Abstract
The release of organic material upon algal cell lyses has a key role in structuring bacterial communities and affects the cycling of biolimiting elements in the marine environment. Here we show that already before cell lysis the leakage or excretion of organic matter by infected yet intact algal cells shaped North Sea bacterial community composition and enhanced bacterial substrate assimilation. Infected algal cultures of Phaeocystis globosa grown in coastal North Sea water contained gamma- and alphaproteobacterial phylotypes that were distinct from those in the non-infected control cultures 5 h after infection. The gammaproteobacterial population at this time mainly consisted of Alteromonas sp. cells that were attached to the infected but still intact host cells. Nano-scale secondary-ion mass spectrometry (nanoSIMS) showed ∼20% transfer of organic matter derived from the infected 13C- and 15N-labelled P. globosa cells to Alteromonas sp. cells. Subsequent, viral lysis of P. globosa resulted in the formation of aggregates that were densely colonised by bacteria. Aggregate dissolution was observed after 2 days, which we attribute to bacteriophage-induced lysis of the attached bacteria. Isotope mass spectrometry analysis showed that 40% of the particulate 13C-organic carbon from the infected P. globosa culture was remineralized to dissolved inorganic carbon after 7 days. These findings reveal a novel role of viruses in the leakage or excretion of algal biomass upon infection, which provides an additional ecological niche for specific bacterial populations and potentially redirects carbon availability.
Keywords: Alteromonas and Roseobacter, carbon remineralisation, nanoSIMS, Phaeocystis globosa, pyrosequencing, marine viruses
Introduction
Marine viruses are the most abundant entities and dynamic components of the microbial loop (Suttle, 2005). Studies conducted in the last years have made it increasingly evident that viruses are significant driving forces in algal (Haaber and Middelboe, 2009) and bacterioplankton populations dynamics (Middelboe et al., 2003). Moreover, through the ‘viral shunt' (Wilhelm and Suttle, 1999), the release of dissolved organic carbon and nutrients from the particulate organic pool is enhanced, leading to increased substrate availability for microbially mediated processes. Thereby, viruses can influence biogeochemical cycling in the world's oceans (Brussaard et al., 2005b; Haaber and Middelboe, 2009).
The prymnesiophyte, Phaeocystis globosa are a dominant algae with the ability to generate high-biomass spring blooms (Brussaard et al., 1996). Viruses infecting P. globosa (PgVs) have recently been brought into culture, and virus–host interactions (for example, latent period) have been well investigated (Baudoux and Brussaard, 2005). Viral-mediated lysis can account for up to 66% of the total mortality of P. globosa single cells (Baudoux et al., 2006) and can even control algal bloom formation (Brussaard et al., 2005a). In a mesocosm study, it was shown that viral-mediated lysis of P. globosa blooms may lead to rapid changes in the microbial community structure and enhanced bacterial carbon utilisation (Brussaard et al., 2005b). Hence, P. globosa is an ideal species to study the impact of viruses structuring bacterial communities and in turn the transfer of algal biomass towards microbial communities affecting coastal biogeochemical element cycling.
Past field observations have shown that the bacterioplankton communities during algal blooms in the coastal North Sea waters are mainly dominated by Alphaproteobacteria, Gammaproteobacteria and Bacteroidetes (Brussaard et al., 2005b; Teeling et al., 2012). The gammaproteobacterial Alteromonadaceae (referred to as Alteromonas cells henceforth) and alphaproteobacterial Rhodobacteriaceae (referred to as Roseobacter cells henceforth) can become very abundant during algal blooms (Pernthaler et al., 2001) and exhibit distinctive temporal patterns most likely in relation to the changes in the organic matter composition during the course of the algal blooms (Eilers et al., 2000).
Viral-mediated algal lysis may induce aggregate formation due to the release of lysis products and can be associated with dense bacterial abundances (Brussaard et al., 2005b). Although the majority of marine pelagic bacteria exists as free-living cells, a substantial portion lives attached to algal surfaces and aggregates (Azam et al., 1983). Aggregate-associated bacteria are often characterized by high cellular abundance, growth rate and enzymatic activity relative to their free-living counterparts (Riemann and Grossart, 2008). It is estimated that about 37% of the aggregate-associated bacteria may be killed by viral lysis because of the density of potential host cells within aggregates (Proctor and Fuhrman, 1991). Moreover, bacterial cell lysis could mediate aggregate dissolution. Consequently, viruses might alter the efficiency of the biological carbon pump by retaining dissolved organic matter (for example, carbon) within the euphotic zone (Brussaard et al., 2008). However, how viral lysis shapes the bacterial composition and diversity and how it influences the bacterial uptake of virally released organic compounds and thereby mediating oceanic biogeochemical cycles, remain poorly understood.
The objective of the current study is to investigate the effects of algal viral infection and subsequent lysis on bacterial community structure, and carbon and nitrogen transfer from the algae to the bacteiroplankton. The utilisation of 13C- and 15N- labelled axenic biomass of the model algae P. globosa biomass by bacterial communities from the coastal North Sea (0.8 μm pre-filtered) during and after algal viral infection was followed by a combination of bulk stable isotopic and molecular analyses as well as novel single-cell analyses. Catalysed reporter deposition – fluorescent in situ hybridisation (CARD-FISH), together with amplicon pyrosequencing, was used to examine the changes in the bacterial composition and diversity. The application of high-resolution single-cell techniques enabled us to visualise the occurrence of aggregates and aggregate-associated and/or free-living cells bacteria (atomic force microscopy imaging, AFM). Single-cell bacterial substrate assimilation was further quantified using nanometer-scale secondary-ion mass spectrometry (nanoSIMS), whereas the organic carbon remineralisation associated with bacteriophage lysis was quantified using isotope ratio mass spectrometry (IRMS).
Materials and methods
Generation of 13C- and 15N-labelled algal biomass
Axenic cultures of P. globosa strain Pg G (A) were obtained from the culture collection of Royal Netherlands Institute for Sea Research (NIOZ). The 13C- and 15N-labelled P. globosa biomass were generated from exponentially growing axenic P. globosa culture grown for 2 days in Erlenmeyer flasks in modified enriched artificial seawater (ESAW; Cottrell and Suttle, 1991) containing 1 mM H13CO3−, 80 μM of 15NO3− (as sodium salts, 99 atom %, ISOTEC) and 5 μM of PO43− (as sodium salts, Sigma-Aldrich Chemie Gmbh, Munich, Germany). The cultures were grown under 95 μmol quanta m−2 s−1 irradiance with a light to dark regime of 16:8 h and at a temperature of 15±1 °C. On day 3, cultures were centrifuged at 1500g (with a swing rotor) for 10 min to remove unincorporated 13C- and 15N-labelled substrates from the media. Algal cell pellets formed after centrifugation were washed twice and re-suspended in an ESAW media without nutrient loadings. The centrifuged and re-suspended algal cultures had similar sensitivity to viral infection when compared with non-centrifuged control cultures, suggesting that physiology of P. globosa was unaltered by centrifugation (data not shown).
P. globosa virus culturing and bacterial inoculum
The lytic P. globosa virus, PgV-07T (Baudoux and Brussaard, 2005) used in this study was produced from exponentially growing P. globosa cultures in ESAW media. The bacterial populations used in this experiment were obtained from Southern North Sea, The Netherlands (December 2008). Before performing experiment, North Sea water was filtered through 0.8-μm pore size filters (GTTP, 45 mm in diameter, Millipore, Eschborn, Germany) to exclude heterotrophic nanoflagellates and other zooplankton (no grazers were found during epifluorescence microscopy). About 25% of the total bacterial cell numbers were excluded after 0.8 μm pre-filtration (data not shown).
Experimental set-up
The centrifuged and re-suspended 13C- and 15N-labelled P. globosa cultures in ESAW media without nutrient loadings were split into four subcultures. Each of these subcultures were transferred (10% v-v) to Erlenmeyer flasks containing 3 l 1:1 mixture of f/2 (Guillard, 1975) and ESAW media. Bacterial inoculation, that is, 0.8-μm-pre-filtered North Sea water (10% v/v) was then added to each of these subcultures. Two of these subcultures were then infected with pre-filtered PgV-07T virus (0.2 μm pore-size, Whatman) at an initial virus to algae ratio of 17:1. The other two cultures served as non-infected control cultures and received medium instead of viral lysate in equal amount. Sampling for flow cytometric algal and viral abundance, bulk particulate 13C- and 15N- measurements, catalysed reporter deposition-fluorescence in situ hybridization analyses (CARD-FISH) and nanoSIMS analyses were taken soon after addition of viral lysate. Samples were taken frequently until day 1 of the experiment (0, 1, 2, 3, 5, 8, 12, 19, 24 and 30 h post addition of viral lysate), followed by daily sampling until day 7 of the experiment.
Control experiments
Independent control experiments were performed in order to investigate the influence of organics derived from algal media and P. globosa viral lysates on the changes in the North Sea bacterial abundance (Supplementary Figure 1). Experiments were conducted in four Erlenmeyer flasks (2 l) containing 1:1 mixture of f/2 and ESAW media with North Sea bacterial inoculation (10% v/v, 0.8 μm pre-filtered). Two of these subcultures received pre-filtered PgV-07T virus (0.2 μm pore-size, Whatman), whereas other two cultures received medium instead of viral lysate in equal amount. Samples for flow cytometric bacterial abundance were taken frequently until day 1 of the experiment (0, 3, 5, 8, 12, 19 and 24 h), followed by daily sampling until day 5 of the experiment.
Abundances
Algal abundance was monitored using flow cytometry with a Beckman Coulter EPICS XL-MCL benchtop flow cytometer, equipped with a 15 mW 488 nm argon laser. One millilitre samples taken at each time point were diluted up to 10-fold in autoclaved seawater (0.2 μm filtered). The flow cytometer trigger was set on the red chlorophyll autofluorescence (emission >630 nm).
The abundance of P. globosa viruses and bacteriophages were enumerated by using a 15 mW 488 nm argon laser Becton-Dickson FACSCalibur flow cytometer according to Brussaard (2004). In short, samples of 1 ml were fixed with 25% glutaraldehyde (0.5% final concentration, EM grade, Sigma- Aldrich, USA) for 15–30 min at 4 °C, flash frozen in liquid nitrogen and stored at −80 °C until analysis. The thawed samples were diluted 50- to 1000-fold in sterile TE-buffer (pH 8.0) and stained with the nucleic acid-specific dye SYBR Green I (Invitrogen-Molecular Probes, USA) at a final concentration of 0.5 × 10−4 of the commercial stock for 10 min at 80 °C. The flow cytometer trigger was set on the green fluorescence, and data files were analysed as described by Brussaard et al. (2010).
CARD-FISH analyses
CARD-FISH analyses were performed to identify and quantify the bacterial populations following the protocol of Pernthaler et al. (2004). Subsamples taken at each time interval were fixed with paraformaldehyde (PFA, 1% final concentration) for 1 h at room temperature or overnight at 4 °C. Subsamples were filtered onto white polycarbonate membrane filters (GTTP, 0.2 μm pore size, Millipore), washed with 5–10 ml of 1 × phosphate buffer saline (PBS), air-dried and stored at −20 °C until analysis. Samples were hybridised with the following probes: Gamma42a for Gammaproteobacteria together with Beta42a as a competitor (Manz et al., 1992), CF319a for Bacteroidetes (Manz et al., 1996), ALF986 for Alphaproteobacteria (Amann et al., 1997), Alt1413 for Alteromonas cells (Eilers et al., 2000), Ros593 for Roseobacter cells (Eilers et al., 2001). Hybridised filters were counterstained with 1 μg ml−1 of 4,6-diamidino-2-phenylindole (DAPI). Subsequently, all DAPI-stained and hybridised cells were quantified using epifluorescence microscopy (Axioplan II Imaging, Zeiss, Jena, Germany). The EUB-I-III probe set was also used as CARD-FISH controls for sample filters and yielded ⩾82% DAPI-stained cells in all samples, but only the counts for the more specific groups are presented here. More than 900 DAPI-stained cells and 450 probe-specific hybridised bacterial cells were evaluated in about 25 randomly chosen microscopic fields.
Bulk carbon and nitrogen measurements
For the determination of bulk particulate 13C- and 15N- measurements, 30–80 ml of the experimental cultures were filtered onto pre-combusted glass fibre filters, (GF/F, Whatman) freeze dried and stored at room temperature until analysis. The C- and N-isotopic composition of particulate organic matter was determined as CO2 and N2 released by flash combustion in an automated elemental analyzer (Thermo Flash EA, 1112 Series; Thermo Fisher Scientific, Schwerte, Germany) coupled to an isotope ratio mass spectrometer (Finnigan Deltaplus XP, Thermo Scientific, Thermo Fisher Scientific).
Carbon remineralisation
Carbon remineralisation was measured as dissolved inorganic 13C-carbon (13C-DIC) from labelled biomass released within the plankton community in our incubation experiments. Subsamples (5 ml) were poisoned with 30 μl of saturated mercury chloride solution. The isotopic component of DIC was determined after acidifying with 1% final concentration of hypo-phosphoric acid as described by Assayag et al. (2006) and was analysed on a gas chromatography-isotope ratio monitoring mass spectrometry (Optima Micromass, Manchester, UK). Any loss of 13C-DIC in the medium because of gaseous exchange with atmospheric CO2 was corrected for as described in Sheik et al. (2013).
Ammonium analysis
Ammonium was measured using a TrAAcs 800 autoanalyzer (detection limit of 0.1 μM) on samples (∼ 5 ml) that were gently filtered through polysulfone filters (0.2-μm pore size, Acrodisc, Gelman Sciences, Zwijndrecht, Netherlands) and stored at −20 °C until analysis.
Atomic force microscopy
Atomic force microscopy (AFM, NT-MDT Co., Moscow, Russia) was performed to visualise surface topography of aggregates (Supplementary Figure 2) on formaldehyde-fixed samples that were filtered on to polycarbonate membrane filters (0.22 μm pore-size; Millipore). AFM analysis was performed in a semi-contact mode as described by Sheik et al. (2013) in air at scan rates between 0.5 and 1 Hz and a spring constant of 11.8 Nm−1. Images were processed with flatten correction function of the software (Nova P9, version 2.1.0.828; NT-MDT Co., Moscow, Russia).
NanoSIMS analyses
Halogen in-situ hybridization assay coupled to nanoSIMS (HISH-SIMS) (Musat et al., 2008) was performed to identify and quantify the substrate assimilation of individual Alteromonas (probe Alt1413) and Roseobacter (probe Ros593) cells. Enrichment of the 13C and 15N in the specific probe-hybridised bacterial cells (Alt1413 and Ros593, as indicated by their respective, high 19F content derived from the fluorine containing Oregon Green-labelled tyramide used), was analysed with NanoSIMS 50 L (CAMECA, Paris, France). The primary ion beam had a nominal size between 50 and 100 nm, and the sample was sputtered with a dwelling time of 2 ms per pixel. The primary current of the Cs+ beam was 20–30 nA during acquisition for most images. For each analysis, we recorded simultaneously secondary-ion images of naturally abundant 12C (measured as 12C−), 14N (measured as 12C14N−) and, similarly, 19F (halogen-labelled HISH-SIMS tyramide) for the identification of specific probe-hybridised bacterial cells, and 13C and 15N for the uptake quantification. NanoSIMS data sets were analysed using the Look@NanoSIMS software (Polerecky et al., 2012). Regions of interest (ROI) around individual bacterial cells were defined manually on on the basis of 19F image. The isotope ratios (r=13C/12C or 15N/14N) were calculated for each ROI on the basis of the total 13C– and 12C– counts for each pixel. Subsequently, the atomic percentage of 13C and 15N were calculated as 13C/(13C+12C) and 15N/(15N+14N), respectively. The 13C and 15N isotopic ratios served as a proxy to quantify the algal biomass that has been transferred towards specific bacterial members.
Calculations of biovolume and single-cell assimilation of 13C and 15N
Biovolume of Alteromonas and Roseobacter cells were calculated in order to estimate the amount of 13C and 15N algal biomass transfer due to viral lysis or non-infected algae. Epifluorescence microscopy images taken during CARD-FISH analyses and before nanoSIMS analyses were used to determine the dimensions of Alteromonas and Roseobacter cells. Assuming these bacterial cells as rotational ellipsoids (Olenina et al., 2006), we deduced the biovolume of Alteromonas and Roseobacter cells at the 5 h (Alteromonas cells only), day 2 and day 7 of the experiment (Supplementary Table 1).
We quantified the 13C and 15N substrate assimilation (fmol per cell−1) within single cells of Alteromonas and Roseobacter due to P. globosa viral lysis, relative to non-infected control cultures. This estimation was on the basis of the 13C and 15N enrichments of Alteromonas and Roseobacter cells and calculated biovolume as described by Musat et al. (2008).
DNA extraction and amplicon pyrosequencing
In order to investigate the community response to viral infection, the same seawater was used in the infected and non-infected cultures, and DNA samples were taken at 6 h, day 2 and day 7 in parallel from both cultures. The DNA samples from the non-infected culture were used as a reference to monitor the changes in microbial community due to viral infection in the infected culture. DNA extraction was performed as described by Zhou et al. (1996) on samples that were filtered (100–200 ml) onto white polycarbonate membrane filters (GTTP, 0.2-μm pore size, Millipore) and stored at −20 °C until analysis. The extracted DNA was further purified using the Wizard DNA Clean-Up System (Promega Corporation, Madison, USA) as per the manufacturer's instructions. The bacterial 16S rRNA genes were amplified and sequenced using amplicon pyrosequencing at the Research and Testing Laboratories (Lubbock, TX, USA). The pyrosequencing was performed at 6 h, day 2 and day 7 of the experiment from the infected and non-infected control cultures targeting Gammaproteobacteria (forward primer 5′-CMATGCCGCGTGTGTGAA-3′, reverse primer 5′-ACTCCCCAGGCGGTCDACTTA-3′), Alphaproteobacteria (forward primer 5′-ARCGAACGCTGGCGGCA-3′, reverse primer 5′-TACGAATTTYACCTCTACA-3′) and Bacteroidetes (forward primer 5′-AACGCTAGCTACAGGCTT-3′, reverse primer 5′-CAATCGGAGTTCTTCGTG-3′). The generated sequences were processed and taxonomically identified as per the company's standard procedure (Sun et al., 2011) to the species level according to the >97% sequence identity of 16S rRNA genes. Thereafter, the species percentage composition of each major bacterial group was based on the relative abundance information within and among the individual samples and relative numbers of reads (Supplementary Tables 2 and 3).
MEGAN 4, a metagenome analysing software, was used to construct the heat plot of 16S rRNA amplicon sequence data set (Supplementary Figure 3, (Huson et al., 2011)). Amplicon sequences were clustered with >97% sequence identity with UCLUST (Edgar, 2010), and BLASTN was used to compare clustered sequences against the SILVA rRNA database (http://www.arb-silva.de). The output of this comparison was then parsed by MEGAN4 (Huson et al., 2011) and mapped onto the NCBI taxonomy. The BLASTN comparison showed that Bacteroidetes 16S rRNA amplicon sequences targeted mostly uncultured Bacteroidetes species and hence is not described further in the text. Comparison tool of MEGAN4 software was used to generate a phylogenetic tree from multiple data sets.
Statistical analyses
Using the Sigmastat version 3.5 software package (Systat Software GmbH, Erkrath, Germany), one-way analyses of variance (ANOVAs) were used to test for differences in the bacterial numbers (∼25 microscopic fields of each replicate, n) and single-cell 13C and 15N enrichments of Alteromonas and Roseobacter cells in infected and non-infected P. globosa cultures at different time intervals. Further, the Pearson product moment correlation was used to determine the correlation between algal cell numbers, particulate 13C-carbon and 13C-DIC.
Results
Microbial abundances
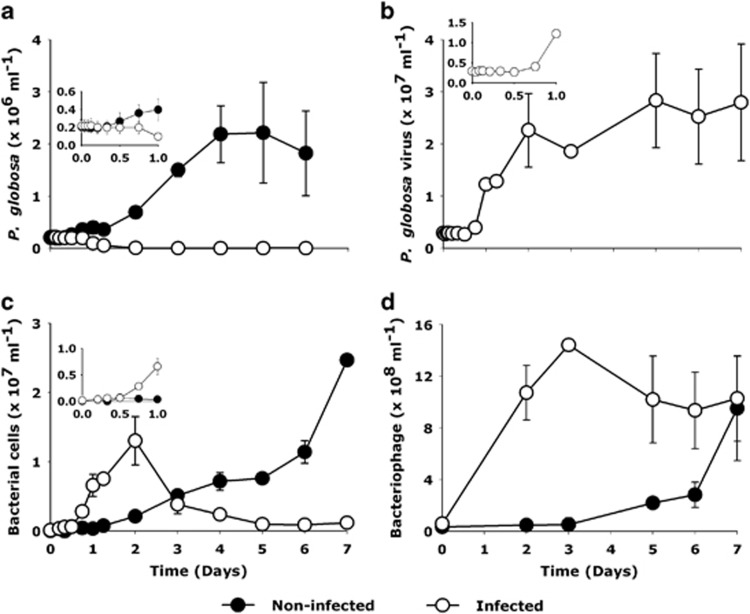
Although the P. globosa cell abundance in non-infected control cultures increased by 10-fold in 4 days (from 0.2 to 2.3 × 106 cells ml−1; Figure 1a), viral infection led to a decline of algal host abundance from 18 h post infection (p.i.) onwards (from 0.2 × 106 to 2.6 × 103 cells ml−1 by day 6). Correspondingly, the abundance of PgV in infected cultures increased between 8 and 12 h p.i. (Figure 1b). The bacterial abundance in non-infected P. globosa cultures increased steadily throughout the 7-day experiment (2.2 × 107 cells ml−1 at day 7; Figure 1c). In infected cultures, bacterial abundance increased rapidly from the point of P. globosa cell lysis (12 h) reaching a maximum by day 2 (2.1 × 107 cells ml−1), after which it dropped sharply (3.7 × 106 cells ml−1, by day 7, Figure 1c). The sharp decline in the bacterial numbers coincided with an increase in the number of bacteriophages (Figure 1d).
Figure 1.
Changes in microbial and viral abundances in response to viral infection of P. globosa. (a) algal abundance, (b) P. globosa virus (PgV-07T) abundance (c) total heterotrophic bacterial abundance and (d) bacteriophage. Closed symbols represent non-infected control and open symbols represent the virally infected cultures. Error bars indicate standard error of mean from duplicate batch cultures (s.e.m.).
Changes in bacterial community composition
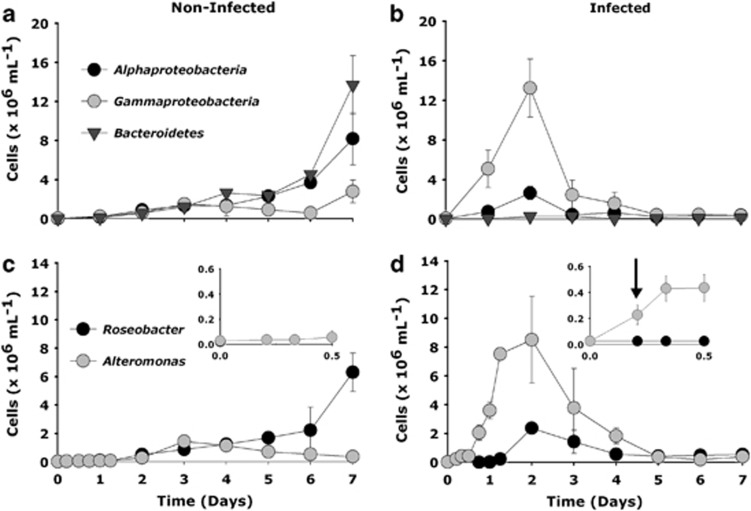
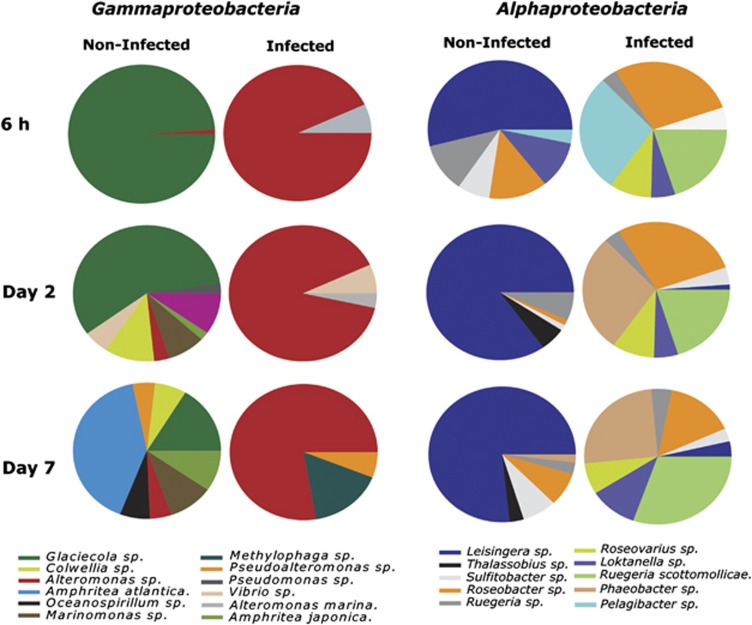
In the non-infected control cultures, catalysed reporter deposition– fluorescence in situ hybridization (CARD-FISH) analysis revealed that Bacteroidetes and Alphaproteobacteria dominated by day 7, accounting for 56 and 34% of the total microbial populations, respectively (Figure 2a). Viral-mediated P. globosa lysis affected most strikingly the gammaproteobacterial populations (Figure 2). Growth of Alteromonas cells was stimulated shortly after P. globosa viral infection, already at 5 and 8 h post infection, yet before cell lysis of P. globosa (ANOVA, number of random microscopic fields analysed (n)=25, P=<0.001). Independent control experiments to test for the effect of the viral lysate added and potential organic metabolites excreted by the algal host showed no significant increase in bacterial abundance until day 2 where a slight increase was observed (Supplementary Figure 1). CARD-FISH and amplicon pyrosequencing analyses showed that Alteromonas dominated the Gammaproteobacteria in the infected cultures with a maximum abundance of 8.5 × 106 cells ml−1 on day 2 (Figures 2c and 3). The Alphaproteobacteria also peaked at day 2 in the infected cultures (2.5 × 106 cells ml−1). The majority of the Alphaproteobacteria phylotypes in the infected cultures, as identified per amplicon pyrosequencing analysis, belonged to diverse community of Roseobacter-related organisms (Figure 3, Supplementary Table 2). In contrast, alphaproteobacterial phylotypes in the non-infected control cultures showed Leisingera sp. to be dominant (Figure 3, Supplementary Table 2). Like the Alphaproteobacteria, the Bacteroidetes population increased towards the end of the experiment in the non-infected algal cultures. In contrast, Bacteroidetes in the infected cultures were hardly affected, maintaining more or less stable but low cell abundance throughout the experiment (Figure 2b). Epifluorescence microscopy revealed that Alteromonas cells from the infected P. globosa cultures showed (micro)aggregate association soon after P. globosa cell lysis (Table 2). The presence of (micro)aggregates was confirmed by atomic force microscopy (AFM) imaging (Supplementary Figure 2). In contrast to Roseobacter, Alteromonas cells showed an increasing degree of aggregation until day 2 of the experiment (Table 1). Roseobacter cells were mainly found in aggregates towards the end of the experiment. Thereafter, gammaproteobacterial populations and similarly Alteromonas cells in infected P. globosa cultures dropped until day 5. Interestingly, amplicon pyrosequencing analysis indicated that only a single phylotype, Alteromonas sp., dominated the gammaproteobacterial populations after P. globosa viral lysis and persisted throughout the experiment (Figure 3, Supplementary Table 3). However, in non-infected control cultures, phylotype Galciecola sp. dominated at 6 h and day 2. Thereafter, at day 7 gammaproteobacterial phylotypes became more diverse (Figure 3, Supplementary Table 3).
Figure 2.
The abundance of major bacterial groups of Alphaproteobacteria, Gammaproteobacteria and the Bacteroidetes (a, b) as determined by CARD-FISH analyses due to viral lysis of P. globosa relative to the non-infected control cultures. Within specific taxonomic groups, P. globosa viral lysis led to rapid changes in the abundance of Alteromonas (Gammaproteobacteria) followed by Roseobacter (Alphaproteobacteria) cells (c, d). Please note the different y-axis scale in the upper and bottom panels. The inlay in the bottom panel (c, d) shows the changes in the abundance of Alteromonas and Roseobacter cells for the first 12 h of the experiment. An initial doubling in the abundance of Alteromonas cells at 5 h post infection is indicated by an arrow (d). Error bars indicate s.e.m.
Figure 3.
Temporal changes in the bacterial community composition of phylotypes belonging to Gamma- and Alphaproteobacteria during and after viral infection of P. globosa relative to non-infected control cultures.
Table 1. Percentage of Alteromonas and Roseobacter cells as free-living and aggregate associated.
| Alteromonas cells |
||||||||
|---|---|---|---|---|---|---|---|---|
|
Non-infected cultures |
Infected cultures |
|||||||
| Time | n | Free-living (mean±s.e.) % | n | Aggregate-associated (mean±s.e.) % | n | Free-living (mean±s.e.) % | n | Aggregate-associated (mean±s.e.) % |
| 0 h | 15 | 100±0 | 0 | 0 | 24 | 100±0 | 0 | 0 |
| 5 h | 21 | 100±0 | 0 | 0 | 35 | 78±2.5 | 26 | 22±3.9 |
| 18 h | 19 | 99±0 | 0 | 1±0 | 324 | 49±3.6 | 335 | 51±2.6 |
| Day 2 | 67 | 81.3±2.7 | 15 | 18.6±2.4 | 168 | 12.2±2.1 | 1196 | 87.7±5.2 |
| Day 4 | 91 | 74.7±2.8 | 31 | 25.8±3.1 | 200 | 40.9±5.2 | 290 | 59.1±6.3 |
| Day 7 | 45 | 83.3±3.5 | 29 | 16.6±3.9 | 69 | 70.8±4.5 | 45 | 29.1±3.5 |
| Roseobacter cells |
||||||||
|---|---|---|---|---|---|---|---|---|
|
Non-infected cultures |
Infected cultures |
|||||||
| Time | n | Free living (mean±s.e.) % | n | Aggregate associated (mean±s.e.) % | n | Free living (mean±s.e.) % | n | Aggregate associated (mean±s.e.) % |
| 0 h | 15 | 100±0 | 0 | 0 | 15 | 100±0 | 0 | 0 |
| 5 h | 17 | 100±0 | 0 | 0 | 35 | 100±0 | 0 | 0 |
| 18 h | 19 | 95.6±1.7 | 6 | 4.4±2.6 | 17 | 98.2±3.2 | 6 | 1.7±2.8 |
| Day 2 | 166 | 83.1±2.5 | 33 | 16.9±5.7 | 172 | 67.8±1.8 | 82 | 32.2±1.2 |
| Day 4 | 214 | 67.3±6.3 | 104 | 32.7±4.6 | 59 | 41.8±4.4 | 90 | 58.1±3.6 |
| Day 7 | 603 | 55.1±3.1 | 459 | 44.9±4.0 | 50 | 27.8±4.6 | 131 | 72.1±4.8 |
n=total number of cells per microscopic field.
Carbon remineralization
There was no significant change in the particulate organic 13C-carbon (13C -POC) and particulate organic 15N-nitrogen (15N -PON) content with time in the non-infected control cultures (Figures 4a and b). However, viral lysis of P. globosa led to a marked decline in the amount of 13C -POC, which correlated strongly with the declining cell abundances (the Pearson product moment correlation, R=−0.947, P⩽0.001, Figure 4b). On the basis of net decline of the 13C-POC by day 7 of the experiment, around 44% of the P. globosa biomass from the infected cultures was converted to dissolved organic forms (Figure 4a, Supplementary Table 4). Meanwhile, there was no significant decline in 15N-PON (ANOVA, P=0.004, Figure 4a). The decrease in the 13C-POC in infected P. globosa cultures strongly correlated with the amount of organic carbon remineralized to 13C-DIC (R=−0.991, P⩽0.001, Figure 4c). By day 1 of the experiment, the amount of carbon remineralised in the infected cultures was already substantially higher compared with the control cultures (ANOVA, P=<0.001). Comparing the net decrease in 13C-POC and the net increase in the 13C-DIC, 22.5 μM 13C, equivalent to 40% of the shunted particulate 13C-organic carbon was remineralized by day 7 (Figure 4c, Supplementary Table 4). In contrast, in the non-infected control cultures, 13C-DIC concentrations did not significantly change until day 6 when a small increase in 13C-DIC was observed. Throughout the experiment, there was no detectable ammonium in both the non-infected and infected P. globosa cultures (data not shown).
Figure 4.
Changes in particulate organic (a) 15N-nitrogen (15N-PON), (b) 13C-carbon (13C-POC) and (c) carbon remineralization due to viral-mediated shunt. Closed symbols represent non-infected control and open symbols represent the viral-infected cultures. Error bars indicate s.e.m.
Effect of viral lysis on algal C and N transfer to specific bacterial groups
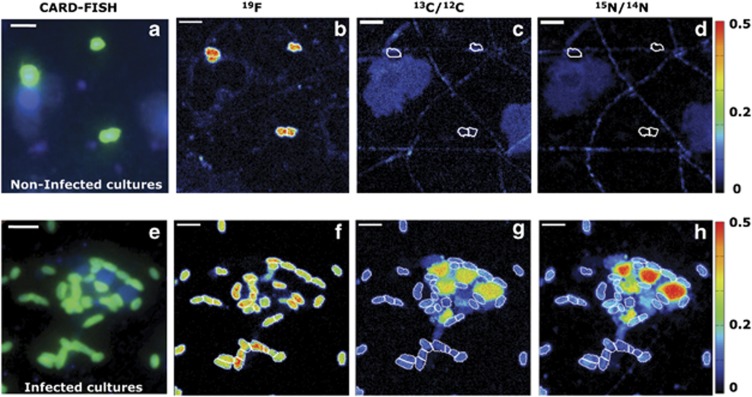
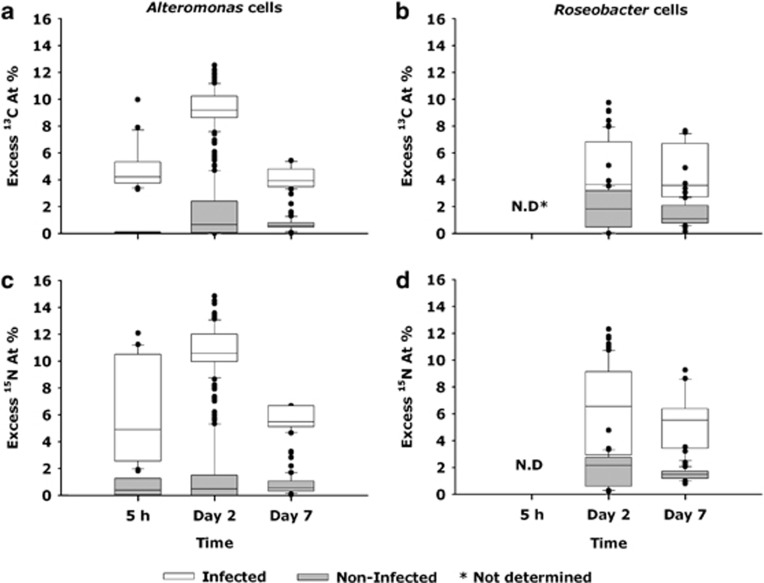
By combining CARD-FISH and HISH-SIMS imaging, we measured the 13C and 15N enrichments of the Alteromonas and Roseobacter cells from the non-infected control and infected P. globosa cultures (Figures 5 and 6). In the non-infected P. globosa control cultures, the 13C and 15N enrichment and hence substrate assimilation of Alteromonas cells remained low within the first 5 h (Figures 5a–d, 6a and c and Table 2, Supplementary Table 4). In contrast, already by 5 h, Alteromonas cells from the infected P. globosa cultures were characterized by significant enrichment in 13C and 15N (Figures 5e–h, and 6a and c), with calculated substrate assimilation rates of 2.14 fmol 13C cell−1 and 0.65 fmol 15N cell−1 (P=<0.001, Table 2). A maximum substrate assimilation of Alteromonas and Roseobacter cells from the non-infected control cultures was observed at day 2 of the experiment. At day 2 of the experiment, in the infected cultures, ∼18% of the bulk 13C-POC was assimilated by the Alteromonas biomass and was about 2.4-fold enhanced relative to Alteromonas biomass in the non-infected cultures (Supplementary Table 4). The 13C and 15N substrate assimilation of Alteromonas from the infected cultures P. globosa cultures decreased from day 2 to day 7, whereas substrate assimilation of Roseobacter cells increased from day 2 to day 7 (Table 2).
Figure 5.
NanoSIMS imaging visualising the transfer of algal biomass towards Alteromonas cells in non-infected control cultures (upper panel) and infected P. globosa cultures (lower panel) at day 2 of the experiment. First column (a, e) illustrates the CARD-FISH image taken before nanoSIMS analyses. The corresponding CARD-FISH hybridised cells were located by the 19F signal during nanoSIMS (b, f) and their respective 13C (c, g) and 15N enrichments (d, h). Scale bar=2 μm.
Figure 6.
The 13C and 15N enrichments as deduced from nanoSIMS analyses indicate the transfer of isotopically labelled biomass to Alteromonas (a, c) and Roseobacter (b, d) cells in both non-infected control and infected P. globosa cultures. The 13C and 15N enrichments of Roseobacter cells at 5 h were not determined due to their low cell abundance.
Table 2. Single cell 13C and 15N substrate assimilation of Alteromonas and Roseobacter cells (f mol per cell) at various temporal stages of P. globosa viral lysis relative to non-infected P. globosa cells.
| Alteromonas cells |
Roseobacter cells |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Non-Infected cultures |
Infected cultures |
Non-Infected cultures |
Infected cultures |
|||||||||
| Time | n | f mol 13C cell −1 (mean±s.e.) | f mol 15 N cell −1 (mean±s.e.) | n | f mol 13C cell −1 (mean±s.e.) | f mol 15 N cell −1 (mean±s.e.) | n | f mol 13C cell −1 (mean±s.e.) | f mol 15 N cell −1 (mean±s.e.) | n | f mol 13C cell −1 (mean±s.e.) | f mol 15 N cell −1 (mean±s.e.) |
| 0 h | 8 | 0 | 0 | 12 | 0 | 0 | ND | — | — | ND | — | — |
| 5 h | 12 | 0.03±0.01 | 0.03±0.02 | 21 | 2.14±0.16 | 0.05±0.04 | ND | — | — | ND | — | — |
| Day 2 | 87 | 0.56±0.08 | 0.07±0.01 | 82 | 1.32±0.02 | 0.22±0.005 | 41 | 0.32±0.04 | 0.05±0.01 | 63 | 0.76±0.06 | 0.16±0.01 |
| Day 7 | 46 | 0.13±0.01 | 0.02±0.002 | 36 | 0.53±0.23 | 0.10±0.004 | 21 | 0.16±0.01 | 0.02±0.002 | 46 | 1.38±0.15 | 0.24±0.02 |
Abbreviation: ND, not determined.
n=number of single cells analysed by nanoSIMS.
Discussion
Role of algal viral lysis in structuring bacterial community composition
The distinct temporal patterns in the abundance of major bacterial groups show that P. globosa viral lysis led to significant and very rapid changes in the bacterial community composition. Further, viral-mediated P. globosa lysis promoted the growth of opportunistic Gamma- and Alphaproteobacteria (r-strategists) relative to slow growing bacteria (for example, Bacteroidetes, K-strategists) (Fuchs et al., 2000). The bacterial community composition also changed in association with aging P. globosa cells from non-infected control cultures. However, these communities were distinct from those in the infected cultures. The predominance of Alphaproteobacteria and Bacteroidetes observed in the non-infected control cultures agrees well with observations from field studies performed during P. globosa blooms (Brussaard et al., 2005b; Alderkamp et al., 2006; Lamy et al., 2009).
Our results show that viral infections of P. globosa led to the rapid growth of Alteromonas cells, which dominated the bacterial community soon after P. globosa viral lysis. In contrast to the development of Alteromonas abundance, the relative contribution of Roseobacter cells rose slowly. The initial rapid growth of Alteromonas cells is consistent with previous phytoplankton incubation studies (Allers et al., 2007, Sandaa et al., 2009). Although the rapid increase in Alteromonas abundance coincided with higher bacteriophage abundance, potential bacteriophage lysis was apparently not able to prevent the initial Alteromonas bloom.
Interestingly, Alteromonas populations in the infected culture mainly consisted of a single phylotype, Alteromonas sp., during the initial doubling of Alteromonas abundance (6 h post infection), and this phylotype remained a major component throughout the remainder of the experiment. Gammaproteobacterial phylotypes in the non-infected control cultures were more diverse. The development of Methylophaga sp., a known dimethylsulfide (DMS) degrader (Schäfer, 2007), by day 7 in the infected cultures could be related to the potential occurrence of P. globosa derived DMS compounds (Liss et al., 1994). Although we did not analyse for DMS, its potential formation could explain the presence of this methylotrophic organism in our cultures. Moreover, viral lysis of P. globosa triggered the development of several alphaproteobacterial phylotypes, most of which belonged to the Roseobacter group. In contrast to Gammaproteobacteria, the alphaproteobacterial phylotypes in the non-infected control cultures consisted mainly of a single phylotype, Leisingera sp. (affiliated to Roseobacter cells), which persisted and remained to be a major phylotype throughout the experiment. Overall, our results indicate that algal viral lysis structures bacterial community composition by favouring distinct bacterial phylotypes.
In the coastal North Sea, the P. globosa blooms are often associated with aggregates that are formed due to the disintegration of P. globosa colonies and/or cell lysis (Brussaard et al., 2005b; Mari et al., 2005). Despite the presence of P. globosa single cells in this study, both the atomic force and epifluorescence microscopy revealed that the initial increasing abundances of Alteromonas and to a lesser extent Roseobacter cells were aggregate associated. AFM imaging visualised that aggregate-associated bacteria appeared to be retained within a flocculant-like material. Aggregate formation might be enhanced due to the impediment in the release of P. globosa star-like structures upon viral infection, which has been suggested to stimulate (micro)aggregate formation (Sheik et al., 2013).
Although aggregation may enhance accessibility to certain substrates and stimulate bacterial growth, the dense bacterial populations within aggregates may also enhance their chance contact with their specific phages, thereby stimulating successful viral (that is, bacteriophage) infection. The declining cell abundance of Alteromonas and Roseobacter cells, which were mostly aggregate associated, in P. globosa-infected cultures, coincided with an increase in ambient bacteriophage abundance. Assuming a burst size (number of viruses produced per cell) of 50 (Parada et al., 2006), the net decrease in the abundances of Alteromonas and Roseobacter cells matched the net increase in the number of ambient bacteriophages.
Viral driven carbon and nitrogen flow
It is generally assumed that viruses stimulate bacterial growth by the release of organic substrates upon host cell lysis. Our nanoSIMS analyses reveal a substantial carbon and nitrogen substrate assimilation by Alteromonas cells in the infected cultures already by 5 h post infection, before the viral-induced cell lysis of P. globosa (latent period 8–12 h, (Baudoux and Brussaard, 2005)). The still intact P. globosa cells and the lack of extracellular P. globosa virus increase also show that viral lysis of P. globosa had not occurred yet. Therefore, the isotopic enrichment in Alteromonas cells would have to come from the leakage or enhanced excretion of organic compounds from the infected but still intact P. globosa host cells. To our knowledge, leakage or enhanced excretion in response to viral infection as an additional substrate source for bacterial growth, has not been reported previously.
Interestingly, at day 2, P. globosa viral lysis specifically enhanced the single-cell 13C and 15N assimilation of Alteromonas cells by about 2.5-fold relative to Roseobacter cellular substrate assimilation. During this time, there was no major change in the bulk 15N-PON content (Figure 4). As Alteromonas and Roseobacter cells are part of the particulate organic matter, the 15N transfer from algae to bacteria does not influence the 15N-PON content. In the control cultures, small increases in 13C and 15N content of both Alteromonas and Roseobacter cells at day 2 (Table 2) indicate that these bacteria at least partly utilised algal derived 13C- and 15N-labelled substrates. The significantly larger contribution of Alteromonas (∼35%, Supplementary Table 4) and to a lesser extent Roseobacter cells (∼6%) to particulate 13C-POC and 15N-PON in the infected cultures suggests an efficient transfer of P. globosa viral lysates towards these specific bacterial members. The observed differences in substrate assimilation were most likely due to different substrate uptake affinities of these two genera. The Alteromonas cells are capable of utilising a high diversity of organic compounds for energy acquisition ranging from low molecular weight organics such as hexoses (Gómez-Consarnau et al., 2012) to complex substrates such as coral mucus (Allers et al., 2008). On the other hand, Roseobacter cells are known to prosper on the phytoplankton-derived material such as algal osmolytes and monomers like amino acids (Zubkov et al., 2001). Thus, the cell abundances attained by Alteromonas and Roseobacter and their 13C and 15N substrate assimilation by day 2 indicate that initial stages of P. globosa viral lysis favour the development of opportunistic bacteria, such as Alteromonas.
Bacterial utilisation might alter the virally released P. globosa organic matter with time, from readily available organic substrates to more refractory compounds (Brussaard et al., 2005b). Given the high enzymatic activity of aggregate-associated bacteria (Proctor and Fuhrman, 1991), we speculate that the bacteriophage-mediated lysis might have facilitated the enzymatic dissolution of aggregates leading to enhanced organic carbon remineralisation rates. On the basis of our estimates, ∼40% of the particulate 13C organic carbon was remineralized to 13C-DIC by day 7 (Supplementary Table 4). The rest likely constituted of less labile particulate organic carbon forms such as cellular P. globosa star-like structures. In fact, the presence of Pseudoalteromonas sp., an algal polysaccharide decomposer (Ivanova et al., 2002) at day 7, might indicate the accumulation of more less labile particulate material.
In the infected P. globosa cultures, the 13C and 15N single-cell substrate assimilation by Alteromonas cells decreased by day 7, even though the particulate 15N-PON remained unchanged. The reduced 13C and 15N labelling of the Alteromonas cells at day 7 might indicate that they utilised less labelled and less labile organic material like P. globosa star-like structures (Sheik et al., 2013). In contrast, the increased substrate assimilation of Roseobacter cells by day 7 suggests that organic material released due to potential Alteromonas phage lysis might have at least partly favoured the observed enhanced growth of Roseobacter cells. The impact of such bacteriophage-mediated lysis of the abundant bacterial species on other bacteria in the marine environment requires further investigation that should include identification of specific bacteriophages.
In contrast, to the particulate 13C content, the particulate 15N-nitrogen content did not show any significant change with time in the infected P. globosa culture (Figure 4a). Assuming Redfield stoichiometry (C/N=6.6; (Redfield, 1934)), the observed 25.5 μM increase in 13C-DIC would mean a release of 3.6 μM of ammonium by day 7 due to remineralization of the isotopically labelled algae (Supplementary Table 4). However, no detectable regeneration of ammonium was observed. The lack of detectable ammonium in our study could have resulted from ammonium adsorption on aggregates as suggested previously (Shanks and Trent, 1979).
Conclusions and implications
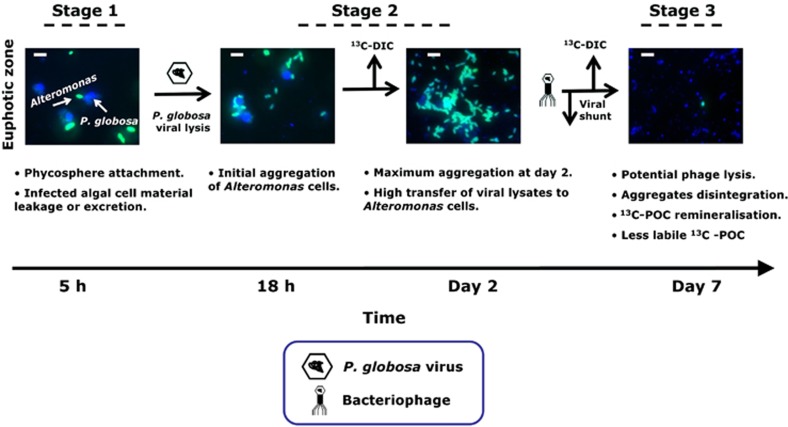
On the basis of our combined results, we propose the following stages of P. globosa viral infections, where the availability of carbon and nitrogen from infected host algae appeared to structure bacterial diversity (Figure 7). During early hours of viral infection, and still before cell lysis, P. globosa cells leaked or excreted ∼20% of organic matter that did not only stimulate substantial substrate assimilation by Alteromonas cells but also triggered its attachment to the infected algal host (stage 1). This association of bacteria with algae is known as the phycosphere (Bell and Mitchell, 1972). Phycosphere formation has been attributed to various environmental factors (Teeling et al., 2012). However, viral infections stimulating algal leakage or excretion and promoting the growth of the associated phycosphere has thus far not been reported.
Figure 7.
Conceptual diagram illustrating the observed temporal microbial regulation and associated biogeochemical processes due to P. globosa viral lysis using Alteromonas CARD-FISH images as an example. Viral infection of P. globosa cells led to leakage or excretion of algal organic matter, stimulating substrate assimilation by Alteromonas cells and also triggered its attachment to the infected algal host (stage 1). Viral lysis of P. globosa single cells resulted in the formation of aggregates that were colonised mostly with Alteromonas cells with an efficient transfer of viral lysates (stage 2). Potential bacteriophage lysis most likely led to aggregate dissolution, leading to regeneration of dissolved inorganic carbon and also less labile organic carbon (stage 3). Scale bar=5 μm.
The bacterial response to the algal viral infections was a rapid temporal succession of bacterial populations consisting of distinct phylotypes. Moreover, our observations indicate that viral lysis of P. globosa single cells resulted in the formation of aggregates that were colonised densely with bacteria (stage 2). Differences in the size of aggregates, bacterial colonisation, time of occurrence and environmental factors (Mari et al., 2005) will determine whether or not aggregate formation enhances or impedes the biological pump. Subsequent aggregate dissolution due to potential bacteriophage lysis appeared to be responsible for the regeneration of dissolved inorganic carbon (stage 3). The sudden appearance of r- strategists (such as Alteromonas cells) and their rapid demise signifies the efficiency of potential bacteriophage-mediated lysis and offers a potential explanation to their apparent rarity in the environment (Pedros-Alio, 2006). Cell lysis due to lytic viral infections is considered to be the simplest explanation of how viruses structure bacterial communities and mediate global biogeochemical cycles. Our results show that already before cell lysis the leakage or excretion of organic matter by infected yet intact algal cells shaped North Sea bacterial community composition and enhanced bacterial substrate assimilation. If these results could be extrapolated to other algal species, loss of organic matter by infected intact algae may have an important but so far unrecognised role in global carbon and nitrogen cycles.
Acknowledgments
We thank Gabriele Klockgether, Daniela Franzke, Thomas Max, Birgit Adam, Anna Noordeloos and Evaline van Weerlee for their technical assistance. We are grateful to two anonymous reviewers for their valuable suggestions, which helped us to improve this manuscript. We thank the Max Planck Society (MPG) and Royal Netherlands Institute for Sea Research for financial support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alderkamp AC, Sintes E, Herndl GJ. Abundance and activity of major groups of prokaryotic plankton in the coastal North Sea during spring and summer. Aquat Microb Ecol. 2006;45:237–246. [Google Scholar]
- Allers E, Gómez-Consarnau L, Pinhassi J, Gasol JM, Šimek K, Pernthaler J. Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ Microbiol. 2007;9:2417–2429. doi: 10.1111/j.1462-2920.2007.01360.x. [DOI] [PubMed] [Google Scholar]
- Allers E, Niesner C, Wild C, Pernthaler J. Microbes enriched in seawater after addition of coral mucus. Appl Environ Microbiol. 2008;74:3274–3278. doi: 10.1128/AEM.01870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, Glöckner FO, Neef A. Modern methods in subsurface microbiology: in situ identification of microorganisms with nucleic acid probes. Fems Microbiol Rev. 1997;20:191–200. [Google Scholar]
- Assayag N, Rivé K, Ader M, Jézéquel D, Agrinier P. Improved method for isotopic and quantitative analysis of dissolved inorganic carbon in natural water samples. Rapid Commun Mass Spectrom. 2006;20:2243–2251. doi: 10.1002/rcm.2585. [DOI] [PubMed] [Google Scholar]
- Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- Baudoux AC, Brussaard CPD. Characterization of different viruses infecting the marine harmful algal bloom species Phaeocystis globosa. Virology. 2005;341:80–90. doi: 10.1016/j.virol.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Baudoux AC, Noordeloos AAM, Veldhuis MJW, Brussaard CPD. Virally induced mortality of Phaeocystis globosa during two spring blooms in temperate coastal waters. Aquat Microb Ecol. 2006;44:207–217. [Google Scholar]
- Bell W, Mitchell R. Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull. 1972;143:265–277. [Google Scholar]
- Brussaard CPD, Gast GJ, vanDuyl FC, Riegman R. Impact of phytoplankton bloom magnitude on a pelagic microbial food web. Mar Ecol Prog Ser. 1996;144:211–221. [Google Scholar]
- Brussaard CPD. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CPD, Kuipers B, Veldhuis MJW. A mesocosm study of Phaeocystis globosa population dynamics: 1. Regulatory role of viruses in bloom. Harmful Algae. 2005a;4:859–874. [Google Scholar]
- Brussaard CPD, Mari X, Van Bleijswijk JDL, Veldhuis MJW. A mesocosm study of Phaeocystis globosa (Prymnesiophyceae) population dynamics—II. Significance for the microbial community. Harmful Algae. 2005b;4:875–893. [Google Scholar]
- Brussaard CPD, Payet JP, Winter C, Weinbauer MG.2010Quantification of aquatic viruses by flow cytometryIn: SW Wilhelm, MG Weinbauer and CA Suttle (eds)Manual of Aquatic Viral Ecology ASLO 102–109.
- Brussaard CPD, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M, et al. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2008;2:575–578. doi: 10.1038/ismej.2008.31. [DOI] [PubMed] [Google Scholar]
- Cottrell MT, Suttle CA. Wide-spread occurrence and clonal variation in viruses which cause lysis of a cosmopolitan, eukaryotic marine phytoplankter, Micromonas pusilla. Mar Ecol Prog Ser. 1991;78:1–9. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Glockner FO, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Applied Environ Microbiol. 2000;66:3044. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Peplies J, Glöckner FO, Gerdts G, Amann R. Isolation of novel pelagic bacteria from the German Bight and their seasonal contributions to surface picoplankton. Appl Environ Microbiol. 2001;67:5134–5142. doi: 10.1128/AEM.67.11.5134-5142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BM, Zubkov MV, Sahm K, Burkill PH, Amann R. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ Microbiol. 2000;2:191–201. doi: 10.1046/j.1462-2920.2000.00092.x. [DOI] [PubMed] [Google Scholar]
- Gómez-Consarnau L, Lindh MV, Gasol JM, Pinhassi J. Structuring of bacterioplankton communities by specific dissolved organic carbon compounds. Environ Microbiol. 2012;14:2361–2378. doi: 10.1111/j.1462-2920.2012.02804.x. [DOI] [PubMed] [Google Scholar]
- Guillard RRL.1975Culture of phytoplankton for feeding marine invertebratesSmith, Walter L, Chanley Matoira H (eds)Culture of Marine Invertebrate Animals Conference, Greenport, NY Oct, 1972 Viii+338p Illus Plenum Press: New York, NY, USA; London, England Isbn 0-306-30804-5; 29–60. [Google Scholar]
- Haaber J, Middelboe M. Viral lysis of Phaeocystis pouchetii: Implications for algal population dynamics and heterotrophic C, N and P cycling. ISME J. 2009;3:430–441. doi: 10.1038/ismej.2008.125. [DOI] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EP, Bakunina IY, Sawabe T, Hayashi K, Alexeeva YV, Zhukova NV, et al. Two species of culturable bacteria associated with degradation of brown algae Fucus evanescens. Microb Ecol. 2002;43:242–249. doi: 10.1007/s00248-001-1011-y. [DOI] [PubMed] [Google Scholar]
- Lamy D, Obernosterer I, Laghdass M, Artigas F, Breton E, Grattepanche JD, et al. Temporal changes of major bacterial groups and bacterial heterotrophic activity during a Phaeocystis globosa bloom in the eastern English Channel. Aquat Microb Ecol. 2009;58:95–107. [Google Scholar]
- Liss PS, Malin G, Turner SM, Holligan PM. Dimethyl sulphide and Phaeocystis: a review. J Marine Syst. 1994;5:41–53. [Google Scholar]
- Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- Mari X, Rassoulzadegan F, Brussaard CPDD, Wassmann P. Dynamics of transparent exopolymeric particles (TEP) production by Phaeocystis globosa under N- or P-limitation: a controlling factor of the retention/export balance. Harmful Algae. 2005;4:895–914. [Google Scholar]
- Middelboe M, Riemann L, Steward GF, Hansen V, Nybroe O. Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat Microb Ecol. 2003;33:1–10. [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci. 2008;105:17861. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olenina I, Hajdu S, Edler L, Andersson A. Biovolumes and size-classes of phytoplankton in the Baltic Sea. HELCOM Baltic Sea Environ Proc. 2006;106:1–144. [Google Scholar]
- Parada V, oacute nica, Herndl GJ, Weinbauer MG. Viral burst size of heterotrophic prokaryotes in aquatic systems. J Marine Biol Assoc UK. 2006;86:613–621. [Google Scholar]
- Pedros-Alio C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms. Mol Microb Ecol Manual. 2004;3:711–726. [Google Scholar]
- Pernthaler A, Pernthaler J, Eilers H, Amann R. Growth patterns of two marine isolates: Adaptations to substrate patchiness. Appl Environ Microbiol. 2001;67:4077. doi: 10.1128/AEM.67.9.4077-4083.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polerecky L, Adam B, Milucka J, Musat N, Vagner T, Kuypers MMM. Look@NanoSIMS—a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- Proctor LM, Fuhrman JA. Roles of viral infection in organic particle flux. Marine Ecol Progress Series Oldendorf. 1991;69:133–142. [Google Scholar]
- Redfield AC. On the proportions of organic derivatives in sea water and their relation to the composition of plankton. James Johnstone Memorial Vol. 1934;176:92. [Google Scholar]
- Riemann L, Grossart HP. Elevated lytic phage production as a consequence of particle colonization by a marine Flavobacterium (Cellulophaga sp.) Microbial Ecol. 2008;56:505–512. doi: 10.1007/s00248-008-9369-8. [DOI] [PubMed] [Google Scholar]
- Sandaa R-A, Gómez-Consarnau L, Pinhassi J, Riemann L, Malits A, Weinbauer MG, et al. Viral control of bacterial biodiversity – evidence from a nutrient-enriched marine mesocosm experiment. Environ Microbiol. 2009;11:2585–2597. doi: 10.1111/j.1462-2920.2009.01983.x. [DOI] [PubMed] [Google Scholar]
- Schäfer H. Isolation of Methylophaga spp. from marine dimethylsulfide-degrading enrichment cultures and identification of polypeptides induced during growth on dimethylsulfide. Appl Environ Microbiol. 2007;73:2580–2591. doi: 10.1128/AEM.02074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks AL, Trent JD. Marine snow: Microscale nutrient patches. Limnol Oceanograph. 1979;24:850–854. [Google Scholar]
- Sheik AR, Brussaard CPD, Lavik G, Foster RA, Musat N, Adam B, et al. Viral infection of Phaeocystis globosa impedes release of chitinous star-like structures: quantification using single cell approaches. Environ Microbiol. 2013;15:1441–1451. doi: 10.1111/j.1462-2920.2012.02838.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wolcott RD, Dowd SE. Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol Biol (Clifton, Nj) 2011;733:129–141. doi: 10.1007/978-1-61779-089-8_9. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea. Bioscience. 1999;49:781–788. [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ Microbiol. 2001;3:304–311. doi: 10.1046/j.1462-2920.2001.00196.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.