Abstract
BACKGROUND
It has been thought that Clostridium difficile infection is transmitted predominantly within health care settings. However, endemic spread has hampered identification of precise sources of infection and the assessment of the efficacy of interventions.
METHODS
From September 2007 through March 2011, we performed whole-genome sequencing on isolates obtained from all symptomatic patients with C. difficile infection identified in health care settings or in the community in Oxfordshire, United Kingdom. We compared single-nucleotide variants (SNVs) between the isolates, using C. difficile evolution rates estimated on the basis of the first and last samples obtained from each of 145 patients, with 0 to 2 SNVs expected between transmitted isolates obtained less than 124 days apart, on the basis of a 95% prediction interval. We then identified plausible epidemiologic links among genetically related cases from data on hospital admissions and community location.
RESULTS
Of 1250 C. difficile cases that were evaluated, 1223 (98%) were successfully sequenced. In a comparison of 957 samples obtained from April 2008 through March 2011 with those obtained from September 2007 onward, a total of 333 isolates (35%) had no more than 2 SNVs from at least 1 earlier case, and 428 isolates (45%) had more than 10 SNVs from all previous cases. Reductions in incidence over time were similar in the two groups, a finding that suggests an effect of interventions targeting the transition from exposure to disease. Of the 333 patients with no more than 2 SNVs (consistent with transmission), 126 patients (38%) had close hospital contact with another patient, and 120 patients (36%) had no hospital or community contact with another patient. Distinct subtypes of infection continued to be identified throughout the study, which suggests a considerable reservoir of C. difficile.
CONCLUSIONS
Over a 3-year period, 45% of C. difficile cases in Oxfordshire were genetically distinct from all previous cases. Genetically diverse sources, in addition to symptomatic patients, play a major part in C. difficile transmission. (Funded by the U.K. Clinical Research Collaboration Translational Infection Research Initiative and others.)
Most episodes of Clostridium difficile infection are believed to result from recent acquisition within a health care setting. Prevention efforts have therefore focused on symptomatic patients, their immediate environment, and judicious use of antimicrobial drugs.1,2 Person-to-person transmission of C. difficile infection and surrounding contamination have been well documented.2-5 However, there are multiple other potential sources, including patients with asymptomatic colonization6,7 and sources in the wider environment, such as water, farm animals or pets, and food.8 The contribution of cases acquired from these sources to the overall burden of disease is unclear, particularly with increasing reports of community-associated C. difficile infection.9
Previous studies combining data from hospital admissions and genotyping have shown that transmission through hospital-based contact with patients with C. difficile infection accounts for less than 25% of new cases.10,11 However, such studies have not definitively described the role of symptomatic patients in transmission, nor do they account for potential spread across hospital wards by the movement of patients, staff, and equipment12 or for potential spread from community contacts. Horizontal transmission from symptomatic patients continues to be viewed as the source of most cases of infection and is the basis for recent prevention guidelines.13
The assessment of hospital-wide transmission with the use of multilocus sequence typing or ribotyping is hampered by the large numbers of patients who share a genotype and hospital-based contact. However, whole-genome sequencing shows that substantial genetic diversity exists, even within isolates of the same genotype.14 To quantify the role of symptomatic patients in the transmission of C. difficile leading to infection and to determine how such transmission has varied over time, we examined whole-genome sequences in isolates obtained from all patients with C. difficile infection in a defined geographic area during a 3.6-year period.
METHODS
POPULATION
The Oxford University Hospitals, comprising four hospitals with a total of approximately 1600 beds (mostly in 4-bed bays within discrete areas of wards containing 20 to 30 beds), provide all acute care and more than 90% of hospital services in Oxfordshire, United Kingdom (approximate population, 600,000). During the study, the infection-control practices in this hospital system were in keeping with published guidelines (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).1,2 All inpatients with diarrhea (defined as ≥3 stools within a 24-hour period that took the shape of a container) underwent testing for the presence of C. difficile. The hospitals’ central microbiology laboratory used enzyme immunoassays for toxins A and B (Meridian Bioscience) to test all samples obtained in the hospitals and the community.
From September 2007 through March 2011, all such samples with positive results on enzyme immunoassay were cultured. Subcultured single colonies from culture-positive isolates underwent multilocus sequence typing10,15 and whole-genome sequencing. Repeat isolates of the same sequence type from the same patient were not sequenced, except for 148 randomly selected sample pairs that were used to estimate rates of within-host diversity and evolution (see the Supplementary Appendix). We sequenced repeat isolates with different sequence types from the same patient, which allowed us to account for the effect of mixed infections and reinfections on transmission.16
Data were available for all patients on hospital admissions, movement throughout the hospital, and home postal-code districts (28 distinct locations) and general medical practices.
SEQUENCING
DNA was extracted and sequenced with the use of Illumina technology.17 Sequence reads were mapped to a reference genome, and base-pair calls at each position in the genome that were identified after quality filtering were used to identify single-nucleotide variants (SNVs) between pairs of sequenced isolates and in phylogenetic comparisons. (Details regarding sequencing, including error rates, are provided in the Supplementary Appendix.)
EPIDEMIOLOGIC ANALYSIS
Epidemiologic relationships between genetically related cases were classified as “ward contact” if cases occurred in two patients who had been on the same hospital ward at the same time and if this period was consistent with between-patient transmission. For such transmission to occur, it was assumed that cases were infectious from 1 week before diagnosis through 8 weeks after diagnosis,10,18,19 with an incubation period (the time from the acquisition of C. difficile to the diagnosis of infection) of 0 to 12 weeks.1,10 Patients from whom multiple samples had been obtained were considered to be infectious for 8 weeks after their last positive test. If no ward contact existed, patients could be linked by time (as above) within the same hospital (hospital-wide contact) or by exposure to the same ward but with an interval of up to 28 days separating the discharge (or end of infectivity) of the first patient and the admission of the second patient (classified as ward contamination). Patients whose cases were classified as community contact received care from the same general medical practice or lived in the same postal-code district.
A sensitivity analysis was performed in which the definition of hospital contact was not restricted to a specified period of infectivity, incubation, or ward exposure. For patients with genetically related infections who did not have hospital contact, we investigated whether we could link patients with undiagnosed infection or asymptomatic colonization using hospital-movement data from patients with negative results on enzyme immunoassay and inpatients without diarrhea (for details, see the Supplementary Appendix).
STUDY OVERSIGHT
The study was approved by the Berkshire Research Ethics Committee and National Information Governance Board, which did not require that patients provide written informed consent. All authors vouch for the completeness and accuracy of the data and analysis.
RESULTS
STUDY SAMPLES
From September 2007 through March 2011, a total of 40,924 fecal samples were submitted for C. difficile testing. Of these samples, 2377 tested positive for C. difficile on enzyme immunoassay, resulting in overall rates of 8.5 positive samples per 10,000 inpatient overnight stays and 13.0 per month for outpatients and patients in the community. Of the samples with positive results on enzyme immunoassay, 2283 (96%) were retrieved for culture. Of these samples, 1714 (75%) were culture-positive for C. difficile, representing 1250 infections after accounting for repeated samples with the same sequence type. Of these isolates, 1223 (98%) were successfully sequenced (Fig. S2 in the Supplementary Appendix): 862 (71%) from Oxford University Hospitals inpatients, 246 (20%) from general medical practices, 60 (5%) from Oxford University Hospitals outpatients, and 55 (4%) from other hospitals.
The median age at the time of the diagnosis of C. difficile infection was 78 years (interquartile range, 66 to 86). The most prevalent sequence types were sequence type 1 (ribotype 027) in 214 samples (17%), sequence type 8 (ribotypes 002 and 159) in 110 samples (9%), and sequence type 2 (ribotypes 014, 020, 076, and 220) in 103 samples (8%).
GENETIC DIVERSITY WITHIN INDIVIDUAL PATIENTS
The analysis of the evolution and genetic diversity of C. difficile within individual patients provides a framework for interpreting differences in SNVs among patients. We evaluated the first and last samples obtained from 145 patients at a median interval of 51 days (interquartile range, 28 to 105; simple range, 0 to 561), using a model based on coalescent theory (in which all alleles of a gene that is shared by all members of a population are traced to a single ancestral copy) (for details, see the Supplementary Appendix).20 On the basis of this evaluation, the estimated evolutionary rate was 0.74 SNVs (95% confidence interval [CI], 0.22 to 1.40) per successfully sequenced genome per year, and the mean within-host diversity was 0.30 SNVs (95% CI, 0.13 to 0.42). Using model-based 95% prediction intervals, we determined that 0 to 2 SNVs would be expected between isolates that were obtained less than 124 days apart, and 0 to 3 SNVs would be expected between isolates that were obtained 124 to 364 days apart (Fig. S3 in the Supplementary Appendix).
GENETIC DIVERSITY AMONG PATIENTS
Of 957 sequenced isolates from patients with C. difficile infection obtained from April 2008 through March 2011, a total of 333 (35%) were genetically related to at least one isolate obtained from a previous case from September 2007 through March 2011. (Genetic relationship was defined as ≤2 SNVs between the two isolates, an association that is close enough to support transmission on the basis of the stated prediction intervals.) The 266 isolates that were obtained from September 2007 to March 2008 were included only as potential sources for later C. difficile infections, since these patients could have acquired C. difficile from a case before the study started. Of the 957 sequenced isolates, 428 (45%) had more than 10 SNVs, as compared with all previous sequenced samples obtained since September 2007 (Fig. 1A). Results were similar in a sensitivity analysis after a basic adjustment for recombination (Table S2 and Fig. S4 in the Supplementary Appendix).
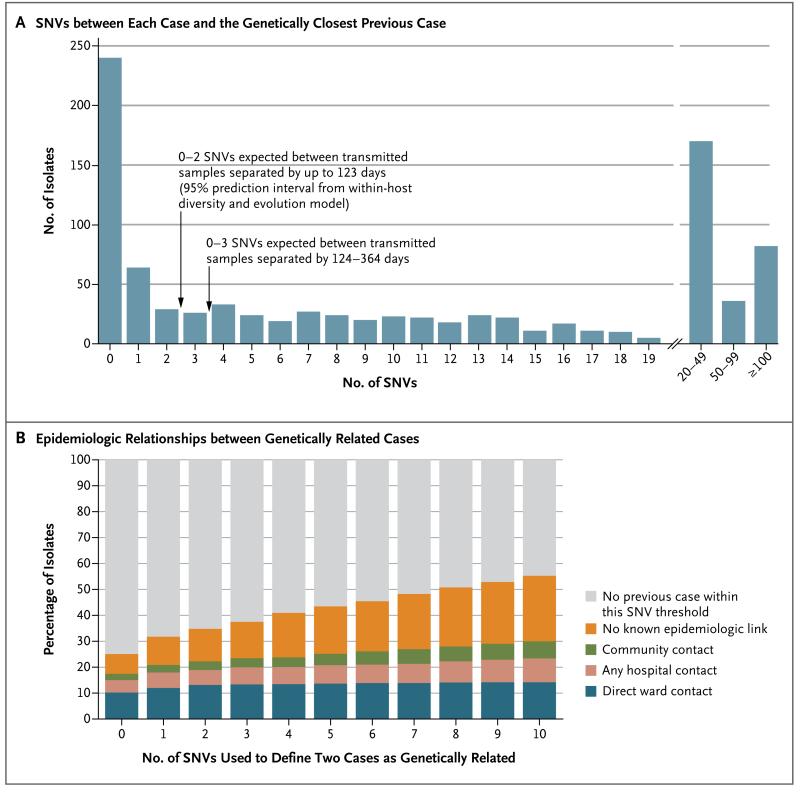
Figure 1. Genetic Variation and Epidemiologic Relationships among 957 Isolates Obtained from Patients with Clostridium difficile Infection.
Panel A shows the number of single-nucleotide variants (SNVs) between each sample obtained during the period from April 1, 2008, through March 31, 2011, and the most closely related previous sample obtained after September 1, 2007. Panel B shows the percentages of isolates that were classified as genetically related, according to the different SNV thresholds, along with the epidemiologic links between related isolates.
Of the 333 patients with C. difficile genetically related to a previous infection, 126 (38%) had ward contact with the previous genetically related case, 5 (2%) were linked only by possible ward-based contamination after the discharge or recovery of an infectious patient, 29 (9%) shared time in the same hospital but were never on the same ward, and 21 (6%) had both ward contamination and hospital-wide contact. For the remaining 152 patients (46%), no hospital-based contact could be established. Of these patients, 15 (10%) were patients at the same general medical practice, and 17 (11%) lived in the same postal-code district. Overall, 120 patients (36%) had no record of any hospital or community contact with a previous genetically related case. Even when the infectious, incubation, and ward-contamination periods were unlimited, 68 patients (20%) had no hospital or community contact (Table S3 in the Supplementary Appendix). The results were consistent when other SNV thresholds for the definition of genetically related cases were used (Table 1 and Fig. 1B).
Table 1.
Classification of 957 Cases of C. difficile Infection According to the SNV Threshold Used to Define a Genetic Relationship.*
| Classification | Main Analysis | Sensitivity Analysis | |
|---|---|---|---|
| 0–2 SNVs | 0 SNVs number of cases (percent) |
0–10 SNVs | |
| Genetically distinct | 624 (65) | 717 (75) | 428 (45) |
| Genetically linked to any previous case | 333 (35) | 240 (25) | 529 (55) |
| Closest genetic link through hospital contact | |||
| Any hospital contact within plausible limits† | 181 (19) | 144 (15) | 224 (23) |
| Ward contact† | 126 (13) | 98 (10) | 136 (14) |
| Ward contamination only‡ | 5 (1) | 7 (1) | 8 (1) |
| Shared medical specialty only† | 17 (2) | 15 (2) | 28 (3) |
| Other hospital-wide contact only† | 12 (1) | 9 (1) | 22 (2) |
| Ward contamination and hospital- wide contact |
21 (2) | 15 (2) | 30 (3) |
| Closest genetic link through community contact, with no hospital contact |
|||
| Any community contact | 32 (3) | 23 (2) | 63 (7) |
| Same general medical practice | 15 (2) | 10 (1) | 37 (4) |
| Same residential postal-code district, but different general medical practice |
17 (2) | 13 (1) | 26 (3) |
| Genetically related but no known hospital or community contact |
120 (13) | 73 (8) | 242 (25) |
Shown are the proportions of cases that would be classified into various epidemiologic groups according to the SNV threshold used to define genetically related infection as compared with previous cases: the threshold used in the main analysis of the study (≤2 SNVs) or two alternative thresholds used in sensitivity analyses (0 SNVs or 0 to 10 SNVs). In the main analysis, isolates that were obtained from April 1, 2008, through March 31, 2011, were classified as genetically distinct from any other isolate obtained after September 1, 2007, if they were separated by more than 10 SNVs. A sensitivity analysis with no restrictions on the duration of infectious, incubation, and ward-contamination periods is provided in Table S3 in the Supplementary Appendix.
This category was defined as contact in which patients shared time and space in the same setting (hospital ward, hospital, or medical specialty) that was consistent with between-patient transmission. It was assumed that cases were infectious from 1 week before diagnosis to 8 weeks after diagnosis, with an incubation period of 0 to 12 weeks.
Ward contamination was defined as contamination persisting for up to 4 weeks after a patient’s discharge from the ward.
EPIDEMIOLOGICALLY UNEXPLAINED CASES
We investigated several possible explanations for the source of the 120 cases for which there was no epidemiologic explanation for the genetic relationship (Table S4 in the Supplementary Appendix). The median time since the most recent previous genetically related case was 113 days (interquartile range, 28 to 281), which suggests that point sources or laboratory contamination (of samples processed in weekly batches) were unlikely. Transmission mediated by health care workers is probably accounted for in the 17 genetically related samples (5%) obtained from patients with physicians in the same medical specialty, which were already excluded from the 120 cases. Of the 120 samples, 32 (27%) were obtained from patients with possible transmission from an intermediate patient who had tested negative for C. difficile on enzyme immunoassay but who had known contact with a symptomatic patient with a genetically related strain. However, 27 of 120 date-matched pairs of genetically distinct (>10 SNVs) cases (22%) also had at least one shared contact who had negative results on enzyme immunoassay, a finding that suggests that many of the intermediate contacts that were found between genetically related cases were the result of chance rather than transmission (P = 0.55). Similarly, 36 patients (30%) had a hospital contact who had not been tested by means of enzyme immunoassay (i.e., a possible asymptomatic carrier) in common with a genetically related case, as compared with 35 pairs of genetically distinct cases (29%) (P = 1.00). Of these 36 patients, 32 also had an enzyme immunoassay–negative contact. Analyses that allowed an unlimited duration of ward contamination (to account for the possible persistence of spores on wards) accounted for an additional 13 cases (11%). No pair of patients who had 2 SNVs or fewer attended the same outpatient clinic on the same day. Overall, none of these potential transmission routes could link 48 patients with genetically related strains who had no hospital or community contact (40%).
WHOLE-GENOME SEQUENCING VERSUS CONVENTIONAL GENOTYPING
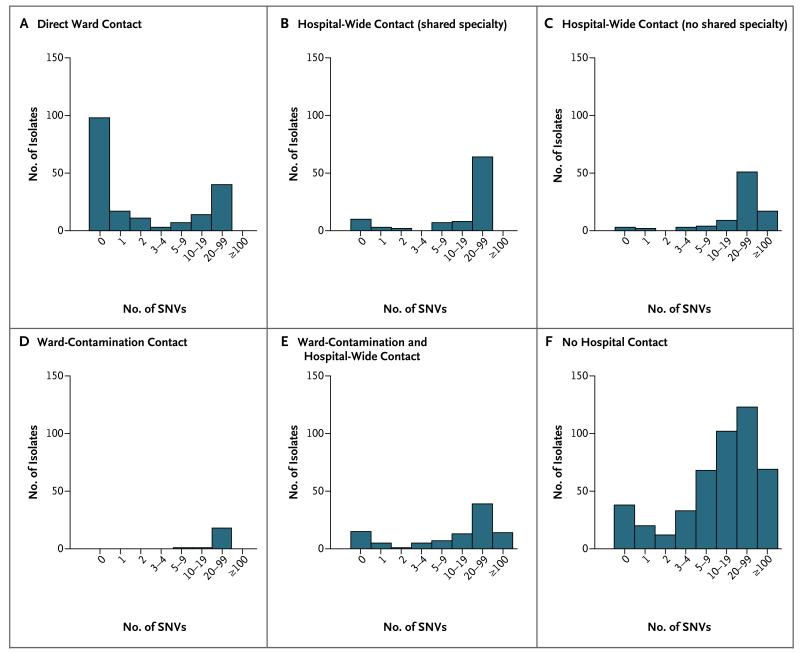
As described above, we classified cases using genetic sequences and then determined the epidemiologic relationships. We also compared the results of previous analyses that were based on combining genotyping (by means of multilocus sequence typing) and epidemiologic data10 with the number of SNVs between patients (Fig. 2). Among 190 pairs of patients with ward contact who had the same sequence type, 126 (66%) had no more than 2 SNVs (consistent with transmission); 54 (28%) had more than 10 SNVs between them. In comparison, of 302 patients who were linked by other forms of hospital contact (predominantly shared time), only 41 (14%) had no more than 2 SNVs, and 230 (76%) had at least 10 SNVs, which ruled out transmission that was previously thought to be plausible.
Figure 2. Number of SNVs between Cases on the Basis of Multilocus Sequence Typing and Epidemiologic Data.
Shown is the breakdown of 957 isolates obtained from patients with C. difficile infection from April 1, 2008, through March 31, 2011. Patients sharing a sequence type and epidemiologic contact with a case since September 1, 2007, are shown in Panels A through E: direct ward contact (Panel A), hospital-wide contact with a shared medical specialty (Panel B), hospital-wide contact with no shared medical specialty (Panel C), only ward-contamination contact (Panel D), and both ward-contamination and hospital-wide contact (Panel E). Patients with no previous case of or hospital contact with a case of the same sequence type are shown in Panel F.10 For multiple contacts of the same type that occurred with different potential sources of infection, the lowest number of SNVs associated with that type of contact is shown.
TEMPORAL PATTERNS IN DIVERSITY
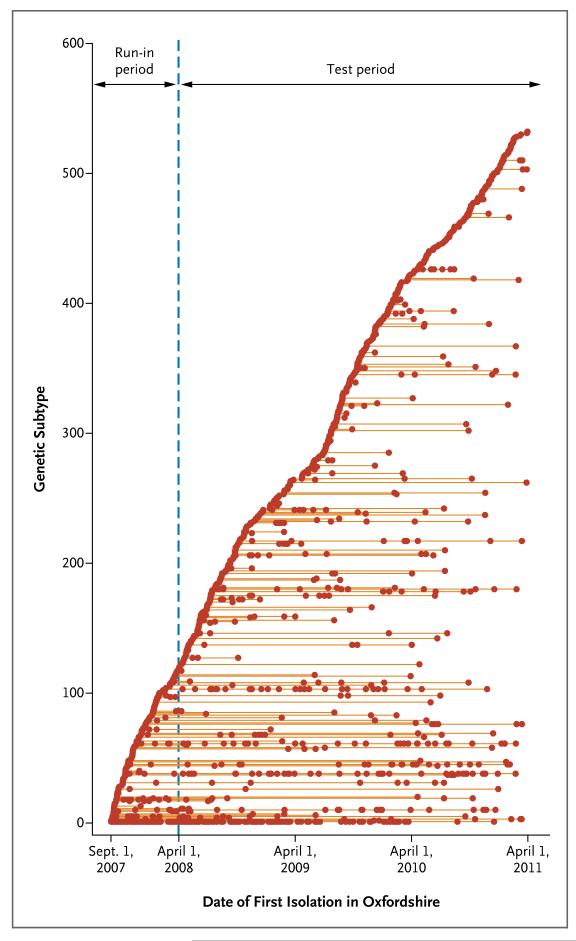
Given the diversity observed within a host over time, we determined that the probability of observing more than 10 SNVs through evolution during the 3.6-year study period was less than 0.001. To assess the number of genetically distinct clusters of cases that occurred during the study, we defined distinct subtypes as separated by more than 10 SNVs from all other cases (Fig. 3). The number of cases of each subtype varied markedly. In addition to clear clustering of cases over time, we found instances in which cases from the same lineage were separated by long periods without a case. In 12 cases, genetically indistinguishable strains were recovered more than a year apart without any intervening case. Distinct subtypes were identified consistently throughout the study, suggesting that the cases arose from a considerable reservoir (Fig. S5 in the Supplementary Appendix).
Figure 3. Timing and Size of C. difficile Genetic Clusters.
Isolates with more than 10 SNVs from any previous sample were defined as distinct genetic subtypes and are plotted on separate horizontal lines, according to the date of the first isolation in Oxfordshire. Isolates that were obtained from September 2007 through March 2008 (run-in period) were included only as potential sources for later C. difficile infections, since patients could have acquired C. difficile from a case before the study started. (Models estimating the total population diversity are provided in Fig. S5 in the Supplementary Appendix.)
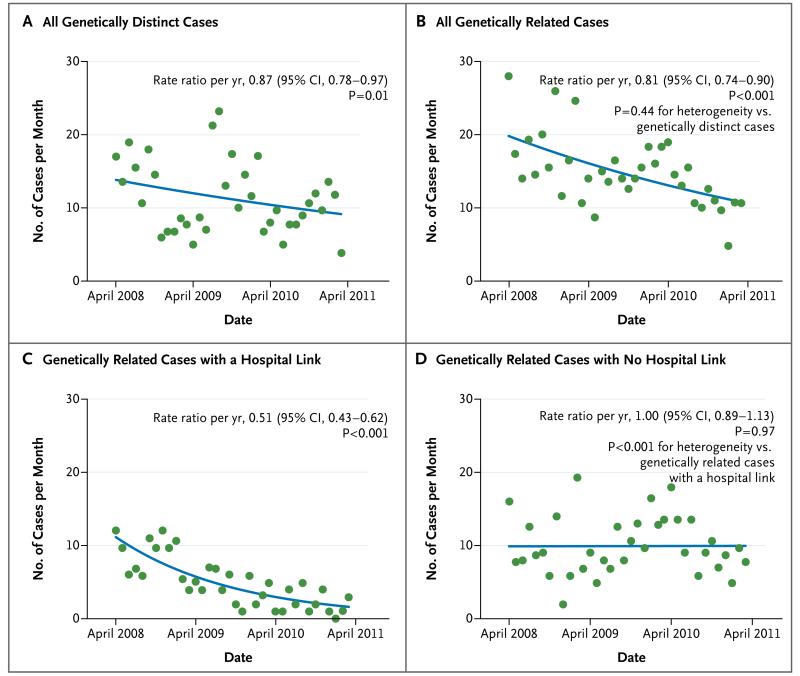
To test the hypothesis that interventions that were aimed at reducing transmission might have preferentially affected genetically related cases, we compared the incidence of cases of C. difficile infection caused by genetically distinct isolates (those with >10 SNVs from any previous case) with the incidence of cases that were genetically related to a previous case (≤2 SNVs). Despite a marked month-to-month variation in the incidence of genetically distinct and related cases, a downward trend was observed in the two types of cases, with no evidence of a significant between-group difference in reduction (P = 0.44 for heterogeneity) (Fig. 4A and 4B). Among the genetically related cases, a marked reduction was seen in cases that involved a known hospital contact, with no change in other cases over time (P<0.001 for heterogeneity) (Fig. 4C and 4D). Genetically related cases that were caused by sequence type (ST) 1 C. difficile known as North American Pulsed Field type 1 (NAP1) and polymerase-chain-reaction (PCR) ribotype 027 (ST1/NAP1/027) accounted for much of this decrease (Fig. S6 in the Supplementary Appendix).
Figure 4. Trends in Genetically Related and Distinct Cases, According to Date.
Shown are cases of C. difficile infection that were genetically distinct (>10 SNVs) from all previous cases (on the basis of samples obtained after September 1, 2007) (Panel A), cases that were genetically related (≤2 SNVs) (Panel B), and cases that were genetically related and either had a hospital link (Panel C) or had no hospital link (Panel D). The study population was 600,000 persons, so the rate of 30 cases per month corresponds to 5 cases per 100,000 population per month. The blue lines indicate the model-based estimates of the per-year rate ratio. (Details regarding the regression models that were used in the analysis and trends in genetically related cases grouped according to the presence or absence of ribotype 027 are provided in the Supplementary Appendix.)
Patients with isolates that were genetically related tended to be older (median age, 81 years; interquartile range, 70 to 87) than those with genetically distinct isolates (median age, 76 years; interquartile range, 64 to 85; P<0.001). On the basis of surveillance definitions,1 253 of 333 genetically related cases (76%) were classified as having a “health care onset” or being “health care associated,” as compared with 282 of 428 genetically distinct cases (66%) (P = 0.003).
DISCUSSION
In a 3.6-year study of whole-genome sequencing of isolates from more than 1200 patients with C. difficile infection who were living in a defined geographic area with a typical incidence of the infection21 and standard infection-control practices,2 we found that a substantial number of patients had acquired C. difficile from sources other than symptomatic patients with positive results for C. difficile toxin on enzyme immunoassay. Only 35% of cases were genetically related to at least one previous case (i.e., ≤2 SNVs). These data show that in a majority of cases, C. difficile infection is not transmitted from another symptomatic patient. This finding is based only on whole-genome data and therefore does not depend on potentially incomplete epidemiologic data that were a key limitation of previous genotyping-based transmission studies10,11 that could assess only ward-based transmission.12 Applying the greater discrimination provided by whole-genome sequencing, we found that 13% of cases were genetically related and involved ward contact, and 19% of cases were genetically related and involved some sort of hospital contact. The remainder of the genetically related cases may represent exposure to at least one intermediate host or source rather than direct contact.
A total of 45% of cases had sufficient genetic diversity (>10 SNVs from any previous cases) to represent transmission originating from sources other than the symptomatic cases that were included in the study. Some patients with genetically distinct isolates may have acquired C. difficile infection from sources outside Oxfordshire, but given the patients’ median age of 78 years, it is more likely that most organisms were acquired from asymptomatic persons22 or some other environmental reservoir. The ongoing identification of genetically distinct isolates throughout the study period points to diverse alternative sources. This finding might be expected, since C. difficile is an ancient organism23 with diverse strains present in humans, animals, and food.24 However, this diversity does not necessarily translate into diverse causes of C. difficile infection. In general, the organism is likely to have been acquired recently,25-27 allowing only a limited window for C. difficile exposure, and many infections are caused by relatively few lineages, in particular ST1/NAP1/027.28 It is therefore striking that we observed diverse subtypes in patients with C. difficile infection, each representing a separate transmission event from a reservoir or asymptomatic carrier. Typically, one or very few cases arose from each exposure, reflecting both limited secondary transmission and the absence of widespread transmission from a few point sources. Therefore, to prevent exposure, numerous sources need to be targeted.
Whole-genome sequencing also identified 13% of cases that were genetically related (≤2 SNVs) but without any evidence of plausible previous contact through a hospital, residential area, or family doctor. Exploring possible explanations for these cases, we found no evidence of substantial contributions from third-party transmission within hospitals by health care workers or patients. Patients with genetically related strains who did not have hospital or community contact could also have been exposed to a common source; the median of 113 days between such cases makes a short-term point source such as fresh food or water unlikely, unless it was persistently contaminated. Further, detailed studies of these patients in real time may provide insights into the location and nature of such sources.
The incidence of genetically distinct C. difficile cases was similar to that of genetically related cases. This finding suggests that interventions to reduce susceptibility to disease in exposed patients (e.g., changes in the use of antibiotics or specific types of antibiotics), rather than just to reduce transmission of C. difficile from symptomatic patients, might have played a major role in reductions in the incidence of C. difficile infection in the region during the past 5 years. Supporting this possibility, the use of fluoroquinolones and cephalosporins fell significantly between 2007 and 2010 in the treatment of 12,603 British patients with C. difficile infection whose cases were reported to the C. difficile Ribotyping Network.29 According to a recent U.K. government report, the rate of use of fluoroquinolones and cephalosporins fell in 175 English hospitals between 2006 and 2009.30 During a similar period, active restriction of the prescribing of fluoroquinolones and cephalosporins in a 450-bed Scottish hospital, without any other change in infection-control procedures, was associated with a relative reduction of 77% in the incidence of C. difficile infection.31 Data from a simulation study also suggested that reducing the susceptibility of exposed patients to infection may be more effective at reducing incidence than lowering transmission rates.32 Interventions to prevent secondary cases in our hospitals from 2007 (Table S1 in the Supplementary Appendix) may underlie the substantially larger decrease in the number of genetically related cases associated with hospital contact, particularly those caused by ST1/NAP1/027, as compared with those not associated with hospital contact. Genotyping and sequencing were performed retrospectively, so these studies did not affect clinical or infection-control practice. Large decreases in the prevalence of this epidemic strain in England have been described recently and are believed to be due to targeted infection-prevention measures.29 Alternatively, ST1/NAP1/027 may have declined independently of infection-control efforts, as has been observed with strains of methicillin-resistant Staphylococcus aureus (MRSA),33 possibly because of changes in strain fitness, host immunity, or changes in antimicrobial use combined with strain-specific antibiotic resistance.34 The unchanged incidence of possible secondary cases with no hospital link is an ongoing challenge.
Our study has several limitations. We ascertained initial cases on the basis of enzyme immunoassay, which has limited sensitivity. However, multiple samples were available from many patients during the study.35 If the missing cases (a minority) were missed at random, our estimates of the diversity and trends in genetically related and distinct cases are likely to be robust. More sensitive or two-step testing may also identify C. difficile carriers with diarrhea, who represent another possible source of transmission.36 The molecular clock that was used to assess potential transmissions is an approximation, particularly since mutation rates among reproducing vegetative forms may be different from such rates among spores. However, slow or arrested evolution in spores would mean that genetically distinct cases were even more distantly related. Genetically quiescent spores may also complicate attempts to reconstruct transmission events, since related cases may be separated by long intervals. However, such separation was seen infrequently, and indistinguishable strains that were separated by at least 1 year were identified in only 12 of 785 samples obtained after the first year of the study. We accounted for mixed infections that were detected by genotyping samples obtained from patients for whom multiple samples were available.16 However, undetected mixed infections that were missed by sequencing a single colony subculture and subsequent reinfections with the same sequence type may provide an explanation for some apparently unlinked cases. The widely reported prevalence of mixed infections (<10%)16 could not explain the diversity that was identified, and minority strains in mixed infections appear to play a limited role in transmission.37 Samples from asymptomatic adults25,38-40 and children41,42 and from the environment8,24 were not available, so we could not assess these potential sources of transmission. Quantifying their contribution to the total incidence of C. difficile infection remains an area for future study.
In conclusion, we found that the transmission of C. difficile infection from symptomatic patients accounted for slightly more than a third of such cases in a region with a typical incidence of infection,21 which suggests that many cases arise from genetically diverse sources. The use of rapid benchtop sequencing allows for the identification of genetically related cases in nearly real time43 so that cases that are clearly linked by hospital or community contact can be targeted to prevent further spread. Such sequencing also permits more sensitive monitoring of institutional infection-control performance through the counting of genetically related cases, rather than all cases. Data from whole-genome sequencing are also sufficiently discriminatory that genetically related cases without a clear epidemiologic link can be investigated in a highly focused way to discover novel routes of transmission. Such investigations are likely to shed new light on the source of currently unexplained infections and the diverse sources of C. difficile infection.
Supplementary Material
Acknowledgments
The views expressed in this article are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research (NIHR), or the Department of Health.
Supported by the NIHR under its Oxford Biomedical Research Centre Infection Theme and the United Kingdom Clinical Research Collaboration (UKCRC) Modernising Medical Microbiology Consortium, the latter funded under the UKCRC Translational Infection Research Initiative through grants from the Medical Research Council, the Biotechnology and Biological Sciences Research Council, and the NIHR on behalf of the Department of Health (G0800778) and the Wellcome Trust (087646/Z/08/Z). Profs. Crook and Peto are NIHR Senior Investigators. Dr. Eyre is an NIHR Doctoral Research Fellow.
Profs. Crook and Peto report receiving grant support through their institution from Optimer Pharmaceuticals and honoraria from Optimer Pharmaceuticals; and Dr. Wilcox, receiving consulting fees from Actelion, Astellas, AstraZeneca, Cerexa, Durata, Cubist, Merck, Nabriva, Novacta, Novartis, Optimer, Pfizer, Roche, Sanofi Pasteur, Summit, the Medicines Company, and VHsquared, lecture fees from Pfizer, AstraZeneca, and Astellas, grant support from Astellas and bioMérieux, and lecture fees from Alere paid to his department.
Footnotes
No other potential conflict of interest relevant to this article was reported.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Cohen SH, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 2.Vonberg R-P, Kuijper EJ, Wilcox MH, et al. Infection control measures to limit the spread of Clostridium difficile. Clin Microbiol Infect. 2008;14(Suppl 5):2–20. doi: 10.1111/j.1469-0691.2008.01992.x. [DOI] [PubMed] [Google Scholar]
- 3.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–10. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 4.Samore MH, DeGirolami PC, Tlucko A, Lichtenberg DA, Melvin ZA, Karchmer AW. Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis. 1994;18:181–7. doi: 10.1093/clinids/18.2.181. [DOI] [PubMed] [Google Scholar]
- 5.Dubberke ER, Reske KA, Olsen MA, et al. Evaluation of Clostridium difficile-associated disease pressure as a risk factor for C. difficile-associated disease. Arch Intern Med. 2007;167:1092–7. doi: 10.1001/archinte.167.10.1092. [DOI] [PubMed] [Google Scholar]
- 6.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. J Infect Dis. 1992;166:561–7. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 7.Muto CA. Asymptomatic Clostridium difficile colonization: is this the tip of another iceberg? Clin Infect Dis. 2007;45:999–1000. doi: 10.1086/521855. [DOI] [PubMed] [Google Scholar]
- 8.Hensgens MPM, Keessen EC, Squire MM, et al. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect. 2012;18:635–45. doi: 10.1111/j.1469-0691.2012.03853.x. [DOI] [PubMed] [Google Scholar]
- 9.Health Protection Agency Quarterly epidemiological commentary: mandatory MRSA, MSSA and E. coli infection data (up to October–December 2012) ( http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1284473407318)
- 10.Walker AS, Eyre DW, Wyllie DH, et al. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med. 2012;9(2):e1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norén T, Akerlund T, Bäck E, et al. Molecular epidemiology of hospital-associated and community-acquired Clostridium difficile infection in a Swedish county. J Clin Microbiol. 2004;42:3635–43. doi: 10.1128/JCM.42.8.3635-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbarth S, Samore MH. Clostridium: transmission difficile? PLoS Med. 2012;9(2):e1001171. doi: 10.1371/journal.pmed.1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surawicz CM, Brandt LJ, Binion DG, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108:478–98. doi: 10.1038/ajg.2013.4. [DOI] [PubMed] [Google Scholar]
- 14.Didelot X, Eyre DW, Cule ML, et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol. 2012;13(12):R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths D, Fawley W, Kachrimanidou M, et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–8. doi: 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyre DW, Walker AS, Griffiths D, et al. Clostridium difficile mixed infection and reinfection. J Clin Microbiol. 2012;50:142–4. doi: 10.1128/JCM.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley DR, Balasubramanian S, Swerdlow HP, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sethi AK, Al-Nassir WN, Nerandzic MM, Bobulsky GS, Donskey CJ. Persistence of skin contamination and environmental shedding of Clostridium difficile during and after treatment of C. difficile infection. Infect Control Hosp Epidemiol. 2010;31:21–7. doi: 10.1086/649016. [DOI] [PubMed] [Google Scholar]
- 19.Jinno S, Kundrapu S, Guerrero DM, Jury LA, Nerandzic MM, Donskey CJ. Potential for transmission of Clostridium difficile by asymptomatic acute care patients and long-term care facility residents with prior C. difficile infection. Infect Control Hosp Epidemiol. 2012;33:638–9. doi: 10.1086/665712. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigo AG, Felsenstein J. Coalescent approaches to HIV population genetics. In: Crandall KA, editor. Evolution of HIV. Johns Hopkins University Press; Baltimore: 1999. pp. 233–72. [Google Scholar]
- 21.Health Protection Agency Results from the mandatory Clostridium difficile reporting scheme. 2012 ( http://www.hpa.org.uk/web/HPAweb&HPAwebStandard/HPAweb_C/1195733750761)
- 22.Bartsch SM, Curry SR, Harrison LH, Lee BY. The potential economic value of screening hospital admissions for Clostridium difficile. Eur J Clin Microbiol Infect Dis. 2012;31:3163–71. doi: 10.1007/s10096-012-1681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He M, Sebaihia M, Lawley TD, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A. 2010;107:7527–32. doi: 10.1073/pnas.0914322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stabler RA, Dawson LF, Valiente E, et al. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One. 2012;7(3):e31559. doi: 10.1371/journal.pone.0031559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo VG, Bourgault A-M, Poirier L, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365:1693–703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 26.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351:633–6. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 27.Johnson S, Clabots CR, Linn FV, Olson MM, Peterson LR, Gerding DN. Nosocomial Clostridium difficile colonisation and disease. Lancet. 1990;336:97–100. doi: 10.1016/0140-6736(90)91605-a. [DOI] [PubMed] [Google Scholar]
- 28.He M, Miyajima F, Roberts P, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–13. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilcox MH, Shetty N, Fawley WN, et al. Changing epidemiology of Clostridium difficile infection following the introduction of a national ribotyping-based surveillance scheme in England. Clin Infect Dis. 2012;55:1056–63. doi: 10.1093/cid/cis614. [DOI] [PubMed] [Google Scholar]
- 30.Annual report of the Chief Medical Officer . Department of Health; London: 2011. ( https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/138331/CMO_Annual_Report_Volume_2_2011.pdf) [Google Scholar]
- 31.Dancer SJ, Kirkpatrick P, Corcoran DS, Christison F, Farmer D, Robertson C. Approaching zero: temporal effects of a restrictive antibiotic policy on hospital-acquired Clostridium difficile, extended-spectrum β-lactamase-producing coliforms and meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2013;41:137–42. doi: 10.1016/j.ijantimicag.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Starr JM, Campbell A, Renshaw E, Poxton IR, Gibson GJ. Spatio-temporal stochastic modelling of Clostridium difficile. J Hosp Infect. 2009;71:49–56. doi: 10.1016/j.jhin.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 33.Wyllie DH, Walker AS, Miller R, et al. Decline of meticillin-resistant Staphylococcus aureus in Oxfordshire hospitals is strain-specific and preceded infection-control intensification. BMJ Open. 2011;1(1):e000160. doi: 10.1136/bmjopen-2011-000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stabler RA, He M, Dawson L, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10(9):R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Updated guidance on the diagnosis and reporting of Clostridium difficile. Department of Health; London: Mar, 2012. ( http://www.dh.gov.uk/en/Publicationsandstatistics/Publications/PublicationsPolicyAndGuidance/DH_132927) [Google Scholar]
- 36.Wilcox MH, Planche T. Summary of research underpinning the Department of Health’s new Clostridium difficile testing guidance. 2011 ( http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1317132979562) [Google Scholar]
- 37.Eyre DW, Cule ML, Griffiths D, et al. Detection of mixed infection from bacterial whole genome sequence data allows assessment of its role in Clostridium difficile transmission. PLoS Comput Biol. 2013;9(5):e1003059. doi: 10.1371/journal.pcbi.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyajima F, Roberts P, Swale A, et al. Characterisation and carriage ratio of Clostridium difficile strains isolated from a community-dwelling elderly population in the United Kingdom. PLoS One. 2011;6(8):e22804. doi: 10.1371/journal.pone.0022804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RLP, Donskey CJ. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45:992–8. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 40.Ozaki E, Kato H, Kita H, et al. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J Med Microbiol. 2004;53:167–72. doi: 10.1099/jmm.0.05376-0. [DOI] [PubMed] [Google Scholar]
- 41.Enoch DA, Butler MJ, Pai S, Aliyu SH, Karas JA. Clostridium difficile in children: colonisation and disease. J Infect. 2011;63:105–13. doi: 10.1016/j.jinf.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Stoesser NE, Martin J, Mawer D, et al. Risk factors for Clostridium difficile acquisition in infants: importance of study design. Clin Infect Dis. 2013;56:1680–1. doi: 10.1093/cid/cit089. [DOI] [PubMed] [Google Scholar]
- 43.Eyre DW, Golubchik T, Gordon NC, et al. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open. 2012;2(3):e001124. doi: 10.1136/bmjopen-2012-001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.