Abstract
The κ-opioid receptor (KOR)-dynorphin system has been implicated in the control of affect, cognition, and motivation, and is thought to be dysregulated in mood and psychotic disorders, as well as in various phases of opioid dependence. KOR agonists exhibit analgesic effects, although the adverse effects produced by some KOR agonists, including sedation, dysphoria, and hallucinations, have limited their clinical use. Interestingly, KOR-mediated dysphoria, assessed in rodents as aversion, has recently been attributed to the activation of the p38 mitogen-activated protein kinase pathway following arrestin recruitment to the activated KOR. Therefore, KOR-selective G protein–biased agonists, which do not recruit arrestin, have been proposed to be more effective analgesics, without the adverse effects triggered by the arrestin pathway. As an initial step toward identifying novel biased KOR agonists, we applied a multifaceted screening strategy utilizing both in silico and parallel screening approaches. We identified several KOR-selective ligand scaffolds with a range of signaling bias in vitro. The arylacetamide-based scaffold includes both G protein– and β-arrestin–biased ligands, while the endogenous peptides and the diterpene scaffolds are G protein biased. Interestingly, we found scaffold screening to be more successful than library screening in identifying biased ligands. Many of the identified functionally selective ligands are potent selective KOR agonists that are reported to be active in the central nervous system. They therefore represent excellent candidates for in vivo studies aiming at determining the behavioral effects mediated by specific KOR-mediated signaling cascades.
Introduction
The κ-opioid receptor (KOR)-dynorphin system has been implicated in the pathogenesis and pathophysiology of affective disorders, drug addiction, and psychotic disorders (Sheffler and Roth, 2003; Bruchas and Chavkin, 2010). KOR and dynorphin are highly expressed in regions of the brain implicated in the modulation of reward, mood, cognition, and perception (ventral tegmental area, nucleus accumbens, prefrontal cortex, hippocampus, striatum, amygdala, and hypothalamus) (Land et al., 2008; Schwarzer, 2009; Knoll and Carlezon, 2010; Tejeda et al., 2012). Accordingly, drugs directed at KOR as antagonists or partial agonists have potential utility for a number of indications, especially as antidepressants and anxiolytics (Carlezon et al., 2009). Additionally, KOR agonists are gaining attention as potential antiaddiction medications and analgesics without a high abuse potential (Tao et al., 2008; Wee and Koob, 2010; Prevatt-Smith et al., 2011). However, the adverse effects produced by many centrally active KOR agonists, including sedation, dysphoria, and hallucinations, have limited their clinical development (Pfeiffer et al., 1986). Dysphoria has been considered the best surrogate marker of KOR agonism, while the hallucinogenic effects of KOR agonists have been relatively unexplored, except in the case of salvinorin A (Roth et al., 2002; White and Roth, 2012).
KOR stimulation leads to the activation of the canonical Gαi signaling cascade, the recruitment of β-arrestin, and activation of p38 mitogen-activated protein kinase and an array of other downstream effectors (Appleyard et al., 1997; Bruchas et al., 2006; Land et al., 2009). It has been hypothesized that the dysphoric effects of KOR agonism is mediated through the arrestin-dependent activation of p38 mitogen-activated protein kinase, while the analgesic effects of KOR agonism are mediated only through G protein signaling (Bruchas et al., 2007). This suggests the potential for functionally selective ligands of KOR as analgesics devoid of dysphoric effects. Ligands that differentially stimulate canonical and noncanonical transduction pathways are considered to be “functionally selective” (Urban et al., 2007), and their differential engagement in signaling is referred to as “biased.” Identifying functionally selective KOR agonists with extreme signaling bias will be useful for determining which signal transduction pathways are important for therapeutic efficacy and which signaling cascades contribute to the side effects (Allen et al., 2011). Due to the diverse structure of KOR ligands, there is the potential to discover a variety of functionally selective ligands that can be used to probe KOR signaling, as well as to improve KOR-based therapeutics. The goal of this study was to identify a range of chemotypes of functionally selective KOR ligands using a parallel in vitro screening approach accompanied by in silico selection.
KOR agonists can be classified into five chemotypes: the endogenous peptides (dynorphins), the benzodiazepines (tifluadom), the benzomorphans (ketazocine), the arylacetamides (U69593; (+)-(5α,7α,8β)-N-methyl-N-[7-(1-pyrrolidinyl)-1-oxaspiro[4.5]dec-8-yl]-benzeneacetamide), and the diterpenes (salvinorin A). Dynorphins (Dyns) have been implicated in addiction and drug-seeking, mood disorders, and the stress response (Bruchas and Chavkin, 2010). The benzomorphans, such as bremazocine, have limited KOR selectivity but show strong analgesic effects. However, despite their low dependence potential, they were removed from clinical development due to psychotomimetic and dysphoric effects (Dortch-Carnes and Potter, 2005). It was originally thought that the negative side effects of KOR agonists were due to off-target effects, and a new class of selective KOR agonists—the arylacetamide derivatives such as U69593—was developed to circumvent these potential shortcomings. However, some arylacetamides are also reported to produce hallucinations and aversion (Millan, 1990). The diterpenes, represented by salvinorin A (which is the main psychoactive compound in Salvia divinorum), represent a novel scaffold of highly potent and selective KOR agonists with no appreciable affinity for any other known neurotransmitter system or receptor (Roth et al., 2002).
Functionally selective ligands at other targets have been identified by screening derivatives of known ligand scaffolds in a parallel fashion, in which libraries of analogs are screened simultaneously against multiple downstream effector pathways (see, for instance, Huang et al., 2009; Allen et al., 2011; Chen et al., 2012; Wacker et al., 2013). The extent of functional selectivity of those compounds, or bias factor, can be quantified using the operational model (Black and Leff, 1983; Kenakin et al., 2012; Kenakin and Christopoulos, 2013; Wacker et al., 2013). Accordingly, we sought to identify and quantify the degree of bias for representative scaffolds that maintain high affinity and selectivity for KOR.
Materials and Methods
Drugs.
The National Institutes of Health Clinical Collection (NCC) library used here is a publicly available library consisting of Food and Drug Administration–approved drugs we have previously used to identify biologically active drugs (Huang et al., 2009, 2011). The synthesis of the RB family of salvinorin derivatives used here has been previously described: 22-chlorosalvinorin A (RB 48), 22-thiocyanatosalvinorin A (RB 64), 22-bromosalvinorin A (RB 50), (22R,S)-22-chloro-22-methylsalvinorin A (RB 55), (22S)-22-chloro-22-methylsalvinorin A (RB 55–1), (22R)-22-chloro-22-methylsalvinorin A (RB 55–2), 22-cyanosalvinorin A (RB 59), and 22-methoxysalvinorin A (RB 65) (Yan et al., 2009). Salvinorin A was isolated from dried leaves of S. divinorum purified as previously reported (Kutrzeba et al., 2009) and hydrolyzed to salvinorin B, which was a starting material for the synthesis of all analogs.
Dynorphin 1–13, dynorphin 1–11, dynorphin 1–9, and dynorphin 1–8 are all obtained from the National Institute on Drug Abuse drug supply program. U69593, (±)-(5α,7α,8β)-3,4-dichloro-N-methyl-N-[7-(1-pyrrolidinyl)-1-oxaspiro[4.5]dec-8-yl]benzeneacetamide mesylate salt (spiradoline, U62066), 17-cyclopropylmethyl-6,7-dehydro-4,5-epoxy-3,14-dihydroxy-6,7,2′,3′-indolomorphinan (naltrindole), L-N-cyclobutylmethyl-3,14-dihydroxymorphinan (+)-tartrate salt (butorphanol), and 17-(cyclobutylmethyl)-4,5-epoxymorphinan-3,6,14-triol hydrochloride hydrate (nalbuphine) were purchased from Sigma-Aldrich (St. Louis, MO). 4-([3,4-Dichlorophenyl]acetyl)-3-(1-pyrrolidinylmethyl)-1-piperazinecarboxylic acid methyl ester fumarate salt (GR89696), 2-(3,4-dichlorophenyl)-N-methyl-N-([1S]-1-phenyl-2-[1-pyrrolidinyl]ethyl)acetamide hydrochloride (ICI 199,441), trans-(−)-3,4-dichloro-N-methyl-N-[2-(1-pyrrolidinyl)cyclohexyl]benzeneacetamide hydrochloride [(−)U50,488], trans-(+)-3,4-dichloro-N-methyl-N-(2-[1-pyrrolidinyl]-cyclohexyl)benzeneacetamide hydrochloride [(+)U50,488], 2-(3,4-dichlorophenyl)-N-methyl-N-([1S]-1-[3-isothiocyanatophenyl]-2-[1-pyrrolidinyl]ethyl)acetamide hydrochloride (DIPPA), (±)-1-(3,4-dichlorophenyl)acetyl-2-(1-pyrrolidinyl)methylpiperidine hydrochloride (BRL 52537), N-methyl-N-([1S]-1-phenyl-2-[1-pyrrolidinyl]ethyl)phenylacetamide hydrochloride (N-MPPP), (RS)-[3-[1-[[(3,4-dichlorophenyl)acetyl]methylamino]-2-(1-pyrrolidinyl)ethyl]phenoxy]acetic acid hydrochloride (ICI 204,448), and dynorphin A were purchased from Tocris Bioscience (Bristol, UK). 3-(Cyclopropylmethyl)-6,11-dimethyl-1,2,3,4,5,6-hexahydro-2,6-methano-3-benzazocin-8-ol (cyclazocine) and (5α,7α)-17-(cyclopropylmethyl)- 4,5-epoxy-18,19-dihydro-3-hydroxy-6-methoxy-α,α-dimethyl-6,14-ethenomorphinan-7-methanol (diprenorphine) were acquired from the drug supply program of the National Institute on Drug Abuse.
The synthesis of N-naphthoyl-β-naltrexamine (β-NNTA), 6′-guanidino-17-(cyclopropylmethyl)-6,7-didehydro-4,5α-epoxy-3,14-dihydroxyindolo[2′,3′:6,7]morphinan (6′-GNTI), and 5′-guanidino-17-(cyclopropylmethyl)-6,7-didehydro-4,5α-epoxy-3,14-dihydroxyindolo[2′,3′:6,7]morphinan (5′-GNTI) (Supplemental Methods).
Measurement of G Protein Activation.
A genetically engineered firefly luciferase cAMP biosensor (GloSensor; Promega, Madison, WI) was used to quantify Gαi-mediated activity as described previously (Allen et al., 2011; Chen et al., 2012; Thompson et al., 2012; Wu et al., 2012; Wacker et al., 2013). Details are available online at the National Institute of Mental Health’s Psychoactive Drug Screening Program site (http://pdsp.med.unc.edu/PDSP%20Protocols%20II%202013-03-28.pdf). In brief, human embryonic kidney cells were transfected with the biosensor and KOR at a 1:1 ratio. The next day, the cells were plated into Greiner white 384-well plates (catalog no. 655098). The cells were incubated with the test compound for 20–30 minutes before addition of the GloSensor reagent (luciferin) and isoproterenol (Allen et al., 2011). Luminescence is quantified 10 minutes after the addition of GloSensor reagent and isoproterenol. The Z′ score for this assay using salvinorin A is 0.89 (Zhang et al., 2000).
Measurement of Arrestin Recruitment.
Two assays were used to assess β-arrestin translocation: the Tango assay as described previously (Barnea et al., 2008; Wu et al., 2012), and a bioluminescence resonance energy transfer (BRET)-based assay as an orthologous confirmatory assay as described previously (Rives et al., 2012). The Tango assay requires the fusion of a transcription factor to the C terminus of KOR via linker that contains a tobacco etch virus (TEV) protease cleavage site. Activation of KOR leads to the recruitment of β-arrestin-2 fused with TEV protease, which releases the transcription factor, making it available for induction of luciferase expression. The BRET assay requires cotransfection of KOR fused with Renilla luciferase, Venus-tagged β-arrestin-2, and G protein–coupled receptor kinase 2. The cells were distributed on 96-well plates 1 day prior to assay. The Z′ scores using salvinorin A are 0.716 and 0.95 for the Tango assay and the BRET assay, respectively.
Virtual Screening for Biased Ligands.
Upon identification of a potential scaffold with signaling bias, we then identified analogs as detailed previously (Huang et al., 2011) using the ZINC database (Irwin and Shoichet, 2005; Irwin et al., 2012). Compounds identified were purchased and screened as described above.
Quantifying Bias.
We used the method developed by Kenakin and Christopolous to quantify the biased signaling of ligands (Kenakin et al., 2012, Kenakin and Christopoulos, 2013). After generating concentration-response curves, we fit the data to a mathematical model based on the Black and Leff operational model to generate log(τ/KA) values. The log(τ/KA) value is a transduction coefficient that represents the affinity and efficacy of a ligand for a specific signaling pathway, in this case either G protein activation or arrestin mobilization. This model also incorporates the receptor density and coupling within a system, and therefore is receptor-expression independent. The log(τ/KA) of each test ligand is then compared with the log(τ/KA) of a reference ligand, in this case salvinorin A, for both G protein activation and arrestin recruitment. Salvinorin A was chosen as the reference ligand because it has very similar EC50 values for both the G protein and arrestin pathways and displays full efficacy at both pathways. Because agonists activate different signaling pathways with different efficacies and potencies, ligand bias is quantified by comparing the activity of an agonist in one assay to the agonist’s relative activity in another assay, using the same reference ligand in both assays. This method reduces observation or assay bias, as well as system bias innate to the assays used (Kenakin and Christopoulos, 2013). Generating a single number that incorporates agonist affinity and efficacy is useful for identifying which ligands to use in future studies.
Results
Screening for Biased Ligands Using G Protein Activation and Arrestin Recruitment Assays.
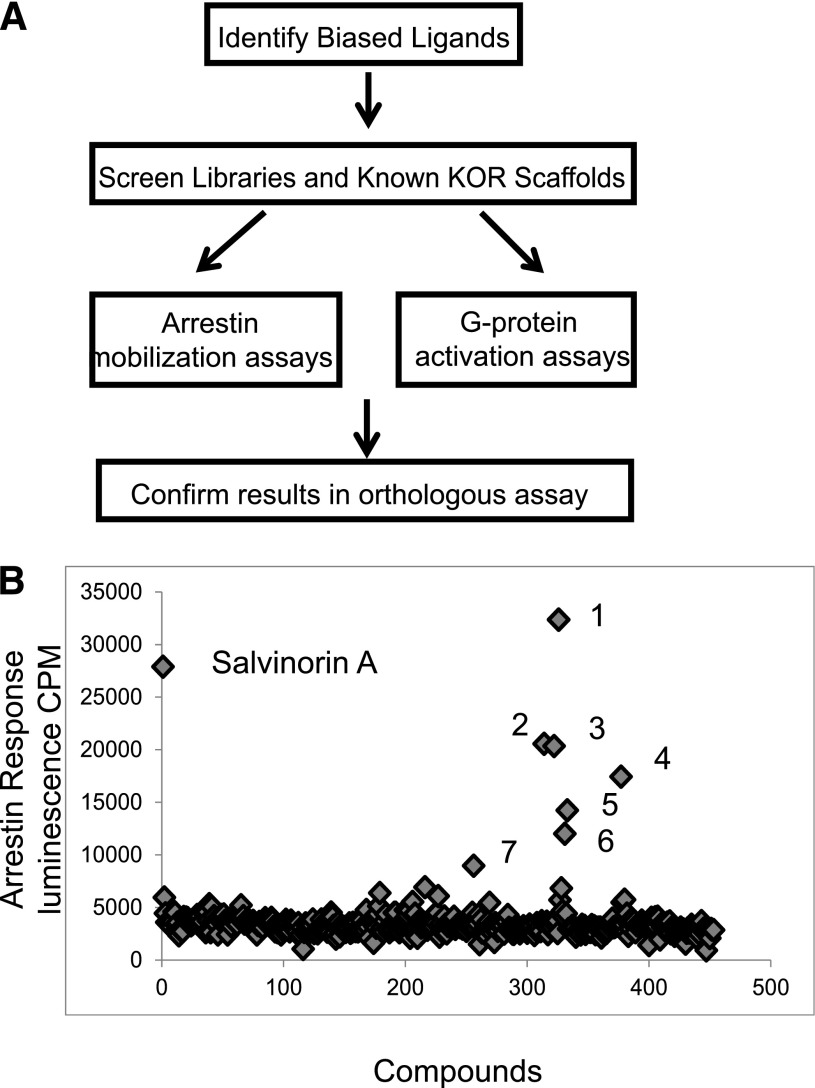
To identify KOR ligands with signaling bias, we screened in parallel the NCC library of approved medications at a concentration of 3 µM using a split luciferase cAMP assay (GloSensor) and a genetically encoded arrestin recruitment assay (Tango). Seven “actives” from this screen were further analyzed by full concentration-response studies (Fig. 1; Supplemental Table 1). GR89696 was the only compound from the NCC library identified as a potent biased ligand for KOR (Supplemental Table 1). The concentration-response analyses of “actives” from the NCC library screen yielded two low-potency agonists: 2-(2-aminoethyl)-pyridine and N-cyano-N′-(1,1-dimethylpropyl)-N′′-3-pyridinylguanidine. Because few compounds in this library were known or predicted to bind to KOR, we continued our screening efforts with scaffolds known to have affinity for KOR. We focused on screening scaffold derivatives of arylacetamides, dynorphins, morphinans, benzomorphans, and salvinorins. Tables 1–5 depict the potencies and efficacies of these ligands for G protein activation and arrestin mobilization (Tango) as well as the calculated bias factors.
Fig. 1.
NCC library screening results. (A) Depiction of the parallel screening approach used. (B) Scatter plot showing the results of the screening of the NCC library in the arrestin assay. 1, Bestatin; 2, GR8969; 3, 2-(2-aminoethyl) pyridine; 4, N-cyano-N′-(1,1-dimethylpropyl)-N′′-3-pyridinylguanidine; 5, brucine; 6, doxapram; 7, diphenoxylate.
TABLE 1.
Affinity and potency values for arylacetamides using GloSensor and Tango
| Arylacetamides | G Protein EC50 | G Protein Emax | Arrestin EC50 | Arrestin Emax | Bias Factor |
|---|---|---|---|---|---|
| nM | nM | ||||
| Salvinorin A | 5.183 (−8.29 ± 0.10) | 99.7 | 5.75 (−8.24 ± 0.06) | 97.2 | 1 |
| ICI 199,441 | 1.63 (−8.79 ± 0.07) | 101 | 0.428 (−9.37 ± 0.05) | 84.8 | 4 Arrestin |
| ICI 204,448 | 4.22 (−8.38 ± 0.09) | 111 | 3.28 (−8.48 ± 0.06) | 77.4 | 2 Arrestin |
| U69593 | 5.89 (−8.23 ± 0.07) | 109 | 6.42 (−8.19 ± 0.09) | 89.3 | 1 |
| GR89696 | 0.970 (−9.01 ± 0.11) | 96.4 | 0.259 (−9.60 ± 0.06) | 92.8 | 5 Arrestin |
| U62066 | 1.01 (−9.00 ± 0.05) | 103 | 6.21 (−8.21 ± 0.10) | 92.7 | 6 G protein |
| (+)U50,488 | 246 (−6.61 ± 0.12) | 102 | 959 (−6.02 ± 0.08) | 92.3 | 8 G protein |
| (−)U50,488 | 0.858 (−9.06 ± 0.07) | 95.5 | 0.822 (−9.09 ± 0.09) | 94.6 | 2 Arrestin |
| DIPPA | 14.5 (−7.84 ± 0.09) | 111 | 8.49 (−8.07 ± 0.07) | 68.5 | 1 |
| N-MPPP | 4.45 (−8.35 ± 0.09) | 109 | 2.41 (−8.62 ± 0.06) | 79.7 | 1 |
| BRL 52537 | 1.85 (−8.73 ± 0.07) | 112 | 1.35 (−8.87 ± 0.05) | 88.9 | 1 |
TABLE 5.
Affinity and potency values for the RB family of salvinorin derivatives using GloSensor and Tango assays
| RB Salvinorins | G Protein EC50 | G Protein Emax | Arrestin EC50 | Arrestin Emax | Bias Factor |
|---|---|---|---|---|---|
| G Protein | |||||
| nM | nM | ||||
| Salvinorin A | 5.183 (−8.29 ± 0.10) | 99.7 | 5.75 (−8.24 ± 0.06) | 97.2 | 1 |
| Salvinorin B | 73.4 (−7.13 ± 0.08) | 95.9 | 428 (−6.37 ± 0.07) | 115 | 4 G protein |
| RB-64 | 5.29 (−8.27 ± 0.06) | 101 | 391 (−6.41 ± 0.05) | 104 | 35 G protein |
| RB-48 | 8.82 (−8.05 ± 0.07) | 101 | 143 (−6.84 ± 0.09) | 63.2 | 25 G protein |
| RB-55_1 | 119 (−6.93 ± 0.07) | 101 | 1492 (−5.83 ± 0.15) | 52.2 | 22 G protein |
| RB-55_2 | 142 (−6.84 ± 0.10) | 105 | 2284 (−5.64 ± 0.09) | 56.8 | 33 G protein |
| RB 55 | 31.3 (−7.50 ± 0.08) | 103 | 229 (−6.64 ± 0.07) | 86.9 | 8 G protein |
| RB 50 | 166 (−6.78 ± 0.10) | 103 | 3812 (−5.42 ± 0.21) | 89.2 | 69 G protein |
| RB 59 | 35.8 (−7.45 ± 0.10) | 95.7 | 4290 (−5.37 ± 0.13) | 76.6 | 95 G protein |
| RB 65 | 145 (−6.83 ± 0.10) | 95.9 | 2767 (−5.56 ± 0.13) | 42.7 | 29 G protein |
All the arylacetamides tested are potent agonists at KOR with varying degrees of bias (Table 1). ICI 204,448 and BRL 52537 were identified from a virtual screen using the ZINC database (Irwin and Shoichet 2005; Irwin et al., 2012) as potentially biased ligands based on the structure of GR89696. GR89696 and ICI 199,441 displayed modest arrestin bias (bias factors 5 and 4, respectively) while ICI 204,448 and (−)U50,488 are only very weakly biased for arrestin (bias factors 2 for each compound). In contrast, U62066 and (+)U50,488 are slightly G protein–biased (bias factors 6 and 8, respectively). Lastly, we found that U69593, DIPPA, N-MPPP, and BRL 52537 are all unbiased agonists.
The dynorphin peptides tested displayed varying degrees of G protein bias (Table 2). Dyn A, Dyn 1-13, and Dyn 1-11 have the highest degree of bias (34, 34, and 44, respectively), while Dyn 1–8 and Dyn 1–9 are more moderately biased (4 and 16, respectively). This represents the first report of endogenous KOR ligands having a biased signaling profile relative to salvinorin A, which equally stimulates G protein and arrestin pathways. Furthermore, the tested morphinans (Table 3) and benzomorphans (Table 4) tested displayed very little bias. Only 6′-GNTI displayed a slight G protein bias (bias factor of 6), consistent with previous studies (Rives et al., 2012; Schmid et al., 2013). Also, we found that the antagonist JDTic has no agonistic activity in either G protein or arrestin assays (Table 4).
TABLE 2.
Affinity and potency values for dynorphin peptides using GloSensor and Tango assays
| Peptides | G Protein EC50 | G Protein Emax | Arrestin EC50 | Arrestin Emax | Bias Factor |
|---|---|---|---|---|---|
| nM | nM | ||||
| Salvinorin A | 5.183 (−8.29 ± 0.10) | 99.7 | 5.75 (−8.24 ± 0.06) | 97.24 | 1 |
| Dynorphin A | 8.12 (−8.09 ± 0.07) | 101 | 268 (−6.57 ± 0.11) | 74.8 | 34 G protein |
| Dyn 1–8 | 57.7 (−7.24 ± 0.05) | 106 | 720 (−6.14 ± 0.11) | 89.9 | 4 G protein |
| Dyn 1–9 | 10.2 (−7.99 ± 0.06) | 101 | 600 (−6.22 ± 0.09) | 64.7 | 16 G protein |
| Dyn 1–11 | 3.26 (−8.49 ± 0.08) | 101 | 450 (−6.35 ± 0.09) | 75.8 | 44 G protein |
| Dyn 1–13 | 2.07 (−8.68 ± 0.07) | 96.6 | 97.8 (−7.01 ± 0.07) | 72.4 | 34 G protein |
TABLE 3.
Affinity and potency values for morphinans using GloSensor and Tango assays
| Morphinans | G Protein EC50 | G Protein Emax | Arrestin EC50 | Arrestin Emax | Bias Factor |
|---|---|---|---|---|---|
| nM | nM | ||||
| Salvinorin A | 5.18 (−8.29 ± 0.10) | 99.7 | 5.75 (−8.24 ± 0.06) | 97.2 | 1 |
| β-NNTA | 0.305 (−9.52 ± 0.12) | 97.0 | 0.268 (−9.57 ± 0.12) | 84.5 | 1 |
| 6′ GNTI | 4.74 (−8.32 ± 0.09) | 96.5 | 7.38 (−8.13 ± 0.12) | 34.7 | 6 G protein |
| 5′ GNTI | Antagonist | — | Antagonist | — |
TABLE 4.
Affinity and potency values for benzomorphans using GloSensor and Tango assays
| Benzomorphans | G Protein EC50 | G Protein Emax | Arrestin EC50 | Arrestin Emax | Bias Factor |
|---|---|---|---|---|---|
| nM | nM | ||||
| Salvinorin A | 3.63 (−8.29 ± 0.10) | 103 | 6.67 (−8.18 ± 0.05) | 99.42 | 1 |
| Naltrindole | Antagonist | — | Antagonist | — | |
| Diprenorphine | 0.960 (−9.02 ± 0.08) | 88.3 | 3.3 (−8.48 ± 0.14) | 87.0 | 2 G protein |
| Nalbuphine | 61.5 (−7.21 ± 0.11) | 81.3 | 47.2 (−7.33 ± 0.08) | 74.1 | 3 Arrestin |
| Butorphanol | 1.82 (−8.74 ± 0.07) | 94.3 | 1.70 (−8.77 ± 0.06) | 59.2 | 2 G protein |
| Cyclazocine | 1.19 (−8.92 ± 0.09) | 102 | 0.806 (−9.09 ± 0.03) | 81.7 | 1 |
| JDTic | Antagonist | — | Antagonist | — |
Additionally, we tested several C-2–modified salvinorin derivatives and found them to display a wide range of G protein bias (Table 5). Of this family, RB 64 and RB 48 are the most potent in activating G protein signaling and have a high degree of bias (35 and 25, respectively). RB 59, RB 55-2, and RB 50 also have high G protein bias factors (95, 33, and 69, respectively). RB 55-1 and RB 65 are lower potency ligands but still have a strong bias (bias factors 22 and 29, respectively). RB 55 has a slight bias factor of 8, while salvinorin B, a metabolite of salvinorin A, has a bias factor of 4.
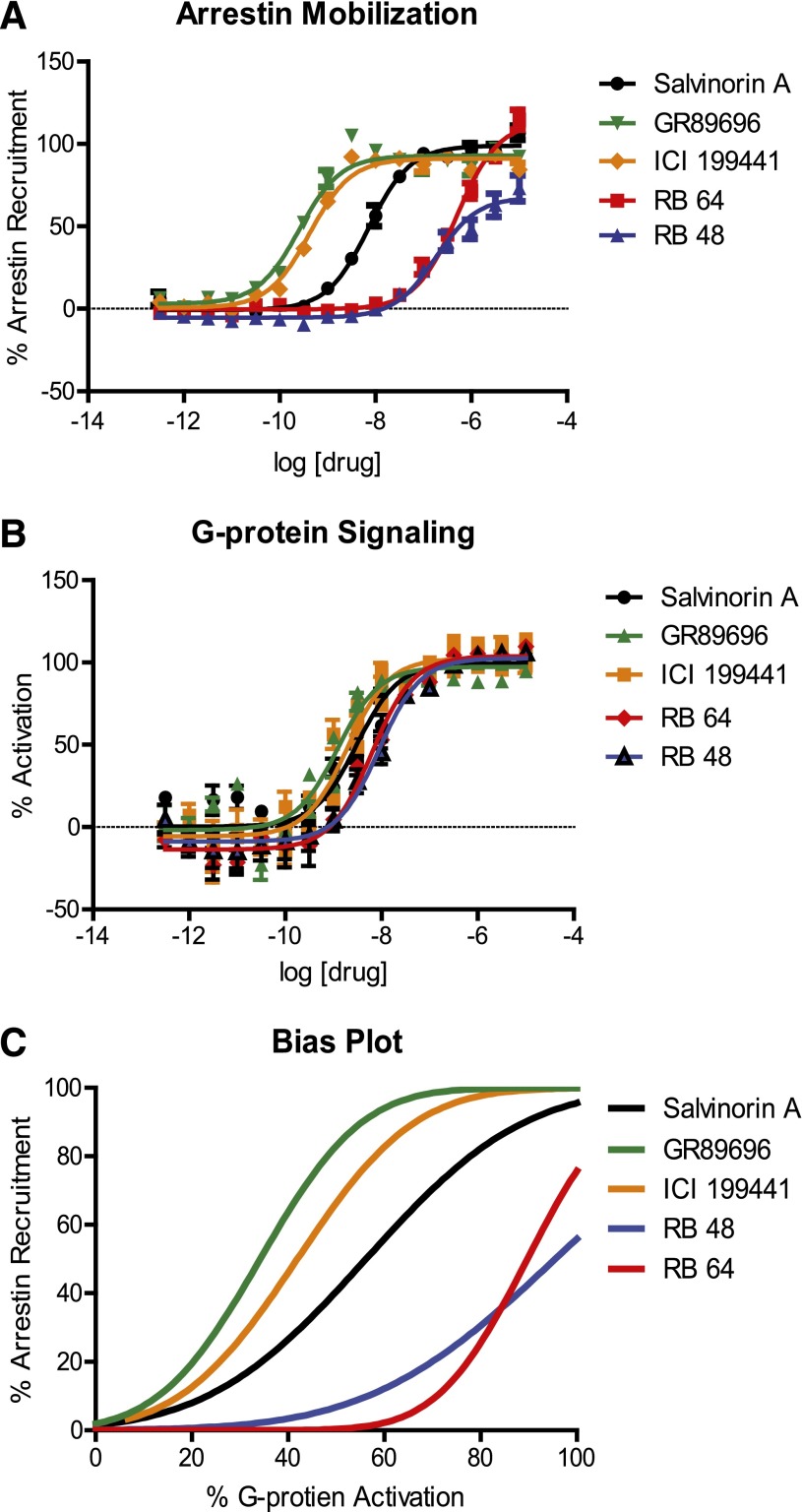
Figure 2 depicts the G protein activation (Fig. 2A) and arrestin mobilization (Fig. 2B) concentration-response curves for the compounds found to be the most potent and the most biased, along with relevant controls. The “bias plot” indicates the signaling bias of each compound by showing the response in the arrestin recruitment assay as a function of the corresponding response in the G protein activation assay (Kenakin and Christopoulos, 2013) (Fig. 2C). Thus, ICI 199,441 and GR89696 are arrestin biased, whereas RB 64 and RB 48 are G protein–biased.
Fig. 2.
Arrestin mobilization and G protein activation dose-response curves of candidates for in vivo studies. The dose-response curves of candidate ligands for arrestin recruitment were measured via Tango (A), G protein activation (B), and the bias plot (C). These ligands all have similar potency and efficacy values for G protein signaling, yet the potency values for arrestin mobilization differ greatly. The bias plot highlights the differences in potency and efficacy values for each ligand in both G protein and arrestin pathways.
Orthologous Arrestin Assay.
To confirm our results from the Tango arrestin recruitment assay, we used a BRET-based arrestin-recruitment assay (Rives et al., 2012) to further analyze the compounds displaying the highest degree of bias. Salvinorin A displayed very similar potency values for the Tango and BRET assays (5.56 and 5.63 nM, respectively) (Tables 1 and 6). Also, the potencies of GR89696 and ICI 199,441 were very similar, based on comparison of results obtained from the Tango and BRET arrestin assays. U62066 has a slightly higher potency in the BRET assay compared with the Tango assay (19.8 and 6.21 nM, respectively). This shift in potency has a modest effect on the bias factor calculated with the BRET data as compared with the Tango data, but both assays suggest a slight G protein bias for U62066 (Supplemental Table 2). Furthermore, RB 64, RB 48, RB 59, RB 55, Dyn 1–13, Dyn 1–9, Dyn 1–11, and Dyn A all have slightly higher potencies in the BRET arrestin assay than the Tango assay, while Dyn 1–8 has a slightly more potent effect in Tango than BRET.
TABLE 6.
BRET arrestin affinity and potency values
| Compound | EC50 | Emax |
|---|---|---|
| nM | ||
| Salvinorin A | 5.55 (−8.25 ± 0.05) | 98.84 |
| GR89896 | 0.265 (−9.58 ± 0.03) | 104 |
| ICI 199,441 | 0.461 (−9.34 ± 0.07) | 100 |
| U62066 | 19.8 (−7.70 ± 0.07) | 101 |
| RB 64 | 118 (−6.93 ± 0.06) | 105 |
| RB 48 | 45.0 (−7.35 ± 0.06) | 101 |
| RB 55 | 196 (−6.71 ± 0.03) | 78.9 |
| RB 59 | 3560 (−5.44 ± 0.18) | 177 |
| Dyn 1–13 | 78.2 (−7.11 ± 0.13) | 86.3 |
| Dyn 1–11 | 132 (−6.87 ± 0.16) | 86.9 |
| Dyn 1–9 | 253 (−6.59 ± 0.11) | 92.8 |
| Dyn 1–8 | 1070 (−5.97 ± 0.11) | 102 |
| Dynorphin A | 112 (−6.95 ± 0.13) | 99.2 |
Despite modest potency differences between the Tango and BRET assays, if a ligand was identified as biased in the Tango assay then it was also identified as biased using the BRET arrestin assay. A comparison of bias factors generated from the BRET arrestin assay and the Tango assay is shown in Supplemental Table 2, and the log(τ/KA) values are listed in Supplemental Table 3.
Discussion
Recent structural evidence suggests that G protein–coupled receptors (GPCRs) adopt multiple conformations and that different ligands can stabilize distinct conformations leading to diverse signaling profiles (Liu et al., 2012; Wacker et al., 2013; Nygaard et al., 2013; Vardy and Roth, 2013; Kenakin, 1995). Additionally, signaling partners including arrestins (Gray et al., 2003) and G proteins (Yan et al., 2008; Nygaard et al., 2013) can allosterically modulate agonist affinities and overall receptor conformations. This bidirectional modulation from both the ligand and the intracellular effector might affect its signaling.
In this study we sought to identify KOR-selective functionally selective ligands, as such ligands have been proposed to potentially function as analgesics with fewer adverse side effects (e.g., sedation and dependence). Our attempts to identify biased KOR agonists were aided by: 1) a wealth of diverse chemical matter reported to be KOR-selective; 2) assays that are both readily available and scalable; and 3) the availability of a KOR crystal structure (Wu et al., 2012). The diverse KOR chemotypes and structural information will be useful as we attempt to further optimize this structurally diverse catalog of biased ligands. Additionally, there is increased interest in developing KOR antagonists for both depression and addiction disorders, and for developing KOR agonists as analgesics with a low abuse potential (Tao et al., 2008; Wee and Koob, 2010; Prevatt-Smith et al., 2011). However, KOR agonists also cause aversion, hallucinations, and psychotomimetic effects (Pfeiffer et al., 1986). To develop KOR agonists that can be used as analgesics, we must understand how KOR mediates these negative side effects and explore the use of functionally selective ligands toward KOR therapies with minimal side effects. Additionally, understanding which KOR-dependent signaling cascades mediate hallucinations will provide insight into how KOR activation affects cognition. Therefore, the first step in understanding the diverse KOR behavioral effects is to identify a range of functionally selective ligands that are potent and selective for KOR. In this study, we identify multiple centrally active KOR-selective biased ligands (RB 64, RB 48, ICI 199,441, and GR89696) that have the potential for probing KOR signaling pathways in vivo (Ravert et al., 2002; Terner et al., 2005; Yan et al., 2009).
Significantly, an unbiased screen of a small library of known drugs yielded only a single KOR-biased ligand (GR89696), although it is possible that larger screens encompassing greater chemical diversity could yield additional scaffolds. Intriguingly, when we focused our investigation on analogs of known KOR ligands, we were able to rapidly identify additional KOR ligands with varying degrees of bias. This suggests that screening scaffold derivatives is a reliable approach for identifying biased ligands, and mirrors our results reported for D2 arrestin–biased drug discovery (Allen et al., 2011). After identifying a scaffold from the NCC screen, for instance, we tested compounds that were similar in structure to the initial arylacetamide hit. Additionally, we performed a similarity search using the ZINC database and found an additional biased ligand possessing the arylacetamide scaffold (ICI 204,448). We found arylacetamide ligands to be either weakly G protein– or arrestin-biased.
We also tested varying lengths of the endogenous KOR peptide ligand, dynorphin, and found them all to be G protein biased. Additionally, we tested the RB family of salvinorin derivatives that were originally synthesized to covalently bind to KOR. Future studies will be needed to investigate how those ligands interact with the receptor and potentially identify residues mediating the signaling bias observed. The RB family of compounds constitutes the first identified KOR G protein–biased ligands that are centrally active and can therefore be used for in vivo probing of KOR-mediated G protein signaling (Yan et al., 2009).
To further investigate our biased ligands, we tested arrestin recruitment in an orthologous assay using bioluminescence resonance energy transfer. In general, ligands tested in the BRET assay displayed similar potencies and efficacies when compared with results obtained with the Tango assay. RB 48 and RB 59, by contrast, possess the largest differences in bias factors quantified using Tango versus BRET assays. Notably, the incubation time is much longer for the Tango assay (16 hours), and proteolysis of the transcription factor, entry into the nucleus, and transcription and translation are required downstream of arrestin recruitment, whereas only arrestin recruitment is assayed in the BRET assay (5 minutes). However, all ligands that we originally found to be biased using the Tango assay were also found to be biased using the BRET assay. Thus, we can infer that these compounds are functionally selective ligands for KOR—at least in human embryonic kidney cells.
This is the first report of KOR-selective biased ligands that may ultimately be useful in vivo to discover which KOR signaling cascades are responsible for various KOR-mediated behavioral effects. Although 6′-GNTI was previously identified as a biased ligand, it has a fixed charge and therefore does not readily cross the blood-brain barrier (Rives et al., 2012). Additionally, while the log(τ/KA) method of quantifying bias is useful for calculating the bias in vitro, further studies are necessary for investigating the in vivo effect of these ligands, as efficacies and potencies in vitro may not correlate with those obtained in other cell types in vivo. Nonetheless, using a similar strategy, we have been able to successfully advance arrestin-biased D2 agonists to in vivo testing and demonstrate that they retain substantial apparent bias in vivo (Allen et al., 2011; Chen et al., 2012).
Finally, the phenomenon of GPCR functional selectivity is not limited to arrestin mobilization and G protein activation. For example, we have identified 5-HT2A inverse agonists which can induce receptor internalization and downregulation in vitro and in vivo without activating either G protein signaling or arrestin translocation (Bhatnagar et al., 2001; Xia et al., 2003; Yadav et al., 2011). In future studies, it will be useful to combine in vivo behavioral studies and a global study of intracellular signaling with functionally selective ligands, to fully understand which signaling cascades contribute to the various behavioral effects of KOR agonism. The present study suggests that simply screening available scaffolds represents a facile method for identifying functionally selective ligands with good drug-like properties. The rapid increase in GPCR structural and dynamic information and our expanded understanding of functional selectivity have enhanced the potential for designing more selective therapies with fewer side effects for a multitude of diseases and conditions. In the future, screening compounds for a more global activation of pathways in addition to those activated by G proteins should allow for a better understanding of how these ligands affect physiology, and how functionally selective compounds might have beneficial therapeutic value.
Supplementary Material
Acknowledgments
The authors thank Dr. Wesely K. Kroeze for critically reading this manuscript and the National Institute on Drug Abuse for the supply of dynorphin peptides. The authors also thank thank Joel Karpiak and Brian Shoichet for assistance with in silico screening.
Abbreviations
- 5′-GNTI
5′-guanidino-17-(cyclopropylmethyl)-6,7-didehydro-4,5α-epoxy-3,14-dihydroxyindolo[2′,3′:6,7]morphinan
- 6′-GNTI
6′-guanidino-17-(cyclopropylmethyl)-6,7-didehydro-4,5α-epoxy-3,14-dihydroxyindolo[2′,3′:6,7]morphinan
- β-NNTA
N-naphthoyl-β-naltrexamine
- BRET
bioluminescence resonance energy transfer
- BRL 52537
(±)-1-(3,4-dichlorophenyl)acetyl-2-(1-pyrrolidinyl)methylpiperidine hydrochloride
- DIPPA
2-(3,4-dichlorophenyl)-N-methyl-N-([1S]-1-[3-isothiocyanatophenyl]-2-[1-pyrrolidinyl]ethyl)acetamide hydrochloride
- Dyn
dynorphin
- GPCR
G protein–coupled receptor
- GR89696
4-([3,4-dichlorophenyl]acetyl)-3-(1-pyrrolidinylmethyl)-1-piperazinecarboxylic acid methyl ester fumarate salt
- ICI 199,441
2-(3,4-dichlorophenyl)-N-methyl-N-([1S]-1-phenyl-2-[1-pyrrolidinyl]ethyl)acetamide hydrochloride
- ICI 204,448
(RS)-[3-[1-[[(3,4-dichlorophenyl)acetyl]methylamino]-2-(1-pyrrolidinyl)ethyl]phenoxy]acetic acid hydrochloride
- KOR
κ−opioid receptor
- NCC
National Institutes of Health Clinical Collection
- N-MPPP
N-methyl-N-([1S]-1-phenyl-2-[1-pyrrolidinyl]ethyl)phenylacetamide hydrochloride
- (+)U50,488
trans-(+)-3,4-dichloro-N-methyl-N-(2-[1-pyrrolidinyl]-cyclohexyl)benzeneacetamide hydrochloride
- (−)U50,488
trans-(−)-3,4-dichloro-N-methyl-N-[2-(1-pyrrolidinyl)cyclohexyl]benzeneacetamide hydrochloride
- U62066
(±)-(5α,7α,8β)-3,4-dichloro-N-methyl-N-[7-(1-pyrrolidinyl)-1-oxaspiro[4.5]dec-8-yl]benzeneacetamide mesylate salt
- U69593
(+)-(5α,7α,8β)-N-methyl-N-[7-(1-pyrrolidinyl)-1-oxaspiro[4.5]dec-8-yl]-benzeneacetamide
Authorship Contributions:
Participated in research design: White, Roth.
Conducted experiments: White.
Contributed new reagents or analytic tools: Bikbulatov, Polepally, Zjawiony, Rives, Javitch, Scopton, Brown.
Performed data analysis: White, Roth, Kenakin, Rives, Javitch.
Wrote or contributed to the writing of the manuscript: White, Roth, Rives, Javitch.
Footnotes
The work was funded by the National Institutes of Health National Institute on Drug Abuse and National Institute of Mental Health [Grants RO1DA01724, U19MH82441, K05DA022413, and R01MH54137]; a National Institute on Drug Abuse EUREKA Grant [R01DA027170]; and the National Institute of Mental Health Psychoactive Drug Screening Program. The Structural Genomics Consortium is a registered charity (No. 1097737) that receives funds from AbbVie, Boehringer Ingelheim, the Canada Foundation for Innovation, the Canadian Institutes for Health Research, Genome Canada through the Ontario Genomics Institute [OGI-055], GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust [092809/Z/10/Z].
Primary laboratory of origin: Roth.
This work was previously presented as follows: White KL, Vardy E, and Roth BL (2013) Utilizing functionally selective ligands to probe specific signaling pathways of the kappa opioid receptor. Second Conference on the Therapeutic Potential of Kappa Opioids in Pain and Addiction; 2013 Apr 24–27; Cambridge, MA. Kappa Therapeutics, Boston, MA.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Allen JA, Yost JM, Setola V, Chen X, Sassano MF, Chen M, Peterson S, Yadav PN, Huang XP, Feng B, et al. (2011) Discovery of β-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc Natl Acad Sci USA 108:18488–18493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard SM, Patterson TA, Jin W, Chavkin C. (1997) Agonist-induced phosphorylation of the kappa-opioid receptor. J Neurochem 69:2405–2412 [DOI] [PubMed] [Google Scholar]
- Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, Lee KJ. (2008) The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci USA 105:64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar A, Willins DL, Gray JA, Woods J, Benovic JL, Roth BL. (2001) The dynamin-dependent, arrestin-independent internalization of 5-hydroxytryptamine 2A (5-HT2A) serotonin receptors reveals differential sorting of arrestins and 5-HT2A receptors during endocytosis. J Biol Chem 276:8269–8277 [DOI] [PubMed] [Google Scholar]
- Black JW, Leff P. (1983) Operational models of pharmacological agonism. Proc R Soc Lond 220:141–162 [DOI] [PubMed] [Google Scholar]
- Bruchas MR, Macey TA, Lowe JD, Chavkin C. (2006) Kappa opioid receptor activation of p38 MAPK is GRK3- and arrestin-dependent in neurons and astrocytes. J Biol Chem 281:18081–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchas MR, Land BB, Aita M, Xu M, Barot SK, Li S, Chavkin C. (2007) Stress-induced p38 mitogen-activated protein kinase activation mediates kappa-opioid-dependent dysphoria. J Neurosci 27:11614–11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchas MR, Chavkin C. (2010) Kinase cascades and ligand-directed signaling at the kappa opioid receptor. Psychopharmacology (Berl) 210:137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA, Jr, Béguin C, Knoll AT, Cohen BM. (2009) Kappa-opioid ligands in the study and treatment of mood disorders. Pharmacol Ther 123:334–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sassano MF, Zheng L, Setola V, Chen M, Bai X, Frye SV, Wetsel WC, Roth BL, Jin J. (2012) Structure-functional selectivity relationship studies of β-arrestin-biased dopamine D₂ receptor agonists. J Med Chem 55:7141–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortch-Carnes J, Potter DE. (2005) Bremazocine: a kappa-opioid agonist with potent analgesic and other pharmacologic properties. CNS Drug Rev 11:195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Bhatnagar A, Gurevich VV, Roth BL. (2003) The interaction of a constitutively active arrestin with the arrestin-insensitive 5-HT(2A) receptor induces agonist-independent internalization. Mol Pharmacol 63:961–972 [DOI] [PubMed] [Google Scholar]
- Huang XP, Setola V, Yadav PN, Allen JA, Rogan SC, Hanson BJ, Revankar C, Robers M, Doucette C, Roth BL. (2009) Parallel functional activity profiling reveals valvulopathogens are potent 5-hydroxytryptamine(2B) receptor agonists: implications for drug safety assessment. Mol Pharmacol 76:710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr, Lee HM, Chen X, et al. (2011) Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK. (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. (2012) ZINC: A free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T. (1995) Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol Sci 16:232–238 [DOI] [PubMed] [Google Scholar]
- Kenakin T, Watson C, Muniz-Medina V, Christopoulos A, Novick S. (2012) A simple method for quantifying functional selectivity and agonist bias. ACS Chem Neurosci 3:193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T, Christopoulos A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov 12:205–216 [DOI] [PubMed] [Google Scholar]
- Knoll AT, Carlezon WA., Jr (2010) Dynorphin, stress, and depression. Brain Res 1314:56–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutrzeba LM, Karamayan VT, Speth RC, Williamson JS, Zjawiony JK. (2009) In vitro studies on metabolism of salvinorin A. Pharm. Biol. 47:1078–1084 [Google Scholar]
- Land BB, Bruchas MR, Lemos JC, Xu M, Melief EJ, Chavkin C. (2008) The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J Neurosci 28:407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land BB, Bruchas MR, Schattauer S, Giardino WJ, Aita M, Messinger D, Hnasko TS, Palmiter RD, Chavkin C. (2009) Activation of the kappa opioid receptor in the dorsal raphe nucleus mediates the aversive effects of stress and reinstates drug seeking. Proc Natl Acad Sci USA 106:19168–19173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wüthrich K. (2012) Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335:1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan MJ. (1990) Kappa-opioid receptors and analgesia. Trends Pharmacol Sci 11:70–76 [DOI] [PubMed] [Google Scholar]
- Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, Manglik A, Pan AC, Liu CW, Fung JJ, Bokoch MP, et al. (2013) The dynamic process of β(2)-adrenergic receptor activation. Cell 152:532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Brantl V, Herz A, Emrich HM. (1986) Psychotomimesis mediated by kappa opiate receptors. Science 233:774–776 [DOI] [PubMed] [Google Scholar]
- Prevatt-Smith KM, Lovell KM, Simpson DS, Day VW, Douglas JT, Bosch P, Dersch CM, Rothman RB, Kivell B, Prisinzano TE. (2011) Potential drug abuse therapeutics derived from the hallucinogenic natural product salvinorin A. Medchemcomm 2:1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravert HT, Scheffel U, Mathews WB, Musachio JL, Dannals RF. (2002) [(11)C]-GR89696, a potent kappa opiate receptor radioligand; in vivo binding of the R and S enantiomers. Nucl Med Biol 29:47–53 [DOI] [PubMed] [Google Scholar]
- Rives ML, Rossillo M, Liu-Chen LY, Javitch JA. (2012) 6′-Guanidinonaltrindole (6′-GNTI) is a G protein-biased κ-opioid receptor agonist that inhibits arrestin recruitment. J Biol Chem 287:27050–27054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Baner K, Westkaemper R, Siebert D, Rice KC, Steinberg S, Ernsberger P, Rothman RB. (2002) Salvinorin A: a potent naturally occurring nonnitrogenous kappa opioid selective agonist. Proc Natl Acad Sci USA 99:11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CL, Streicher JM, Groer CE, Munro TA, Zhou L, Bohn LM. (2013) Functional selectivity of 6'-guanidinonaltrindole (6'-GNTI) at κ-opioid receptors in striatal neurons. J Biol Chem 288:22387–22398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer C. (2009) 30 years of dynorphins—new insights on their functions in neuropsychiatric diseases. Pharmacol Ther 123:353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler DJ, Roth BL. (2003) Salvinorin A: the “magic mint” hallucinogen finds a molecular target in the kappa opioid receptor. Trends Pharmacol Sci 24:107–109 [DOI] [PubMed] [Google Scholar]
- Tejeda HA, Shippenberg TS, Henriksson R. (2012) The dynorphin/κ-opioid receptor system and its role in psychiatric disorders. Cell Mol Life Sci 69:857–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y-M, Li Q-L, Zhang C-F, Xu X-J, Chen J, Ju Y-W, Chi Z-Q, Long Y-Q, Liu J-G. (2008) LPK-26, a novel kappa-opioid receptor agonist with potent antinociceptive effects and low dependence potential. Eur J Pharmacol 584:306–311 [DOI] [PubMed] [Google Scholar]
- Terner JM, Lomas LM, Lewis JW, Husbands SM, Picker MJ. (2005) Effects of the long-lasting kappa opioid 2-(3,4-dichlorophenyl)-N-methyl-N-[(1S)-1-(3-isothiocyanatophenyl)-2-(1-pyrrolidinyl) ethyl] acetamide in a drug discrimination and warm water tail-withdrawal procedure. Behav Pharmacol 16:665–670 [DOI] [PubMed] [Google Scholar]
- Thompson AA, Liu W, Chun E, Katritch V, Wu H, Vardy E, Huang XP, Trapella C, Guerrini R, Calo G, et al. (2012) Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature 485:395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL, Christopoulos A, Sexton PM, et al. (2007) Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther 320:1–13 [DOI] [PubMed] [Google Scholar]
- Vardy E, Roth BL. (2013) Conformational ensembles in GPCR activation. Cell 152:385–386 [DOI] [PubMed] [Google Scholar]
- Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, McCorvy JD, Jiang Y, Chu M, Siu FY, et al. (2013) Structural features for functional selectivity at serotonin receptors. Science 340:615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Koob GF. (2010) The role of the dynorphin-kappa opioid system in the reinforcing effects of drugs of abuse. Psychopharmacology (Berl) 210:121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KL, Roth BL. (2012) Psychotomimetic effects of kappa opioid receptor agonists. Biol Psychiatry 72:797–798 [DOI] [PubMed] [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, et al. (2012) Structure of the human κ-opioid receptor in complex with JDTic. Nature 485:327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PN, Kroeze WK, Farrell MS, Roth BL. (2011) Antagonist functional selectivity: 5-HT2A serotonin receptor antagonists differentially regulate 5-HT2A receptor protein level in vivo. J Pharmacol Exp Ther 339:99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Mosier PD, Westkaemper RB, Roth BL. (2008) Galpha-subunits differentially alter the conformation and agonist affinity of kappa-opioid receptors. Biochemistry 47:1567–1578 [DOI] [PubMed] [Google Scholar]
- Yan F, Bikbulatov RV, Mocanu V, Dicheva N, Parker CE, Wetsel WC, Mosier PD, Westkaemper RB, Allen JA, Zjawiony JK, et al. (2009) Structure-based design, synthesis, and biochemical and pharmacological characterization of novel salvinorin A analogues as active state probes of the kappa-opioid receptor. Biochemistry 48:6898–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Gray JA, Compton-Toth BA, Roth BL. (2003) A direct interaction of PSD-95 with 5-HT2A serotonin receptors regulates receptor trafficking and signal transduction. J Biol Chem 278:21901–21908 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TDY, Oldenburg KR. (2000) Confirmation of primary active substances from high throughput screening of chemical and biological populations: a statistical approach and practical considerations. J Comb Chem 2:258–265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.