Abstract
Microbial life in the nutrient-limited and low-permeability continental crystalline crust is abundant but remains relatively unexplored. Using high-throughput sequencing to assess the 16S rRNA gene diversity, we found diverse bacterial and archaeal communities along a 2516-m-deep drill hole in continental crystalline crust in Outokumpu, Finland. These communities varied at different sampling depths in response to prevailing lithology and hydrogeochemistry. Further analysis by shotgun metagenomic sequencing revealed variable carbon and nutrient utilization strategies as well as specific functional and physiological adaptations uniquely associated with specific environmental conditions. Altogether, our results show that predominant geological and hydrogeochemical conditions, including the existence and connectivity of fracture systems and the low amounts of available energy, have a key role in controlling microbial ecology and evolution in the nutrient and energy-poor deep crustal biosphere.
Keywords: deep subsurface, crystalline rock, metagenomic sequencing, metabolism, bacteria, archaea
Introduction
Independent of photosynthetic carbon fixation and energy production, formation waters in low-porosity continental crust form a nutrient-poor environment where autogenic chemical energy sources are also scarce. Yet, studies conducted in mines and deep drill holes have documented that microbial life extends several kilometers into the Earth's crystalline crust with diverse microbial communities detected in a variety of continental areas (Havemann et al., 1999; Zhang et al., 2005; Gihring et al., 2006; Sahl et al., 2008; Onstott et al., 2009; Itävaara et al., 2011a; Nyyssönen et al., 2012). Culture-based studies under laboratory conditions show that these microbial communities include both autotrophic and heterotrophic members with variable modes of metabolism including sulfate and iron reduction, methanogenesis and acetogenesis (Haveman et al., 1999; Haveman and Pedersen, 2002). However, low rates of microbial metabolism in situ and inability to culture most of these microbes using conventional techniques limit our understanding on the mechanisms that drive and sustain such diversity in this nutrient- and energy-limited environment.
Recent advances in culture-independent techniques such as metagenomics have enabled more straightforward investigation of the deep crustal microbiome in its geological and geobiochemical context (Chivian et al., 2008; Brazelton et al., 2012). It is now postulated that the long-term survival of these microbial communities in the deep subsurface relies on diverse modes of hydrogen-driven carbon cycling, reduced carbon compounds and complex microbial interactions (Lin et al., 2006; Pedersen, 2006; Chivian et al., 2008; Brazelton et al., 2012). However, our understanding on the metabolic diversity, activity and extent of these processes continues to be limited by the scarcity of suitable study sites, technical difficulties in sampling the deep crystalline rock environment and lack of comprehensive data sets combining metagenomic sequence data with geochemical features.
The Outokumpu deep drill hole located in Finland provides a unique environment for studying the deep subsurface microbiome in low-permeability crystalline rock. The drill hole is 2516 m deep and provides direct access to a Paleoproterozoic sequence of metasediments, pegmatitic granodiorite and an ophiolitic rock assemblage where old, highly saline, poorly buffered alkaline formation water originates from long-term water–rock interactions in igneous rock aquifers (Ahonen et al., 2011; Västi, 2011; Kukkonen, 2011). A large amount of geological and hydrogeochemical data has been collected from the formation through drill core analyses, fluid samplings and in situ loggings. Recently, a diverse bacterial community was also detected down to 1500 m depth in the drill hole (Itävaara et al., 2011a, 2011b). By using 16S rRNA gene cloning, the authors demonstrated that the bacterial communities varied with depth and suggested a relationship between the diversity of these communities and geochemical composition of formation water. In this study, we extended the drill hole sampling down to 2300 m depth and used high-throughput sequencing of both bacterial and archaeal 16S rRNA genes to gain deeper insight into the microbial community structuring in the Outokumpu deep subsurface. By combining the comprehensive geological and hydrogeochemical data sets, that are available from the site with the high-throughput sequencing data, we then evaluated how environmental conditions that prevail in this environment shape the structure and functionality of these microbial communities. Finally, we used shotgun-metagenomic sequencing to develop plausible models of metabolism and functional adaptation that enable the long-term stability of these microbial communities in this low-permeability, nutrient-poor environment.
Materials and methods
Study site
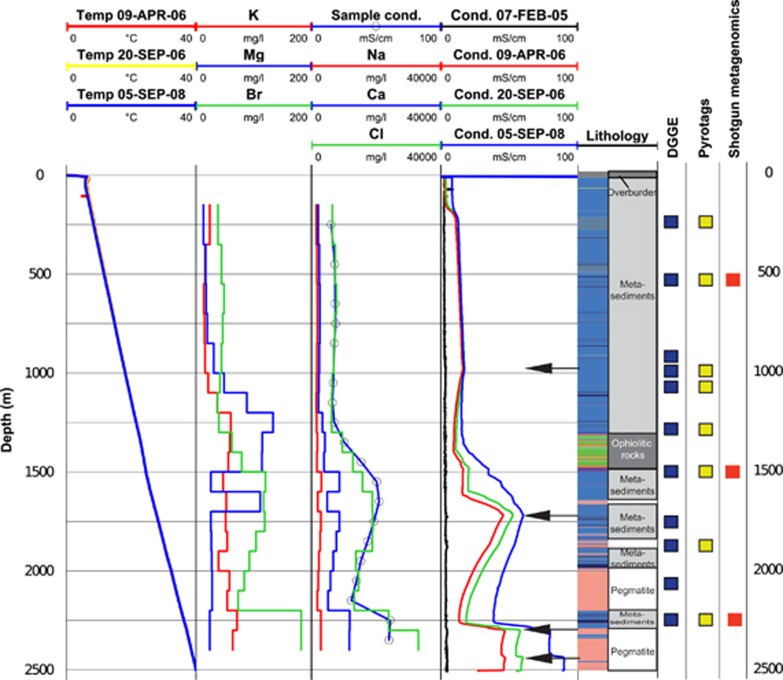
The Outokumpu deep drill hole, located at Outokumpu, Finland (E29.062°, N62.717°), extends 2516 m down from the surface (Ahonen et al., 2010; Kukkonen, 2011; Västi, 2011). The geothermal gradient in the drill hole is 13–16 mK m−1, the temperature ranging from 5 °C at the surface to 40 °C at the bottom (Figure 1) (Kukkonen et al., 2011). The upper 1300 m of the drill hole is predominantly mica schist in which thin layers of black schist represent organic marine sediments (Kontinen et al., 2006; Peltonen et al., 2008). The ophiolitic rock sequence at 1300–1500 m is mainly composed of serpentinized, mantle-derived ultramafic rock, which was likely an early seabed at the time of deposition. Other typical components of the Outokumpu assemblage include serpentinite, black schist, carbonate (skarn) and quartz. Below the assemblage the drill hole is coarse-grained intrusive pegmatitic granodiorite. The low-permeability crystalline rock is intersected by conductive fracture zones with minimal hydraulic connections (Ahonen et al., 2011). Below 39 m, the drill hole with a diameter of 22 cm provides direct access to the bedrock.
Figure 1.
The Outokumpu deep drill hole. Temperature and fluid electrical conductivity data are based on four separate post-drilling logging sessions (first and fourth column from left). Sample conductivity and chemical analyses were measured from water samples recovered in this study (second and third column from left). Lithological rock types intersected by the hole are shown in the lithology column (blue: metasedimentary mica schists; green and orange: ophiolite-derived serpentinites, skarn rock and quartz rock; pink: pegmatitic granite). The arrows indicate the most important inflow points of saline fluid to the drill hole. Analyses performed in this study are shown on the right.
The drill hole was rotary-drilled in 2004–2005. During drilling, purified drinking water from the municipal water system was used for cooling the bit and flushing the drill hole. The water was labeled with sodium fluoresceine (500 mg m−3) in order to trace the flushing water during future sampling campaigns.
Repeated EC logs in 2005–2008 and drill hole water sampling in 2007 and 2008 document the gradual replacement of fresh drilling fluids by saline formation waters (Figure 1). Inflow of formation waters occurs at 967, 1400, 1460, 1600, 1720, 2250 and 2300 m, of which the three most important ones occur at 967, 1720 and 2250 m. Fluorescein was below the detection limit in all samples analyzed in this study, indicating that formation water has replaced the cooling and flushing fluids used during and after drilling (Ahonen et al., 2011).
Sample collection
Sampling of drill hole water was performed with a sterile pressure-tight tubing from August 20 to 21, 2008, according to the method described by Nurmi and Kukkonen (1986). A plastic tube with a back-pressure check valve at the lower end was lowered into the drill hole and filled with water. When lifted, the valve activates and the contents were held in the tube. The polyamide tube consisted of 50-m-long sections connected by ball valves and/or tube fittings. During retrieval of the tube, ball valves were closed as they emerged from the drill hole. A 100 m section of the tube was treated as one sample representing a 100 m interval in the drill hole.
The gas-impermeable polyamide (PA 11) tube (outer/inner diameter 12/9 mm, Toppi Oy, Espoo, Finland) used was obtained directly from the manufacturing process and sterilized by autoclaving in 50 m lengths. The valves, connections and tools for connecting the tubes were washed with 70% (v/v) ethanol and autoclaved. The 50 m tubes were connected to the valves and to each other as they were lowered into the drill hole, and care was taken to maintain surface sterility. These precautions ensured that surface microbes were not introduced to the drill hole during sampling. Negative controls for molecular biological and microbiological sampling were taken in the beginning and at the end of the sampling from a 50-m section of tube that was sterilized and filled with distilled, autoclaved water. A total of 2350 m of tubing was inserted into the drill hole. The tube was estimated to stretch approximately 5% of the total length due to its weight.
Samples for molecular biological and microbiological analyses were recovered at 11 depths. Sampling was performed directly from the tube through a flame-sterilized, pressure-tight valve. Water samples for nucleic acid analyses (500 ml) were filtered directly with Sterivex filter units (Sterivex GP 0.22 μm, Millipore, Billerica, MA, USA) and immediately frozen on dry ice. Samples for microbial cell counting were placed in sterile 100-ml anaerobic head-space vials and transported in the dark at 4 °C to the laboratory for fluorescence staining the same day. Negative controls were taken as described above from a 50 m tube that was filled with autoclaved water and analyzed in a similar manner as those containing deep groundwater. Samples for geochemical analyses were taken from 19 depths.
Geochemistry
Electrical conductivity and pH were measured directly in the field with a field analyzer (WTW GmbH, Weilheim, Germany). Chemical analyses were conducted by Labtium Oy (Espoo, Finland) and are described in detail in Supplementary Information.
Microbial density
The number of microorganisms was determined from five milliliters of sample water by staining with the BacLight Bacterial Viability Kit (Molecular Probes, Invitrogen Corp., USA) as described in Supplementary Information.
DNA isolation
Total DNA was isolated directly from the frozen Sterivex filters in a laminar flow hood. DNA was isolated from the filter pieces with the PowerSoil DNA Isolation kit (MoBio Laboratories, Carlsbad, CA, USA) according to the manufacturer's instructions with the exception that bead beating was performed in Ribolyser (Thermo Scientific, Waltham, MA, USA) at 6 m s−1 for 30 s. The entire filter was used for DNA isolation in all samples except for the 1100 m sample, which used half. To provide a negative control, clean Sterivex filters were processed in the same way. DNA was eluted in a final volume of 50 μl. Because DNA concentration in all samples was below the detection limit of a Nanodrop ND-1000 (Thermo Scientific, Waltham, MA, USA), an equal volume of each sample was used in PCR (2–4 μl) and MDA (1 μl) amplifications.
PCR-DGGE analysis of bacterial and archaeal communities
In order to select samples for high-throughput sequencing, denaturing gradient gel electrophoresis (DGGE) was used for preliminary characterization of the bacterial and archaeal communities at the 11 sampling depths. A 193 bp fragment covering the V3 hypervariable region of the bacterial 16S rRNA gene was PCR amplified with the P2 and P3 primers (Muyzer et al., 1993).
The archaeal 16S rRNA genes were amplified with nested PCR. First, a 806 bp fragment of the archaeal 16S rRNA gene was PCR amplified with A109f and Arch915R primers (Stahl and Amann, 1991; Grosskopf et al., 1998). Second, PCR amplification of a 227 bp fragment flanking the V3 hypervariable region of the 16S rRNA gene was performed using A344FGC and 519RP primers (Lane, 1991; Bano et al., 2004). PCR conditions are described in Supplementary Information.
Amplification products were separated on a denaturing gradient gel containing 8% acrylamide and 20–65% denaturing gradient at 60 V (bacteria) or 65 V (archaea) and 60 °C for 18–20 h, and visualized with SYBR Green I staining. Similarities between banding patterns were analyzed with BioNumerics software version 4.6 (Applied Maths, Austin, TX, USA) using Dice's coefficient of similarity, the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) clustering algorithm and 0.5% optimization.
High-throughput sequencing and analysis of 16S rRNA genes
In order to obtain more comprehensive sequence coverage for further microbial community analyses, bacterial and archaeal 16S rRNA gene pyrotags were then prepared from eight sample depths by PCR as described above except that sequencing adapters and depth-specific barcode tags were attached to the 5′ ends of primers. The library preparation, emulsion PCR and pyrosequencing were conducted with a Genome Sequencer FLX 454 System according to manufacturer's protocol (454 Life Sciences/Roche Applied Biosystems, Branford, CT, USA) as described in Supplementary Information. The sequence reads were quality filtered and analyzed with mothur (Schloss et al., 2009). Reads were assigned to operational taxonomic units (OTUs) at 97% similarity and OTUs were assigned to taxonomy using BLASTN (Altschul et al., 1990) against the Greengenes database (version 4 February 2011, DeSantis et al., 2006) with the best hit taken into account with an e-value of<0.001. For the rarefaction analysis, Venn diagrams and alpha diversity estimates, a number of sequences were chosen randomly to match the same number as the sample yielding the fewest sequence (461 in bacteria and 1936 in archaea).
Non-metric multidimensional scaling analysis
The relative abundance of bacterial and archaeal OTUs at the genus level were exposed to non-metric multidimensional scaling with the R software package vegan (Oksanen et al., 2006). A Bray–Curtis distance matrix was calculated with 1000 permutations.
Metagenomic library preparation for shotgun sequencing
In order to gain further insight to the metabolic potential of the microbial communities, metagenomic libraries for shotgun sequencing were prepared from drill hole water samples recovered from 600, 1500 and 2300 m. In order to obtain a sufficient amount of DNA for sequencing library production, whole genome amplification of the total DNA was performed with the Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Pollards Wood, UK) according to the manufacturer's recommendations. In order to reduce amplification bias the amplification was performed in three separate reactions that were pooled for sequencing. Negative controls included sampling controls, DNA isolation controls and amplification controls that contained PCR-grade water instead of template DNA. The amplified DNA was processed as described in Supplementary Information and libraries were sequenced with the 454 Genome Sequencer FLX using GS FLX Titanium series reagents (454 Life Sciences/Roche Applied Biosystems). Due to the challenging nature of sample retrieval, limited amount of sample water and low amount of recovered DNA, replication of the metagenomes was not possible.
Metagenomic sequence analysis
Redundant reads potentially produced during the whole genome amplification were removed from metagenomic sequences using mothur (Schloss et al., 2009). Unique sequence reads were annotated in the metagenomics RAST server (MG-RAST) and the annotations were imported to STAMP for pair-wise statistical comparisons (Meyer et al., 2008; Parks and Beiko, (2010). Samples were compared using Fisher's exact two-sided test with Newcombe–Wilson confidence interval and Holm–Bonferroni correction. Results with a P-value below 0.05 and effect size of 0.5 between proportions were analyzed further. The effect size was used in order to focus on the biologically relevant genes.
In order to compare the composition of microbial communities detected by 16S rRNA gene sequencing and metagenomic sequencing, 16S rRNA sequences were analyzed from metagenomes by comparing all metagenomic sequences to the Greengenes database using BLASTN.
Similarity between metagenomic samples was analyzed by pairwise BLAST that was performed for individual sequences. All sample pairs were compared together with best hit for each sequence taken into account with a bit score of >40. Distance matrix was built based on the BLAST hits. Principal component analysis was done with the R function prcomp. The first two principal components were used to generate 2-dimensional scatter plots.
Material availability
The unfiltered 16S rRNA sequence reads were submitted to the short read archive under accession numbers ERS225199-ERS225214. The annotated metagenomes are available at MG-RAST (http://metagenomics.anl.gov; MG-RAST IDs 4453297.3, 4451761.3, 4451759.3).
Results and discussion
Geochemical composition and source of formation water
Water in the Outokumpu drill hole was saline below the uppermost 200 m (Figure 1). Total dissolved solids (TDS) increased with depth to about 50 g l−1 at the bottom of the drill hole (Table 1 and Supplementary Table S1). Sulfate concentrations of 13–17 mg l−1 were observed at 1200–1500 m, whereas its concentration in all other samples remained below the detection limit of 10 mg l−1. Phosphate concentrations of 0.03–0.16 mg l−1 were detected within the upper 1200 m. Nitrate was below the limit of detection (20 mg l−1) in all studied samples. The most important components of the gas phase were methane and nitrogen (average 74 vol% and 20 vol%), followed by 2 vol% helium. The stable isotope (δ2H and δ13C) composition of methane differed from its typical composition in both microbiologically and thermogenetically produced methane (Supplementary Table S2).
Table 1. Hydrogeochemical characteristics of the drill hole sections investigated with DGGE (•), pyrotag sequencing (○) and shotgun metagenomic sequencing (○) in this study.
| 200 m •○ |
600 m •○
|
900 m• | 1000 m•○ | 1100 m•○ | 1300 m•○ |
1500 m•○
|
1700 m• | 1900 m•○ | 2100 m• |
2300 m•○
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample depth | m | 150–250 | 550–650 | 850–950 | 950–1000 | 1050–1150 | 1250–1350 | 1450–1550 | 1650–1750 | 1850–1950 | 2050–2150 | 2250–2350 |
| Main rock types | Mica schist, biotite gneiss | Mica schist, biotite gneiss | Biotite gneiss, mica schist | Biotite gneiss, mica schist | Biotite gneiss, mica schist | Black schist, mica schist, skarn, serpentinite | Serpentinite, skarn, black schist | Biotite gneis, mica schist | Pegmatitic granite, mica schist, biotite gneiss | Pegmatitic granite | Pegmatitic granite, mica schist | |
| Pressure | bar | 20 | 60 | 90 | 100 | 110 | 130 | 150 | 170 | 190 | 210 | 230 |
| Hydraulic conductivity | km s−1 | n.d. | n.d. | n.d. | 5.3 × 10−7 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Permeability | km2 | n.d. | n.d. | n.d. | 4.8 × 10−14 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Ca:Na | 0.80 | 0.83 | 0.81 | 0.81 | 0.87 | 1.36 | 1.65 | 1.68 | 1.67 | 1.65 | 2.31 | |
| Br:Cl | 0.0026 | 0.0027 | 0.0029 | 0.0028 | 0.0026 | 0.0029 | 0.0034 | 0.0028 | 0.0030 | 0.0025 | 0.0024 | |
| Temperature | oC | 7.75 | 12.91 | 16. | 18.14 | 19.48 | 22.22 | 24.56 | 27.55 | 30.67 | 32.31 | 37.07 |
| pH | 9.7 | 7.2 | 8.3 | 7.9 | 7.3 | 8.2 | 8.2 | 8.5 | 8.4 | 8.2 | 8.9 | |
| Ca | mg l−1 | 2070 | 2600 | 2450 | 2280 | 2200 | 4210 | 8640 | 8740 | 6760 | 5190 | 11700 |
| Mg | mg l−1 | 12.6 | 17.5 | 30 | 48.2 | 88.1 | 115 | 25.2 | 25.5 | 27.1 | 27.8 | 23.4 |
| Na | mg l−1 | 1490 | 1800 | 1740 | 1620 | 1450 | 1780 | 3010 | 2990 | 2320 | 1800 | 2910 |
| Br | mg l−1 | 38 | 48 | 44 | 41 | 37 | 62 | 120 | 119 | 92 | 73 | 182 |
| Cl | mg l−1 | 6430 | 7760 | 6830 | 6410 | 6250 | 9590 | 15800 | 19000 | 13700 | 13200 | 33000 |
| SO4 | mg l−1 | <10 | <10 | <10 | <10 | <10 | 16 | 15 | <10 | <10 | <10 | <10 |
| TDS | g l−1 | 10 | 12 | 11 | 10 | 10 | 16 | 28 | 31 | 23 | 20 | 48 |
| EC | mS m−1 | 1640 | 1940 | 1760 | 1760 | 1700 | 2940 | 5400 | 5220 | 4110 | 3330 | 6450 |
| Alk | mmol l−1 | 0.48 | 0.25 | 0.23 | 0.32 | 0.25 | 0.34 | 0.37 | 0.77 | 2.55 | 1.47 | 0.39 |
| NO3 | mg l−1 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| PO4 | mg l−1 | <0.02 | 0.07 | 0.10 | 0.09 | 0.16 | <0.2 | <0.2 | <0.2 | <0.2 | <0.2 | <0.2 |
Abbreviations: DGGE, denaturing gradient gel electrophoresis; n.d., not detected; TDS, total dissolved solids.
On the basis of molar Ca:Na ratio, processes affecting the salinity varied with depth (Table 1). The Ca:Na ratio was 0.8 in the upper 1200 m, 1.7 at 1500–2200 m and 2.4 in the deepest and most saline samples. The molar ratio of bromide vs chloride was uniform throughout the upper 1200 m, suggesting a common source. Stable isotope composition of water (2H, 18O) indicated that saline water in the drill hole deviates distinctly from the isotopic composition of meteoric waters and suggested that formation fluids can be attributed to a long-term interaction between water and rock, and likely represent different sources (Supplementary Table S3) (Kietäväinen et al., 2013).
Microbial diversity and community composition in Outokumpu deep subsurface
Microbial diversity
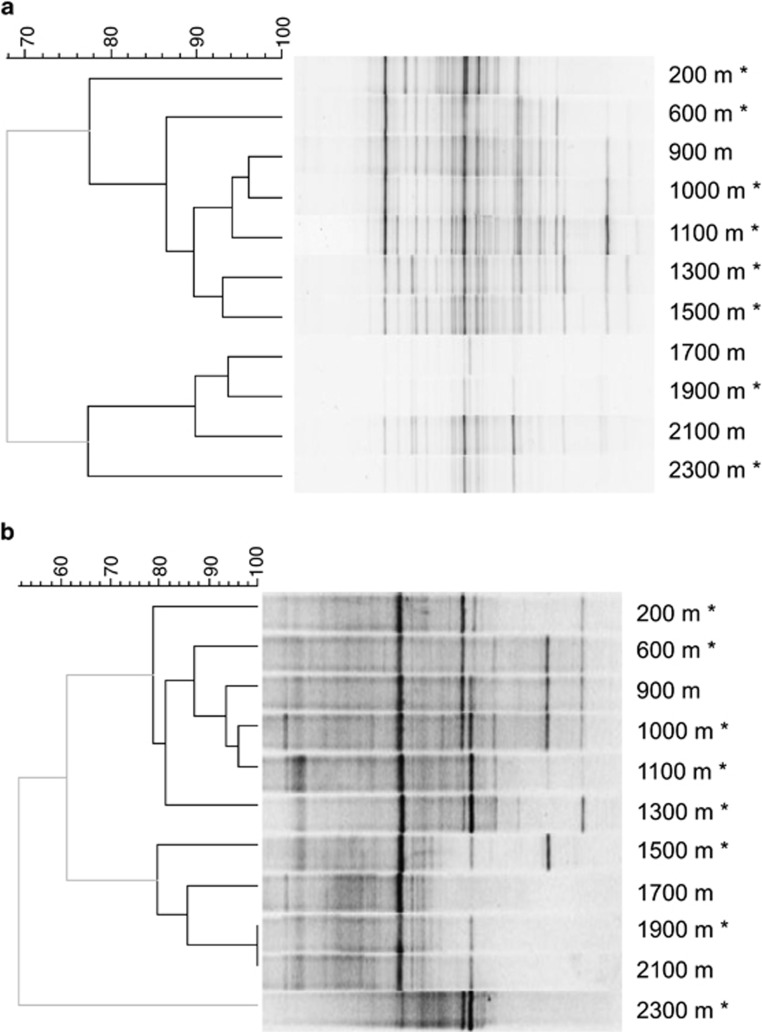
Preliminary comparison of the 11 water samples with 16S rRNA gene-based DGGE revealed diverse bacterial and archaeal communities along the 2300 m sampling section, which varied with sample depth and geochemical and isotopic composition of water (Figure 2). On the basis of their DGGE profiles and hydrogeological characteristics, eight samples were selected for further investigation by high-throughput sequencing of the hypervariable V3 region of the 16S rRNA gene. The 11 616 bacterial and 53 748 archaeal sequences, obtained after quality filtering (Huse et al., 2007), represented 26–60% of the estimated bacterial diversity and 21–60% of the estimated archaeal diversity (Supplementary Figure S2, Supplementary Table S4).
Figure 2.
Bacterial (a) and archaeal (b) community composition in the Outokumpu drill hole determined with DGGE. Similarity of DGGE profiles was calculated with the Dice's coefficient of similarity using 0.5% optimization. Clustering was performed with the UPGMA algorithm. Significant clusters (black lines) were determined by the cluster cut-off method. The scale bar represents the percentage similarity between DGGE banding patterns. Samples analyzed by 16S rRNA gene sequencing are indicated with asterisks.
The additional resolution achieved with the pyrosequencing revealed an unexpectedly high number of bacterial and archaeal OTUs (97% sequence similarity). Previous studies using 16S rRNA gene cloning have reported up to 42 bacterial OTUs in continental crystalline rocks (Gihring et al., 2006; Sahl et al., 2008; Onstott et al., 2009), whereas analysis of rarefied pyrotag data obtained from Outokumpu in this study inferred 48–110 bacterial OTUs per sample (Supplementary Table S4). Archaeal OTU richness also greatly exceeded expectations based on earlier studies, with up to 112 OTUs detected in samples recovered above 1500 m (Takai et al., 2001; Sahl et al., 2008). Bacterial OTU richness peaked at 1300 m depth, which coincides with ophiolitic sequence of altered ultramafic rocks (that is, serpentinite, skarn and quartz). This depth also contains graphite- and sulphide-rich layers derived from ancient organic material deposits, is characterized by a higher concentration of permeable fracture zones as well as an active fracture system at 1460 m, and was shown to harbor high bacterial diversity during the first characterization of drill hole microbial communities and in DGGE analysis performed in this study (Itävaara et al., 2011a). The highest number of archaeal OTUs was observed at 1000 m and at 1300 m. At 1000 m, the drill hole hydrogeochemistry is affected by methane-rich saline water that discharges from an active fracture zone at 967 m.
Microbial community composition
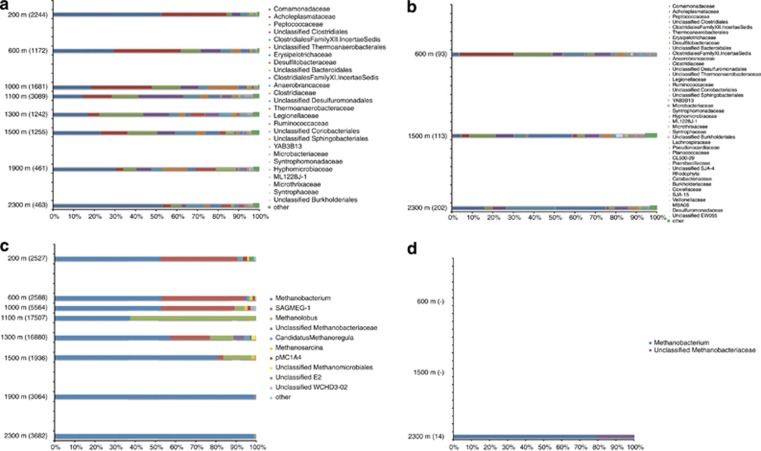
Most abundant bacterial phyla included Proteobacteria, Firmicutes and Tenericutes (Figure 3). Proteobacteria were represented by Comamonadaceae, which at 200 m accounted for 51.7% of all sequences. Chemoheterotrophic Acholeplasma were also most prominent at 200 and 600 m at 31.7 and 32.5% relative abundance, respectively, and decreased with increasing sampling depth. The highest Clostridiales diversity was detected between 1000–1500 m, and associated with genera such as the chemolithoheterotrophic Dehalobacter, thiosulfate-reducing Dethiosulfatibacter, fermentative or sulfite- and thiosulfate-reducing Desulfitibacter and heterotrophic Alkaliphilus. This high diversity occurred where saline water is discharged to the drill hole at 976 m and also between 1300–1500 m, where a higher sulfate concentration occurs within the graphite- and sulphide-rich black schist and ophiolitic rocks. At 1900 and 2300 m, sequences identified as Fusibacter, and unclassified Comamonadaceae and Thermoanaerobacterales were detected in larger numbers, accounting for 44–62% of all sequences. Archaeal communities were dominated by hydrogenotrophic Methanobacterium and other Methanobacteriaceae in almost all samples (Figure 3). At 1100 m, 38% of reads clustered to Methanobacterium and 62% to methylotrophic Methanolobus. At this depth, the drill hole water is mixed by methane-rich saline water that discharges from a fracture zone at 967 m and flows both downwards and upwards. South Africa gold mine euryarchaeotic group 1 was detected at 200–1000 m and at 1300–1500 m. At 1900 and 2300 m, over 99% of the sequences were classified to Methanobacteriaceae family, causing a reduction in community evenness, diversity and richness (Supplementary Table S4). The prevalence of Proteobacteria, Firmicutes and methanogenic archaea in Outokumpu was consistent with the detection of these taxa in geographically distant continental crustal areas (Gihring et al., 2006; Sahl et al., 2008).
Figure 3.
Bacterial (a, b) and archaeal (c, d) community composition in the Outokumpu deep drill hole as determined by 16S rRNA gene sequencing (a, c) and metagenomic sequencing (b, d). Clusters denote taxonomically assigned reads on family (bacteria) and genus (archaea) level and include taxa that represent more than 0.5% of bacterial sequences and 0.3% of archaeal sequences. The remaining sequences are grouped to ‘others.' The number of sequences used in the classification is shown in parenthesis. No archaeal 16S rRNA gene sequences were identified from metagenomic sequences obtained from 600 and 1500 m depths.
Sample validation and endemic Outokumpu deep biosphere
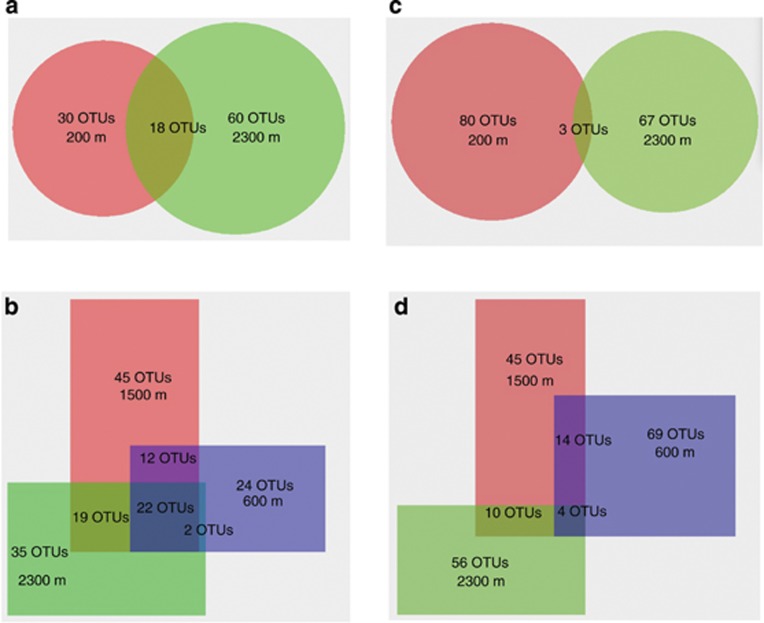
The low proportion of shared bacterial (35–55%) and archaeal (11–44%) OTUs between samples (Figure 4), the high number of OTUs unique to each sample and the correspondence between microbial community composition and depth (Supplementary Figure S3) demonstrated that the detected microbial communities describe the true diversity of the endemic microbiome at Outokumpu. This was also supported by the consistent detection of high proportion of Proteobacteria close to the surface and higher clostridial diversity between 1000–1500 m in this study (Figure 3) and by Itävaara et al., (2011a, 2011b). Together with the similar electrical conductivity measurements obtained in situ and from the sampled water, our results verify that the tube sampling method provided representative samples at each depth without cross-contamination.
Figure 4.
Venn diagrams representing shared bacterial (a, b) and archaeal (c, d) OTUs between water samples recovered close to ground surface (200 m) and toward the bottom of the drill hole (2300 m) (a, c) as well as between the three samples used for metagenomic sequencing (600, 1500 and 2300 m) (b, d). 16S rRNA reads were assigned to OTUs at 97% similarity.
Drilling required to access the deep biosphere always involves a risk of surface-derived contamination. In Outokumpu, drill hole contamination with drilling fluids and surface-derived microbes can be assumed negligible due to several reasons. Repeated in situ loggings and water sampling campaigns document the replacement of fresh drilling fluids by saline formation waters (Figure 1). The isotopic composition of sample water deviates distinctly from the isotopic composition of meteoric waters (Kietäväinen et al., 2013), and no tritium or drilling water tracer (sodium fluorescein) could be detected in the water samples (Ahonen et al., 2011). Distinct and separate bacterial and archaeal communities that were independent of changes in cell density were also detected at different depths of the drill hole.
Linking microbial community composition to hydrogeochemistry
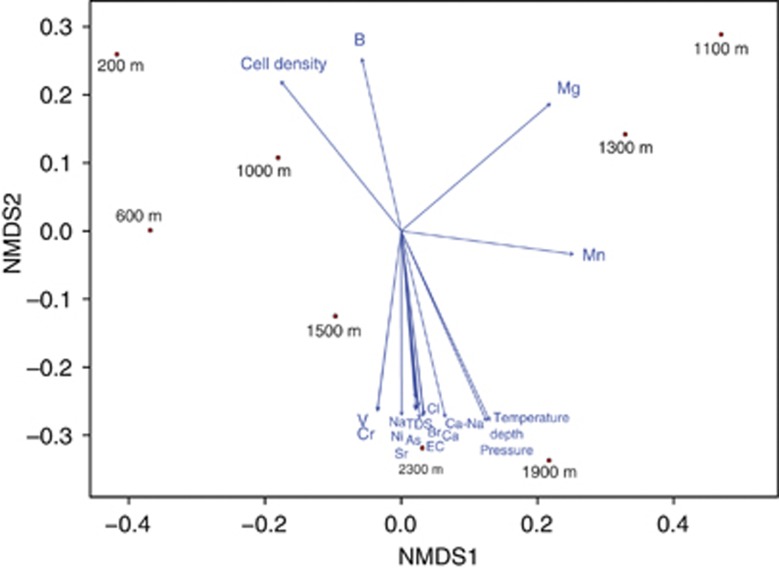
Comparison of bacterial and archaeal OTU clustering with environmental variables via non-metric multidimensional scaling analysis grouped the samples into three distinct clusters based on the sample depth (Figure 5 and Supplementary Table S5), in accordance with the UPGMA clustering of DGGE profiles and 16S rRNA gene sequences (Figure 2 and Supplementary Figure S3). At 200 m, ten-times higher cell density than other samples correlated positively (P<0.01) with OTU clustering. At this depth, an active fracture system mixes drill hole water and potentially stimulates the locally abundant chemoorganotrophic Proteobacteria. At 1100 and 1300 m, a positive correlation was observed with an increased concentration of manganese and magnesium, whereas below 1500 m temperature (25–37 °C) and pressure (150–230 bar) had a significant effect (P<0.01) on the structure of microbial communities. High salinity (TDS 28 and 48 g l−1) and Ca:Na molar ratio also correlated significantly (P<0.05) with OTU clustering in the three deepest samples. In general, bacterial OTU clustering correlated more strongly with chemical composition of drill hole water, whereas archaeal OTU clustering was more dependent on sampling depth and changes in pressure and temperature (data not shown). This indicates that bacterial communities in the Outokumpu deep subsurface respond to distinct environmental factors than the archaeal communities. In addition, although decreasing cell density with increasing sampling depth with the concomitant increase in osmotic stress due to pressure and salinity has an important role in structuring bacterial and archaeal communities in the Outokumpu subsurface, the composition of the communities is also dependent of the source of drill hole water and its formation mechanisms, which are determined by local geology. For instance, according to Ca:Na and Br:Cl molar ratios and stable isotope composition, water below 1500 m originates from a different source than that above 1200 m, where methane-rich saline water that discharges from a fracture zone at 967 m controls the water composition. At 1300 m, increased levels of manganese and magnesium coincide with manganese- and magnesium-bearing ophiolitic rocks.
Figure 5.
Microbial community structure in relation to sample depth and its associated environmental factors in the Outokumpu drill hole. A non-metric multidimensional scaling analysis of Bray–Curtis distances was performed to examine the relative abundance of bacterial and archaeal OTUs clustering at the genus level. Environmental variables with P-values >0.05 are noted on the ordination. The stress for the Bray–Curtis distance matrix was 1.26 × 10−14 for bacteria and 2.99 × 10−14 for archaea.
Most importantly, microbial diversity did not correlate with cell density or sampling depth (Supplementary Figure S4). Instead, bacterial and archaeal communities remained diverse and even along the drill hole despite a 100-fold decrease in the cell density (Supplementary Figure S4). A possible explanation for this is that more diverse communities can utilize the limited amount of available nutrients more efficiently via niche partitioning (Cardinale 2011). Mutualistic interactions among community members or emergent properties resulting from slow reproduction rates may also promote diversity and resilience of the entire community.
Functional diversity and adaptation of the drill hole microbial communities
To investigate the factors that sustain such microbial diversity in the nutrient-limited crystalline rock, shotgun-metagenomic sequencing with 454 technology was used to examine the functional, potential and survival strategies of the microbial communities at 600, 1500 and 2300 m, that is, at depths where sample water distinctly deviated from meteoric water but no significant trends between community composition and local hydrogeology were observed (600 m), where high microbial diversity coincided with graphite- and sulfide-rich rock layers derived from ancient organic material deposits (1500 m), and where the communities were structured by high salinity (2300 m).
Due to low yield, DNA isolated from the water samples was amplified using whole genome amplification. Although amplification of nanogram quantities of DNA with whole genome amplification has made it possible to study microbial communities in environments where the amount of microbial biomass would otherwise be limiting (for example, Mason et al., 2010), the technique can introduce bias in the distribution of the amplified DNA (Pinard et al., 2006). In order to minimize this bias, whole genome amplification was performed in replicate reactions and the metagenomic sequences were filtered for redundant sequences (Supplementary Table S6). Comparison of the taxonomic distribution in 16S rRNA gene amplicons and metagenomic sequences showed that the most abundant bacterial families were detected with both sequencing approaches (Figure 3). However, 16S rRNA gene sequencing detected higher relative abundance of Comamonadaceae and lower relative abundance of Clostridiales than metagenomic sequencing at all three sample depths, whereas according to metagenomic sequencing, Peptococcaceae were more abundant at 600 m, and Erysipelotrichaceae at 1500 and 2300 m. These differences in relative abundance are most likely due to different number of 16S rRNA gene sequences analyzed and coverage of sampling. 461–2244 sequences were obtained from bacterial 16S rRNA gene sequencing but only 93–202 16S rRNA gene sequences were identified from the three metagenomic libraries. In addition, while the 16S rRNA gene fragments were PCR amplified from V3 variable region of the gene, the 16S rRNA genes identified from metagenomic sequences can cover any region of the 16S rRNA gene. This can result in differences in taxonomic resolution (Ward et al., 2012). Possible PCR bias through primer selection should also not be ignored. Archaeal 16S rRNA gene sequences were detected only in the 2300-m metagenome, and were all assigned to Methanobacteriaceae in agreement with the 16S rRNA gene sequence data obtained from the same depth (Figure 3). Altogether, these results showed that the metagenomic libraries obtained in this study are representative of the endemic Outokumpu microbial communities.
In order to further demonstrate that the metagenomic libraries represented distinct microbial communities, metagenomic sequences from the three sample depths were compared using pair-wise BLAST and principal component analysis. All three samples clustered separately confirming that there was minimal overlap in gene content between the analyzed depths (Supplementary Figure S5).
Individual metagenomic sequences were then annotated in MG-RAST. Based on their phylogenetic affinities, over 78% were of bacterial origin at all sample depths, whereas the proportion of archaeal sequences increased from <2% above 1500 m to 12% at 2300 m (Supplementary Table S7).
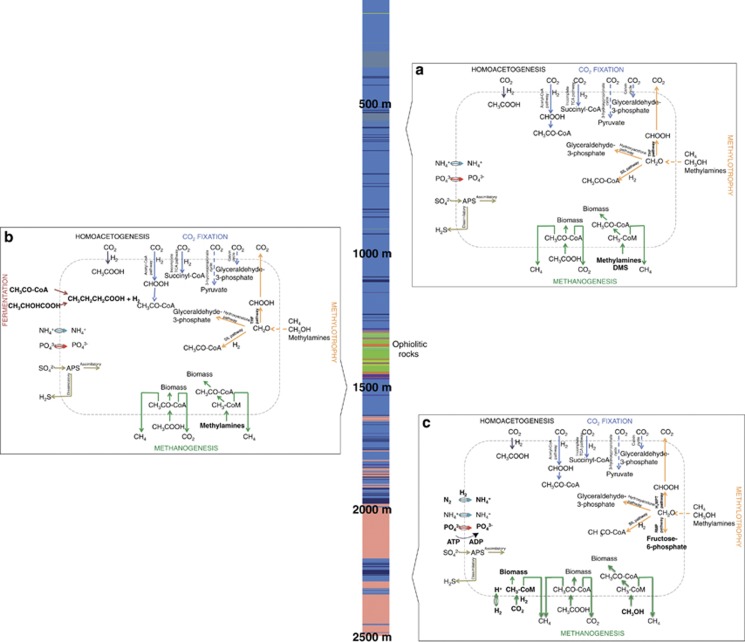
Carbon cycling
Annotation of the metagenomic sequences to functional subsystems indicated that microbial communities in Outokumpu have various mechanisms of carbon assimilation and cycling (Figure 6). Identification of the incomplete reductive TCA cycle and the reductive acetyl-CoA cycle in addition to several genes from Calvin and 3-hydroxypropionate cycles in all three metagenomes demonstrated that the drill hole microbes can fix CO2 in several ways. The reductive TCA cycle is often found in methanogenic archaea that use this pathway for producing biosynthetic precursors. Detection of this pathway is in agreement with 16S rRNA gene sequencing, which displayed a dominance of methanogenic archaea in Outokumpu. Evidence of autotrophic acetate production via the reduction CO2 and oxidation of hydrogen was also present in all metagenomes, whereas autotrophic methanogenesis from CO2 and H2 was detected only at 2300 m. The metagenome from 2300 m was also significantly enriched with hydrogenases that couple hydrogen oxidation to autotrophic CO2 fixation (Major et al., 2010).
Figure 6.
Microbial carbon and nutrient utilization strategies in the Outokumpu drill hole at (a) 600, (b) 1500 and (c) 2300 m depth predicted from MG-RAST annotations of 454 shotgun metagenomes. Pathways that differed statistically significantly (P<0.05) between metagenomes are indicated in bold. Solid lines indicate pathways from which all genes were detected in the respective metagenomes. The lithology column represents the drill hole and shows rock types intersected by the drill hole (blue: metasedimentary mica schists; green and orange: ophiolite-derived serpentinites, skarn rock and quartz rock; pink: pegmatitic granite).
The resulting reduced C-1 compounds can provide a carbon source for a variety of methylotrophic microbes (Figure 6). Genes required for both methylotrophic and acetoclastic methanogenesis were detected at all three sample depths. However, substrates for methylotrophic methanogenesis changed from methylamines and dimethylsulfide at 600 and 1500 m to methanol at 2300 m. Reduced C-1 compounds are also metabolized via formaldehyde that is assimilated through serine and hydroxyacetone pathways at all three depths. Mechanisms of formaldehyde oxidation, in contrast, depend on sample depth. At 600 and 1500 m, formaldehyde is converted to formate via tetrahydrofolate pathway, whereas at 2300 m tetrahydromethanopterin and ribulose monophosphate pathways are used. The drill hole microbes also have the ability to assimilate more complex carbon substrates through fermentation. Genes involved in lactate fermentation were detected in all three metagenomes, whereas genes involved in the fermentation of acetyl-CoA to butyrate, acetone–butanol–ethanol synthesis and butanol biosynthesis were found to be more abundant at 1500 m where black schist forms a typical component of ophiolitic rocks. High microbial diversity with a particularly high proportion of chemoorganoheterotophic Clostridia (38%) in comparison with the other two depths (21–23%), many of which ferment ethanol and acetate (Kim and Gadd, 2008), were also detected at this depth.
Hydrogen
In line with the earlier proposals (Pedersen 1997), our results demonstrate that the deep hydrogen-driven biosphere of the Fennoscandian shield relies on homoacetogenesis and autotrophic methanogenesis. Up to 3.1 mmol l−1 of hydrogen has been detected in the Outokumpu deep drill hole water (Kietäväinen et al., 2013), but its primary origin and availability are unclear. It has been suggested that hydrogen in deep crystalline rocks could be generated by the radiolysis of water as a result of uranium decay (Lin et al., 2005; Pratt et al., 2006). Hydrogen may also be produced as a result of mineral reactions including serpentinization or oxidation of sulfide-containing minerals such as black schist (Hoffman 1992; Sleep et al., 2004).
Nutrient uptake and metabolism
Nutrients are mainly obtained via active uptake (Figure 6). The main source of nitrogen is ammonium that is assimilated via glutamine and glutamate. Denitrification and nitrate respiration are negligible in all three samples, which is in accordance with the low levels or absence of nitrate in the sample water. Also nitrogen fixation has a minor role at 600 and 1500 m. However, at 2300 m the number of nitrogenase reads increased 10- to 100-fold indicating that the reduction of dissolved nitrogen gas has a key role in ammonium production at this depth.
Phosphate is obtained through organic and inorganic phosphate mineralization and phosphate uptake at all three sample depths despite dissimilar phosphate concentrations measured in the drill hole water (Figure 6). However, phosphate uptake occurs through different mechanisms at various depths. Sodium-dependent phosphate transporters are especially abundant at 1500 m, whereas at 2300 m phosphate uptake occurs via active transport that requires adenosine triphosphate.
Genes involved in the assimilation of organic sulfur and complete pathways for both assimilatory and dissimilatory sulfate reduction were present in all three metagenomes (Figure 6). However, there was no difference in the relative abundance of sequence reads between the three samples despite different amounts of sulfur. Because many sulfur-cycling bacteria are also fermentative, it is possible that these bacteria are not dependent on sulfur availability.
Iron reduction was not apparent in any of the metagenomes, in agreement with the low amount of iron in all samples.
Adaptation
In accordance with a significant positive correlation between water salinity and both bacterial and archaeal community structure in non-metric multidimensional scaling, the metagenomic-shotgun sequencing revealed functional adaptation of the drill hole microbial communities to high salinity. Genes involved in the uptake and biosynthesis of compatible solutes such as proline and glycine betaine were significantly more abundant at 2300 m, where high salinity correlates positively with both bacterial and archaeal OTU clustering (Figure 5). Significantly higher amounts of cation efflux and metal resistance systems were also detected at 2300 m. These systems may increase the resilience of the deep microbial communities to high osmotic stress.
Viral communities and CRISPRs
We also detected small amounts of various phage-related sequences and clustered regularly interspaced short palindromic repeat (CRISPR)-associated genes in the three metagenomes (Supplementary Table S8 and S9) (Makarova et al., 2011). Although the low abundance of viral sequences (<0.0002% of sequences) is most likely caused by our sampling method that was not designed to capture intact virus particles, the presence of prokaryotic viral defense systems suggests that host–phage interactions may have a role in the diversification of microbial life in deep granitic groundwater (Kyle et al., 2008). In soil, fluctuating selection dynamics resulting from such host–phage interactions have been proposed to sustain microbial community diversity (Gomez and Buckling, 2011).
Conclusions
Geological and hydrogeochemical conditions have been proposed to have a key role in the structuring and functional adaptation of microbial communities in deep continental crystalline rock (Pedersen, 1997). Our analyses show that, in the deep subsurface at Outokumpu, the lack of hydraulic connections, very low or negligible exchange of fluids among adjacent fracture systems, long residence times, unique geochemical characteristics and low amount of available energy have resulted in highly diverse bacterial, archaeal and viral communities with various metabolic strategies for hydrogen-driven carbon cycling, assimilation of reduced carbon compounds and nutrient cycling. Distinct signatures at the community level, predicted metabolic pathways and adaptations at different sample depths suggest that the phylogenetic and functional diversity of microbial communities confer them with a selective advantage and resilience over geological time scales. Co-evolution with viruses may have an important role in the long-term survival and stability of these communities possibly also contributing to genomic plasticity.
Acknowledgments
We thank Angelito Ramos, Aura Nousiainen, Marjo Öster and Sarah Scholtz for their assistance during the sampling campaign at Outokumpu, and Kirsi Lipponen, Eeva-Marja Turkki and Marjo Öster for sample preparation. We thank Olivia U. Mason and Eoin L. Brodie for their comments on an earlier version of this article, and Michael Hardman for revision of the language. This work was funded by the Finnish Funding Agency for Technology and Innovation (grant METAGENO), the Academy of Finland (grant 133348, DEEP LIFE), the European Regional Development Fund (grant YMLI) and the Finnish Research Program on Nuclear Waste management 2007–2010 (grants KABIO and GEOMOL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ahonen L, Kietäväinen R, Kortelainen N, Kukkonen IT, Pullinen A, Toppi T, et al. 2011Hydrogeological characteristics of the Outokumpu deep drill holeIn: Kukkonen IT (eds).Outokumpu Deep Drilling Project 2003-2010. Special Paper 51. Geological Survey of Finland: Espoo; 151–168. [Google Scholar]
- Bano N, Ruffin S, Ransom B, Hollibaugh JT. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl Environ Microbiol. 2004;70:781–789. doi: 10.1128/AEM.70.2.781-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Nelson B, Schrenk MO. Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front Microbiol. 2012;2:268. doi: 10.3389/fmicb.2011.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale BJ. Biodiversity improves water quality through niche partitioning. Nature. 2011;472:86–89. doi: 10.1038/nature09904. [DOI] [PubMed] [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, et al. Environmental genomics reveals a single-species ecosystem deep within Earth. Science. 2008;322:275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gihring TM, Moser DP, Lin LH, Davidson M, Onstott TC, Morgan L, et al. The distribution of microbial taxa in the subsurface water of the Kalahari shield, South Africa. Geomicrobiol J. 2006;23:415–430. [Google Scholar]
- Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- Grosskopf R, Janssen PH, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemann SA, Pedersen K, Ruotsalainen P. Distribution and metabolic diversity of microorganisms in deep igneous rock aquifers of Finland. Geomicrobiol J. 1999;16:277–294. [Google Scholar]
- Haveman SA, Pedersen K. Distribution of culturable anaerobic microorganisms in Fennoscandian shield groundwater. FEMS Microbiol Ecol. 2002;39:129–137. doi: 10.1111/j.1574-6941.2002.tb00914.x. [DOI] [PubMed] [Google Scholar]
- Hoffman BA. Distribution of culturable anaerobic microorganisms in Fennoscandian shield groundwater. FEMS Microbiol Eco. 1992;39:129–137. doi: 10.1111/j.1574-6941.2002.tb00914.x. [DOI] [PubMed] [Google Scholar]
- Hoffman BA.1992Isolated reduction phenomena in red-beds: a result of porewater radiolysisIn: Kharaka YK, Maest AS (eds)Proceedings of the 7th International Symposium on Water–Rock Interaction 503–506.
- Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itävaara M, Nyyssönen M, Kapanen A, Nousiainen A, Ahonen L, Kukkonen I. Characterization of bacterial diversity to a depth of 1500 m in the Outokumpu deep borehole, Fennoscandian Shield. FEMS Microbiol Ecol. 2011a;77:295–309. doi: 10.1111/j.1574-6941.2011.01111.x. [DOI] [PubMed] [Google Scholar]
- Itävaara M, Nyyssönen M, Bomberg M, Kapanen A, Nousiainen A, Ahonen L, et al. 2011bMicrobiological sampling and analysis of the Outokumpu deep drill biosphere in 2007-2009In: Kukkonen IT (eds)Outokumpu Deep Drilling Project 2003-2010, Geological Survey of Finland, Espoo, Special Paper 51199–206. [Google Scholar]
- Kietäväinen R, Ahonen L, Kukkonen IT, Hendriksson N, Nyyssönen M, Itävaara M. Characterisation and isotopic evolution of saline waters of the Outokumpu Deep Drill Hole, Finland—implications for water origin and deep terrestrial biosphere. Appl Geochem. 2013;32:37–51. [Google Scholar]
- Kim BK, Gadd GM. Bacterial Physiology and Metabolism. Cambridge University Press: Cambridge, UK; 2008. [Google Scholar]
- Kukkonen IT.(ed.). (2011Outokumpu Deep Drilling Project 2003-2010Special Paper 51.Geological Survey of Finland: Espoo; 252 [Google Scholar]
- Kukkonen IT, Rath V, Kivekäs L, Safanda J, Cermak V. Geothermal studies of the Outokumpu Deep Drill Hole: Vertical variation in heat flow and palaeoclimatic implications. Phys Earth Planet Inter. 2011;188:9–25. [Google Scholar]
- Kyle JE, Eydal HS, Ferris FG, Pedersen K. Viruses in granitic groundwater from 69 to 450 m depth of the Aspo hard rock laboratory, Sweden. ISME J. 2008;2:571–574. doi: 10.1038/ismej.2008.18. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn Stackebrandt E, Goodfellow M (eds)Nucleic Acid Techniques in Bacterial Systematics John Wiley and Sons: New York, USA; 115–175. [Google Scholar]
- Lin LH, Slater GF, Sherwood Lollar B, Lacrampe-Couloume G, Onstott TC. The yield and isotopic composition of radiolytic H2, a potential energy source for the deep subsurface biosphere. Geochim Cosmochim Acta. 2005;69:893–903. [Google Scholar]
- Lin LH, Wang PL, Rumble D, Lippmann-Pipke J, Boice E, Pratt LM, et al. Long-term sustainability of a high-energy, low-diversity crustal biome. Science. 2006;314:479–482. doi: 10.1126/science.1127376. [DOI] [PubMed] [Google Scholar]
- Mason OU, Nakagawa T, Rosner M, Van Nostrand JD, Zhou J, Maruyama A, et al. First investigation of the microbiology of the deepest layer of ocean crust. PLoS One. 2010;5:e15399. doi: 10.1371/journal.pone.0015399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major TA, Liu Y, Whitman WB. Characterization of energy-conserving hydrogenase B in Methanococcus maripaludis. J Bacteriol. 2010;192:4022–4030. doi: 10.1128/JB.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmi P, Kukkonen I. A new technique for sampling water and gas from deep drill holes. Can J Earth Sci. 1986;23:1450–1454. [Google Scholar]
- Nyyssönen M, Bomberg M, Kapanen A, Nousiainen A, Pitkänen P, Itävaara M. Methanogenic and sulphate-reducing microbial communities in deep groundwater of crystallinen rock fractures in Olkiluoto, Finland. Geomicrobiol J. 2012;29:863–878. [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara RB. Vegan: Community Ecology Package. Version 1.8-2 2006.
- Onstott TC, McGown DJ, Bakermans C, Ruskeeniemi T, Ahonen L, Telling J, et al. Microbial communities in subpermafrost saline fracture water at the Lupin Au mine, Nunavut, Canada. Microb Ecol. 2009;58:786–807. doi: 10.1007/s00248-009-9553-5. [DOI] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Pedersen K. Microbial life in deep granitic rock. FEMS Microbiol Rev. 1997;20:399–414. [Google Scholar]
- Pedersen K. Microbial life in deep granitic rock. FEMS Microbiol Rev. 2006;3–4:399–414. [Google Scholar]
- Peltonen P, Kontinen A, Huhma H, Kuronen U. Outokumpu revisited: new mineral deposit model for the mantle peridodite-associated Cu-Co-Zn-Ni-Ag-Au sulphide deposits. Ore Geol Rev. 2008;33:559–617. [Google Scholar]
- Pinard R, de Winter A, Sarkis GJ, Gerstein MB, Tartaro KR, Plant RN, et al. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics. 2006;7:216–236. doi: 10.1186/1471-2164-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LM, Lefticariu L, Ripley EM, Onstott TC. Radiolysis of water as a source of bioavailable chemical energy. Geochim Cosmochim Acta. 2006;70:A503. [Google Scholar]
- Sahl JW, Schmidt R, Swanner ED, Mandernack KW, Templeton AS, Kieft TL, et al. Subsurface microbial diversity in deep-granitic-fracture water in Colorado. Appl Environ Microbiol. 2008;74:143–152. doi: 10.1128/AEM.01133-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleep NH, Meibom A, Fridriksson T, Coleman RG, Bird DK. H2-rich fluids from serpentinization: geochemical and biotic implications. Proc Natl Acad Sci USA. 2004;101:12818–12823. doi: 10.1073/pnas.0405289101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, Amann R.1991Development and application of nucleic acid probesIn Stackebrandt E, Goodfellow M (eds).Nucleic Acid Techniques in Bacterial Systematics John Wiley and Sons: New York, USA; 205–248. [Google Scholar]
- Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol. 2001;67:5750–5760. doi: 10.1128/AEM.67.21.5750-5760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Västi K.2011Petrology of the drill hole R2500 at Outokumpu, eastern Finland–the deepest drill hole ever drilled in FinlandIn: Kukkonen IT (ed)Outokumpu Deep Drilling Project 2003-2010, Special paper 51. Geological Survey of Finland: Espoo, Paper 5117–46. [Google Scholar]
- Ward DV, Gevers D, Giannoukos G, Earl AM, Methé BA, Sodergren E, et al. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Dong H, Xu Z, Zhao D, Zhang C. Microbial diversity in ultra-high-pressure rocks and fluids from the Chinese continental scientific drilling project in China. Appl Environ Microbiol. 2005;71:3213–3227. doi: 10.1128/AEM.71.6.3213-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.