Abstract
Virioplankton have a significant role in marine ecosystems, yet we know little of the predominant biological characteristics of aquatic viruses that influence the flow of nutrients and energy through microbial communities. Family A DNA polymerases, critical to DNA replication and repair in prokaryotes, are found in many tailed bacteriophages. The essential role of DNA polymerase in viral replication makes it a useful target for connecting viral diversity with an important biological feature of viruses. Capturing the full diversity of this polymorphic gene by targeted approaches has been difficult; thus, full-length DNA polymerase genes were assembled out of virioplankton shotgun metagenomic sequence libraries (viromes). Within the viromes novel DNA polymerases were common and found in both double-stranded (ds) DNA and single-stranded (ss) DNA libraries. Finding DNA polymerase genes in ssDNA viral libraries was unexpected, as no such genes have been previously reported from ssDNA phage. Surprisingly, the most common virioplankton DNA polymerases were related to a siphovirus infecting an α-proteobacterial symbiont of a marine sponge and not the podoviral T7-like polymerases seen in many other studies. Amino acids predictive of catalytic efficiency and fidelity linked perfectly to the environmental clades, indicating that most DNA polymerase-carrying virioplankton utilize a lower efficiency, higher fidelity enzyme. Comparisons with previously reported, PCR-amplified DNA polymerase sequences indicated that the most common virioplankton metagenomic DNA polymerases formed a new group that included siphoviruses. These data indicate that slower-replicating, lytic or lysogenic phage populations rather than fast-replicating, highly lytic phages may predominate within the virioplankton.
Keywords: viral ecology, metagenomics, phage diversity
Introduction
Measurements of viral production within marine ecosystems indicate that a significant proportion of the bacterioplankton standing stock is lost to viral lysis (Winget et al., 2011) and that virioplankton populations turn over rapidly, often in less than a day for productive coastal marine ecosystems (Winget et al., 2011). The lytic release of viruses and nutrients is a key mechanism shaping the flow of C and energy through marine ecosystems (Poorvin et al., 2004; Suttle, 2005) and influencing the productivity and composition of marine microbial communities. Although appreciation of the importance of viral processes to ecosystem function has grown, we have only a cursory understanding of the predominant biological characteristics of abundant viral populations that are driving viral effects within the ocean. Such information is crucial to a deeper, mechanistic understanding of the virus–host relationships and how these relationships shape the microbial activities critical to global biogeochemical cycles.
One impediment to these investigations has been the lack of a universal genetic marker that can draw connections between the evolutionary history, diversity and biological characteristics of viruses. By analogy, the use of the 16S ribosomal RNA gene as a universal genetic marker among prokaryotic life has provided a means to investigate links between the phylogeny and population biology of prokaryotic groups. Nevertheless, as many viruses and bacteriophages carry informational protein genes (that is, genes involved in maintenance of genetic information), these genes have been used in studies examining the diversity, biogeography and population biology of viruses. In particular, because polymerases are critical to viral replication, these genes can have a disproportionately major role in shaping the evolutionary history and fitness (Gimenes et al., 2011) of the viruses that carry them.
For example, DNA polymerase sequences have been critical to constructing phylogenetic relationships between viruses infecting eukaryotic microalgal host groups (Brussaard et al., 2004), whereas RNA-dependent RNA polymerase gene sequences revealed a remarkable diversity of picornaviruses and other RNA viruses within the virioplankton (Culley et al., 2003, 2006). In the case of bacteriophages, previous studies have utilized DNA polymerase sequences to examine the distribution and diversity of phages related to coliphage T7 belonging to the Podoviridae morphological family (Breitbart et al., 2004; Labonté et al., 2009). The ubiquity of T7-like DNA polymerase genes has led some to propose that highly lytic podoviruses are key factors in the virioplankton (Labonté et al., 2009). However, a critical shortcoming of these previous examinations has been the limited ability of PCR approaches to detect novel viral groups on the basis of DNA polymerase gene sequences. A telling example of this shortcoming is the fact that sequences related to polA genes from known siphoviruses and myoviruses have not been detected within environmental samples, despite the fact that polA is known to be carried by phages within these morphological families (Scarlato and Gargano, 1992; Buechen-Osmond and Dallwitz, 1996; Lohr et al., 2005).
Today, the use of high-throughput DNA sequencing to assess the genetic diversity and composition of natural viral assemblages—shotgun viral metagenomics—enables much broader access to the extant viral genetic diversity and thus deeper investigations of viral population genetics through phylogenetic analysis of viral genes. This study used just such an approach through characterization of virioplankton metagenomic sequences homologous to full-length family A DNA polymerases. The family A DNA polymerases, encoded by the polA gene, are a large and varied group of polymerases that includes all bacterial Pol I. In bacteria, Pol I primarily functions as a DNA proofreading enzyme and includes a polymerase domain, a 3′–5′ exonuclease domain, and a 5′–3′ exonuclease domain (Ollis et al., 1985; Beese et al., 1998; Li et al., 1998). However, the polA gene is also common in tailed dsDNA phages (Breitbart et al., 2004). Among phages, the protein does not include a 5′-3′ exonuclease and is generally the DNA polymerase primarily responsible for phage genome replication (Tabor and Richardson, 1987; Doublie et al., 1998; Naryshkina et al., 2006). Here we report that well-known structural features of DNA polymerases, which shape its enzymatic activities, provide a framework for understanding the prevalent biological features of phage populations within the virioplankton.
Materials and methods
Viral metagenome sequence libraries (viromes)
Details of library preparation are available in the supplementary material. Sequences from 10 virioplankton metagenomic libraries collected at three sampling sites (Table 1 and Supplementary Figure S1) were analyzed. Libraries from the Dry Tortugas (Andrews-Pfannkoch et al., 2010) and the Chesapeake Bay (Bench et al., 2007; Rusch et al., 2007; Andrews-Pfannkoch et al., 2010) were constructed from environmental viral nucleic acids that had been separated into three fractions: dsDNA, ssDNA and RNA (Andrews-Pfannkoch et al., 2010). Only dsDNA virioplankton were analyzed from the Gulf of Maine sample (Tully et al., 2012). After separation, the ssDNA and RNA fractions were transformed into dsDNA copies and subsequently all dsDNA fragment libraries were constructed using the linker-amplified shotgun library procedure (Andrews-Pfannkoch et al., 2010). Insert DNA from randomly selected clones was sequenced using the Sanger dideoxy-chain terminator method (Sanger et al., 1977) to provide ∼64–117 thousand sequence reads (Figure 1), with read lengths of ca. 750 bp. The longer read lengths provided by Sanger sequencing were critical to unambiguous assembly of putative full-length DNA polymerase genes (Wommack et al., 2008). Environmental metadata, sequences and bioinformatic analyses for these libraries are available on the Viral Informatics Resource for Metagenome Exploration (VIROME) website (http://virome.dbi.udel.edu) (Wommack et al., 2012).
Table 1. DNA polymerase A reads and contigs by library.
| VIROME a Library Name | Location | Library type | polA reads (%b) | polA contigsc |
|---|---|---|---|---|
| CFA-Dd | Chesapeake | dsDNA | 239 (0.29) | 12 |
| CIA-Bd | Chesapeake | dsDNA | 10 (0.18) | 0 |
| CBB | Chesapeake | dsDNA | 39 (0.69) | 1 |
| CBJ | Chesapeake | dsDNA | 80 (0.70) | 2 |
| CBS | Chesapeake | ssDNA | 122 (2.13) | 2 |
| CBR | Chesapeake | RNA | 12 (0.21) | 0 |
| Total | Chesapeake | 502 (0.43) | 17 | |
| DTF | Dry Tortugas | dsDNA | 573 (0.88) | 37 |
| DTS | Dry Tortugas | ssDNA | 47 (0.82) | 0 |
| DTR | Dry Tortugas | RNA | 33 (0.60) | 0 |
| Total | Dry Tortugas | 653 (0.86) | 37 | |
| GMF | Gulf of Maine | dsDNA | 680 (1.06) | 33 |
Library identifier in VIROME. Additional library details available at (http://virome.dbi.udel.edu).
polA reads per total number of reads in library.
Contigs were assembled with up to 2% gaps and 3% mismatches, and the consensus sequences were translated and used only if they were longer than 300 amino acids and had a conserved domain hit to the DNA polymerase A domain.
Multiple libraries collected at the same site were combined for the purpose of this study.
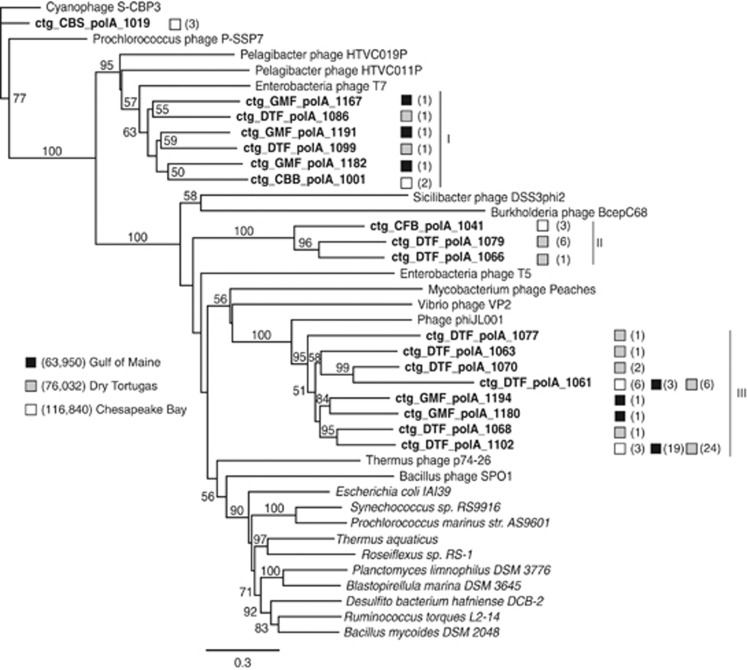
Figure 1.
Unrooted maximum likelihood tree of representative peptide sequences from translated metagenomic DNA polymerase A contig clusters with known phage and bacterial Pol I sequences. Known phages are shown in normal text, bacteria in italics and metagenomic sequences in bold. Sequences were trimmed to the polymerase domain. Each metagenomic sequence is the representative sequence of a peptide cluster and numbers in parentheses indicate the number of contigs in the cluster. Shaded boxes indicate the location of virioplankton metagenomic libraries. Roman numerals indicate clades of environmental sequences. Numbers in parentheses in the legend are total sequences from each location. Bootstrap values below 50% are not shown. Scale bar represents 0.3 amino acid substitutions per site.
Metagenomic and phylogenetic analysis
Open reading frames (ORFs) were predicted from all virome reads using MetageneAnnotator (Noguchi et al., 2006, 2008), translated and subsequently clustered (supplementary methods) at a similarity cutoff of 40% (Edgar, 2010). Representative sequences from all of the peptide clusters were searched in the VIROME database to determine which clusters contained a representative peptide sequence with significant homology (BLASTP E-score ⩽10−3) to a known DNA polymerase. Subsequently, the ORFs within putative DNA polymerase clusters were retrieved from each viral metagenome library. These ORFs were separated by library and assembled with a minimum overlap of 50 bp and maximum of 2% gaps and 3% mismatches (Drummond et al., 2011). Contig consensus sequences were translated into amino acids according to the ORF call, and a conserved domain BLAST search (Marchler-Bauer and Bryant, 2004; Marchler-Bauer et al., 2009, 2011) was run on translated consensus sequences of ⩾300 amino acids and only those sequences with hits to the polymerase domain were used. Putative DNA polymerase I sequences identified in this study have been deposited in GenBank Acc. KF514434–KF514521.
Each virioplankton DNA polymerase contig translation was clustered using the nearest neighbor algorithm of DOTUR with a minimum similarity of 40%, producing 18 clusters (Schloss and Handelsman, 2005) (Supplementary Table S1). A representative translated contig sequence from each cluster was aligned (MUSCLE, 8 iterations, gap extension penalty: -2) with known phage and bacterial DNA polymerase A sequences (Edgar, 2004), and the alignment was trimmed to exclude all exonuclease domains. A maximum likelihood tree of this alignment (Figure 1) was constructed using the JTT substitution model of PHYML (Guindon et al., 2005) with 100 bootstrap replicates.
The genetic content of dsDNA and ssDNA virioplankton from the Chesapeake Bay were compared using four dsDNA viral metagenome libraries (CFA, CFB, CFC and CFD) and one ssDNA viral metagenome library (CBS) (Figure 2). For the purpose of this comparative analysis the four dsDNA libraries were considered as one library. In actuality, each of these libraries was from a single station in the Chesapeake Bay sampled over a 24 h period.
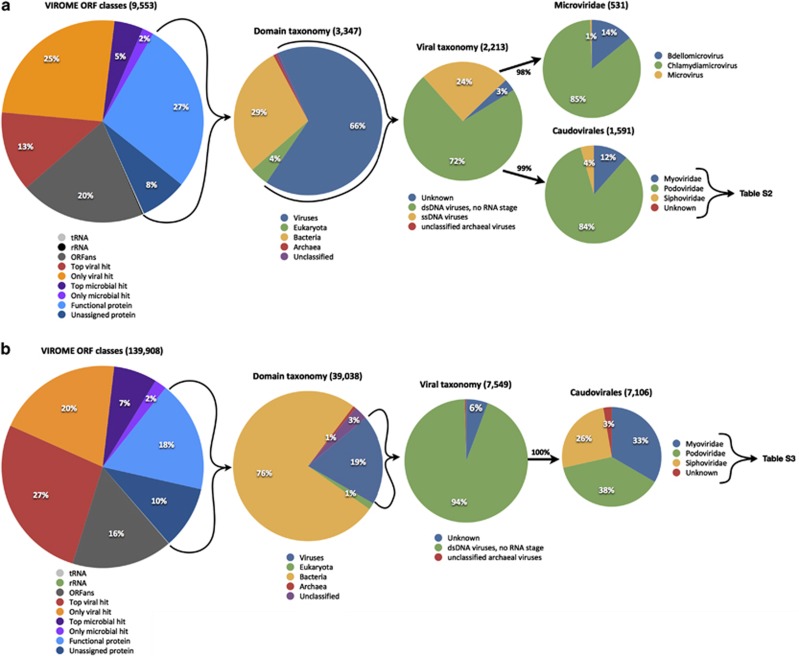
Figure 2.
Distribution of BLAST homologs to predicted ORFs from shotgun metagenome sequence libraries of: (a) single-stranded DNA; and (b) double-stranded DNA virioplankton in the Chesapeake Bay water samples. Virioplankton ORFs within VIROME ORF classes; top viral hit, only viral hit, top microbial hit and only microbial hit showed homology to only environmental peptides. Virioplankton ORFs within the VIROME ORF classes; functional protein or unassigned protein showed homology to a peptide in the UniRef 100 database. ORFans were defined as virioplankton ORFs having no significant homology (E⩽0.001) to either an environmental peptide or a UniRef peptide. Subsequent pie charts based on taxonomy of BLAST homologs are based on virioplankton ORFs showing significant homology to a UniRef peptide. Details of virioplankton ORFs with homology to known phages within the Caudovirales are shown in Supplementary Table S2 (ssDNA virioplankton) and Supplementary Table S3 (dsDNA virioplankton).
Phylogenetic relationships between assembled virioplankton DNA polymerases and PCR-amplified virioplankton DNA polymerases were investigated by aligning (MUSCLE, 8 iterations, gap extension penalty: -2) (Edgar, 2004) the representative consensus sequences with a large collection of environmental DNA polymerase sequences obtained using degenerate PCR primers (Labonté et al., 2009). All metagenomic sequences were trimmed to match the correct amplicon length, and a maximum likelihood tree (Figure 3) was constructed as described above.
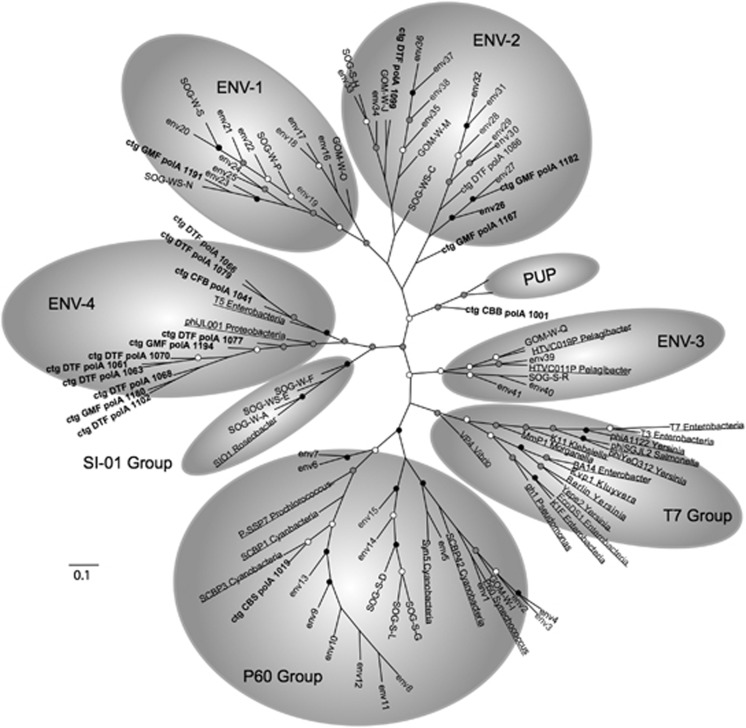
Figure 3.
Unrooted maximum likelihood tree based on the alignment of translated metagenomic contigs (bold) with translated DNA Pol A amplicons from degenerate primers from Labonté et al. (2009) and Breitbart et al. (2004) and known phage (underlined). All metagenomic contigs were trimmed to the amplicon length. Black, gray and white circles indicate nodes with at least 100%, 75% and 50% bootstrap support, respectively. PUP clade as identified in Breitbart et al. (2004). ENV clade 1-3 are as designated in Labonté et al. (2009) and include all sequences from clade I of Figure 1. ENV-4 is newly identified in this study and contains all sequences from clade II and III of Figure 1. Scale bar represents amino acid substitutions per site.
Structural prediction of DNA polymerase from ssDNA virome
Because no known ssDNA viruses have been shown to carry a DNA polymerase gene, the structure of a DNA polymerase A peptide from the CBS ssDNA viral library (ctg_CBS_polA_1019, Supplementary Table S1) was examined using the first approach, automated mode of homology modeling in Swiss-Model Workspace (Peitsch et al., 1995; Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006; Kiefer et al., 2009). The amino acid sequence of ctg_CBS_polA_1019 was modeled onto structure 1 × 9WA of T7 DNA polymerase (Dutta et al., 2004).
Results
Contig assembly and homology to DNA polymerase
Sequence reads showing significant homology (BLASTP E-score⩽10−3) to known DNA polymerase I sequences were present in all libraries, although only 6 of the 10 libraries contained contigs that were longer than 300 amino acids and showed homology to the polA domain (Table 1). Among dsDNA libraries, sequences from the Dry Tortugas (DTF) yielded the greatest number of usable contigs. With the exception of the CFA-D and CIA-B libraries, all dsDNA libraries showed a frequency of polA-encoding reads between 1.1 and 0.7%, making this one of the most abundant functional proteins detected within the libraries (Supplementary Table S3). The highest observed frequency of pol A reads was from the Chesapeake Bay ssDNA library (CBS), but only two viable full-length contigs were assembled from the 122 sequences showing homology to DNA polymerase A (Table 1). The frequency of polA reads in the Dry Tortugas RNA library (DTR) was nearly three times that of the Chesapeake RNA library (CBR), but no viable contigs were assembled from either library.
Multiple sequence alignment of translated metagenomic contigs with known DNA polymerase I sequences showed that these putative DNA polymerases contained many conserved residues critical to metal and DNA binding and enzymatic function (Figure 4). Similar to family A DNA polymerases from known phage, contigs from dsDNA libraries had the 3′–5′ exonuclease and DNA polymerase domains but lacked the 5′–3′ exonuclease domain. Contigs from the ssDNA libraries contained the DNA polymerase domain but neither of the exonucleases at the N-terminus (Figure 4).
Figure 4.
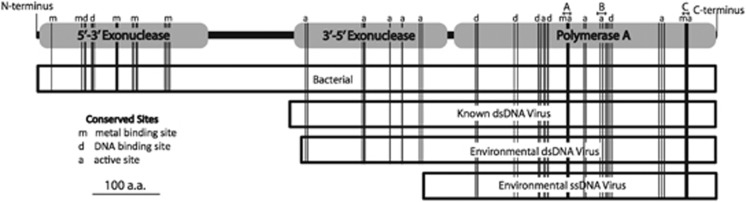
Schematic of typical bacterial, phage and putative environmental viral DNA polymerase I proteins. Lengths in amino acids based on averages from multiple sequence alignments. Capital letters A-C indicate motifs conserved across DNA polymerases (Loh and Loeb, 2005). Vertical lines represent conserved amino acids. All contigs of environmental viral putative DNA polymerase I proteins contained these sites.
All virioplankton metagenomic DNA polymerases contained three motifs that were conserved throughout DNA polymerases (Figure 4) (Loh and Loeb, 2005). Except where noted, residue number refers to its position in the E. coli Pol I. In motif A, Asp705 is immutable because of its binding of catalytic magnesium (Patel and Loeb, 2000). Also highly conserved within motif A are Glu710, which stabilizes the closed form of the enzyme and prevents the incorporation of ribonucleotides (Loh and Loeb, 2005), and Arg712. All of these residues were conserved in the virioplankton DNA polymerases. Motif B, which contacts the nascent base pair, has the key residues Arg754, Lys758, Phe762 and Tyr766 (Loh and Loeb, 2005). These residues were universally conserved in the viroplankton contigs except for Phe762. Of the metagenomic sequences, 12% had phenylalanine, 13% had tyrosine and 75% had leucine in this position. Residues 881-883 of motif C were highly conserved across the reference and metagenomic sequences. His881 coordinates to the sugar of the primer terminus. Asp882 binds to catalytic magnesium and coordinates with Glu883 via a water molecule (Loh and Loeb 2005). In the crystal structure of phage T7 DNA polymerase, Glu883 is proximal to the C-terminal histidine, which is critical to T7 DNA polymerase function (Doublie et al., 1998). However, histidine is not the terminal residue in a number of phage and bacteria including T. aquaticus, Enterobacteria phage T5 and 85% of the metagenomic sequences. In motif C, Val880 is evolutionarily conserved in bacteria (Loh and Loeb, 2005) but is substituted with threonine and isoleucine in cultured phage and in the metagenomic DNA polymerases.
Phylogeny of metagenomic DNA polymerases
Phylogenetic analysis showed that bacterial polymerases form a single, well-supported clade that was less diverse than the environmental and known phage sequences (Figure 1). Larger, well-supported clades of virioplankton metagenomic sequences each contained representatives from at least two of the three locations. This analysis confirmed the ubiquity of T7-like podoviruses observed in previous PCR-based studies (Breitbart et al., 2004; Labonté et al., 2009), as full-length polymerase sequences from all three environments claded with T7 DNA Polymerase I (Clade I, Figure 1). Recently identified T7-like Pelagibacter phages HTVC011P and HTVC019P were also part of this clade. However, this was not the largest clade of environmental sequences and contained only six clusters and a total of seven metagenomic polymerases. In total, the 18 virioplankton DNA polymerase clusters contained 88 contigs assembled from 320 sequence reads (Supplementary Table S1). Thus, T7-like DNA polymerase I sequences accounted for one-third of the virioplankton DNA polymerase clusters, but only 8% of sequenced contigs. Astonishingly, only 5% of the reads contributing to full-length DNA polymerase contigs occurred in the T7-like Clade I (Supplementary Figure S2). These data illustrate that the T7-like DNA polymerases are polymorphic and ubiquitous, yet the phage carrying these genes appeared to be less abundant than other phage groups within the virioplankton.
The second largest group of virioplankton DNA polymerase sequences (Clade II, Figure 1) was distantly related to the DNA polymerase of Enterobacteriophage T5. Although this clade contained only three clusters from the Dry Tortugas and Chesapeake Bay, it comprised a greater number of contigs (10) than the T7-like viruses (Supplementary Table S1), and a larger proportion of the reads (10%) contributing to full-length DNA polymerase contigs (Supplementary Figure S2). Thus polymerases from phages in Clade II were less diverse across the sampling sites than T7-like viruses but more abundant within the virioplankton. The largest group of virioplankton DNA polymerase sequences belonged to Clade III, a clade containing only metagenomic sequences that was distantly related to Proteobacteria phage phiJL001, a siphovirus infecting an α-proteobacterial symbiont of a marine sponge (Lohr et al., 2005). This clade contained 44% of all DNA polymerase clusters, 77% of contigs and 83% of reads contributing to full-length DNA polymerase contigs (Supplementary Figure S2). Of the eight clusters within Clade III, six contained one or two contigs and were only found in a single environment. However, the two largest DNA polymerase clusters in this clade (represented by ctg_DTF_polA_1061 & ctg_DTF_polA_1102 (Figure 1)) contained contigs from all three locations and accounted for 76% of reads contributing to full-length DNA polymerase contigs (15% for cluster-ctg_DTF_polA_1061 and 61% for cluster-ctg_DTF_polA_1102). Therefore, a large majority of DNA polymerase-carrying phages within the virioplankton have a DNA polymerase within Clade III.
The final clade of virioplankton DNA polymerase sequences contained a cluster of ssDNA Pol A sequences represented by ctg_CBS_polA_1019 and was distantly related to podoviruses infecting cyanobacteria, Cyanophage S-CBP3 and Prochlorococcus phage P-SSP7 (Figure 1). This clade contained 2% of reads contributing to full-length DNA polymerase contigs (Supplementary Figure S2). Although much shorter than the sequence of T7 DNA polymerase, ctg_CBS_polA_1019 aligned well with the polymerase domain of this protein, forming the critical thumb, palm and finger regions of the DNA polymerase structure (Figure 5). As the contig had all the key residues discussed above, this predicted polymerase from the Chesapeake Bay ssDNA virome library would make the necessary contacts with the template DNA molecule. As expected from the conserved domain BLAST and alignment (Figure 4), the ssDNA viral metagenomic sequence did not have either exonuclease structure.
Figure 5.
(a) Predicted structure of DNA polymerase from a ssDNA virus based on aligning the sequence of ctg_CBS_pola_1019 with the known structure of Enterobacteria Phage T7. DNA Polymerase DNA molecule within the enzyme structure is shown in green; (b) Structure of T7 DNA polymerase with the polymerase region in red and the exonuclease region in blue; (c) DNA polymerase from ctg_CBS_pola_1019 (ribbon model) overlaid with T7 DNA polymerase (carbon backbone).
Confirmation of purity of ssDNA library
Comparisons of the Chesapeake ssDNA and dsDNA libraries showed that these viromes had similar proportions – 35% and 28%, respectively – of predicted peptides showing significant BLAST homology (E⩽10−3) to a peptide in the UniRef 100 database (Figure 2). These peptides comprised the ‘functional protein' and ‘unassigned protein' VIROME classes (Wommack et al., 2012) and could be further assessed according to the taxonomic origin and function of the top UniRef BLAST homolog. The taxonomic origin of top BLAST hits against UniRef 100 peptides differed sharply between the viromes. At the domain level, most of the ssDNA virioplankton peptides showed significant homology to other known viral proteins (66%); whereas, only 19% of dsDNA virioplankton peptides hit another known viral peptide. For the ssDNA library, 24% of these viral hits were against known proteins from ssDNA viruses with the majority (85%) belonging to phages within the Chlamydiamicrovirus genus. No predicted peptides from the dsDNA virioplankton library had a top BLAST hit to a ssDNA viral peptide.
Most of the viral hits to dsDNA virioplankton peptides (94%) were from phages within the order Caudovirales (tailed phages). Surprisingly, three quarters of the viral BLAST hits to peptides in the ssDNA virome belonged to tailed phages. The family-level distribution of Caudovirales hits between the ssDNA and dsDNA virioplankton libraries was very different. For the ssDNA library the majority of Caudovirales hits (84%) were to phages within the Podoviridae. For the dsDNA virioplankton, hits to phage within the Caudovirales were distributed almost evenly among the three tailed phage families: Myo-, Sipho- and Podoviridae.
The presence of Caudovirales homologs within the ssDNA virioplankton library warranted further investigation to determine the possible functional characteristics of the genes that appear to bridge the evolutionary divide between ssDNA and dsDNA phages. For the ssDNA virome, 85% of peptides with a top UR100 hit in the Caudovirales fell within only seven functional genes, nearly all of which were related to nucleic acid metabolism ((RNA polymerase, endonuclease, uncharacterized protein, DNA polymerase, ssDNA binding protein, DNA primase/helicase and exonuclease) (Supplementary Table S2 and Supplementary Figure S6)). In contrast, the top 85% of Caudovirales hits to the dsDNA viromes included 106 gene descriptions across numerous categories of functional genes (Supplementary Table S3). By and large, only four cyanophages (P-SSP7, P-SSM2, S-CBP3 and Syn5) accounted for the lion's share of top UniRef hits to the ssDNA virome, with podovirus P-SSP7 being the most common (Supplementary Table S2 and Supplementary Figure S6). Recruitment of the Chesapeake Bay and Dry Tortugas ssDNA virome reads to the P-SSP7 genome showed that homology to P-SSP7 genes was largely restricted to genes involved in nucleotide metabolism (Supplementary Figure S4).
Discussion
Key mutations cause biochemical changes relevant to phage lifestyle
The conservation of residues critical to the function of DNA polymerase indicates that the metagenomic contigs likely encoded functional polymerases. However, many of the evolutionarily conserved residues in bacterial polymerases were not conserved in known phage or metagenomic sequences, suggesting that they are less critical to polymerase function or may serve to uniquely alter enzymatic characteristics of this enzyme within bacteria. Of particular interest was residue Phe762, where both known phage and metagenomic sequences frequently encoded tyrosine or leucine instead. At this position, the ssDNA cluster and Clade I (T7-like DNA polymerases) exclusively contained tyrosine, whereas Clades II and III were exclusive for phenylalanine and leucine, respectively (Figure 1).
Mutations in Phe762 are well studied because the site has important roles in discrimination against dideoxynucleotide (ddNTP) incorporation, polymerase activity and fidelity. Site-directed mutagenesis of Phe762 to tyrosine in DNA polymerase I from E. coli and Thermus aquaticus (Taq polymerase) increases the incorporation of ddNTPs 1000-fold or more than the native enzymes (Tabor and Richardson, 1995), a feature essential for DNA sequencing by chain-termination (Tabor and Richardson, 1987). Tyrosine occurs naturally at this position in Mycobacterium spp. and some phage polymerases, including that from phage T7 (Tyr526) (Tabor and Richardson, 1987; Doublie et al., 1998). Despite a low dNTP:ddNTP incorporation ratio of three (Tabor and Richardson 1995), T7 polymerase contains a strong 3′–5′ exonuclease capable of degrading ddNTPs (Tabor and Richardson, 1987). Thus, the selective pressure of tyrosine at this position in phage T7 is likely not the incorporation of ddNTPs but rather increased efficiency. A Phe762Tyr mutation in the Klenow fragment of E. coli decreases the Km fivefold, resulting in an approximately fourfold increase in catalytic efficiency (kcat/Km) over wild type (Astatke et al., 1998). Similarly, conversion of tyrosine at the corresponding residue in Mycobacterium tuberculosis (Tyr737) to phenylalanine results in a sixfold reduction of polymerase activity (Mizrahi and Huberts 1996).
The incorporation of a tyrosine residue at this location might be especially advantageous for highly lytic phage with large burst sizes such as T7-like podoviruses. Indeed, all of the cultured phages with the tyrosine mutation at this position were lytic (Supplementary Figure S3). This included the lytic Pelagibacter phages HTVC011P and HTVC019P, which infect abundant and ubiquitous SAR11 hosts. Together, these data indicate that the members of Clade I are virulent phages.
Like the sequences in Clade III, all cultured lysogenic phages carrying the polA gene contain the leucine substitution in the site corresponding to Phe762 or Phe667 (T. aquaticus) (Supplementary Figure S3). This suggests a link between this particular substitution and the biological requirements for a lysogenic life cycle. The induced mutation Phe667Leu in T. aquaticus increases the accuracy of Taq DNA polymerase threefold compared with the wild type, but simultaneously decreases the specific activity and catalytic efficiency (Vmax/Km) (Suzuki et al., 2000), whereas the T7 Tyr526Leu mutant polymerase displays a 1000-fold decrease in polymerase activity (Tabor and Richardson, 1987). A more accurate phage polymerase could slow the background mutation rate in these phages and have implications for phage evasion of host resistance through high mutation rates. The decreased efficiency of this polymerase also suggests that the phages that possess it replicate more slowly and may produce fewer progeny per burst. Presumably, such a polymerase would be more suitable to a temperate rather than a virulent phage. Alternatively, this mutation could be an adaptation for replication within hosts with low growth rates, although the presence of leucine in cultivated lysogenic phages with DNA polymerase A spanning a broad diversity of hosts suggests the mutation is directly linked to phage lifestyle. It is important to note that although leucine occurs in the Phe762 position in known phage DNA polymerases and is most frequent among marine phages, the biochemistry of this substitution has only been studied in the context of an induced mutation in T. aquaticus Pol I (Suzuki et al., 2000) and bacteriophage T7 (Tabor and Richardson, 1987). These cultured and metagenomic phage polymerases offer a new avenue for biochemists to study this mutation in its naturally occurring form.
The unusual case of Pol I in ssDNA virioplankton
Surprisingly, the metagenomic ssDNA libraries showed similar or higher frequencies of polA reads than the dsDNA libraries. The two CBS contigs had the essential active sites and necessary predicated shape to be functional polymerases despite their shorter coding length. As ssDNA phages typically have smaller genomes than their dsDNA counterparts, it is logical that the ssDNA phage proteins would minimize gene length while maintaining necessary structural domains for protein function. Also, the fact that ssDNA phages have higher mutation rates than dsDNA phages (Duffy et al., 2008) is consistent with the observation that the ssDNA virioplankton DNA polymerase did not include a proofreading exonuclease.
Several lines of evidence indicate that the presence of family A DNA polymerases in the ssDNA virioplankton libraries was not the result of contamination with DNA from dsDNA viruses. First, in the hydroxyapatite chromatography method used to separate the virioplankton nucleic acid fractions, the ssDNA fraction elutes first followed by RNA and finally dsDNA. Thus, the ssDNA and dsDNA fractions are well resolved from one another. Tests on a known mixture of viral nucleic acid types found that each viral nucleic acid eluted in the expected fraction without cross contamination (Andrews-Pfannkoch et al., 2010). Second, sequences from the ssDNA Chesapeake Bay library (CBS) were frequently most similar to DNA polymerase genes from podoviruses S-CBP3 and P-SSP7 in BLAST searches. All metagenomic sequence reads within the CBS library were tested for recruitment to the P-SSP7 genome. Contrary to what would be expected in the event of contamination, the ssDNA fragments recruited at high frequency to only a few sites of the P-SSP7 genome, most notably those regions related to genes encoding proteins involved in DNA replication (Supplementary Figure S4 and Supplementary Table S2). Finally, the distribution of BLASTP homologs among taxa and functional gene groups was substantially different for the dsDNA and ssDNA viromes from the Chesapeake Bay water samples (Supplementary Tables S2–S5).
One possible explanation why DNA pol I and other nucleic acid metabolism genes were so readily detected in the Chesapeake Bay and, to a lesser extent, the Dry Tortugas ssDNA viromes may be the procedures used in the preparation of these samples. Other ssDNA virome studies have relied on the propensity of the phi29 DNA polymerase to preferentially amplify small circular DNA molecules to selectively enrich DNA samples with amplified ssDNA viral genomes (Kim et al., 2008). Indeed, because of this preferential bias, all viromes in which multiple displacement amplification has been used to amplify environmental viral genomic DNA are enriched for the presence of ssDNA viral sequences (Angly et al., 2006; Tucker et al., 2011). To our knowledge, this is the first viral metagenomics study to avoid the use of MDA (and the phi29 DNA polymerase) in the process of preparing environmental viral DNA for sequencing. The combination of hydroxyapatite chromatography for selective isolation of ssDNA molecules along with linker amplification may have enabled detection of ssDNA viral groups that have evaded detection in MDA-based library preparation techniques. Recent work has demonstrated that viral metagenomes prepared using linker amplification more accurately preserve the underlying distributions of viruses within a community (Duhaime et al., 2012).
The critical role of virome data
Assessing viral diversity remains a challenge because viruses lack any universal marker genes. To date, studies that have used marker gene polymorphism as a means for investigating viral diversity and population ecology within natural environments have largely used PCR-based approaches that rely on primers designed from known phage genome sequences. For example, bacteriophage structural proteins g20 (portal vertex protein) (Short and Suttle 2005; Sullivan et al., 2008) and gp23 (major capsid protein) (Filee et al., 2005; Jamindar et al., 2012) have been used to identify the diversity and distribution of cyanomyoviruses and T4-like phages. Functional genes like DNA polymerase A (Labonté et al., 2009; Huang et al., 2010) and photosystem genes psbA and psbD (Bench et al., 2007; Chenard and Suttle 2008) have been used as proxies of T7-like podoviruses and cyanophage diversity, respectively. These marker genes, which have been examined in both cultivated phages and environmental amplicon sequence data, have yielded key insights into the diversity and distribution of their respective phage targets. However, in each case these studies have relied on a priori approaches for the design of PCR primers and the degree to which this reliance has limited our view of viral diversity is not well appreciated. The abundant, novel DNA polymerase A sequences identified in this study using a metagenomic approach highlight the limitations of PCR-based approaches for investigations of viral diversity and population ecology.
Adding our representative metagenomic DNA polymerase sequences to a preexisting alignment of T7-like DNA polymerase PCR amplicons (Labonté et al., 2009) did not alter previously defined groups but instead produced an additional clade labeled ENV-4 (Figure 3). Metagenomic contig sequences that were the closest to T7 DNA polymerase on the full-length sequence tree (Figure 1) surprisingly did not fall into the T7 group on the PCR-amplicon tree (Figure 3), but rather claded with the amplicons in the Labonté et al. (Labonté et al., 2009) ENV groups and with the amplicons within the PUP clade (Breitbart et al., 2004). The ssDNA virus sequence claded within the cyanophage P60 group, close to cyanophages P-SSP7 and S-CBP3 as on the full-length sequence tree (Figure 1). The remaining 11 representative DNA Pol sequences did not fall into any of the previously described groups, but formed a new clade along with Enterobacteria phage T5 and Proteobacteria phage phiJL001 labeled ENV-4 (Figure 3). This new clade corresponded to Clade II and III in Figure 1 and represented a total of 78 out of 88 contigs. It also included all three sampling locations and 93% of all sequence reads contributing to the assembly of full-length virioplankton DNA polymerases. It is important to note that the degenerate PCR primers used to obtain the T7-like environmental sequences would not have amplified the majority of virioplankton polymerase gene sequences found in this study. As a consequence, an a priori approach based on any of the previously reported DNA pol I primer sets would have missed most of the diversity of viruses carrying the DNA polA gene.
Both the full-length (Figure 1) and PCR-amplicon length (Figure 3) analyses of DNA polymerase I from viral metagenomic data indicate that the most abundant DNA polymerase-carrying viruses in coastal and estuarine environments may be similar to siphoviruses, like proteobacteria phage phiJL001. This prediction is supported by observations that myo- and siphoviruses are most frequently isolated from marine environments (Breitbart et al., 2004; Labonté et al., 2009). Moreover, Pol I sequences from cultivated siphoviruses grouped closely with the abundant metagenomic sequences in Clade III (Supplementary Figure S3). These phages are more likely to be lysogenic or pseudo-lysogenic and have slow replication rates as compared with the highly lytic podoviruses such as coliphage T7, and cyanophages P-SSP7 and P60 (Liu et al., 2004; Lohr et al., 2005; Sabehi and Lindell, 2012).
Relationship with DNA polymerase γ
Recent studies have reported that genes with homology to mitochondrial DNA polymerase γ are abundant within the virioplankton have been found in cyanophage genomes and the Global Ocean Survey data set (Chan et al., 2011; Sabehi et al., 2012). To determine the prevalence of these polymerases in our metagenomic libraries the VIROME databank was queried with the set of S15 DNA pol γ sequences from the mitochondria, cyanophage and the Global Ocean Survey (Rusch et al., 2007; Yooseph et al., 2007) and BroadPhage (John et al., 2011) data sets (BLASTP E-score ⩽10−10). This gene does not appear to be abundant in our metagenomic libraries, as only seven ORFs with homology to DNA pol γ were found in the VIROME database.
A maximum likelihood tree encompassing all the polymerase groups described in (Filee et al., 2002) and trimmed to their specified conserved regions confirmed that metagenomic Clade I–III are not related to the group of phages encoding DNA pol γ-like polymerases (Supplementary Figure S5). The clades containing metagenomic representative sequences described in Figure 1 remained distinct and did not disrupt previously established relationships, further supporting our finding that these clades are valid groupings linked to polymerase functionality. Because Clade I–III (Figure 1) contained a greater number of sequences than those identified as DNA pol γ across all VIROME libraries, phages carrying these DNA polA genes likely have a larger impact on aquatic ecosystems. These data should ignite subsequent investigations on the biochemistry of unique phage enzymes and how this biochemistry shapes the mechanistic details behind viral impacts on ecosystems.
Acknowledgments
The authors are grateful for the support of a NSF Microbial Genome Sequencing Program grant (EF 0626826) to SJW and KEW and a Gordon and Betty Moore Foundation grant to KEW and SWP. HFS was supported through summer undergraduate research internships from the Delaware EPSCoR program and the Howard Hughes Medical Institute. EGS was supported through a graduate fellowship from the Delaware Water Resources Center. Usage of the computational infrastructure of the University of Delaware Center for Bioinformatics and Computational Biology Core Facility was made possible by support from Delaware INBRE (NIH NCRR 2 P20 RR016472-09) and Delaware EPSCoR (NSF EPS-0814251). The authors are grateful to Curtis Suttle and Jessica Labonté for providing their alignment of DNA polymerase amplicon sequences.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andrews-Pfannkoch C, Fadrosh DW, Thorpe J, Williamson SJ. Hydroxyapatite-mediated separation of double-stranded DNA, single-stranded DNA, and RNA genomes from natural viral assemblages. Appl Environ Microb. 2010;76:5039–5045. doi: 10.1128/AEM.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:2121–2131. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Astatke M, Grindley ND, Joyce CM. How E. coli DNA polymerase I (Klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J Mol Biol. 1998;278:147–165. doi: 10.1006/jmbi.1998.1672. [DOI] [PubMed] [Google Scholar]
- Beese LS, Kiefer JR, Mao C, Braman JC. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, et al. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microb. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Miyake JH, Rohwer F. Global distribution of nearly identical phage-encoded DNA sequences. Fems Microbiol Lett. 2004;236:249–256. doi: 10.1016/j.femsle.2004.05.042. [DOI] [PubMed] [Google Scholar]
- Brussaard CP, Short SM, Frederickson CM, Suttle CA. Isolation and phylogenetic analysis of novel viruses infecting the phytoplankton Phaeocystis globosa (Prymnesiophyceae) Appl Environ Microb. 2004;70:3700–3705. doi: 10.1128/AEM.70.6.3700-3705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechen-Osmond C, Dallwitz M. Towards a universal virus database - progress in the ICTVdB. Arch Virol. 1996;141:392–399. doi: 10.1007/BF01718409. [DOI] [PubMed] [Google Scholar]
- Chan YW, Mohr R, Millard AD, Holmes AB, Larkum AW, Whitworth AL, et al. Discovery of cyanophage genomes which contain mitochondrial DNA polymerase. Mol Biol Evol. 2011;28:2269–2274. doi: 10.1093/molbev/msr041. [DOI] [PubMed] [Google Scholar]
- Chenard C, Suttle CA. Phylogenetic diversity of sequences of cyanophage photosynthetic gene psbA in marine and freshwaters. Appl Environ Microbiol. 2008;74:5317–5324. doi: 10.1128/AEM.02480-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. High diversity of unknown picorna-like viruses in the sea. Nature. 2003;424:1054–1057. doi: 10.1038/nature01886. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. 2011. Geneious v5.4. http://www.geneious.com/ .
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Duhaime MB, Deng L, Poulos BT, Sullivan MB. Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method. Environ Microbiol. 2012;14:2526–2537. doi: 10.1111/j.1462-2920.2012.02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Li Y, Johnson D, Dzantiev L, Richardson CC, Romano LJ, et al. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc Natl Acad Sci USA. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Filee J, Forterre P, Sen-Lin T, Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J Mol Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- Filee J, Tetart F, Suttle CA, Krisch HM. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci USA. 2005;102:12471–12476. doi: 10.1073/pnas.0503404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenes MV, Zanotto PMdA, Suttle CA, da Cunha HB, Mehnert DU. Phylodynamics and movement of Phycodnaviruses among aquatic environments. ISME J. 2011;6:237–247. doi: 10.1038/ismej.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online - a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wilhelm SW, Jiao N, Chen F. Ubiquitous cyanobacterial podoviruses in the global oceans unveiled through viral DNA polymerase gene sequences. ISME J. 2010;4:1243–1251. doi: 10.1038/ismej.2010.56. [DOI] [PubMed] [Google Scholar]
- Jamindar S, Polson SW, Srinivasiah S, Waidner L, Wommack KE. Evaluation of two approaches for assessing the genetic similarity of virioplankton populations as defined by genome size. Appl Environ Microbiol. 2012;78:8773–8783. doi: 10.1128/AEM.02432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John SG, Mendez CB, Deng L, Poulos B, Kauffman AK, Kern S, et al. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ Microbiol Rep. 2011;3:195–202. doi: 10.1111/j.1758-2229.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, et al. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol. 2008;74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté JM, Reid KE, Suttle CA. Phylogenetic analysis indicates evolutionary diversity and environmental segregation of marine Podovirus DNA polymerase gene sequences. Appl Environ Microb. 2009;75:3634–3640. doi: 10.1128/AEM.02317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Gingery M, Doulatov SR, Liu Y, Hodes A, Baker S, et al. Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J Bacteriol. 2004;186:1503–1517. doi: 10.1128/JB.186.5.1503-1517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Loeb LA. Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair. 2005;4:1390–1398. doi: 10.1016/j.dnarep.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Lohr JE, Chen F, Hill RT. Genomic analysis of bacteriophage Phi JL001: Insights into its interaction with a sponge-associated alpha-proteobacterium. Appl Environ Microb. 2005;71:1598–1609. doi: 10.1128/AEM.71.3.1598-1609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi V, Huberts P. Deoxy- and dideoxynucleotide discrimination and identification of critical 5' nuclease domain residues of the DNA polymerase I from Mycobacterium tuberculosis. Nucleic Acids Res. 1996;24:4845–4852. doi: 10.1093/nar/24.24.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryshkina T, Liu J, Florens L, Swanson SK, Pavlov AR, Pavlova NV, et al. Thermus thermophilus bacteriophage phi YS40 genome and proteomic characterization of virions. J Mol Biol. 2006;364:667–677. doi: 10.1016/j.jmb.2006.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H, Taniguchi T, Itoh T. Metageneannotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA-polymerase-I complexed with Dtmp. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Patel PH, Loeb LA. DNA polymerase active site is highly mutable: evolutionary consequences. Proc Natl Acad Sci USA. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch MC, Wells TNC, Stampf DR, Sussman JL. The Swiss-3dimage Collection and Pdb-Browser on the Worldwide Web. Trends Biochem Sci. 1995;20:82–84. doi: 10.1016/s0968-0004(00)88963-x. [DOI] [PubMed] [Google Scholar]
- Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. Viral release of iron and its bioavailability to marine plankton. Limnol Oceanogr. 2004;49:1734–1741. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G, Lindell D. The P-SSP7 cyanophage has a linear genome with direct terminal repeats. PLoS One. 2012;7:e36710. doi: 10.1371/journal.pone.0036710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, Lindell D. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc Natl Acad Sci USA. 2012;109:2037–2042. doi: 10.1073/pnas.1115467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlato V, Gargano S. The DNA polymerase-encoding gene of Bacillus subtilis bacteriophage SPO1. Gene. 1992;118:109–113. doi: 10.1016/0378-1119(92)90256-o. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microb. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short CM, Suttle CA. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl Environ Microbiol. 2005;71:480–486. doi: 10.1128/AEM.71.1.480-486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Coleman ML, Quinlivan V, Rosenkrantz JE, Defrancesco AS, Tan G, et al. Portal protein diversity and phage ecology. Environ Microbiol. 2008;10:2810–2823. doi: 10.1111/j.1462-2920.2008.01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Yoshida S, Adman ET, Blank A, Loeb LA. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J Biol Chem. 2000;275:32728–32735. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S, Richardson CC. A single residue in DNA-polymerases of the Escherichia Coli DNA-polymerase-I family is critical for distinguishing between deoxyribonucleotides and dideoxyribonucleotides. Proc Natl Acad Sci USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KP, Parsons R, Symonds EM, Breitbart M. Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J. 2011;5:822–830. doi: 10.1038/ismej.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully BJ, Nelson WC, Heidelberg JF. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol. 2012;14:254–267. doi: 10.1111/j.1462-2920.2011.02628.x. [DOI] [PubMed] [Google Scholar]
- Winget DM, Helton RR, Williamson KE, Bench SR, Williamson SJ, Wommack KE. Repeating patterns of virioplankton production within an estuarine ecosystem. Proc Natl Acad Sci USA. 2011;108:11506–11511. doi: 10.1073/pnas.1101907108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Polson SW, Chen J, Dumas M, Srinivasiah S, et al. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand Genomic Sci. 2012;6:427–439. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.