Abstract
Diverse associations between methanotrophic archaea (ANME) and sulfate-reducing bacterial groups (SRB) often co-occur in marine methane seeps; however, the ecophysiology of these different symbiotic associations has not been examined. Here, we applied a combination of molecular, geochemical and Fluorescence in situ hybridization (FISH) coupled to nanoscale secondary ion mass spectrometry (FISH-NanoSIMS) analyses of in situ seep sediments and methane-amended sediment incubations from diverse locations (Eel River Basin, Hydrate Ridge and Costa Rican Margin seeps) to investigate the distribution and physiology of a newly identified subgroup of the Desulfobulbaceae (seepDBB) found in consortia with ANME-2c archaea, and compared these with the more commonly observed associations between the same ANME partner and the Desulfobacteraceae (DSS). FISH analyses revealed aggregates of seepDBB cells in association with ANME-2 from both environmental samples and laboratory incubations that are distinct in their structure relative to co-occurring ANME/DSS consortia. ANME/seepDBB aggregates were most abundant in shallow sediment depths below sulfide-oxidizing microbial mats. Depth profiles of ANME/seepDBB aggregate abundance revealed a positive correlation with elevated porewater nitrate relative to ANME/DSS aggregates in all seep sites examined. This relationship with nitrate was supported by sediment microcosm experiments, in which the abundance of ANME/seepDBB was greater in nitrate-amended incubations relative to the unamended control. FISH-NanoSIMS additionally revealed significantly higher 15N-nitrate incorporation levels in individual aggregates of ANME/seepDBB relative to ANME/DSS aggregates from the same incubation. These combined results suggest that nitrate is a geochemical effector of ANME/seepDBB aggregate distribution, and provides a unique niche for these consortia through their utilization of a greater range of nitrogen substrates than the ANME/DSS.
Keywords: niche differentiation, nitrate assimilation, Desulfobulbaceae, methane seep, symbiosis
Introduction
The coexistence of physiologically related strains or species may be explained by niche partitioning (Gause, 1934), in which coexisting strains or species have differing resource use within an environment. A classic example of this is the low- and high-light strains of Prochlorococcus, co-isolates from the same habitat that have differing light intensity requirements (Moore et al., 1998). Similar ecophysiological differentiation driven by different energy source and electron acceptor utilization has also been described between Desulfovibrio species from sludge bed reactors (Dar et al., 2007). Niche diversification has also been shown between coexisting marine microorganisms based on metatranscriptome analyses, which revealed preferential expression of genes involved in organic compound usage (Gifford et al., 2013). This kind of niche differentiation based on ecophysiological differences is likely more common in environments than was previously appreciated.
Molecular and fluorescence in situ hybridization (FISH) microscopy investigations of methane-seep habitats indicate there is significant diversity within the sulfate-reducing bacterial (SRB) partners found in physical association with the methanotrophic ANME archaea (Orphan et al., 2002; Knittel et al., 2003, 2005; Losekann et al., 2007; Pernthaler et al., 2008; Schreiber et al., 2010; Holler et al., 2011; Kleindienst et al., 2012). SeepSRB1a members of the Desulfobacteraceae (DSS) family are frequently the dominant partner of ANME-2 (Schreiber et al., 2010), however, there appears to be flexibility in partner selection; aggregates of ANME-3 and seepSRB1 cells (Schreiber et al., 2010) and ANME-3 with members of the Desulfobulbaceae (Losekann et al., 2007) have been reported, novel ANME-1 consortia have been shown to associate with Deltaproteobacteria from the HotSeep-1 cluster (Holler et al., 2011) and ANME-2c (a subgroup of the ANME-2 lineage) were found in association with seepSRB2 (Kleindienst et al., 2012), seepSRB1a, Desulfobulbaceae and other bacteria (Pernthaler et al., 2008). Interestingly, in both of the latter cases these alternative aggregate forms were recovered from the same sediment sample along with the dominant consortia type (ANME/DSS), suggesting the different SRB partners may occupy distinct niches across small spatial distances. These studies raise the question of what sustains diversity of seemingly functionally similar organisms, in this case two families within the SRB guild.
Cultured members of the DSS and Desulfobulbaceae families differ in several key metabolic pathways related to carbon assimilation and energy harvesting (Kuever et al., 2005b), suggesting uncultured syntrophic SRB lineages belonging to these families may also have different ecophysiologies. A review of published 16S rRNA and FISH-based studies reporting the presence of Desulfobulbaceae in methane-seep sediment reveals an increase in Desulfobulbaceae-affiliated cells and sequences in shallow horizons beneath sulfur-oxidizing microbial mats (Orphan et al., 2001; Knittel et al., 2003; Niemann et al., 2006; Losekann et al., 2007; Pernthaler et al., 2008), some of which are known to store nitrate (McHatton et al., 1996). Nitrate has a complicated role with regards to its effect on SRB activity from stimulating guilds that can outcompete SRB to leading to an accumulation of byproducts toxic to SRB (Greene et al., 2003). This creates a precedent for differential adaptation and utilization of nitrate by SRB, and sediment depth horizons containing both SRB and complex nitrogen sources are an ideal habitat for studying such differences.
Although very little is known about which factors lead to differences in syntrophic SRB distribution, it is possible that these same factors are important to the symbiosis as a whole, presenting a unique opportunity to uncover additional environmental regulators of the anaerobic oxidation of methane (AOM) via single-cell comparative physiology and distribution of two very distinct syntrophic SRB. FISH-NanoSIMS (nanoscale secondary ion mass spectrometry) analyses of sediments incubated with stable isotope-labeled substrates allow the simultaneous detection of phylogenetic identity and metabolic activity at single-cell resolution. This provides a unique opportunity to investigate potential ecophysiological differences between ANME/DSS and ANME/seepDBB aggregates. Owing to the observed spatial distribution of Desulfobulbaceae we used FISH-NanoSIMS to investigate the potential role of nitrogen substrates in defining unique niches for ANME/seepDBB, focusing on nitrate as it is known to be dynamic in methane-seep sediments (Bowles and Joye, 2010). Using a combination of molecular, geochemical and NanoSIMS analyses of environmental and incubation samples from diverse methane seeps (Eel River Basin, Hydrate Ridge and Costa Rican Margin), we investigated the role of nitrate in ANME/seepDBB (versus ANME/DSS) aggregate distribution and metabolism.
Materials and methods
Site selection, sampling and processing:
Detailed information for all samples used in this study can also be found in Table 1.
Table 1. Summary of samples used in this study.
| Sample name | Location | Date | Cruise | Dive | Latitude and longitude | Depth (mbsl) | Push core | Sediment depths (cmbsf) | Habitat | Sample first described | Analysis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eel River Basin | October-06 | AT 15–11 | |||||||||
| Clam1 | Northern Ridge | AD4256 | 40°N 48.6 124°W 36.6 | 520 | PC17 | 3–12 | Clam bed | This study | Agg. counts, geochemistry | ||
| Low-methane | Northern Ridge | AD4256 | 40°N 48.6 124°W 36.6 | 520 | PC20 | 3–12 | Low methane | This study | Agg. counts, geochemistry | ||
| Clam2 | Northern Ridge | AD4256 | 40°N 48.6 124°W 36.6 | 520 | PC23 | 3–12 | Clam bed | This study | Agg. counts, geochemistry | ||
| Mat | Northern Ridge | AD4256 | 40°N 48.6 124°W 36.6 | 520 | PC29 | 3–12 | Microbial mat | Pernthaler et al., 2008 | DNA, agg. counts, geochemistry | ||
| Incubation, PC11 | Southern Ridge | AD4254 | 40°N 47.2 124°W 35.7 | 520 | PC11 | 0–12 | Clam bed | Dekas et al., 2009 | FISH-nanoSIMS, agg. counts | ||
| Incubation, PC14 | Southern Ridge | AD4254 | 40°N 47.2 124°W 35.7 | 520 | PC14 | 0–15 | Microbial mat | Dekas et al., 2009 | FISH-nanoSIMS | ||
| Costa Rica Margin | February-09 | AT 15–44 | |||||||||
| Costa Rica Mat 1 | Jaco Summit | AD4510 | 9°N 10.3 84°W 47.9 | 745 | PC6 | 0–9 | Microbial mat | Dekas et al., in press | DNA, agg. counts, geochemistry | ||
| Costa Rica Mat 2 | Jaco Summit | AD4510 | 9°N 10.3 84°W 47.9 | 745 | PC1 | 0–9 | Microbial mat | Dekas et al., in press | Agg. counts, geochemistry | ||
| Hydrate Ridge | August-10 | AT 15–68 | |||||||||
| Hydrate Ridge Mat 1 | Southeast Knoll | AD4633 | 44°N 27.0 125°W 1.7 | 625 | PC2 | 0–9 | Microbial mat | This study | Agg. counts, geochemistry | ||
| Hydrate Ridge Mat 2 | Hydrate Ridge South | AD4635 | 44°N 34.1 125°W 9.1 | 775 | PC18 | 0–6 | Microbial mat | This study | Agg. counts, geochemistry | ||
| Hydrate Ridge Mat 3 | Hydrate Ridge South | AD4636 | 44°N 34.1 125°W 9.1 | 772 | PC19 | 0–9 | Microbial mat | This study | Agg. counts, geochemistry | ||
| Hydrate Ridge | September-11 | AT 18–10 | |||||||||
| DNA sample | Hydrate Ridge North | J2 593 E3 | 44°N 40.0 125°W 6.0 | 600 | PC47 | 0–9 | Microbial mat | This study | DNA |
Abbreviations: agg., aggregate; cmbsf, centimeters below sea floor; FISH, fluorescence in situ hybridization; mbsl, meters below sea level.
Eel River Basin (AT 15–11) October 2006
Samples from the Northern Ridge of Eel River Basin (40°N 48.6 124°W 36.6; 520 m water depth), an active methane-seep off the coast of Northern California (described by Orphan et al. (2004)), were collected by manned submersible Alvin in October of 2006 using push cores. Four, 30-cm long push cores were collected during dive AD4256 along a transect, which spanned two habitats defined by distinct chemosynthetic communities residing at the sediment surface in a ‘bulls-eye' pattern (governed by sulfide concentration gradients). Microbial mats were present in the center (PC29:mat), surrounded by clam beds (PC17:clam1 and PC23:clam2), which decrease in abundance towards the outer rim of the ‘bulls-eye', which has lower methane flux and a low concentration of sulfide (PC20:low methane). Two additional cores, AD4254 PC11 and AD4254 PC14, were collected from a clam bed (40°N 47.2 124°W 35.7) and microbial mat (40°N 47.2 124°W 35.7), respectively, for incubation experiments. Cores were processed shipboard (as described by Pernthaler et al. (2008)).
Costa Rica Margin (AT 15–44) February 2009, Hydrate Ridge (AT 15–68) August 2010, Hydrate Ridge (AT 18–10) September 2011
Push core samples were also collected from active methane seeps in the Costa Rica Margin (Mau et al., 2006; Sahling et al., 2008) and Hydrate Ridge (Boetius and Suess, 2004) off the coast of Oregon using manned submersible Alvin and remotely operated vehicle Jason (AT 18–10 only). These push cores were collected through three microbial mats (AD4633 PC2: Hydrate Ridge Mat 1: SE Knoll, 44°N 26.99 125°W 01.69, 625 m water depth, AD4635 PC18: Hydrate Ridge Mat 2: Hydrate Ridge South, 44°N 34.09 125°W 9.14, 775 m water depth; and AD4636 PC19: Hydrate Ridge Mat 3: Hydrate Ridge South, 44°N 34.09 125°W 9.14, 772 m water depth) from Hydrate Ridge and two microbial mats (AD4510: Jaco Summit, 9°N 10.29 84°W 47.92, 745 m water depth; PC6: Costa Rica Mat 1 and PC1: Costa Rica Mat 2) from Costa Rica Margin. Samples for DNA extractions were also collected from Hydrate Ridge AT18-10 (J2 593 E3 PC47: Hydrate Ridge North, 44°N 40.0 125°W 6.0, 600 m water depth 0–9 cm horizon below microbial mat). All cores for this study were processed shipboard (as described by Pernthaler et al. (2008)).
Microcosm experiments
The microcosm experiments used in this study have been previously described by Dekas et al. (2009). Briefly, sediments from Eel River Basin clam bed core AD4254 PC11 (top 12 cm) and microbial mat core AD4254 PC14 (top 15 cm) were mixed ∼1:1 with filtered seawater sparged with Argon. The sediment slurries were amended to 0 or 2 mM 15N-nitrate (PC11) or 2 mM 15N-ammonium (PC14) and incubated anoxically with a headspace of methane (overpressed to 30 PSI) in glass bottles with butyl stoppers at 4–8 °C. 15N2 (comprising 5.2% of the headspace) was also present in the 15N-ammonium incubation. 15N-uptake from 15N2 is not considered a significant contributor to the overall 15N-uptake observed in this incubation owing to the relatively low levels of 15N-uptake in a 15N2-only incubation conducted in parallel (Dekas et al., 2009), as well as the possibility of an ammonium switch-off mechanism that would cease all 15N2 fixation in the presence of 2 mM ammonium (pure cultures of methanogenic archaea cease N2 fixation around 100 μm ammonium, Kessler et al., 2001). Sediment samples were taken anoxically via syringe at 3 (nitrate incubations) and 6 months (ammonium incubations). Sediment samples were fixed in 4% formaldehyde for 1 h, washed with 1 × phosphate-buffered saline (PBS; Pernthaler et al., 2008; 145 mM NaCl, 1.4 mM NaH2PO4, 8 mM Na2HPO4 (pH 7.4)), then PBS and EtOH (1:1), and then resuspended in EtOH, and stored at −20 °C.
DNA extraction and clone library analysis
DNA was extracted from methane-seep sediment collected from Costa Rica (AT15-44 AD4510 PC6: 0–1 cm below a microbial mat), and from magneto-FISH-captured aggregates (see below for details on magneto-FISH) from Eel River Basin (AT15-11 AD4256 PC29: 3–6 cm horizon below microbial mat) and Hydrate Ridge (AT18-10 J2 593 E3 PC47: Hydrate Ridge North, 44°N 40.0 125°W 6.0, 600 m water depth 0–9 cm horizon below microbial mat) using probes seepDBB653 (this study) and ANME_2c_760 (Knittel et al., 2005), respectively. Sediment extractions were conducted using the MoBio Ultraclean soil kit (MO BIO Laboratories Inc., Carlsbad, CA, USA) following a previously published protocol (Orphan et al., 2001). DNA extraction from magneto-FISH-captured aggregates was conducted as described by Pernthaler et al. (2008). Following extraction, magneto-FISH DNAs from Eel River Basin were amplified using Multiple Displacement Amplification (MDA performed using REPLI-g Mini Kit from Qiagen, Valencia, CA, USA) before PCR amplification.
Bacterial 16S rRNA genes were amplified from Hydrate Ridge, Eel River Basin and Costa Rica Margin samples using bacteria specific forward primer BAC-27F and universal reverse primer U-1492R (Lane, 1991). Thermocycling conditions consisted of an initial 94 °C denaturing step for 3 min followed by 30 cycles of 94 °C for 45 s, 54 °C for 45 s and 72 °C for 1 min 20 s, and then a final 72 °C elongation step for 7 min. Amplification reactions followed published PCR mixtures and conditions (Harrison et al., 2009) with 0.5 μl of Hotmaster Taq polymerase (Eppendorf AG, Hamburg, Germany).
Sequencing and phylogenetic analysis
The amplified 16S rRNA gene products were cleaned using a Multiscreen HTS plate (Millipore, Billerica, MA, USA). The purified amplicons were ligated into pCR 4.0 TOPO TA (Invitrogen Corp., Carlsbad, CA, USA) vectors and used to transform One-Shot TOP10 (Invitrogen Corp) chemically competent cells according to the manufacturer's instructions. A minimum of 10 clones were cleaned using Multiscreen HTS plates (Millipore) and sequenced either in house with a CEQ 8800 capillary sequencer according to the DTCS protocol (Beckman Coulter, Fullerton, CA, USA), at the ASGPB DNA Sequencing Facility of the University of Hawai'i at Manoa or at the Laragen sequencing facility (www.laragen.com).
The quality of sequence chromatograms and base calls were manually verified using Sequencher 4.5 software (Gene Codes, Ann Arbor, MI, USA) and sequences minus the vector and priming sites (750 bp) were subsequently aligned using the SILVA online aligner (SINA; http://www.arb-silva.de/aligner) and imported into ARB using the Silva 108 full-length 16S rRNA gene alignment (http://www.arb-silva.de/; ARB software package version 7.12.07org, ARB_EDIT4; Ludwig et al., 2004). Distance values between seepDBB and cultured representatives were computed in ARB using the distance matrix function. Aligned sequences were exported from ARB and phylogenies computed using the MrBayes software program (Ronquist et al., 2012), with inverse gamma rates and default recommendations from Hall for all other parameters (Hall, 2004). Sequences Acidobacterium capsulatum (CP0001472), Terriglobus roseus (DQ660892), Acanthopleuribacter pedis (AB303221) and Geothrix fermentans (AB303221) served as outgroups to root the tree. Genbank accession numbers for sequences reported in this study are KC598077, KC598080 and KC598082.
Probe design
An alignment of pure culture and putative Desulfobulbaceae 16S rRNA gene sequences retrieved from Eel River Basin by Pernthaler et al. (2008) was used to design oligonucleotide probe seepDBB653 (5′-CTTTCCCCTCCGATACTCA-3′). This 19 bp probe contains one mismatch, at position 660, to sequences (EU622284, EU622288 and EU622294) retrieved in this earlier study that makes the probe less homologous to DSS and more homologous to pure culture Desulfobulbaceae reference sequences. This probe also contains one mismatch, in a different position, to the Desulfobulbaceae ANME-3 partner sequence (AM404331; reported by Losekann et al. (2007)). Non-target organisms affiliated with Gammaproteobacteria may also hybridize with this probe based on sequence similarity.
CARD-FISH reactions were performed, on the original sample (Eel River Basin, PC29, 6–9 cm horizon; Pernthaler et al., 2008) from which seepDBB cells were identified, at formamide concentrations ranging from 15–60% with probe seepDBB653. An optimal signal was obtained at 15% formamide. The seepDBB653 probe was also observed to occasionally result in weak cross-hybridization with DSS cells, but could be clearly discerned from the strong fluorescence signal of the seepDBB cells when dual hybridizations using the seepDBB653 and Desulfosarcina-targeted DSS658 probes were used. The specificity of seepDBB653 was further verified via magneto-FISH using this probe to target seepDBB-associated aggregates. A screen of the recovered bacterial 16S rRNA gene sequences (n=9 randomly selected clones) were all found to cluster within the seepDBB group initially recovered by Pernthaler et al. (2008).
Catalyzed reporter deposition FISH (CARD-FISH)
Sediment samples were fixed in 2% formaldehyde for ∼1.5 h at room temperature, washed twice with 1 × PBS, once with 1:1 PBS: ethanol, resuspended in 100% ethanol and stored at −20 °C. For CARD-FISH analyses, 40–75 μl fixed sediment collected from each depth horizon added to a TE (10 mM Tris-HCl and 1 mM EDTA (pH 9.0)), 0.01 M pyrophosphate solution for a final volume of 1.5 ml, heated in a histological microwave oven (Microwave Research and Applications, Carol Stream, IL, USA) for 3 min at 60 °C, cooled to room temperature and incubated in 0.1% hydrogen peroxide for 10 min. The solution was then sonicated on ice for three 10 s bursts with a Vibra Cell sonicating wand (Sonics and Materials, Danbury, CT, USA) at an amplitude setting of 3.0 and overlaid on a percoll density gradient before centrifugation and filtration (Orphan et al., 2002) onto a 3.0-μm pore filter (Millipore). Percoll density gradients were used in this study to concentrate aggregates from fixed sediments before CARD-FISH analyses and are likely necessary for ANME/seepDBB detection in many methane-seep habitats due to their lower abundance, which may explain the lack of their detection in previous studies (for example, the study by Schreiber et al. (2010)). Cells on the resulting filters were permeabilized in sequential HCl (0.01 M), SDS and lysozyme solutions as described by Pernthaler et al. (2004). Horseradish peroxidase-labeled probes (Biomers, Ulm, Germany) targeting seep Desulfobulbaceae (seepDBB653, 15% formamide; this study) and either DSS (DSS_658, targets Desulfosarcina spp./Desulfococcus spp./Desulfofrigus spp. and Desulfofaba spp; Manz et al., 1998) or anaerobic methane-oxidizing archaeal clade ANME-2 (Eel_MS_932; Boetius et al., 2000) were then used in a dual-hybridization CARD-FISH reaction (Pernthaler et al., 2008). The first hybridization reaction was conducted in a histological microwave oven, enabling the hybridization time to be reduced to 30 min at 46 °C as opposed to the standard 2 h incubation, followed by an amplification reaction with fluorescein-conjugated tyramides. The second CARD-FISH hybridization reaction was carried out in a standard hybridization oven to avoid bleaching of the initial probe fluorescence. This hybridization was conducted for 2.5 h at 46 °C followed by an amplification reaction using Alexa Fluor 546-labeled tyramides. Samples were then counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; 5 μg ml−1 in mounting medium at least 30 min before examination). Micrograph images were taken with a Deltavision RT microscope system (Applied Precision, Issaquah, WA, USA).
Magneto-FISH
Magneto-FISH was performed on 75 μl of fixed sediment as described by Pernthaler et al. (2008), using HRP-labeled oligonucleotide probes Eel_MS_932 (Boetius et al., 2000) and seepDBB653 with the following modifications. During the amplification reactions, 0.1% blocking reagent (Roche Diagnostics Corporation, Indianapolis, IN, USA) prepared in maleic acid buffer (100 mM maleic acid, 150 mM NaCl; Ph 7.5) was used instead of BSA. Following the CARD-FISH reaction, monoclonal mouse anti-fluorescein-antibodies (Molecular Probes, Life Technologies Corporation, Grand Island, NY, USA) were applied directly to the sediment (∼1 μg per 106 cells), incubated for 10 min on ice and washed via two centrifugation steps at 300 g for 8 min with re-suspension in PBS (containing 0.1% BSA; pH 7.4) in 1.5 ml tubes. Sediment was then incubated with pan-mouse paramagnetic beads (5 μm diameter; ∼25 μl per 107 cells) (Dynal, AS, Norway) at 4 °C, rotating, for 1 h. Tubes of sediment were then washed 15 times by placing near a magnet (Dynal MPC-E) for 2 min, removing supernatant and re-suspending in PBS (containing 0.1% BSA; pH 7.4), with a final re-suspension in TE before DNA extraction.
Morphological data and aggregate counts
Morphological data were collected from ANME/seepDBB aggregates (using probes seepDBB653 probe and Eel_MS_932) in sediment samples from four push cores collected along a transect within an Eel River Basin methane-seep. A total of 86 positively hybridized aggregates were imaged and characterized as one of the following morphotypes: shell, partial shell, clumped or mixed (Supplementary Figure S2). Morphological data were also collected from ANME/seepDBB and ANME/DSS aggregates (using probes seepDBB653 probe and DSS_658, respectively) in Eel River Basin sediment incubated with 2 mM 15N-nitrate or 15N-ammonium and sampled at 3 or 6 months, respectively. A total of 84 aggregates were imaged and characterized as one of the four morphotypes (shell, partial shell, clumped or mixed).
Seep porewater profiles of nitrate were used to identify samples with low (<50 μM nitrate) and high (>50 μM nitrate) nitrate concentrations to examine the distribution and abundance of seepDBB-associated aggregates via CARD-FISH. Depth profiles of the relative DAPI/seepDBB versus DAPI/DSS aggregate abundance were generated from five seep-associated push cores collected within three microbial mats (AD4633 PC2: Hydrate Ridge Mat 1, AD4635 PC18: Hydrate Ridge Mat 2 and AD4636 PC19: Hydrate Ridge Mat 3) from Hydrate Ridge and two microbial mats (AD4510 PC6: Costa Rica Mat 1 and AD4510 PC1: Costa Rica Mat 2) from the Costa Rica Margin. Samples for aggregate counts were obtained from 1 cm (Hydrate Ridge Mat 2 and Costa Rica Mat 1) or 3 cm (Hydrate Ridge Mats 1 and 3 and Costa Rica Mat 2) core slices and hybridized with probes seepDBB653 and DSS_658. DAPI/seepDBB and DAPI/DSS aggregates were counted from a total of 50 aggregate-containing fields per sample. Relative numbers of DAPI/seepDBB aggregates are expressed as percent DAPI/seepDBB of total DAPI/SRB aggregates.
Samples for Eel River Basin aggregate counts were obtained from 3 cm core slices and hybridized with probes seepDBB653 and Eel_MS_932. A total of approximately 100 ANME-containing aggregates were counted per sample. Relative numbers of ANME/seepDBB aggregates are expressed as percent ANME/seepDBB of total hybridized aggregates. Total aggregate counts were also done via epifluorescent microscopy after staining the sediment with DAPI. Briefly, 0.1–0.5 μl of fixed and washed sample, diluted in PBS, was filtered onto 0.22 μm pore filters (Millipore) and enumerated according to Turley (1993).
Samples for incubation aggregate counts came from a previously described push core collected through a clam bed in Eel River Basin (PC11) and incubated with and with out 2 mM nitrate under a methane headspace as described by Dekas et al. (2009). After performing a percoll density separation as describe above, samples were hybridized with probes seepDBB653 and DSS_658 (Manz et al., 1998). DAPI/seepDBB and DAPI/DSS aggregates were counted from a total of fifty aggregate-containing fields per sample. Due to sample limitation, counts were made from three replicate incubations from the control (no nitrate) and from three filter portions obtained from one nitrate-amended incubation. The relative numbers of DAPI/seepDBB aggregates are expressed as percent DAPI/seepDBB of total DAPI/SRB aggregates.
Geochemical analyses
Push cores were processed shipboard and porewater depth profiles of methane, sulfate and sulfide concentrations were obtained at 3 cm resolution from push cores collected at Eel River Basin, CA, USA following protocols outlined in Orphan et al. (2004). Specifically, methane and sulfate were measured via ion and gas chromatography and sulfide was quantified using the Cline Assay (Cline, 1969) as described by Dekas et al. (2009).
Nitrate and nitrite concentrations for Costa Rica Margin samples (2009) were analyzed with an Antek chemiluminescence detector at the University of Georgia, Athens and reported in (Dekas et al., in press). Ammonium concentrations for these samples were also analyzed at the University of Georgia, Athens via the indol-phenol method in filtered sample water (500 μl) preserved with 200 μl phenol-EtOH solution (5.5 ml phenol, 49 ml EtOH, 2 ml H2O) and as reported in (Dekas et al., in press). Nitrate and ammonium concentrations for Hydrate Ridge samples (2010) were measured as follows. Porewater was collected from 1 to 3 cm sediment intervals using a Reeburgh style squeezer (Reeburgh, 1967; KC Denmark A/S Research Equipment, Silkenborg, Denmark) immediately after collection and filtered with a 0.2-μm syringe filter into 1.5 ml epitubes and stored at −20 °C until analysis. Ion chromatography systems operated in parallel (Dionex, Sunnyvale, CA, USA) DX-500, Environmental Analysis Center, Caltech) were used to measure cations and anions in the porewater samples (ammonium, nitrate, nitrite and sulfate). A single autosampler loaded both systems' sample loops serially, with the 10 μl sample loop on the anion IC system loaded first, followed by a 5-μl sample loop on the cation IC system. Temperatures of the columns and detectors were not controlled.
Nitrite, nitrate and sulfate were resolved from other anionic components in the sample using a Dionex AS-19 separator column (4 × 250 mm) protected by an AG-19 guard column (4 × 50 mm). A hydroxide gradient was produced using a potassium hydroxide eluent generator cartridge and pumped at 1 ml min−1. The gradient began with a 10 mM hold for 5 min, increased linearly to 48.5 mM at 27 min and finally to 50 mM at 41 min. 10 min were allowed between analyses to return the column to initial conditions. Nitrite and nitrate were determined by UV absorption at 214 nm using a Dionex AD25 Absorbance detector downstream from the conductivity detection system. Suppressed conductivity detection using a Dionex ASRS-300 4 mm suppressor operated in eluent recycle mode with an applied current of 100 mA was applied to detect all other anions, including redundant measurement of nitrite and nitrate. A carbonate removal device (Dionex CRD 200 4 mm) was installed between the suppressor eluent out and the conductivity detector eluent in ports.
Ammonium was resolved from other cationic components using a Dionex CS-16 separator column (5 × 250 mm) protected by a CG-16 guard column (5 × 50). A methylsulfonate gradient was produced using a methylsulfonic acid based eluent generator cartridge and pumped at 1 ml min−1. The gradient began with a 10 mM methylsulfonate hold for 5 min, then increased to 20 mM at 20 min following a non-linear curve (Chromeleon curve 7, concave up), increased further to 40 mM at 41 min following a non-linear curve (Chromeleon curve 1, concave down). Ten minutes was allowed between analyses to return the column to initial conditions. Suppressed conductivity detection using a Dionex CSRS-300 4 mm suppressor operated in eluent recycle mode with an applied current of 100 mA.
Standard curves were generated for each species. For nitrate, nitrite and sulfate, standard measurements were fitted to a linear curve; for ammonium, standard measurements were fitted to a quadratic curve. Standard ranges were 10 μM–2 mM (nitrate, nitrite and ammonium) and 500 μM–32 mM (sulfate). s.d. of repeated injections of a standard (250 μm ammonium, nitrate and nitrite, 8000 μM sulfate) throughout the analysis were 5.0 μM (ammonium), 4.2 μM (nitrate), 5.8 μM (nitrate) and 113 μM (sulfate).
Linear regression analyses for Figure 3 were performed using SigmaPlot version 11 (Systat Software, Inc., San Jose, CA, USA). Measured nitrate concentrations were log transformed to adjust for a skewed distribution due to very high concentrations in one sample (Hydrate Ridge Mat 2). Linear regression analyses were performed on the transformed nitrate values versus percent seepDBB (seepDBB%=3.341+(3.382*ln(Nitrate))). The original depth profiles of nitrate and %seepDBB are provided in Supplementary Figure S3. Linear regression analyses were also performed on the log transformed ammonium values versus percent seepDBB (seepDBB%=−34.021+(8.989*ln(Ammonium))).
FISH nanoscale secondary ion mass spectrometry (FISH-NanoSIMS)
Thirteen ANME/SRB aggregates (seven ANME/seepDBB and six ANME/DSS) were examined from an ammonium-amended incubation (∼2 mM 15N-ammonium, sampled at 6 months) inoculated with methane-seep sediment slurries from a push core collected through a microbial mat in Eel River Basin (PC14; Dekas et al., 2009). Fourteen ANME/SRB aggregates (six ANME/seepDBB and eight ANME/DSS) were examined from a nitrate-amended incubation (2 mM 15N-nitrate, sampled at 3 months) inoculated with methane-seep sediment slurries from a push core collected through a clam bed in Eel River Basin (PC11; Dekas et al., 2009).
All samples were deposited onto 1′′ diameter round microprobe slide (Lakeside city, IL, USA) and hybridized with HRP-labeled probes seepDBB653 and DSS_658 (using the above-described CARD-FISH method); DAPI/seepDBB and DAPI/DSS aggregates were then mapped for NanoSIMS analysis (Orphan et al., 2002; Dekas and Orphan, 2011). Clostridia spores (with known δ13C and δ15N) were spotted onto a blank section of the glass and used as standards during the analysis. Samples were then gold-coated and analyzed using a CAMECA NanoSIMS 50 l housed at Caltech, using a mass resolving power ∼5,000. A primary Cs+ ion beam (4.3–22 pA) was used to raster over target cells, with a raster size ranging from 8–25 μm. Secondary ion images were collected at 256 × 256 pixel resolution with a dwell time of 14 000 ct/pixel over a period of 4–20 h, resulting in 7–97 cycles, depending on target size. This range of ion beam current was used to maximize counts with no offset in 15N observed in standards run before and after the analysis. We used a high-intensity beam for the NanoSIMS analyses, sacrificing some spatial resolution in order to sputter through as much biomass as possible, therefore correlation between FISH and NanoSIMS micrographs is approximate. Clostridia spores were measured periodically as a standard throughout the analysis using the same range in ion beam current. Several masses were collected in parallel including: 12C14N−, and 12C15N− using electron multiplier detectors. Resulting ion images were processed using the L'Image software (developed by L. Nittler, Carnegie Institution of Washington, Washington DC). Each cycle was manually checked and removed from the analysis if streaks were present in the ion image due to instrument charging. The reported isotope ratio for each aggregate was extracted from the isotope image by identifying regions of interest (ROI). The edge of each ROI was standardized by setting a lower threshold of 35% of the maximum value of 12C15N/12C14N counts within a given cycle in L'image. The ratio from the cycle with the highest 12C15N/12C14N was then collected from each aggregate. A second analysis in which the entire aggregate (ANME and SRB) was defined as the region of interest by setting a lower threshold of 5% of the maximum value of 12C14N− counts within a given cycle was also performed. The ratio from the cycle with the highest 12C15N/12C14N was then collected from each aggregate in this second analysis. For simplicity, we refer to the ratio of 12C15N− to 12C14N− as the 15N/14N ratio throughout the text.
Results
Phylogenetic characterization of Desulfobulbaceae from multiple seeps
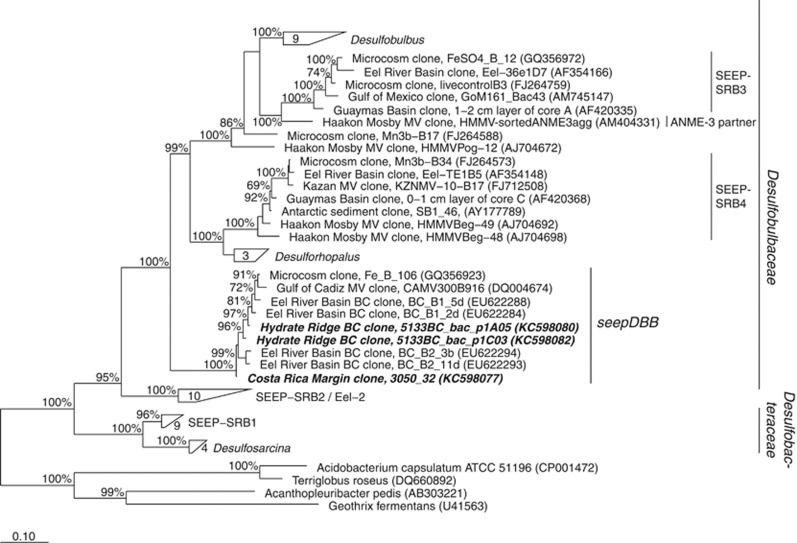
Bacterial 16S rRNA gene sequences distantly related to cultured Desulfobulbaceae sequences were recovered from methane-seep sediments collected from Costa Rica and Hydrate Ridge. These sequences formed a well-supported clade putatively within the Desulfobulbaceae family, along with seepDBB sequences previously retrieved from magneto-FISH enriched ANME-2c aggregates from Eel River Basin (Pernthaler et al., 2008), and distinct from the ANME-3 partners and previously described seepSRB3 and seepSRB4 clades (Knittel et al., 2005); (Figure 1). SeepDBB 16S rRNA sequences shared a similarity of 86–89% to the nearest cultured Desulfobulbaceae sequences (Desulforhopalus species) and 82–83% to the nearest DSS sequences (Desulfosarcina species) in the phylogeny. Initial Eel River Basin sequences from the seepDBB clade were used to design an oligonucleotide probe for CARD-FISH analyses of ANME/seepDBB consortia in situ.
Figure 1.
16S rRNA gene phylogeny of pure culture representatives and sulfate-reducing Deltaproteobacterial sequences retrieved from methane seeps inferred by Bayesian analysis (MrBayes software; Ronquist et al., 2012). Sequences from the current study are in bold italices. Scale bar represents 0.10 substitutions per site. Bootstrap values greater than 60% are reported. BC, bead-captured (that is, originating from magneto-FISH). MV, mud volcano.
Aggregate characterization
Environmental data
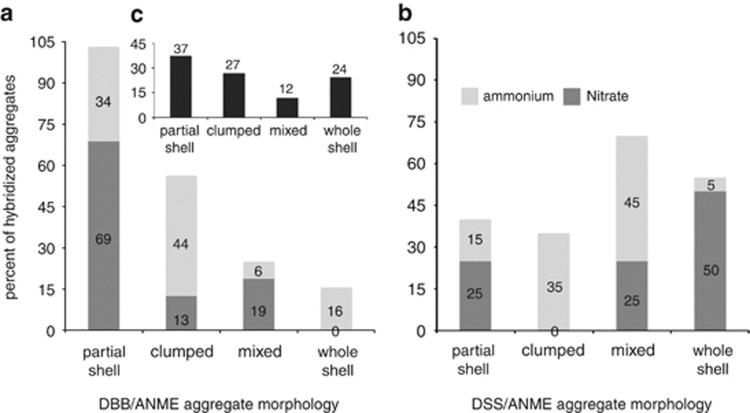
A total of 86 positively hybridized ANME/seepDBB aggregates from Eel River Basin samples were characterized by aggregate morphology, with the majority of Desulfobulbaceae aggregates consisting of partial shell (37%) followed by whole shells and clumped aggregates (24% and 27%, respectively); mixed aggregates represented 12% (Figure 2c, Supplementary Figure S2). The majority (75%) of examined aggregates were 2–6 μm in diameter, with smaller percentages forming aggregates greater than 6 μm.
Figure 2.
Relative proportions of total DBB/ANME or DSS/ANME aggregate morphologies from (a, b) nitrogen-amended incubations or (c) push core sediments collected from Eel River Basin.
Incubation data
Similar to the in situ observations, the dominant ANME/seepDBB morphology in the nitrate incubation was also partial shell (69%), followed by mixed (19%) and clumped aggregates (13% Figure 2a). The ANME/seepDBB aggregates in the ammonium incubation were dominated by clumped morphology (44%), followed by partial shell (34%), whole shell (16%) then mixed (19% Figure 2a). The average ANME/seepDBB aggregate diameter was 6.6 μm in the nitrate incubation and 4.5 μm in the ammonium incubation.
The dominant ANME/DSS morphology in the nitrate incubation was whole shell (50%), followed by equal proportions of mixed and partial shells (25%), with no clumped aggregates detected (Figure 2b). The ammonium incubation in contrast, was dominated by mixed ANME/DSS morphology (45%), followed by clumped (35%), partial shell (15%) and whole shell (5%).
Geochemistry and ANME/seepDBB distribution in diverse methane-seep environments
Eel river basin (AT 15–11)
The seepDBB653 probe along with Eel_MS_932 (targeting ANME cells, Boetius et al., 2000) was initially used to calculate abundance of aggregates associated with the three main seep habitats (clam, mat and low-methane flux periphery) and with increasing sediment depth. Four cores were selected along a transect (mat, clam1, clam2 and low-methane) containing one central mat, two flanking clam beds and the surrounding sediment.
The relative ANME/seepDBB aggregate abundance decreased with depth in 3 of 4 cores (mat, clam2 and low-methane flux site; Supplementary Figure S1). The geochemical profiles of clam1 indicate relatively low levels of sulfate depletion compared with clam2 and mat, perhaps resulting from lower methane flux along the periphery of the clam bed. The apparent correlation between relative ANME/seepDBB aggregate abundance and depth seen in mat, clam2 and low-methane did not appear to be related to sulfate, sulfide or methane concentrations.
Costa rican margin (AT 15–44) and hydrate ridge (AT 15–68)
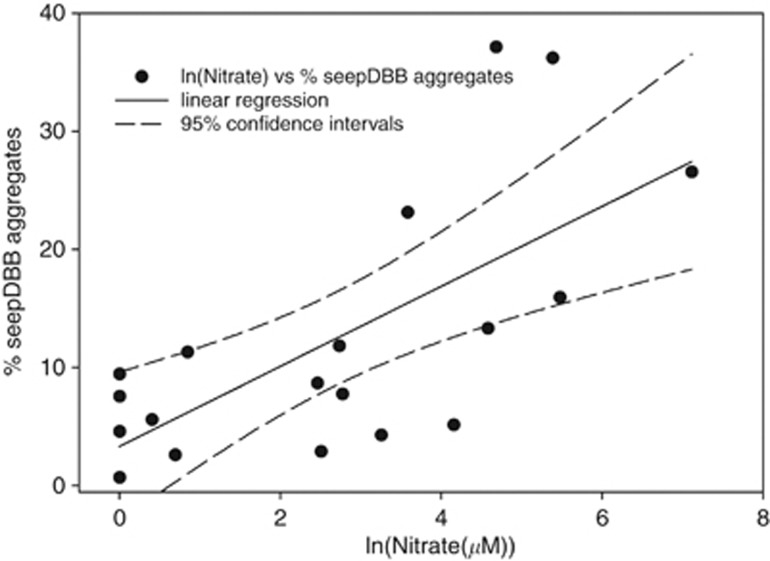
Porewater nitrate concentration profiles were used to select cores containing greater than 50 μM nitrate for further analysis. Nitrate profiles from Costa Rica Margin cores are previously described in Dekas et al. (in press). Cores collected through microbial mats had the highest levels of porewater nitrate of the habitats examined, with the greatest concentrations associated with sediments just below microbial mats, similar to previous reports (Bowles and Joye, 2010). The cores examined in this study contained nitrate ranging from 97 to 1227 μM in the shallowest depth horizon (0–3 cmbsf in Hydrate Ridge Mat 1, 0–1 cmbsf in Costa Rica Mat and Hydrate Ridge Mat 2) that decreased below the detection limit in the deeper depth horizons (>7 cmbsf; Supplementary Figure S3). Depth profiles of relative DAPI/seepDBB (versus DAPI/DSS) aggregate abundance positively correlated with those of nitrate in the resulting cores in both Hydrate Ridge and the Costa Rican Margin (n=3 cores). Low-nitrate (<50 μM nitrate) cores were also examined (n=2 cores), revealing consistently low (DAPI/seepDBB aggregates<10% of total aggregates) relative DAPI/seepDBB (versus DAPI/DSS) aggregate abundance. Overall, a significant correlation was observed between the relative seepDBB aggregate abundance and porewater nitrate concentrations (Figure 3; r2-value=0.469 (t-test P=0.001)). The same linear regression of ammonium and ANME/seepDBB abundance had an r2-value of 0.275 (t-test P=0.021), suggesting ammonium is less correlated with ANME/seepDBB abundance than is nitrate.
Figure 3.
Relative abundance of ANME/seepDBB (to total ANME/SRB) aggregates versus log transformed nitrate (μM) concentrations from depth horizons of five push cores collected from Hydrate Ridge and Costa Rica margin methane seeps. Linear regression analyses were performed on the transformed nitrate values versus percent seepDBB (seepDBB%=3.341+(3.382 * ln(Nitrate))) using SigmaPlot version 11 (Systat Software, Inc.). The r2-value is 0.469 (t-test P=0.001). Measured nitrate concentrations were log transformed to adjust for a skewed distribution due to very high concentrations in one sample (Hydrate Ridge Mat 1). The dashed lines show 95% confidence intervals.
Microcosm analyses via FISH-NanoSIMS
CARD-FISH analyses using probes seepDBB653 and DSS658 were employed on previously prepared methane-amended incubations of seep sediment from the Eel River Basin supplemented with 2 mM nitrate, 2 mM ammonium or no amendment (Dekas et al., 2009). The relative abundance of ANME/seepDBB aggregates (represented as a fraction of total DAPI/SRB aggregates) at 3 months was greater in the nitrate-amended incubation (0.146; s.d.=0.047) compared with the non-amended control (0.087; s.d.=0.018).
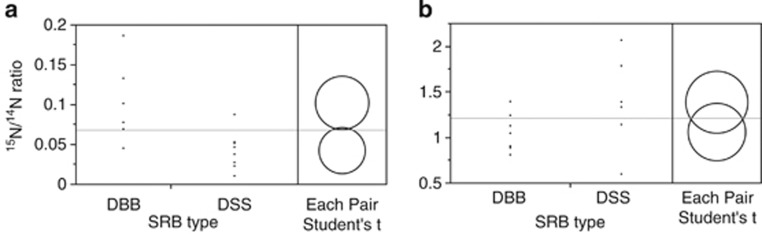
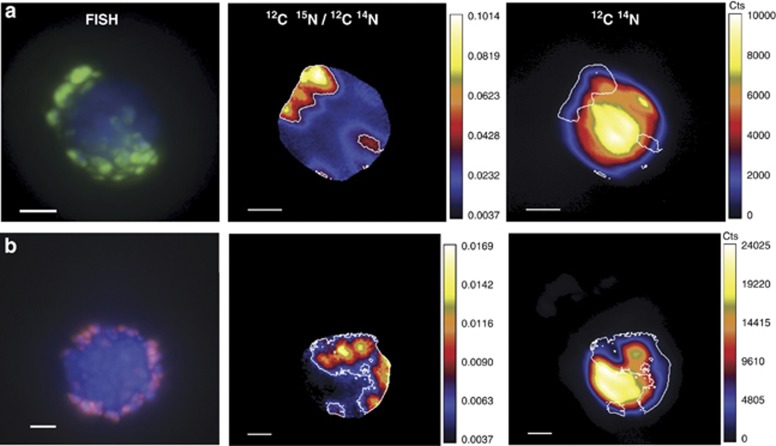
A total of fourteen ANME/SRB aggregates (six ANME/seepDBB and eight ANME/DSS) were examined via FISH-NanoSIMS from the same nitrate-amended microcosm at 3 months. Significantly higher maximum 15N incorporation levels were observed in ANME/seepDBB (versus ANME/DSS) aggregates where 15N/14N ratios ranged from 0.05 to 0.19 in ANME/seepDBB aggregates and from 0.01 to 0.09 in ANME/DSS aggregates (Figure 4; nonparametric Wilcoxon P-val=0.024) when the ROI specified in the data analyses was automatically defined in L'image by setting a lower threshold of 35% of the maximum value of 12C15N/12C14N counts within a given cycle. When the entire aggregate was defined as the ROI by setting a lower threshold of 5% of the maximum value of 12C14N counts within a given cycle, a similar, but not significant trend was observed (nonparametric Wilcoxon P-val=0.081), likely resulting from there being more concentrated regions of 15N incorporation in the ANME/seepDBB aggregates. This may be due to the differences in aggregate morphology, with ANME/seepDBB aggregates typically having half shells or less of seepDBB cells, relative to the ANME/DSS aggregates, frequently observed as whole shell or heterogeneously mixed consortia (Figure 2).
Figure 4.
15N enrichment measured via NanoSIMS in ANME/seepDBB and ANME/DSS aggregates from incubations of Eel River Basin methane-seep sediment and amended with either (a) 15N-nitrate or (b) 15N-ammonium. Circles represent results of t-test.
Overall levels of 15N enrichment were lower in the nitrate-amended microcosms relative to ammonium-amended microcosms. This may be attributed to differences in the sediment source, sampling times and ability of these microorganisms to assimilate the two nitrogen sources, as previously observed in Dekas et al. (2009). Isotope ratio images revealed that several of the aggregates (n=5) from the 15N-nitrate-amended incubation exhibited the highest 15N enrichment in the region corresponding to SRB cells (Figure 5, Supplementary Figure S4).
Figure 5.
FISH and corresponding 12C15N/12C14N and 12C14N ion images of an (a) ANME/seepDBB and (b) ANME/DSS aggregate showing the distribution of 15N incorporation from 15N-nitrate. The edge of each ROI (shown traced in white) was standardized by setting a lower threshold of 35% of the maximum value of 12C15N/12C14N counts within a given cycle. All scale bars represent 2 μm. For FISH images, aggregates were hybridized with probes seepDBB653 (green; targeting methane seep Desulfobulbaceae; this study) and DSS_658 (red; targeting Desulfosarcina/Desulfococcus/Desulfofrigus/Desulfofaba; Manz et al., 1998). All cells were counter stained (blue) using DAPI.
To compare relative uptake of 15N-ammonium, a total of 13 ANME/SRB aggregates (seven ANME/seepDBB and six ANME/DSS) were examined via FISH-NanoSIMS from the ammonium-amended microcosm (sampled at 6 months). There was no significant difference in maximum 15N incorporation levels between ANME/seepDBB and ANME/DSS aggregates (Figure 4; nonparametric Wilcoxon P-value=0.175). 15N/14N ratios for ranged from 0.81–1.39 in ANME/seepDBB aggregates and from 0.60–2.07 in ANME/DSS aggregates. At 6 months, the level of 15N enrichment in ammonium-amended incubations was too high (15N/14N ratios ranged from 0.60–2.07) to distinguish higher incorporation levels in SRB regions versus ANME regions of the aggregate.
Discussion
Characterization of seepDBB partner
Very little is known about the potential physiologies or habitat preferences of the various groups of SRB forming physical associations with the methanotrophic ANME archaea (Knittel and Boetius, 2009). Members of the sulfate-reducing seepSRB1a group are frequently observed as the dominant partner of ANME-2 in many seep habitats (Schreiber et al., 2010), however, other SRB groups and additional bacterial partners have been observed to form physical associations with the ANME-2 lineage (Orphan et al., 2002; Knittel et al., 2005; Pernthaler et al., 2008; Schreiber et al., 2010; Kleindienst et al., 2012). Using an immuno-magnetic cell capture technique (magneto-FISH) to enrich for ANME-2c aggregates, Pernthaler et al. (2008) reported the detection of ANME/Desulfobulbaceae coexisting with ANME/DSS aggregates. These ANME-2c-associated Desulfobulbaceae (seepDBB) are phylogenetically distinct from the Desulfobulbaceae group previously described in association with ANME-3 (Niemann et al., 2006; Losekann et al., 2007; Figure 1). Here, we studied the ecophysiology of co-occurring SRB/ANME-2 consortia, as well as expanded the known distribution of ANME-associated seepDBB cells.
The seepDBB lineage was first described from a sediment sample collected from a methane-seep site in the Eel River Basin (Pernthaler et al., 2008); in the present study CARD-FISH surveys were used to better characterize the depth and habitat distribution of the ANME/seepDBB consortia. We analyzed samples from push cores from a transect spanning three habitats (a sulfur-oxidizing microbial mat, a Calyptogena clam bed and the peripheral sediments with lower methane flux) within this methane seep. Incubations of Eel River Basin sediment amended with either 2 mM nitrate or ammonium were also examined. Interestingly, the dominant morphology was different between ANME/seepDBB versus ANME/DSS consortia in both environmental and incubation data suggesting different dynamics may exist between the partners comprising these consortia (Figure 2).
Although ANME/seepDBB aggregates were found in all Eel River Basin habitats examined, as well as below microbial mat habitats in Hydrate Ridge and Costa Rica margin methane seeps, they consistently occurred as a lower proportion of the total ANME/SRB aggregates relative to ANME/DSS (Supplementary Figures S1 and S3). Despite the relative difference in abundance, the consistent coexistence of two types of ANME/SRB aggregates could result from niche partitioning, which has been demonstrated in cultured species of SRB within the same class (Dar et al., 2007). ANME-associated DSS and seepDBB belong to distinct families (Desulfobacteraceae and Desulbulbaceae, respectively) whose cultured representatives differ in multiple key metabolic pathways (Kuever et al., 2005a, 2005b), suggesting these syntrophic SRB lineages may also have distinct ecophysiologies, which we first explored by comparing their relative distribution in diverse methane seeps to the geochemical gradients in these habitats.
Geochemical profiles and ANME/seepDBB distribution in diverse methane-seep environments
Investigated cores from Eel River Basin were collected along a transect spanning multiple seep habitats (Supplementary Figure S1). ANME/seepDBB aggregates were typically most abundant in the shallower depth horizons of the Eel River Basin transect, with the greatest relative proportions documented below a sulfur-oxidizing microbial mat (Supplementary Figure S1). Available depth profiles of methane, sulfate and sulfide did not appear to explain this distribution. A review of published 16S rRNA and FISH-based studies reporting the presence of Desulfobulbaceae in methane-seep sediment also revealed an increase in seepDBB-affiliated cells and sequences in shallow horizons beneath sulfur-oxidizing microbial mats (Orphan et al., 2001; Knittel et al., 2003; Niemann et al., 2006; Losekann et al., 2007; Pernthaler et al., 2008). Although geochemical porewater profiles for methane, sulfate and sulfide in the 0–10 cm sediment horizons are highly variable between methane-seep sites (Valentine and Reeburgh, 2000; Linke et al., 2005; Niemann et al., 2006; Knittel and Boetius, 2009; Lloyd et al., 2010; Bowles et al., 2011), nitrate levels from methane seeps are typically highest just below microbial mats (Linke et al., 2005; Priesler et al., 2007; Bowles and Joye, 2010; Lichtschlag et al., 2010). We hypothesized nitrate to be a potential geochemical effector of ANME/seepDBB aggregate distribution, and focused subsequent analyses on sediment cores varying in nitrate concentration.
Environmental trends in seepDBB aggregate abundance from Costa Rican margin and Hydrate Ridge methane-seep sites showed a significant correlation with nitrate (Figure 3; r2-value=0.469 (t-test P=0.001)). The highest proportions of ANME/seepDBB (>35% of all ANME/SRB aggregates) were recorded in the shallow horizons of the Costa Rica Margin core where two maxima of ANME/seepDBB aggregate abundance were observed at both 0–1 and 2–3 cm depth. Interestingly, this was the only core that correspondingly contained two peaks of increased porewater nitrate concentrations, which roughly matched the increase in ANME/seepDBB aggregates (Supplementary Figure S3d). To understand the relationship between seepDBB cells and nitrate, we established seep sediment microcosm experiments with 15N-labeled nitrate to measure the effects of nitrate-amendment on the anabolic activity of ANME/seepDBB and ANME/DSS aggregates.
Nitrate utilization by ANME/seepDBB aggregates
Methane-seep sediment previously collected from Eel River Basin and amended with 2 mM 15N-labeled nitrate or ammonium (under methane headspace; Dekas et al., 2009) was used in the current study for CARD-FISH and NanoSIMS analyses. Active sulfate reduction, inferred from sulfide production, was previously measured in both incubations (Dekas et al., 2009). After 3 months, the relative abundance of ANME/seepDBB (represented as a fraction of total DAPI/SRB aggregates) was greater in the nitrate-amended incubation (0.146) than the non-amended control (0.087).
Higher maximum 15N incorporation levels were observed in ANME/seepDBB aggregates versus ANME/DSS aggregates from the 15N-nitrate incubation (Figure 4a), while there was no significant difference in maximum 15N incorporation levels between ANME/seepDBB and ANME/DSS aggregates from the 15N-ammonium incubation (Figure 4b). These data suggest similar assimilation rates for ANME/seepDBB and ANME/DSS in the presence of ammonium, and control for the possible artifact of overall higher growth rates in ANME/seepDBB (versus ANME/DSS) aggregates leading to increased incorporation of any labeled nutrient. Previous FISH-SIMS studies using 15N-labeled ammonium- and N2-amended sediment incubations showed the greatest 15N assimilation by the ANME archaea in intact consortia (Dekas et al., 2009; Orphan and House, 2009). In contrast, several ANME/SRB aggregates analyzed from the labeled nitrate incubation showed clear 15N enrichment in the region associated with SRB cells (Figure 5, Supplementary Figure S4), suggesting that the SRB partner may be responsible for the majority of the nitrogen incorporation from nitrate in these aggregates. This trend was seen in both ANME/SRB aggregate types but was not as pronounced in ANME/DSS aggregates (Figure 5). As expected, 15N enrichment is also seen in the region corresponding to ANME cells, likely resulting from either the SRB cells sharing the enriched nitrogen source with the ANME cells or the ANME cells assimilating it directly.
Although ANME/seepDBB aggregates were consistently less abundant than ANME/DSS, their role in nitrate processing may afford them a more prominent role in marine methane-seep ecosystems than their numbers suggest. It is possible for the impact of an organism to have a disproportionate effect on the environment relative to its abundance. For example, in a FISH-NanoSIMS based study by Musat et al. (2008) the purple sulfur bacterium Chromatium okenii, representing 0.3% of total microbial cell numbers, was found to be responsible for over 40% of total ammonium uptake and 70% of total carbon fixation in an oligotrophic, meromictic Lake Cadagno. Further, Pester et al. (2010) found that a population of Desulfosporosinus, comprising only 0.006% of total 16S rRNA genes, showed potential in situ sulfate reduction rates that could account for a considerable part of sulfate reduction in the peatland soil investigated (Pester et al., 2010).
Our data suggest a role for nitrate as a nutrient source, which is primarily utilized by ANME/seepDBB relative to ANME/DSS aggregates. Although it is not possible to exclude dissimilatory reduction of nitrate to ammonium that can then be incorporated in to biomass (Rabus et al., 2006), the low amount of 15N-nitrate seen assimilated into cells relative to 15N-ammonium still suggests nitrate is a nutrient source secondary to ammonium.
Nitrate has been shown to inhibit sulfate reduction by stimulating resident nitrate-reducing bacteria that outcompete SRB and also produce nitrite, which is toxic to SRB likely due to its inhibition of their dissimilatory sulfite reductase enzyme (DsrAB; Haveman et al., 2004). However, nitrate does not appear to inhibit the SRB population in our study as seen catabolically through sulfide production in nitrate-amended incubations and anabolically through incorporation of 15N-nitrate measured via FISH-NanoSIMS. Greene et al. (2003) showed that different species of SRB, as well as certain strains of the same species, are able to prevent nitrite inhibition by reducing it to ammonium via a nitrite reductase (Nrf). Given that certain species of SRB and nitrate-reducing sulfur-oxidizing bacteria (NR-SOB, which also produce nitrite as a metabolic intermediate of nitrate reduction) coexist in marine methane-seep sediment, this evolution of at least certain species of SRB to overcome nitrite toxicity would be favorable. These SRB would also potentially be able to take advantage of available nitrate as a nitrogen source. Future studies focusing on the genomes of SRB partners involved in AOM may confirm these capabilities, specifically in seepDBB.
The coexistence of physiologically related species may be explained by niche partitioning (Gause, 1934). Complex environments, such as those encountered in seep sediments, are defined by steep chemical gradients, which can lead to distinct microniches, and, in turn, can result in diversification of species harbored in these habitats (Gray et al., 1999; Torsvik et al., 2002). The observed preference for nitrate by ANME/seepDBB versus ANME/DSS aggregates may be one such mechanism by which two apparently functionally redundant consortia can coexist via partitioning the environment into niches defined by nitrogen source.
Conclusions
Very little is known about factors influencing the distribution and fitness of distinct sulfate-reducing bacteria partnered with methanotrophic ANME archaea. Poorly constrained ecological and physico-chemical factors are almost certainly important to the AOM symbiosis as a whole, and present a unique opportunity to uncover additional environmental regulators of sulfate-dependent methane oxidation. Most studies to date have focused on the dynamics of carbon and sulfur metabolism by the AOM symbiosis. Here, we demonstrate a role for nitrate as a geochemical effector influencing the distribution of Desulfobulbaceae-ANME-2 consortia within methane seeps. Although bulk geochemical and molecular analyses provide information on community level diversity and activity, complementary single-cell techniques, like the FISH-NanoSIMS method used in this study, provide direct information on the metabolic function of phylogenetically identified microorganisms in situ and allow for the assessment of ecophysiological differences among coexisting microbial species.
Acknowledgments
We would like to thank Grayson Chadwick for his help with image analysis, Elizabeth Trembath-Reichert for contributing clone sequences, Ankur Saxena for figure design, Yunbin Guan NanoSIMS technical assistance and Joshua Steele for statistical analyses. We would like to also acknowledge Tsege Embaye for methane and sulfate measurements from AT 15–11 and the science party of cruises AT 15–11, AT 15–44, AT 15–68 and AT 18–10 and pilots of the DSRV Alvin and ROV Jason for their assistance with various aspects of this work. Funding for this work was provided by the Department of Energy Division of Biological Research (DE-SC0004949; to VJO), and a National Science Foundation Graduate Research Fellowship (to AG-S). The editing of this work by AD was partially performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. Samples were collected with funding from the National Science Foundation (BIO-OCE 0825791; to VJO).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- Boetius A, Suess E. Hydrate Ridge: a natural laboratory for the study of microbial life fueled by methane from near-surface gas hydrates. Chem Geol. 2004;205:291–310. [Google Scholar]
- Bowles M, Joye S. High rates of denitrification and nitrate removal in cold seep sediments. ISME J. 2010;5:565–567. doi: 10.1038/ismej.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles MW, Samarkin VA, Bowles KM, Joye SB. Weak coupling between sulfate reduction and the anaerobic oxidation of methane in methane-rich seafloor sediments during ex situ incubation. Geochimica et Cosmochimica Acta. 2011;75:500–519. [Google Scholar]
- Cline JD. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanograph. 1969;14:454–458. [Google Scholar]
- Dar S, Stams A, Kuenen J, Muyzer G. Co-existence of physiologically similar sulfate-reducing bacteria in a full-scale sulfidogenic bioreactor fed with a single organic electron donor. Appl Microbiol Biotechnol. 2007;75:1463–1472. doi: 10.1007/s00253-007-0968-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekas AD, Poretsky RS, Orphan VJ. Deep-sea archaea fix and share nitrogen in methane-consuming microbial consortia. Science. 2009;326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- Dekas AE, Chadwick GL, Bowles MW, Joye SB, Orphan VJ. (in press) Spatial distribution of nitrogen fixation in methane seep sediment and the role of the ANME Archaea. Environ Microbiol. [DOI] [PubMed]
- Dekas AE, Orphan VJ. Identification of diazotrophic microorganisms in marine sediment via fluorescence in situ hybridization coupled to nanoscale secondary ion mass spectrometry (FISH-NanoSIMS) Methods Enzymol. 2011;486:281–305. doi: 10.1016/B978-0-12-381294-0.00012-2. [DOI] [PubMed] [Google Scholar]
- Gause G. The Struggle for Existence. Williams and Wilkins: Baltimore, MD, USA; 1934. [Google Scholar]
- Gifford SM, Sharma S, Booth M, Moran MA. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 2013;7:281–298. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N, Howarth R, Rowan A, Pickup R, Jones JG, Head I. Natural communities of Achromatium oxaliferum comprise genetically, morphologically, and ecologically distinct subpopulations. Appl Environ Microbiol. 1999;65:5089–5099. doi: 10.1128/aem.65.11.5089-5099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E, Hubert C, Nemati M, Jenneman G, Voordouw G. Nitrite reductase activity of sulphate-reducing bacteria prevents their inhibition by nitrate-reducing, sulphide-oxidizing bacteria. Environ Microbiol. 2003;5:607–617. doi: 10.1046/j.1462-2920.2003.00446.x. [DOI] [PubMed] [Google Scholar]
- Hall BG. Phylogenetic Trees Made Easy: A How-to Manual. Sinauer Associates: Sunderland, MA, USA; 2004. [Google Scholar]
- Harrison BK, Zhang H, Berelson W, Orphan VJ. Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California) Appl Environ Microbiol. 2009;75:1487–1499. doi: 10.1128/AEM.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveman SA, Greene EA, Stilwell CP, Voordouw JK, Voordouw G. Physiological and gene expression analysis of inhibition of Desulfovibrio vulgaris Hildenborough by nitrite. J Bacteriol. 2004;186:7944–7950. doi: 10.1128/JB.186.23.7944-7950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler T, Widdel F, Knittel K, Amann R, Kellermann MY, Hinrichs K-W, et al. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 2011;5:1946–1956. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler PS, Daniel C, Leigh JA. Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GLnB homologues, NifI(1) and NifI(2) J Bacteriol. 2001;183:882–889. doi: 10.1128/JB.183.3.882-889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleindienst S, Ramette A, Amann R, Knittel K. Distribution and in situ abundance of sulfate,Äêreducing bacteria in diverse marine hydrocarbon seep sediments. Environ Microbiol. 2012;14:2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- Knittel K, Boetius A, Lemke A, Eilers H, Lochte K, Pfannkuche O, et al. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon) Geomicrobiol J. 2003;20:269–294. [Google Scholar]
- Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Knittel K, Losekann T, Boetius A, Kort R, Amann R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol. 2005;71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuever J, Rainey F, Widdel F.2005aFamily I. Desulfobulbaceae family, novIn: Brenner DJ, Krieg NR, Garrity GM, et al (eds)Bergey's Manual® of Systematic Bacteriology Springer: NY, USA; 960–962. [Google Scholar]
- Kuever J, Rainey F, Widdel F.2005bFamily II. Desulfobacteraceae family, NovIn: Brenner DJ, Krieg NR, Garrity GM, et al (eds)Bergey's Manual® of Systematic Bacteriology Springer: NY, USA; 988–992. [Google Scholar]
- Lane D.199116S/23S rRNA sequencingIn Stackebrandt E, Goodfellow M edsNucleic Acid Techniques in Bacterial Systematics Wiley and Sons: Chichester, UK; 115–175. [Google Scholar]
- Lichtschlag A, Felden J, Brüchert V, Boetius A, De Beer D. Geochemical processes and chemosynthetic primary production in different thiotrophic mats of the Håkon Mosby Mud Volcano (Barents Sea) Limnol Oceanograph. 2010;55:931–949. [Google Scholar]
- Linke P, Wallmann K, Suess E, Hensen C, Rehder G. In situ benthic fluxes from an intermittently active mud volcano at the Costa Rica convergent margin. Earth Plane Sci Letters. 2005;235:79–95. [Google Scholar]
- Lloyd KG, Albert DB, Biddle JF, Chanton JP, Pizarro O, Teske A. Spatial structure and activity of sedimentary microbial communities underlying a Beggiatoa spp. Mat in a Gulf of Mexico Hydrocarbon Seep. PLoS One. 2010;5:e8738. doi: 10.1371/journal.pone.0008738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losekann T, Knittel K, Nadalig T, Fuchs B, Niemann H, Boetius A, Amann R. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby mud volcano, Barents Sea. Appl Environ Microbiol. 2007;73:3348–3362. doi: 10.1128/AEM.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A, et al. ARB: a software environment for sequence data. Nucleic acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W, Eisenbrecher M, Neu TR, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. Fems Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- Mau S, Sahling H, Rehder G, Suess E, Linke P, Soeding E. Estimates of methane output from mud extrusions at the erosive convergent margin off Costa Rica. Marine Geol. 2006;225:129–144. [Google Scholar]
- McHatton SC, Barry JP, Jannasch HW, Nelson DC. High nitrate concentrations in vacuolate, autotrophic Marine Beggiatoa spp. Appl Environ Microbiol. 1996;62:954–958. doi: 10.1128/aem.62.3.954-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natil Acad Sciences. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann H, Lösekann T, De Beer D, Elvert M, Nadalig T, Knittel K, et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature. 2006;443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci. 2002;99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- Orphan VJ, House CH. Geobiological investigations using secondary ion mass spectrometry (SIMS): microanalysis of extant and paleo-microbial processes. Geobiology. 2009;7:360–372. doi: 10.1111/j.1472-4669.2009.00201.x. [DOI] [PubMed] [Google Scholar]
- Orphan VJ, Ussler W, Naehr TH, House CH, Hinrichs KU, Paull CK. Geological, geochemical, and microbiological heterogeneity of the seafloor around methane vents in the Eel River Basin, offshore California. Chem Geol. 2004;205:265–289. [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R.2004Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms Molecular Microbial Ecology Manual2nd edn, Vol. 3.11Kluwer Academic Publishers: Netherlands; 711–726. [Google Scholar]
- Pester M, Bittner N, Deevong P, Wagner M, Loy A. A ‘rare biosphere' microorganism contributes to sulfate reduction in a peatland. ISME J. 2010;4:1591–1602. doi: 10.1038/ismej.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisler A, de Beer D, Lichtschlag A, Lavik G, Boetius A, Jorgensen BB. Biological and chemical sulfide oxidation in a Beggiatoa inhabited marine sediment. ISME J. 2007;1:341–353. doi: 10.1038/ismej.2007.50. [DOI] [PubMed] [Google Scholar]
- Rabus R, Hansen T, Widdel F. Dissimilatory sulfate-and sulfur-reducing prokaryotes. Prokaryotes. 2006;2:659–768. [Google Scholar]
- Reeburgh W. An improved interstitial water sampler. Limnol Oceanogr. 1967;12:163–165. [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahling H, Masson DG, Ranero CR, Hühnerbach V, Weinrebe W, Klauke I, et al. Fluid seepage at the continental margin offshore Costa Rica and southern Nicaragua. Geochemistry Geophysics Geosystems. 2008;9:Q05S05. [Google Scholar]
- Schreiber L, Holler T, Knittel K, Meyerdierks A, Amann R. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ Microbiol. 2010;12:2327–2340. doi: 10.1111/j.1462-2920.2010.02275.x. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Sci Signal. 2002;296:1064. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- Turley CM.1993Direct estimates of bacterial numbers in seawater samples without incurring cell loss due to sample storageIn Kemp PF, Sherr BF, Sherr EB, Cole JJ (eds)Handbook of Methods in Aquatic Microbial Ecology Lewis Publishers: Boca Raton, Fla; 143–147. [Google Scholar]
- Valentine DL, Reeburgh WS. New perspectives on anaerobic methane oxidation. Environ Microbiol. 2000;2:477–484. doi: 10.1046/j.1462-2920.2000.00135.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.