed mice.
Rhythmic expression of SIRT1 was disrupted by CS exposure, resulting in increased acetylation of BMAL1 in lungs
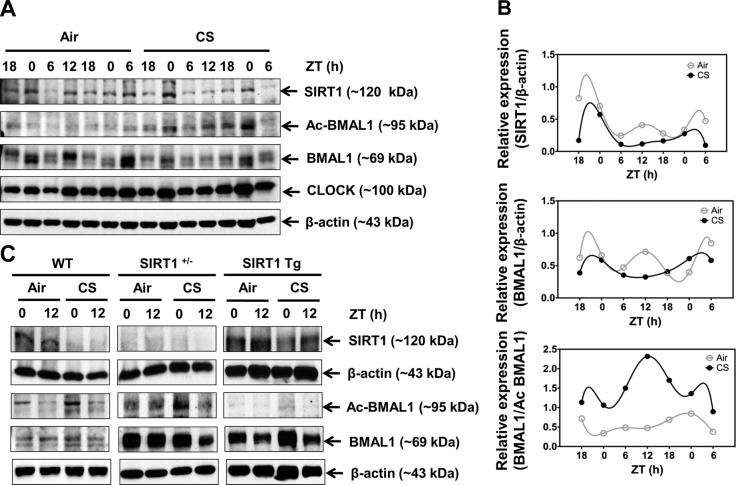
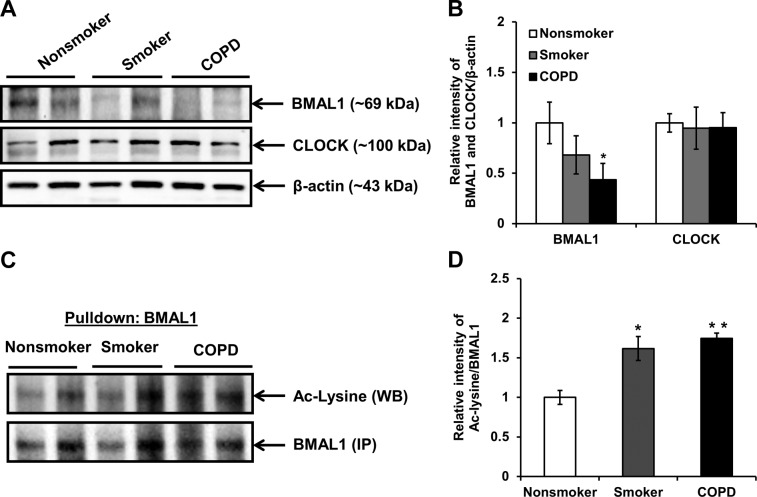
To investigate the mechanisms of CS-induced disruption of the lung inflammatory response, we determined the levels of BMAL1 acetylation and the abundance of SIRT1 in mouse lung after an acute (10 d) CS exposure. In the lungs of the control air-exposed mice, the protein abundance of SIRT1 and BMAL1 showed clear rhythmic oscillations that peaked in the night (Fig. 7A, B). However, CS reduced the abundance of SIRT1 and BMAL1 across the entire day, which was consistent with gene expression data (Fig. 1A and Supplemental Fig. S1A). We observed a similar effect of CS on the rhythm of sirt1 gene expression in the lung and brain tissues (Fig. 1C and Supplemental Fig. S3C). Furthermore, the mice with CS exposure showed a reduction in BMAL1 levels (Fig. 7A). Similarly, the BMAL1 levels were modestly reduced in the smokers and significantly reduced in lungs of the patients with COPD, compared with levels in lungs of the nonsmokers (Fig. 8A, B). CS caused increased BMAL1 acetylation in the mouse lungs (Fig. 7A, B), and a similar increase in BMAL1 acetylation was observed in the lungs of the patients with COPD and the smokers, compared to those of the nonsmokers (Fig. 8C, D). Increased BMAL1 acetylation in the CS-exposed mice was associated with reduced locomotor activity (Figs. 9 and 10). Given that SIRT1 modulates the circadian clock mechanism by controlling BMAL1 acetylation, we hypothesize that acetylation of BMAL1 is increased by the reduction of SIRT1 levels after CS exposure.
Figure 7.
Circadian rhythm of SIRT1 expression in lung tissue was disrupted by CS exposure, resulting in elevated BMAL1 acetylation. A) Immunoblot analysis of SIRT1, total BMAL1, acetylated BMAL1 (Ac BMAL1), and CLOCK in lung tissue homogenates from mice after 10 d of air or CS exposure. Images are representative of ≥2 separate experiments. B) Oscillation patterns of SIRT1, BMAL1, and Ac BMAL1 protein as shown by immunoblot analysis. After densitometric analysis, levels of SIRT1 and BMAL1 were both normalized against β-actin, and Ac BMAL1 was normalized against total BMAL1. Data from air- and CS-exposed samples are representative of the immunoblot data, and were analyzed with nonlinear regression, as described in Materials and Methods (R2=1 for all data). C) Genetic manipulation of SIRT1 regulated BMAL1 acetylation in response to CS in the lungs. Immunoblot analysis of SIRT1, BMAL1, and acetylated BMAL1 performed in lung tissue homogenates from SIRT1 heterozygous knockout (SIRT1+/−) and SIRT1-overexpressing (SIRT1 Tg) mice following 3 d of air or CS exposure. Images are representative of ≥2 separate experiments. Reassembly of noncontiguous gel lanes is demarcated by white spaces and boxes, aligned so as to reflect the overall representative data. Data from air- and CS-exposed tissue are shown as means ± se (n=2–3/group) for each time point.
Figure 8.
BMAL1 was down-regulated in lungs of smokers and patients with COPD. A) Immunoblot analysis of BMAL1 and CLOCK in whole-tissue lysates extracted from lung tissue of nonsmokers, smokers, and patients with COPD. B) After densitometric analysis, the levels of BMAL1 and CLOCK in lung tissue were normalized against β-actin (loading control). Relative level of BMAL1 was significantly decreased in lung tissue from smokers and patients with COPD when compared with that of nonsmokers. C) Whole-tissue lysates extracted from lung tissues (collected at midmorning and midday) of nonsmokers, smokers, and patients with COPD (26, 27, 34) were immunoprecipitated with anti-BMAL1 antibody and probed with anti-acetylated lysine antibody. d) Relative intensity of acetylated lysine/BMAL1 represents the increased acetylation of BMAL1 in lungs of smokers and patients with COPD as compared to nonsmokers. Images and data represent means ± se (n=3–4/group). *P < 0.05, **P < 0.01 vs. nonsmokers.
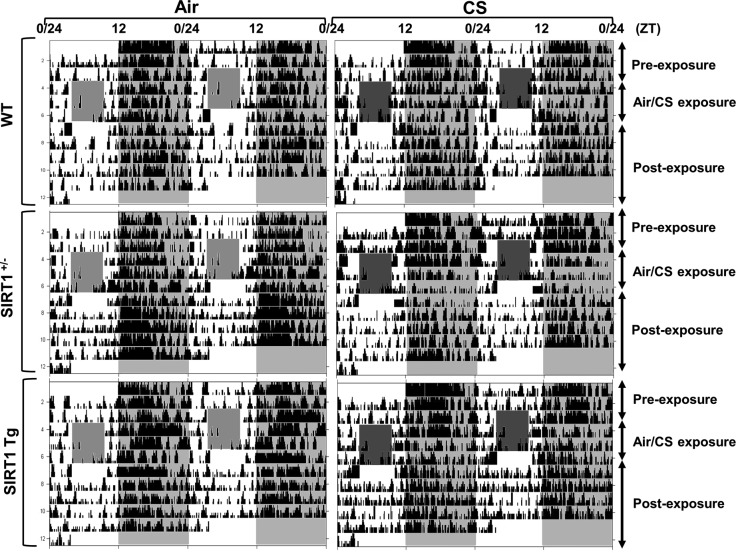
Figure 9.
Locomotor activity of WT, heterozygous SIRT1-knockout (SIRT+/−), and SIRT1-overexpressing (SIRT1 Tg) mice during acute CS exposure. Wild-type (WT), SIRT1+/−, and SIRT1 Tg mice were kept in a 12:12 L:D cycle throughout the experiment. Mice were not exposed to CS for the first 3 d (preexposure); were exposed to CS during the light phase (ZT4-8) for 3 consecutive days (CS exposure); and then were kept in room air without CS exposure for 4 d (postexposure). Representative actograms of locomotor activity from mice exposed to air or CS for 3 d. During the period of CS exposure, activity was not recorded (light-gray or dark-gray shaded area during the 12-h light phase). Locomotor activity during the dark phase was plotted as ambulatory counts. Shading indicates the days of CS exposure.
Figure 10.
Ambulatory counts (A) and activity onset time (ZT) by acute CS exposure (B) in WT, heterozygous SIRT1-knockout (SIRT+/−), and SIRT1-overexpressing (SIRT1 Tg) mice. Activity onset relative to lights off (ZT12; phase angle of entrainment) during the period of preexposure (d 1–3), CS exposure (d 4–6), and postexposure (d 7–10). Gray shading indicates the dark phase (ZT12-24). Data from air- and CS-exposed mice represent means ± se (n=3–4 mice/group) for each time point.
To determine whether SIRT1 reduction is responsible for increased BMAL1 acetylation, SIRT1-deficient (SIRT1+/−) and SIRT1-overexpressing (Tg) mice were exposed to acute CS for 3 d, and the level of BMAL1 acetylation was measured in lung tissue. Acute CS exposure significantly increased the acetylation of BMAL1 in the SIRT1-deficient mice relative to both the air-exposed SIRT1-deficient mice and the CS-exposed WT mice (Fig. 7C). This response was attenuated in the SIRT1 Tg mice, most likely because of the persistently elevated level of SIRT1 deacetylase activity (Fig. 7C). Moreover, locomotor activity levels were significantly reduced during acute (3 d) CS exposure in SIRT1-deficient but not in SIRT1-overexpressing mice (Figs. 9 and 10). These results suggest that SIRT1 reduction in response to CS exposure leads to an increase in BMAL1 acetylation and reduced levels of BMAL1, causing altered molecular clock function and increased proinflammatory responses in the lungs.
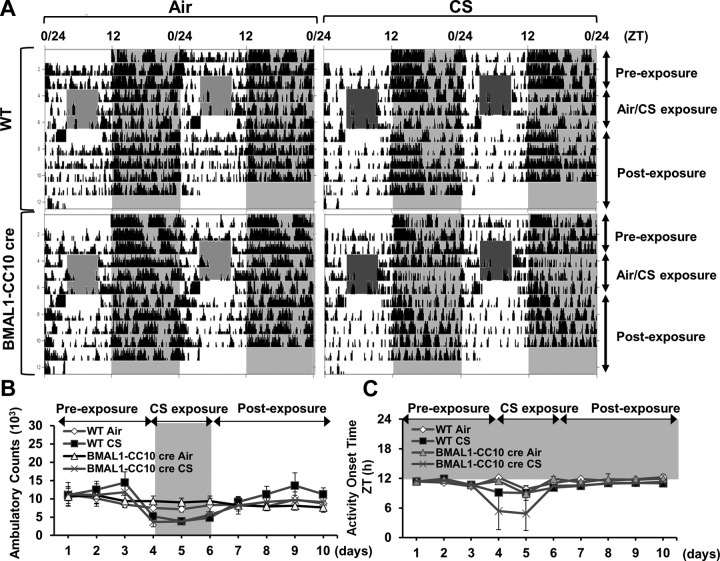
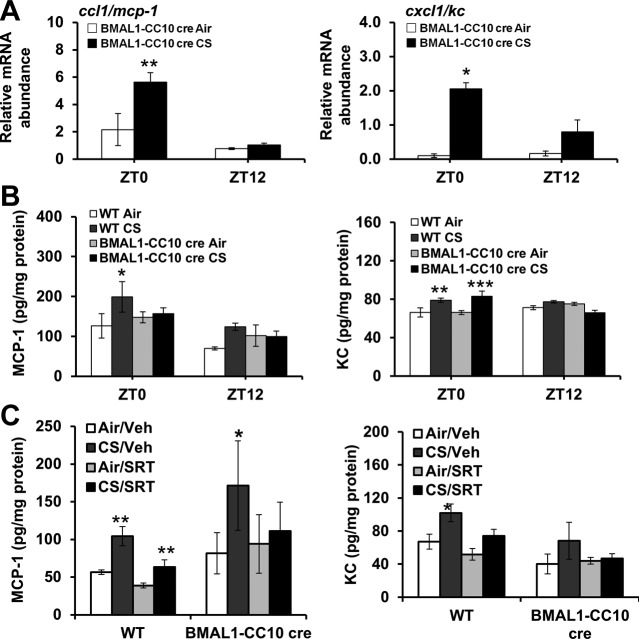
Lung epithelial cell–specific BMAL1 deletion increased lung inflammation, which was not attenuated by treatment with a selective pharmacological SIRT1 activator
To determine the role of the SIRT1-BMAL1 pathway in CS-induced circadian disruption and inflammatory responses, we established lung epithelial cell–specific BMAL1−/− (BMAL1-CC10 cre) mice and exposed them to an acute period (3 d) of CS. The BMAL1-CC10 cre mice exhibited normal rhythms of locomotor activity (ambulatory count and onset time; Fig. 11) and, as in the WT mice, the activity in these mice was significantly reduced by CS exposure (Fig. 11A, B). As in the WT mice, we detected rhythmic expression of proinflammatory cytokine genes, including ccl1/mcp-1 and cxcl1/kc, in the BMAL1-CC10 cre mice that were affected by CS, such that the expression of ccl1/mcp-1 was increased at ZT0 but remained unchanged at ZT12 (Fig. 12A, B). Cxcl1/kc gene expression was elevated at both times in the CS-exposed animals. We found similar effects of lung-specific BMAL1 deletion on rhythms of proinflammatory cytokine release (Fig. 12B). Overall, lung epithelial cell–specific clock disruption altered the inflammatory response to CS. To further examine the involvement of SIRT1 and BMAL1 in these proinflammatory responses, we exposed BMAL1-CC10 cre mice to CS (3 d), followed by administration of the selective pharmacologic SIRT1 activator SRT1720. The BMAL1-CC10 cre, global SIRT1-deficient, and SIRT1 Tg mice exhibited normal diurnal patterns of locomotor activity (Figs. 9–11). As expected, locomotor activity during the night was reduced in these mice during the acute CS exposure and returned to normal levels after 4 d of clean-air recovery (Figs. 9–11). Treatment with SRT1720 attenuated proinflammatory cytokine release in the CS-exposed WT mice, but not in the CS-exposed BMAL1-CC10 cre mice (Fig. 12C). These data strongly suggest that epithelial BMAL1 plays a critical role in the regulation of CS-induced inflammatory responses and that the effect of BMAL1 is modulated by SIRT1.
Figure 11.
Locomotor activity of BMAL1-CC10 cre mice in response to acute CS exposure. BMAL1 CC10 cre (epithelial BMAL1 knockout) mice were kept in a 12:12 L:D cycle throughout the experiment. Mice were not exposed to CS for the first 3 d (preexposure), were exposed to CS during the light phase (ZT4-8) for 3 consecutive days (CS exposure), and then were kept in room air without CS exposure for 4 d (postexposure). A) Representative actograms of locomotor activity from WT or BMAL1-CC10 cre mice exposed to air/CS for 3 d. During the period of CS exposure, activity was not recorded (light-gray or dark-gray shaded area during the 12-h light phase). B) Locomotor activity during the dark phase was plotted as ambulatory counts. Shading indicates the days of CS exposure. C) Activity onset relative to lights off (ZT12; phase angle of entrainment) during the period of preexposure (d 1–3), CS exposure (d 4–6) and postexposure (d 7–10). Gray shading indicates the dark phase (ZT12-24). Data from air- and CS-exposed mice represent means ± se (n=3–4 mice/group) for each time point.
Figure 12.
Lung epithelial cell-specific BMAL1 knockout enhanced CS-induced lung inflammation. A) Expression of proinflammatory cytokine genes (ccl1/mcp-1, and cxcl1/kc) was performed by qPCR. CircWave analysis confirmed circadian rhythms of each proinflammatory cytokine in air-exposed mice. *P < 0.05, **P < 0.01 vs. corresponding air-exposed mice. B) Levels of proinflammatory mediators, including CCL1/MCP-1 and CXCL1/KC, were measured in lung homogenates obtained from air- or CS-exposed WT and BMAL1 CC10 cre mice. Data are shown as means ± se (n=3/group) for each time point. *P < 0.05, **P < 0.01, ***P < 0.001 vs. corresponding air-exposed mice. C) WT and BMAL1-CC10 cre mice were treated with pharmacologic SIRT1 activator SRT1720 (SRT) or vehicle (Veh) during CS exposure for 3 d. Levels of proinflammatory mediators, such as CCL1/MCP-1 and CXCL1/KC, were measured in lung homogenates obtained from air- or CS-exposed WT and BMAL1-CC10 cre mice. Data are shown as means ± se (n=3 mice/group) for each time point. *P < 0.05, **P < 0.01 vs. corresponding air-exposed mice.
DISCUSSION
Patients with COPD experience circadian disruption (14, 17–20). Hence, CS exposure may affect circadian clock function in the lung, leading to inflammatory and injurious responses. We examined the putative mechanisms of clock disruption in response to chronic environmental CS exposure in lung and brain tissues in a mouse model of COPD and emphysema. We determined the effect of COPD and emphysema on molecular clock function in the lung. We show for the first time that CS exposure alters circadian clock gene expression in both the lungs and brain. Our results show that both acute and chronic CS exposure significantly reduced the amplitude of locomotor activity in mice. This effect persisted even after CS exposure ended, with locomotor activity levels not fully recovering until 14–30 d after smoking cessation in the chronic-exposure model. Despite this effect on locomotor activity levels, the phase angle of entrainment and consolidation of activity in the dark phase were not disrupted following CS exposure. Further, acute CS exposure had a small but significant effect on the free-running period of wheel-running activity, suggesting that CS may alter the timing of clock gene expression in brain regions, like the SCN, that drive behavioral rhythms. This interpretation is strengthened by our observation of altered clock gene expression in whole brain following acute CS in vivo and the period of PER2::LUC expression in SCN explants ex vivo. Further, it is worth noting that the shortened period of wheel-running after acute CS paralleled the effects of CSE on SCN tissue explants. Together, these data suggest that CS exposure affects both lung and brain clock function. Although the effects of CS as an agent of circadian disruption may not be as severe as the effects of jet lag or shift work, it is reasonable to speculate that even subtle alteration of clock function can have a substantial long-term effect on clock-dependent physiology. The differential and opposing effects of CSE on the period and phase of clock gene expression in the lung and SCN suggest that internal circadian disruption due to phase dissociation between these central and peripheral clocks is a factor in the development of diseases such as COPD.
Emerging evidence indicates that inflammation and immune functions are governed, in part, by the circadian timing system (41–43). However, it is not known to what extent circadian rhythms in the immune inflammatory system are modulated by timing cues delivered to this system from sources such as the SCN or are regulated locally by peripheral clocks. Circadian desynchrony or environmental circadian disruption during experimental jet lag increases the susceptibility to inflammation through dysregulation of innate immune responses (44). Consistent with other peripheral tissues, circadian timing in the lung, although able to oscillate independently (14–16), is synchronized with other tissues and the external environment by the central clock in the SCN (1). We have shown that CS exposure disrupts circadian expression of clock and clock-controlled genes in the lung, resulting in altered patterns of inflammatory cytokine gene expression and secretion. It is possible that the effects of acute CS exposure (10 d) in mice were amplified by the fact that CS exposure was conducted during the active phase in mice housed in an inverted L:D cycle (lights on from 6 PM to 6 AM). Because of the limitations of our facility, it was not feasible to conduct long-term (chronic CS) exposure studies in an inverted L:D cycle to confirm this possibility. Hence, we do not see a strong correlation in the effects of CS on clock gene expression between the acute and chronic treatment groups. Although certainly adequate, a 6-h time resolution is a less than ideal sampling frequency for detection of subtle variation in gene expression and protein levels across the 24-h day. That said, we were able to detect significant changes in the phase of several clock genes after both acute and chronic CS exposure with that sampling frequency. Studies designed to interrogate subtle changes in clock gene expression following in vivo CS exposure at more frequent sampling rates are warranted. Similar effects of CS on circadian rhythms of cardiac function and lung tumorigenesis have been reported (45–47). Given the parallel nature of the responses, it is reasonable to define CS exposure as a form of environmental circadian disruption, capable of altering the molecular clock and clock-dependent processes in a manner similar to repeated jet lag or rotating shift work. These effects may be more adverse, given that the disruption occurs in the presence of a regular 12:12 L:D cycle and apparently normal entrainment. Furthermore, it is possible that CS-induced proinflammatory cytokines contribute to circadian disruption via TLRs/NF-κB (48–50), as CS components may recognize TLRs and activate NF-κB for proinflammatory responses (12, 51). In acute CS exposure, neutrophil influx was observed at ZT24, but not at ZT0, which is in fact the same time point. However, neutrophil influx in the lung usually increases 16 h after the last CS exposure. Thus, we expected to see a delayed increase in neutrophil influx. Earlier reports from our laboratory have clearly demonstrated an increase in proinflammatory cytokine release and neutrophil influx after acute CS exposure at a concentration of 300 mg/m3 TPM, which was achieved with a mainstream CS exposure system (Baumgartner-Jaeger CSM2072i, CH Technologies; ref. 33). This response is not attainable with the Teague smoke exposure system [a mixture of sidestream (89%) and mainstream (11%) smoke] used in the current study (∼100 mg/m3 TPM). Furthermore, in our protocol, there was no influx of neutrophils into the BAL fluid with the use of the Teague machine for CS exposure.
The role of clock proteins in increased susceptibility to oxidative stress–mediated inflammatory and injurious responses is implicated. For example, PER2 mutant mice are known to be sensitive to oxidative stress and to be susceptible or prone to the development of cancer (9). Recently, it was reported that BMAL1-deficient mice have an increased sensitivity to oxidative stress that correlates with cellular senescence (11). Although the nuclear receptor REV-ERBα, which is known as a negative regulator of BMAL1, has been reported to have a role in the regulation of inflammatory mediators (41, 44, 52), evidence supporting the role of BMAL1 in oxidative stress-induced inflammation remains elusive.
Cigarette smoking not only induces oxidative stress, but is also associated with clinical depression (53, 54) and sleep disorders (55). Patients with COPD have an early morning surge in respiratory symptoms (cough, sputum production, and wheezing) and sleep abnormalities (13, 14, 17, 19, 20). Disrupted sleep in patients with COPD correlates with respiratory symptoms (cough, sputum production, and wheezing), nocturnal oxygen desaturation, hypercapnia, and circadian changes in airway caliber and resistance (20, 56). Each of these effects of chronic tobacco/CS is closely related to a significant deterioration in quality of life in smokers and contributes to the development of COPD. In these patients depression often lasts even after smoking cessation (57) and is a frequent comorbidity of COPD (58–61). However, the underlying molecular and cellular mechanisms for CS-induced circadian abnormalities are not fully understood. In this study, both acute and chronic CS exposure reduced nighttime activity in mice. Surprisingly, chronically exposed mice were less active only through 3 mo of CS exposure. Contrary to our expectations, locomotor activity was similar in the air- and CS-exposure groups after 6 mo. When we compared the overall activity levels in mice from the beginning (1st mo) of the experiment until completion (6th mo), there was a clear age-dependent decline in total activity levels. Hence, the overall reduction in activity in aged mice may have masked the effects of chronic CS exposure. Furthermore, it is possible that the effects of chronic CS exposure on the brain are less pronounced because of the habituation of neural responses to CS. Regardless, our data suggest that reduced sleep quality in patients with COPD is a direct consequence of altered clock function in sleep-regulatory centers of the brain, including the central pacemaker in the SCN.
SIRT1 is an NAD+-dependent deacetylase involved in regulation of various biological processes. The levels of SIRT1 are reduced through protein carbonylation in response to CS exposure in the lung (26, 62). A reduction in the levels and activity of SIRT1 in response to CS leads to lung inflammation, emphysema, and senescence (27). Recently, it has been reported that SIRT1 has an important role in the regulation of circadian clock mechanisms by affecting circadian clock gene transcription in an NAD+-dependent manner (24, 25, 63). Another interesting report showed that SIRT1 in the brain governs central circadian control and activates transcription of the BMAL1:CLOCK transactivator complex by amplifying the SIRT1-PGC-1α-Nampt axis (64). Altered SIRT1 levels in the brain also exert moderate changes in the intrinsic circadian periods of young and old mice (64). SIRT1 plays an essential role in the age-dependent decline in central clock function, and targeting these circadian regulated loops in the brain (SIRT1, PGC-1α, and Nampt) may provide novel strategies for ameliorating the negative effects of aging on circadian clock function (64). SIRT1 affects the molecular clock by binding with CLOCK:BMAL1 complexes and deacetylating the BMAL1 and PER2 proteins (21–24). Thus, CS could affect clock function by reducing SIRT1, leading to increased inflammatory responses in the lung. Our preliminary observations showed that PER2 levels, which are CLOCK:BMAL1 dependent, are reduced in lungs of patients with COPD. CS also decreased abundance of PER2, which may be acetylated and regulated by SIRT1 in lungs of mice exposed to CS. Our results show that SIRT1 expression is rhythmic in the lungs of air-exposed mice, which is consistent with a previous report (24). CS significantly reduced SIRT1 protein abundance, concomitant with a reduction in BMAL1 levels, and increased acetylation on lysine residue K537 of the remaining BMAL1 protein. These results were confirmed in the lungs of SIRT1-deficient and SIRT1-overexpressing mice that were exposed to CS and paralleled a report showing that acetylation of BMAL1 on lysine residue K537 is regulated by SIRT1 (25). We found a modest decrease in BMAL1 levels in lung tissue from patients with COPD, although it was still significant when compared with that in lung tissue of nonsmokers. We also observed a considerable variation in the expression levels of BMAL1 in the lungs of patients with COPD, compared to those of nonsmokers, which is not unexpected, given a large variation in the intensities and grades of COPD severity (GOLD stages I–IV), timing of sample collections, the duration and amount of cigarette inhalation and exposure, and the possible influence of prescribed drugs, which varies greatly between patients. To our knowledge, this is the first study showing that SIRT1 is expressed with a circadian rhythm under clock control in lung tissue. Further, we report for the first time that BMAL1 acetylation in the lung in vivo is due to a CS-dependent reduction in SIRT1 levels. There is emerging evidence that circadian rhythms govern proinflammatory cytokine gene expression (41, 44, 50). It is also known that inflammation and alteration in clock gene expression (BMAL1 and PER2) are intimately associated with fatigue and diminished locomotor activity (65–67). Hence, it is possible that SIRT1 acts as a critical link between core clock function and inflammatory response in the lung.
BMAL1, as part of the primary enhancer complex with CLOCK, has a pivotal role in regulation of circadian clock function through transactivation of clock components and downstream clock-controlled genes. Acetylation is linked to BMAL1 phosphorylation (25, 67, 68), possibly leading to its nuclear accumulation (68) and subsequent degradation (67). The results presented here reveal that CS exposure induced increased acetylation of BMAL1 and its loss of stability. Intriguingly, as shown in a previous study (25), we observed that the level of BMAL1 was greater in the lungs of SIRT1-deficient mice, suggesting the SIRT1 is involved in BMAL1 stability. Further study is needed to investigate SIRT1-regulated mechanisms involved in BMAL1 stability, shuttling, phosphorylation, and acetylation in response to CS exposure and in the development of COPD.
BMAL1-deficient mice show signs of advanced aging and an age-related phenotype, correlating with increased levels of ROS and cellular senescence (69). CS exposure reduced the levels of BMAL1 mRNA and protein, and simultaneously evoked lung inflammatory responses, suggesting that the loss of BMAL1 in response to CS exposure increases inflammatory responses through deregulation of CS-induced oxidative stress. It has been shown that BMAL1 interacts with the CCL1/MCP1 promoter, enhancing CCL1/MCP1 gene expression. We found a sharp surge of CCL1/MCP1 in the lungs of CS-exposed WT mice, which was augmented in the lungs of CS-exposed mice harboring lung epithelium-specific deletion of bmal1. Although the specific mechanisms remain unknown, it is likely that BMAL1 has a role in the regulation of oxidative stress and could be a target of SIRT1 in oxidative stress–induced inflammatory responses. A recent study showed a similar pharmacologic activation of SIRT1 in modulation of circadian rhythms in liver via a reduction in H3K9/K14 acetylation (70). Furthermore, augmented inflammatory responses observed in CS-exposed mice harboring a lung epithelium–specific bmal1 deletion compared to WT mice suggest the involvement of the epithelial molecular clock in regulation of CS-induced lung inflammation.
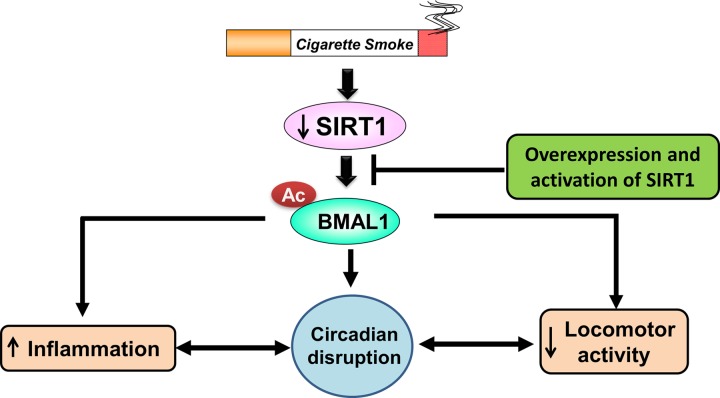
In conclusion, our data show that environmental CS exposure in mice caused alterations in circadian clock gene expression in brain and lung tissue, altered rhythms of locomotor activity, and increased lung inflammation and emphysema. BMAL1 levels were reduced in lung tissue from patients with COPD, most likely owing to increased turnover mediated by enhanced acetylation of BMAL1 by SIRT1. These data clearly indicate that circadian rhythms of lung function are dampened in patients with COPD, which is associated with abnormal airway inflammation. We also report that BMAL1 regulated CS-induced lung inflammatory responses through SIRT1-controlled acetylation and stability of BMAL1 in lung epithelium (Fig. 13). Thus, our results highlight the importance of the molecular clock in the regulation of lung inflammation and injurious responses caused by CS. Overall, CS-mediated disruption of circadian clock function has implications in the pathogenesis of COPD. Understanding molecular clock function in the lung and its association with daily lung physiological function, particularly as it relates to the response to tobacco/CS, may strengthen the rationale for chronotherapy in COPD management.
Figure 13.
Environmental circadian disruption after CS exposure is SIRT1-BMAL1 dependent, and is associated with increased inflammation and reduced locomotor activity. CS exposure affects SIRT1 levels in the lung, which leads to BMAL1 acetylation/degradation, culminating in increased inflammation, circadian disruption (altered gene expression of clock and clock-controlled genes), and reduced locomotor activity. Overexpression or pharmacologic activation of SIRT1 in cell-specific BMAL1-knockout mice does not attenuate lung inflammation, suggesting that the effects of CS on circadian clock function and inflammatory responses are mediated almost entirely by SIRT1-BMAL1-dependent mechanism.
Supplementary Material
Acknowledgments
The authors thank Dr. Thomas J. Mariani (University of Rochester, Rochester, NY) for providing the Cre recombinase transgenic mice with CC10 promoter, Dr. Michael McBurney (University of Ottawa, Ottawa, ON, Canada) for providing SIRT1-knockout mice (SIRT1-deficient mice), Dr. Leonard Guarente (Massachusetts Institutes of Technology, Cambridge, MA, USA) and Dr. Wei Gu (Columbia University, New York, NY, USA) for providing Tg mice overexpressing Sirt1 (SIRT1 Tg mice), and Dr. Sangwoon Chung, Katherine Bachmann, Lindsay Marchetti, Zachary Murphy, Drew Phillips, Suzanne Bellanca, and Stephanie Uhrinek (University of Rochester, Rochester, NY, USA) for technical assistance.
This study was supported by grants from the U.S. National Institutes of Health (1R01HL097751, 1R01HL092842) to I.R. and a grant from the National Institute of Environmental Health Sciences (NIEHS) Environmental Health Science Center (P30-ES01247).
The authors declare no conflicts of interest. Author contributions: J.H. performed mouse studies, cell counts, immunoblotting, real-time PCR analysis, ELISA, and manuscript writing; I.S.K. performed cell counts, real-time PCR analysis, ELISA, mean linear intercept (Lm) analysis, immunoblotting, immunoprecipitation, and overall data collection and analysis; J.H. and I.S.K. performed: data analysis for circadian experiments and cell counts; H.Y., I.S.K., M.T.S., and I.R. performed: interpretation of results and manuscript editing; M.T.S. performed wheel running activity assay and, PER2::luciferase assays; I.R. performed study design and manuscript writing.
Dedication:
The authors dedicate this work to the loving memory of Dr. Vuokko L. Kinnula (Pulmonary Division, Department of Medicine and Pathology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland), who provided the human tissue samples and left us suddenly during the preparation of this article.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ANOVA
- analysis of variance
- BAL
- bronchoalveolar lavage
- BW
- body weight
- COG
- center of gravity
- COPD
- chronic obstructive pulmonary disease
- CRY
- cryptochrome
- CS
- cigarette smoke
- CSE
- cigarette smoke extract
- E
- tissue elastance
- H&E
- hematoxylin and eosin
- IP-10
- CXCL10/interferon-γ inducible protein 10
- KC
- CXCL1/keratinocyte-derived chemokine
- Lm
- mean linear intercept
- MCP-1
- CCL2/monocyte chemotactic protein
- PER
- Period
- qPCR
- quantitative real-time PCR
- QsC
- quasi-static compliance
- R
- lung resistance
- SCN
- suprachiasmatic nucleus
- SIRT1
- Sirtuin1
- Tg
- transgenic
- TPM
- total particulate matter
- ZT
- zeitgeber time
REFERENCES
- 1. Mohawk J. A., Green C. B., Takahashi J. S. (2012) Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dibner C., Schibler U., Albrecht U. (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 [DOI] [PubMed] [Google Scholar]
- 3. Bechtold D. A., Gibbs J. E., Loudon A. S. (2010) Circadian dysfunction in disease. Trends Pharmacol. Sci. 31, 191–198 [DOI] [PubMed] [Google Scholar]
- 4. Mortola J. P., Seifert E. L. (2002) Circadian patterns of breathing. Respir. Physiol. Neurobiol. 131, 91–100 [DOI] [PubMed] [Google Scholar]
- 5. Hadden H., Soldin S. J., Massaro D. (2012) Circadian disruption alters mouse lung clock gene expression and lung mechanics. J. Appl. Physiol. 113, 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boysen P. G., Block A. J., Wynne J. W., Hunt L. A., Flick M. R. (1979) Nocturnal pulmonary hypertension in patients with chronic obstructive pulmonary disease. Chest 76, 536–542 [DOI] [PubMed] [Google Scholar]
- 7. Spengler C. M., Shea S. A. (2000) Endogenous circadian rhythm of pulmonary function in healthy humans. Am. J. Respir. Crit. Care Med. 162, 1038–1046 [DOI] [PubMed] [Google Scholar]
- 8. Sukumaran S., Jusko W. J., Dubois D. C., Almon R. R. (2011) Light-dark oscillations in the lung transcriptome: implications for lung homeostasis, repair, metabolism, disease, and drug action. J. Appl. Physiol. 110, 1732–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu L., Pelicano H., Liu J., Huang P., Lee C. (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50 [DOI] [PubMed] [Google Scholar]
- 10. Zheng X., Yang Z., Yue Z., Alvarez J. D., Sehgal A. (2007) FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 104, 15899–15904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khapre R. V., Kondratova A. A., Susova O., Kondratov R. V. (2011) Circadian clock protein BMAL1 regulates cellular senescence in vivo. Cell Cycle 10, 4162–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yao H., Rahman I. (2011) Current concepts on oxidative/carbonyl stress, inflammation and epigenetics in pathogenesis of chronic obstructive pulmonary disease. Toxicol. Appl. Pharmacol. 254, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoegh S. V., Sorensen G. L., Tornoe I., Lottenburger T., Ytting H., Nielsen H. J., Junker P., Holmskov U. (2010) Long-term stability and circadian variation in circulating levels of surfactant protein D. Immunobiology 215, 314–320 [DOI] [PubMed] [Google Scholar]
- 14. Thomas A., Petro W., Konietzko N. (1993) [The circadian rhythm of ciliary beat frequency of human nasal cilia in probands with healthy lungs and in patients with chronic obstructive lung disease: includes adrenergic stimulation by terbutaline]. Pneumologie 47, 526–530; German [PubMed] [Google Scholar]
- 15. Gebel S., Gerstmayer B., Kuhl P., Borlak J., Meurrens K., Muller T. (2006) The kinetics of transcriptomic changes induced by cigarette smoke in rat lungs reveals a specific program of defense, inflammation, and circadian clock gene expression. Toxicol. Sci. 93, 422–431 [DOI] [PubMed] [Google Scholar]
- 16. Gibbs J. E., Beesley S., Plumb J., Singh D., Farrow S., Ray D. W., Loudon A. S. (2009) Circadian timing in the lung: a specific role for bronchiolar epithelial cells. Endocrinology 150, 268–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Casale R., Pasqualetti P. (1997) Cosinor analysis of circadian peak expiratory flow variability in normal subjects, passive smokers, heavy smokers, patients with chronic obstructive pulmonary disease and patients with interstitial lung disease. Respiration 64, 251–256 [DOI] [PubMed] [Google Scholar]
- 18. Petty T. L. (1988) Circadian variations in chronic asthma and chronic obstructive pulmonary disease. Am. J. Med. 85, 21–23 [DOI] [PubMed] [Google Scholar]
- 19. Tsai C. L., Brenner B. E., Camargo C. A., Jr. (2007) Circadian-rhythm differences among emergency department patients with chronic obstructive pulmonary disease exacerbation. Chronobiol. Int. 24, 699–713 [DOI] [PubMed] [Google Scholar]
- 20. Agusti A., Hedner J., Marin J. M., Barbe F., Cazzola M., Rennard S. (2011) Night-time symptoms: a forgotten dimension of COPD. Eur. Respir. Rev. 20, 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Belden W. J., Dunlap J. C. (2008) SIRT1 is a circadian deacetylase for core clock components. Cell 134, 212–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 23. Grimaldi B., Nakahata Y., Kaluzova M., Masubuchi S., Sassone-Corsi P. (2009) Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int. J. Biochem. Cell Biol. 41, 81–86 [DOI] [PubMed] [Google Scholar]
- 24. Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W., Schibler U. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 [DOI] [PubMed] [Google Scholar]
- 25. Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rajendrasozhan S., Yang S. R., Kinnula V. L., Rahman I. (2008) SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 177, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao H., Chung S., Hwang J. W., Rajendrasozhan S., Sundar I. K., Dean D. A., McBurney M. W., Guarente L., Gu W., Ronty M., Rahman I. (2012) SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J. Clin. Invest. 122, 2032–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang S. R., Wright J., Bauter M., Seweryniak K., Kode A., Rahman I. (2007) Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L567–L576 [DOI] [PubMed] [Google Scholar]
- 29. Simon D. M., Arikan M. C., Srisuma S., Bhattacharya S., Tsai L. W., Ingenito E. P., Gonzalez F., Shapiro S. D., Mariani T. J. (2006) Epithelial cell PPARγ contributes to normal lung maturation. FASEB J. 20, 1507–1509 [DOI] [PubMed] [Google Scholar]
- 30. McBurney M. W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J. R., Lansdorp P. M., Lemieux M. (2003) The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23, 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bordone L., Cohen D., Robinson A., Motta M. C., van Veen E., Czopik A., Steele A. D., Crowe H., Marmor S., Luo J., Gu W., Guarente L. (2007) SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6, 759–767 [DOI] [PubMed] [Google Scholar]
- 32. Yao H., Arunachalam G., Hwang J. W., Chung S., Sundar I. K., Kinnula V. L., Crapo J. D., Rahman I. (2010) Extracellular superoxide dismutase protects against pulmonary emphysema by attenuating oxidative fragmentation of ECM. Proc. Natl. Acad. Sci. U. S. A. 107, 15571–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yao H., Edirisinghe I., Rajendrasozhan S., Yang S. R., Caito S., Adenuga D., Rahman I. (2008) Cigarette smoke-mediated inflammatory and oxidative responses are strain-dependent in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L1174–L1186 [DOI] [PubMed] [Google Scholar]
- 34. Hwang J. W., Rajendrasozhan S., Yao H., Chung S., Sundar I. K., Huyck H. L., Pryhuber G. S., Kinnula V. L., Rahman I. (2011) FOXO3 deficiency leads to increased susceptibility to cigarette smoke-induced inflammation, airspace enlargement, and chronic obstructive pulmonary disease. J. Immunol. 187, 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoon J. A., Han D. H., Noh J. Y., Kim M. H., Son G. H., Kim K., Kim C. J., Pak Y. K., Cho S. (2012) Meal time shift disturbs circadian rhythmicity along with metabolic and behavioral alterations in mice. PLoS One 7, e44053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) Period2: :luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U. S. A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Foronjy R. F., Mercer B. A., Maxfield M. W., Powell C. A., D'Armiento J., Okada Y. (2005) Structural emphysema does not correlate with lung compliance: lessons from the mouse smoking model. Exp. Lung Res. 31, 547–562 [DOI] [PubMed] [Google Scholar]
- 38. Guerassimov A., Hoshino Y., Takubo Y., Turcotte A., Yamamoto M., Ghezzo H., Triantafillopoulos A., Whittaker K., Hoidal J. R., Cosio M. G. (2004) The development of emphysema in cigarette smoke-exposed mice is strain dependent. Am. J. Respir. Crit. Care Med. 170, 974–980 [DOI] [PubMed] [Google Scholar]
- 39. Oster H., Damerow S., Hut R. A., Eichele G. (2006) Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J. Biol. Rhythms 21, 350–361 [DOI] [PubMed] [Google Scholar]
- 40. Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. (2007) Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keller M., Mazuch J., Abraham U., Eom G. D., Herzog E. D., Volk H. D., Kramer A., Maier B. (2009) A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. U. S. A. 106, 21407–21412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Logan R. W., Sarkar D. K. (2012) Circadian nature of immune function. Mol. Cell. Endocrinol. 349, 82–90 [DOI] [PubMed] [Google Scholar]
- 43. Narasimamurthy R., Hatori M., Nayak S. K., Liu F., Panda S., Verma I. M. (2012) Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc. Natl. Acad. Sci. U. S. A. 109, 12662–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Castanon-Cervantes O., Wu M., Ehlen J. C., Paul K., Gamble K. L., Johnson R. L., Besing R. C., Menaker M., Gewirtz A. T., Davidson A. J. (2010) Dysregulation of inflammatory responses by chronic circadian disruption. J. Immunol. 185, 5796–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen C. Y., Chow D., Chiamvimonvat N., Glatter K. A., Li N., He Y., Pinkerton K. E., Bonham A. C. (2008) Short-term secondhand smoke exposure decreases heart rate variability and increases arrhythmia susceptibility in mice. Am. J. Physiol. Heart Circ. Physiol. 295, H632–H639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gentner N. J., Weber L. P. (2012) Secondhand tobacco smoke, arterial stiffness, and altered circadian blood pressure patterns are associated with lung inflammation and oxidative stress in rats. Am. J. Physiol. Heart Circ. Physiol. 302, H818–H825 [DOI] [PubMed] [Google Scholar]
- 47. Vasu V. T., Cross C. E., Gohil K. (2009) Nr1d1, an important circadian pathway regulatory gene, is suppressed by cigarette smoke in murine lungs. Integr. Cancer Ther. 8, 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silver A. C., Arjona A., Walker W. E., Fikrig E. (2012) The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36, 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leone M. J., Marpegan L., Duhart J. M., Golombek D. A. (2012) Role of proinflammatory cytokines on lipopolysaccharide-induced phase shifts in locomotor activity circadian rhythm. Chronobiol. Int. 29, 715–723 [DOI] [PubMed] [Google Scholar]
- 50. Spengler C. M., Shea S. A. (2012) Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc. Natl. Acad. Sci. U. S. A. 109, E2457–E2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yao H., Edirisinghe I., Yang S. R., Rajendrasozhan S., Kode A., Caito S., Adenuga D., Rahman I. (2008) Genetic ablation of NADPH oxidase enhances susceptibility to cigarette smoke-induced lung inflammation and emphysema in mice. Am. J. Pathol. 172, 1222–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gibbs J. E., Blaikley J., Beesley S., Matthews L., Simpson K. D., Boyce S. H., Farrow S. N., Else K. J., Singh D., Ray D. W., Loudon A. S. (2012) The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. U. S. A. 109, 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murphy J. M., Horton N. J., Monson R. R., Laird N. M., Sobol A. M., Leighton A. H. (2003) Cigarette smoking in relation to depression: historical trends from the Stirling County Study. Am. J. Psychiatry 160, 1663–1669 [DOI] [PubMed] [Google Scholar]
- 54. Mykletun A., Overland S., Aaro L. E., Liabo H. M., Stewart R. (2008) Smoking in relation to anxiety and depression: evidence from a large population survey: the HUNT study. Eur. Psychiatry 23, 77–84 [DOI] [PubMed] [Google Scholar]
- 55. Hida A., Kitamura S., Mishima K. (2012) Pathophysiology and pathogenesis of circadian rhythm sleep disorders. J. Physiol. Anthropol. 31, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lewis D. A. (2001) Sleep in patients with asthma and chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 7, 105–112 [DOI] [PubMed] [Google Scholar]
- 57. Gierisch J. M., Bastian L. A., Calhoun P. S., McDuffie J. R., Williams J. W., Jr. (2012) Smoking cessation interventions for patients with depression: a systematic review and meta-analysis. J. Gen. Intern. Med. 27, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Antoniu S. A. (2011) Predictors of depression in chronic obstructive pulmonary disease patients. Expert Rev. Respir. Med. 5, 333–335 [DOI] [PubMed] [Google Scholar]
- 59. Divo M., Cote C., de Torres J. P., Casanova C., Marin J. M., Pinto-Plata V., Zulueta J., Cabrera C., Zagaceta J., Hunninghake G., Celli B. (2012) Comorbidities and risk of mortality in patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 186, 155–161 [DOI] [PubMed] [Google Scholar]
- 60. Hanania N. A., Mullerova H., Locantore N. W., Vestbo J., Watkins M. L., Wouters E. F., Rennard S. I., Sharafkhaneh A. (2011) Determinants of depression in the ECLIPSE chronic obstructive pulmonary disease cohort. Am. J. Respir. Crit. Care Med. 183, 604–611 [DOI] [PubMed] [Google Scholar]
- 61. Laurin C., Moullec G., Bacon S. L., Lavoie K. L. (2012) Impact of anxiety and depression on chronic obstructive pulmonary disease exacerbation risk. Am. J. Respir. Crit. Care Med. 185, 918–923 [DOI] [PubMed] [Google Scholar]
- 62. Caito S., Rajendrasozhan S., Cook S., Chung S., Yao H., Friedman A. E., Brookes P. S., Rahman I. (2010) SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 24, 3145–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chang H. C., Guarente L. (2013) SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153, 1448–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cavadini G., Petrzilka S., Kohler P., Jud C., Tobler I., Birchler T., Fontana A. (2007) TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc. Natl. Acad. Sci. U. S. A. 104, 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Okada K., Yano M., Doki Y., Azama T., Iwanaga H., Miki H., Nakayama M., Miyata H., Takiguchi S., Fujiwara Y., Yasuda T., Ishida N., Monden M. (2008) Injection of LPS causes transient suppression of biological clock genes in rats. J. Surg. Res. 145, 5–12 [DOI] [PubMed] [Google Scholar]
- 67. Kwon I., Lee J., Chang S. H., Jung N. C., Lee B. J., Son G. H., Kim K., Lee K. H. (2006) BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell. Biol. 26, 7318–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tamaru T., Isojima Y., van der Horst G. T., Takei K., Nagai K., Takamatsu K. (2003) Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells 8, 973–983 [DOI] [PubMed] [Google Scholar]
- 69. Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P. (2006) Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20, 1868–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bellet M. M., Nakahata Y., Boudjelal M., Watts E., Mossakowska D. E., Edwards K. A., Cervantes M., Astarita G., Loh C., Ellis J. L., Vlasuk G. P., Sassone-Corsi P. (2013) Pharmacological modulation of circadian rhythms by synthetic activators of the deacetylase SIRT1. Proc. Natl. Acad. Sci. U. S. A. 110, 3333–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.