Abstract
Responses to host amyloids and curli amyloid fibrils of Escherichia coli and Salmonella enterica serotype Typhimurium are mediated through Toll-like receptor (TLR) 2. Here we show that TLR2 alone was not sufficient for mediating responses to curli. Instead, transfection experiments with human cervical cancer (HeLa) cells and antibody-mediated inhibition of TLR signaling in human macrophage-like (THP-1) cells suggested that TLR2 interacts with TLR1 to recognize curli amyloid fibrils. TLR1/TLR2 also serves as a receptor for tri-acylated lipoproteins, which are produced by E. coli and other Gram-negative bacteria. Despite the presence of multiple TLR1/TLR2 ligands on intact bacterial cells, an inability to produce curli amyloid fibrils markedly reduced the ability of E. coli to induce TLR2-dependent responses in vitro and in vivo. Collectively, our data suggest that curli amyloid fibrils from enterobacterial biofilms significantly contribute to TLR1/TLR2-mediated host responses against intact bacterial cells.
Introduction
Amyloids are protein deposits with a fibrillar cross β-sheet quaternary structure, which exhibit a starch (amylose)-like ability to stain with iodine. Host amyloid deposition in tissue is a result of protein misfolding and is associated with a number of illnesses, such as Alzheimer’s disease and prion diseases. In contrast, many bacteria produce functional amyloid deposits, which are an important component of their extracellular biofilm matrix (Larsen et al., 2007, Jordal et al., 2009). Perhaps the best-characterized bacterial amyloid is encoded by the csgDEFG csgBA gene cluster of Escherichia coli and Salmonella enterica serotype Typhimurium (S. Typhimurium) (Olsen et al., 1989, Hammar et al., 1995, Romling et al., 1998), which directs the biosynthesis of extracellular amyloid fibrils, termed curli, composed of the CsgA protein (Chapman et al., 2002). Curli fibrils are a major extracellular matrix component contributing to biofilm formation in E. coli (Prigent-Combaret et al., 2000, Vidal et al., 1998) and S. Typhimurium (Romling et al., 2003).
Amyloids of host and bacterial origin share a number of characteristics, including an ability to trigger innate immune responses. Host amyloid deposits induce chronic inflammation, which in turn results in the tissue injuries responsible for neurodegenerative diseases, such as Alzheimer’s disease (Akiyama et al., 2000). Similarly, expression of bacterial amyloid fibrils during infection is associated with the induction of inflammatory responses (Bian et al., 2000, Bian et al., 2001, Tukel et al., 2005, Tukel et al., 2009). Recent research on the mechanisms by which amyloids initiate innate immune responses has revealed some diversity. For example, receptors implicated in inducing responses to β-amyloid from plaques of Alzheimer’s patients include Toll-like receptor (TLR)2 (Richard et al., 2008, Tukel et al., 2009, Reed-Geaghan et al., 2009, Udan et al., 2008, Jana et al., 2008), TLR4 (Reed-Geaghan et al., 2009, Tang et al., 2008, Udan et al., 2008) and a TLR4/TLR6 receptor complex (Stewart et al.). Acute phase serum amyloid A, a host amyloid, stimulates innate responses through TLR2 (He et al., 2009, Chen et al.), while proinflammatory cytokine expression is further enhanced when cells express a TLR2/TLR1 receptor complex (Cheng et al., 2008). The multitude of different receptors and receptor complexes implicated in mediating responses to host amyloids illustrates that it is not straightforward to predict by which mechanisms bacterial amyloids stimulate innate immunity.
Recent studies show that responses to amyloid fibrils formed by the S. Typhimurium CsgA protein are mediated through TLR2 (Tukel et al., 2005, Tukel et al., 2009). Since both TLR1 and TLR6 have been implicated as co-receptors for different host amyloids (Stewart et al., Cheng et al., 2008), we explored the possibility that TLR2 forms functional hetero-dimers with TLR1 or TLR6 to mediate responses against bacterial amyloid fibrils formed by the CsgA protein. In addition to amyloid fibrils, TLR2 mediates responses against other pathogen associated molecular patterns (PAMPs) present in bacterial cells. For example, lipoproteins of Gram-negative bacteria contain conserved tri-acylated cysteines at their N termini (Hantke & Braun, 1973), which stimulate innate immune responses through a receptor complex containing TLR2 and TLR1 (Takeuchi et al., 2002). Lipoproteins of Mycoplasma spp. contain di-acylated cysteines (Shibata et al., 2000), a conserved structure recognized by TLR2 cooperatively with TLR6 (Takeuchi et al., 2001). To test the biological relevance of recognizing bacterial amyloids, we investigated whether formation of amyloid fibrils contributes significantly to TLR2-mediated responses against whole bacterial cells.
Results
Cooperativity between TLR1 and TLR2 in initiating responses to CsgA
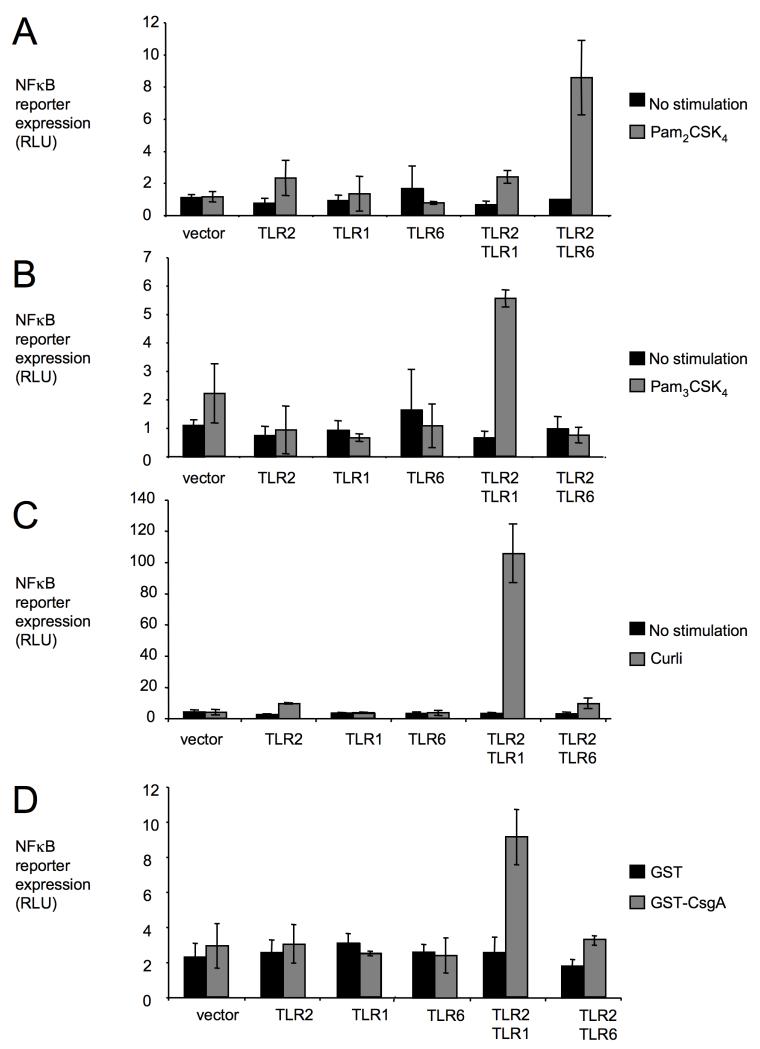
TLR2 is necessary for initiating responses against amyloids in HEK293 cells (Tukel et al., 2005, Tukel et al., 2009), a cell line intrinsically producing TLR1 and TLR6 (Buwitt-Beckmann et al., 2006, Kurt-Jones et al., 2004). To determine whether TLR2 cooperates with other receptors to initiate these responses, transfection experiments were performed with human cervical cancer (HeLa) cells, which lack TLR expression. HeLa cells carrying a NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells)-dependent luciferase reporter fusion were transfected transiently with the empty cloning vector (mock transfection) alone or in combination with constructs encoding human TLR1, human TLR6 or human TLR2 (Figure 1). Stimulation with di-acylated synthetic lipopeptide (Pam2CSK4) resulted in marked induction of the NFκB luciferase reporter only when HeLa cells were transfected with both TLR2 and TLR6 (Figure 1A). In contrast, stimulation with tri-acylated synthetic lipopeptide (Pam3CSK4) induced the NFκB luciferase reporter only when HeLa cells were transfected with both TLR2 and TLR1 (Figure 1B). These data validated our transfection approach and confirmed that tri-acylated lipopeptides stimulate a receptor complex containing TLR2 and TLR1 (Takeuchi et al., 2002), while di-acylated lipopeptides are recognized by TLR2 cooperatively with TLR6 (Takeuchi et al., 2001).
Figure 1. Transfection of HeLa cells suggests that responses to curli require an interaction between TLR2 and TLR1.
HeLa cells carrying a NFκB luciferase reporter were mock-transfected (vector) or transfected with the indicated human TLRs. Cells were stimulated with synthetic bi-acylated lipopeptide (Pam2CSK4) (A), synthetic tri-acylated lipopeptide (Pam3CSK4) (B), purified curli fibrils (C) or GST-CsgA fusion protein (D). Non stimulated cells (A, B and C) or cells stimulated with GST protein (D) served as negative controls. Activity of the NFκB luciferase reporter was monitored by measuring relative luminescence units (RLU). Bars represent averages from at least three independent experiments ± standard error.
To determine whether TLR2 cooperates with other receptors to mediate responses to amyloids present in bacterial biofilm, curli fibrils were purified from S. Typhimurium biofilm and used for stimulation of HeLa cells. Transfection with TLR2 alone did not confer responsiveness to curli, suggesting that TLR2 alone was not sufficient for mediating responses to amyloid fibrils purified from bacterial biofilm (Figure 1C). A marked induction of the NFκB luciferase reporter was observed only with HeLa cells transfected with both TLR1 and TLR2. The main component of curli fibrils is the CsgA protein (Olsen et al., 1998). To further investigate the responsiveness of HeLa cells to curli, a fusion between Gluthathione S transferase (GST) and the S. Typhimurium CsgA protein was affinity purified from the E. coli cytosol. Contamination with lipoprotein cannot account for the agonist activity of this fusion protein, because TLR2-mediated responses to GST-CsgA, but not to Pam3CSK4, can be abrogated by pretreatment with protease K (Tukel et al., 2005). Stimulation with GST-CsgA induced expression of the NFκB luciferase reporter only in HeLa cells transfected with both TLR1 and TLR2 (Figure 1D). HeLa cells did not respond to control stimulation with GST protein purified from E. coli by the same method. Collectively, these data suggested that a TLR1/TLR2 receptor complex is sufficient for mediating responses to the CsgA protein from curli fibrils.
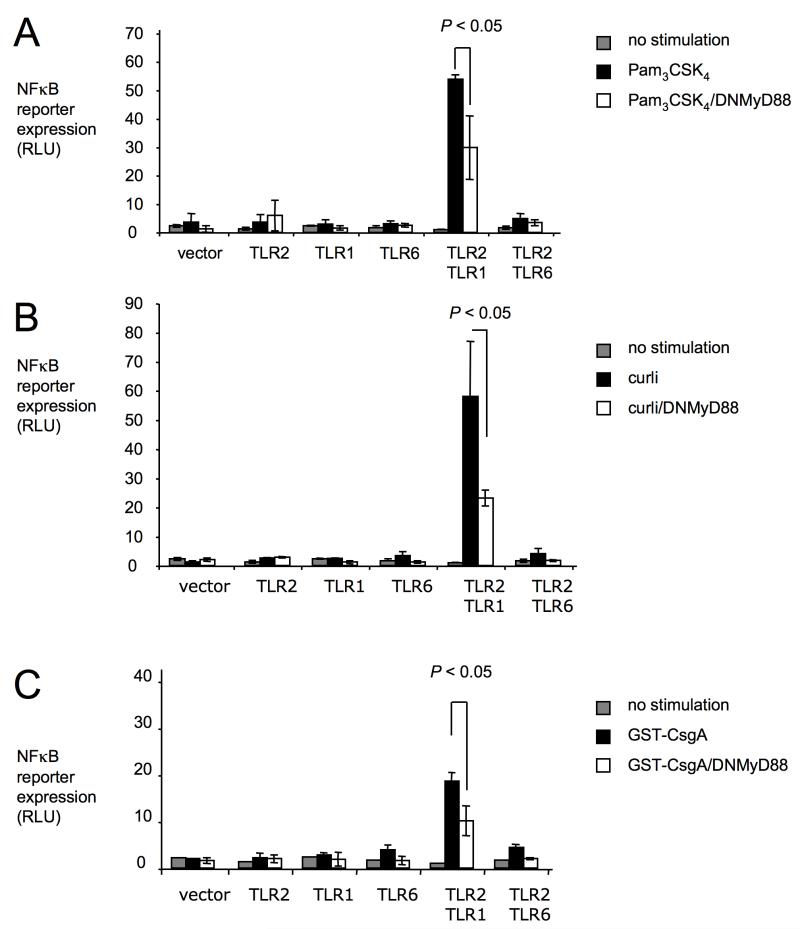
TLR1 and TLR2 initiate signaling cascades leading to NFκB translocation into the nucleus by engaging an intracellular adaptor protein, termed myeloid differentiation primary response protein 88 (MyD88) (Underhill et al., 1999, Wang et al., 2001). In HeLa cells transfected with both TLR1 and TLR2, expression of a dominant negative form of MyD88 resulted in a significantly (P < 0.05) blunted expression of the NFκB luciferase reporter in response to stimulation with Pam3CSK4, purified curli fibrils or GST-CsgA protein (Figure 2). Our data were consistent with a role of MyD88 in initiating TLR1/TLR2-mediated responses to tri-acylated lipopeptide and to CsgA.
Figure 2. Responses to curli are MyD88-dependent.
HeLa cells carrying a NFκB luciferase reporter were mock-transfected (vector) or transfected with the indicated human TLRs. In some cases, cells were transfected in addition with a dominant negative form of MyD88 (DNMyD88, open bars). Cells were stimulated with synthetic tri-acylated lipopeptide (Pam3CSK4) (A), purified curli fibrils (B) or GST-CsgA fusion protein (C). Non stimulated cells served as negative controls. Activity of the NFκB luciferase reporter was monitored by measuring RLU. Bars represent averages from at least three independent experiments ± standard error.
TLR1 and TLR2 are necessary for responses to CsgA in human macrophage-like cells
While transfection of HeLa cells represents a convenient model for investigating the contribution of individual TLRs to host responses (Figure 1), drawbacks of this approach include artificial levels of receptor expression and the questionable relevance of using cervical cancer cells to study responses to E. coli or S. Typhimurium. We therefore investigated responses to curli fibrils using THP-1 cells, a human macrophage-like cell line naturally expressing TLR1 and TLR2.
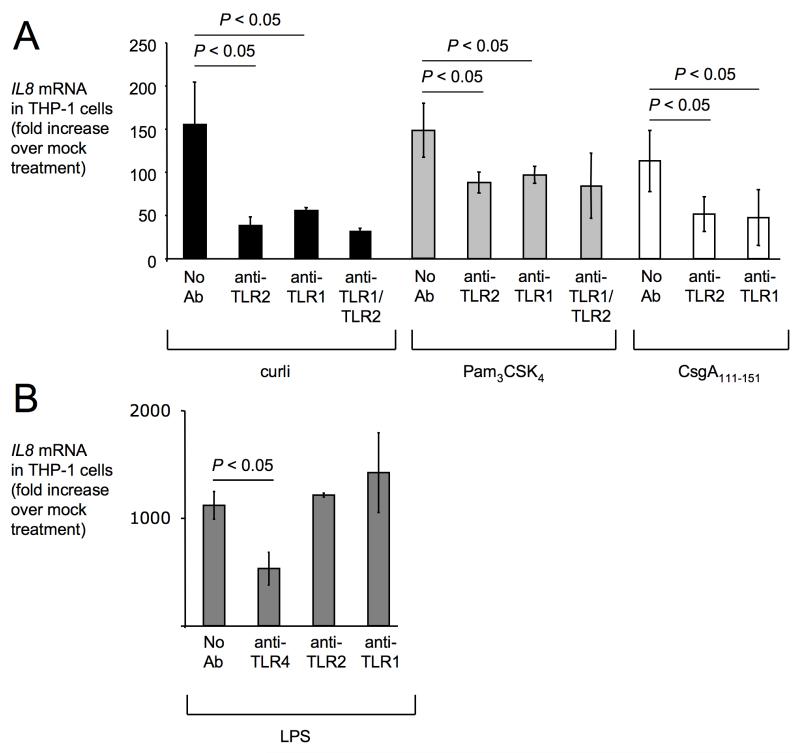
Stimulation of THP-1 cells with purified curli fibrils resulted in increased mRNA levels of IL8, the gene encoding CXCL8 (formerly known as interleukin-8) (Figure 3A). IL8 mRNA levels were significantly (P < 0.05) reduced when the stimulation was performed in the presence of blocking anti-TLR2 and/or anti-TLR1 antibodies. These data were consistent with the idea that responses to curli fibrils are mediated though TLR1/TLR2 in THP-1 cells. Similarly, THP-1 cells responded to stimulation with synthetic tri-acylated lipopeptide (Pam3CSK4) by producing increased mRNA levels of IL8. Induction of IL8 expression by Pam3CSK4 was significantly (P < 0.05) blunted when stimulation was performed in the presence of blocking anti-TLR2 and/or anti-TLR1 antibodies. These data were consistent with previous reports implicating a TLR1/TLR2 receptor complex in responses to tri-acylated lipoproteins (Takeuchi et al., 2002). Next, THP-1 cells were stimulated with a synthetic peptide containing residues 111-151 of S. Typhimurium CsgA (CsgA111-151), a region of the protein involved amyloid formation and sufficient for stimulating TLR2-mediated responses (Tukel et al., 2009). Compared to mock infected cells, expression of IL8 was markedly upregulated in THP-1 cells upon stimulation with CsgA111-151. Incubation with blocking anti-TLR1 or anti-TLR2 antibodies significantly (P < 0.05) blunted IL8 expression in response to CsgA111-151. In contrast, anti-TLR2 and anti-TLR1 antibodies did not block induction of IL8 mRNA THP-1 cells treated with lipopolysaccharide (LPS) (Figure 3B). The use of the synthetic Pam3CSK4 and CsgA111-151 peptides (Figure 3A) excluded the possibility that responses of THP-1 cells were due to contamination with other pathogen associated molecular patterns (PAMPs). We conclude that the TLR1/TLR2 receptor complex mediates responses against both tri-acylated lipoprotein and curli amyloid fibrils.
Figure 3. Responses to curli in human macrophage-like (THP-1) cells are mediated through TLR1 and TLR2.
(A) THP-1 cells were stimulated with purified curli fibrils (black bars), synthetic tri-acylated lipopeptide (Pam3CSK4) (gray bars), or synthetic CsgA111-151 peptide (open bars) in the presence or absence (No Ab) of blocking antibodies against TLR2 (anti-TLR2), TLR1 (anti-TLR1) or a combination of anti-TLR1 and anti-TLR2 antibodies (anti-TLR1/TLR2). Mock-treated THP-1 cells served as a negative control. (B) THP-1 cells were stimulated with LPS (gray bars) in the presence or absence of blocking antibodies against TLR4 (anti-TLR4), TLR2 or TLR1. Expression of IL8 in (A) and (B) was determined by quantitative real-time PCR. Data are expressed as fold increases over mRNA levels detected in mock-treated cells. Bars represent averages from at least three independent experiments ± standard error.
TLR2-mediated responses to whole bacterial cells are markedly influenced by expression of curli fibrils
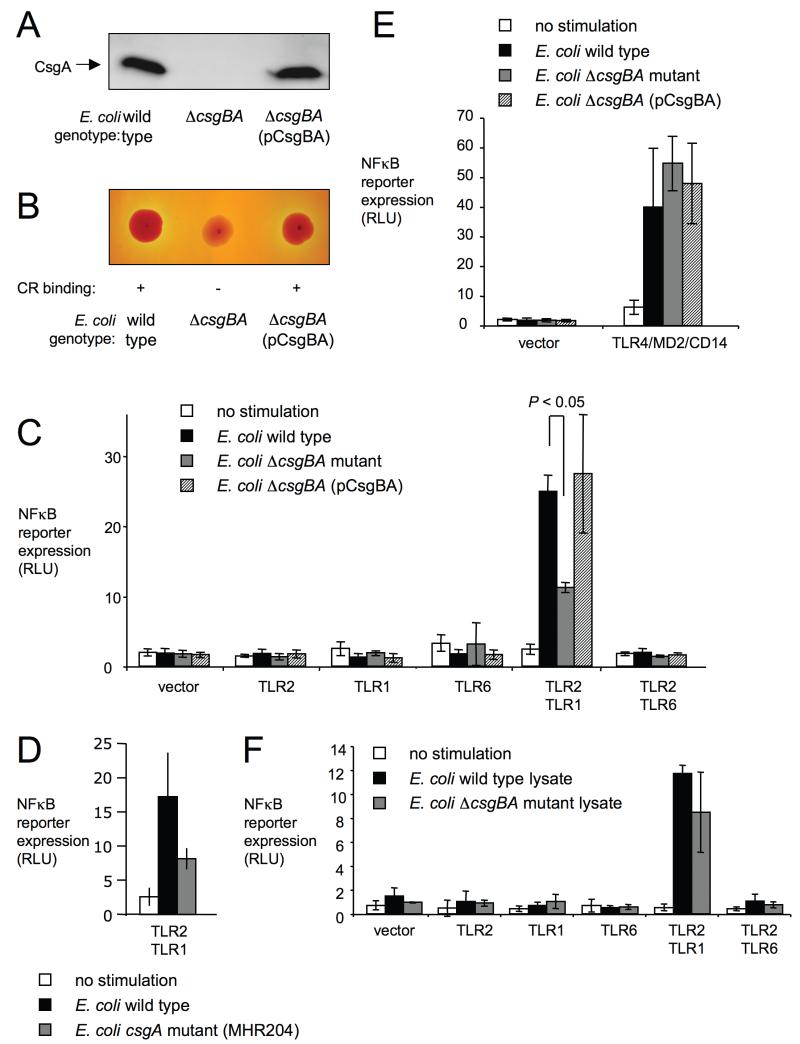
In addition to curli fibrils, intact E. coli cells contain tri-acylated lipoproteins (Hantke & Braun, 1973), which stimulate responses through a TLR2/TLR1 receptor complex (Takeuchi et al., 2002). Due to the presence of multiple agonists for TLR2/TLR1, the biological relevance of curli in mediating responses to whole bacterial cells is not apparent from stimulating host cells with purified ligands (Figure 1). To address this issue, we analyzed responses to the E. coli wild type (MC4100), an isogenic csgBA mutant (LSR13) and the csgBA mutant complemented with the cloned csgBA genes from S. Typhimurium (LSR13[pCsgBA]). Expression of CsgA was detected by Western blot in whole cell extracts from the E. coli wild type (MC4100) and the complemented csgBA mutant (LSR13[pCsgBA]), but not in the csgBA mutant (LSR13) (Figure 4A). Biofilm formation of the E. coli wild type (MC4100) on agar plates results in secretion of extracellular matrix, which contains curli amyloid fibrils (Hammar et al., 1995). Bacterial colonies formed under this growth condition bind congo red, a dye which stains amyloid deposits (Hammar et al., 1995). Consistent with the elaboration of curli fibrils, colonies of the E. coli wild type (MC4100) and the complemented csgBA mutant (LSR13[pCsgBA]) bound congo red (Figure 4B). In contrast, colonies of the csgBA mutant did not exhibit congo red binding.
Figure 4. Curli contribute to recognition of whole E. coli cells through TLR1/TLR2.
(A) Detection of CsgA expression in the indicated E. coli strains using Western blot with rabbit anti-CsgA serum. (B) Congo red binding activity of bacterial colonies formed by the indicated E. coli strains. (C, D, E and F) HeLa cells carrying a NFκB luciferase reporter were mock-transfected (vector) or transfected with the indicated pathogen recognition receptors. Cells were stimulated with the indicated E. coli strains (C, D and E) or with whole cell lysates of the indicated E. coli strains (F). Non-stimulated cells served as negative controls. Activity of the NFκB luciferase reporter was monitored by measuring RLU. Bars represent averages from at least three independent experiments ± standard error.
Stimulation with whole bacterial cells induced expression of the NFκB luciferase reporter only in HeLa cells transfected with TLR1/TLR2, but not is cells transfected with either TLR1, TLR2, TLR6 or TLR2/TLR6 (Figure 4C). These data suggested that E. coli cells contain agonists for TLR1/TLR2, but not for any of the other receptors tested. Expression of the NFκB luciferase reporter was significantly (P < 0.05) higher after stimulation with the E. coli wild type (MC4100) than with the csgBA mutant (LSR13), which illustrated that curli production contributed markedly to TLR1/TLR2-mediated responses induced by whole bacterial cells. Complementation (LSR13[pCsgBA]) restored the level of NFκB luciferase reporter expression to levels observed after stimulating cells with E. coli wild type (MC4100). A single mutation in csgA (E. coli strain MHR204) also reduced NFκB luciferase reporter expression in HeLa cells transfected with TLR1/TLR2 (Figure 4D).
Expression of the NFκB luciferase reporter induced by stimulation with the csgBA mutant was significantly higher (P < 0.05) than expression in mock-stimulated HeLa cells (Figure 4C). This stimulation of the NFκB luciferase reporter by the csgBA mutant was likely due the induction of TLR1/TLR2-mediated responses by other ligands, such as tri-acylated lipoproteins (Hantke & Braun, 1973). We next investigated responses to LPS, a TLR4 agonist that was predicted to be expressed equally by all three bacterial strains. The E. coli wild type (MC4100), the csgBA mutant (LSR13) and the complemented csgBA mutant (LSR13[pCsgBA]) induced similar expression levels of the NFκB luciferase reporter in HeLa cells transfected with TLR4, MD2 and CD14 (Figure 4E). Thus, the reduced ability of the csgBA mutant to induce the NFκB luciferase reporter in HeLa cells transfected with TLR1/TLR2 (Figure 4C) was not due to a general defect of the bacterial strain in inducing host responses in HeLa cells (Figure 4E), but to its inability to produce curli (Figure 4A and B).
The important role of amyloids in stimulating TLR1/TLR2-mediated responses against whole bacterial cells may relate to the accessibility of curli on the cell surface. In contrast, the incorporation of other TLR1/TLR2 ligands into the cell envelope may make these less accessible to pattern recognition receptors in intact bacterial cells. To test this idea, HeLa cells were stimulated with whole bacterial cell lysates to release internal PAMPs. Whole cell lysates of the E. coli wild type (MC4100) and the csgBA mutant (LSR13) elicited similar NFκB luciferase reporter expression levels (Figure 3F), suggesting that after a substantial release of other PAMPs from bacterial cells, curli is no longer the major ligand responsible for stimulating the TLR1/TLR2 receptor complex. Collectively, our results provided evidence that curli fibrils contributed markedly to TLR1/TLR2-mediated responses to intact bacterial cells in vitro.
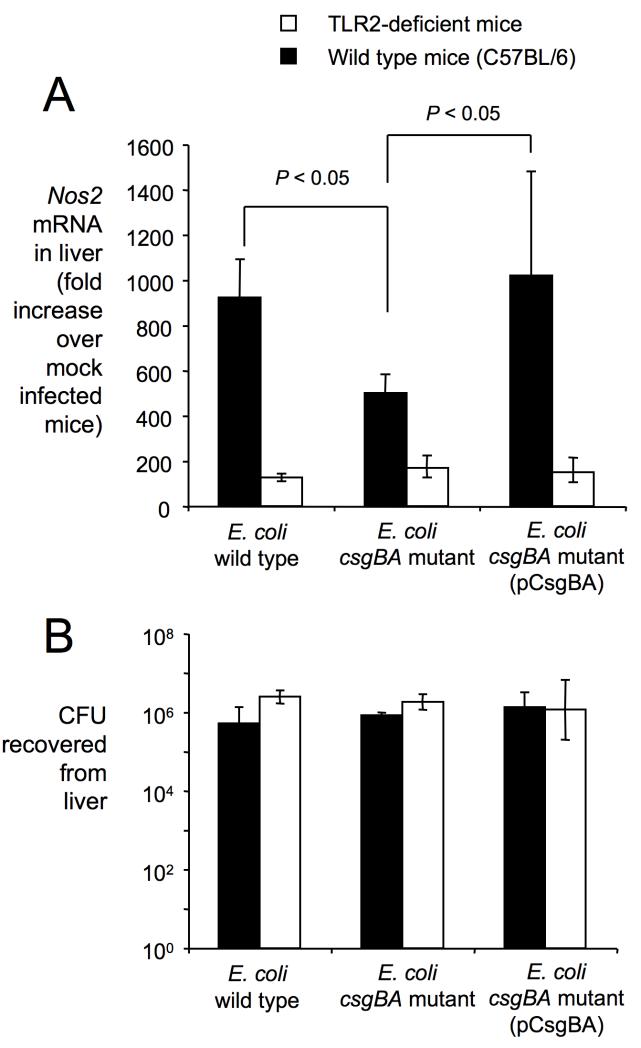
While HeLa cells are not able to kill bacterial cells, which would result in the release of PAMPs, lyses of E. coli cells may occur in vivo during an infection. To determine whether curli are an important contributor to TLR2-mediated responses generated by bacteria in vivo, mice were injected intraperitoneally with the E. coli wild type (MC4100), the csgBA mutant (LSR13) or the complemented csgBA mutant (LSR13[pCsgBA]) and transcript levels of Nos2, encoding inducible nitric oxide synthase (iNOS), were quantified 8 hours after infection using quantitative real-time PCR (Figure 5). Expression of Nos2 in the liver was significantly (P < 0.05) higher after stimulation with the E. coli wild type (MC4100) than with the csgBA mutant (LSR13) (Figure 5A) although both bacterial strains were recovered in similar numbers from this organ (Figure 5B). Reduced Nos2 production elicited by the csgBA mutant could be complemented in vivo by introducing the cloned csgBA genes from S. Typhimurium (LSR13[pCsgBA]). Differences between the E. coli wild type (MC4100) and the csgBA mutant (LSR13) in inducing Nos2 expression were TLR2-dependent, because both strains elicited similar Nos2 mRNA levels when the experiment was repeated with TLR2-deficient mice (Figure 5A). These data show that curli fibrils contribute significantly to TLR2-mediated responses against bacterial cells in vivo.
Figure 5. Contribution of curli and TLR2 to inducing Nos2 expression during E. coli sepsis.
(A) Nos2 mRNA levels observed in the liver 8 hours after intraperitoneal infection of wild type mice (black bars) or TLR2-deficient mice (open bars) with 108 colony forming units (CFU) of the indicated E. coli strains. Each bar represents the average fold increases in Nos2 expression of E. coli-infected mice (n=4) compared to mock-infected mice (n=4) ± standard error. Statistical significance of differences is indicated by a bracket. (B) Bacterial numbers recovered from the liver 8 hours after intraperitoneal infection of wild type mice (black bars) or TLR2 deficient mice (open bars) with 108 colony forming units (CFU) of the indicated E. coli strains.
Discussion
Biofilms of E. coli and S. Typhimurium contain extracellular amyloid deposits, termed curli (Prigent-Combaret et al., 2000, Vidal et al., 1998, Romling et al., 2003), which can elicit inflammatory responses in the host (Bian et al., 2000, Bian et al., 2001, Tukel et al., 2005, Tukel et al., 2009). While TLR2 is necessary for initiating these responses (Tukel et al., 2005, Tukel et al., 2009), here we show that this receptor was not sufficient for initiating inflammatory gene expression after stimulation with curli. Instead, a receptor complex containing TLR1 and TLR2 was sufficient for mediating responses to curli amyloid fibrils in HeLa cells. In addition to curli, TLR2 has been implicated in recognizing other PAMPs present in E. coli, including tri-acylated lipoproteins (Takeuchi et al., 2002), peptidoglycan (Schwandner et al., 1999) and B subunits of type II heat-labile enterotoxins (Hajishengallis et al., 2005, Liang et al., 2007). The E. coli isolate used in this study does not express a heat-labile enterotoxin. Furthermore, ultra pure peptidoglycan preparations do not activate TLR2, thus calling into question whether peptidoglycan is indeed a genuine TLR2-ligand (Travassos et al., 2004). Thus, bacteria used in this study expressed at least two classes of TLR1/TLR2 ligands, curli amyloid fibrils and tri-acylated lipoproteins. The latter group consists of 96 distinct lipoproteins, some of which, such as the Braun lipoprotein (Hantke & Braun, 1973), are highly expressed in E. coli (Brokx et al., 2004).
Despite the presence of multiple TLR2 ligands, an inability to produce curli fibrils markedly reduced the ability of HeLa cells transfected with TLR1/TLR2 to respond to stimulation with intact bacterial cells. Elaboration of curli fibrils on the surface of E. coli induces Nos2 expression in a mouse sepsis model (Bian et al., 2001) and here we show that this effect was TLR2-dependent. Curli fibrils might serve as an important PAMP because these structures are secreted, thereby being more readily accessible to detection by pattern recognition receptors than lipoproteins, which are buried in the cell envelope of intact bacterial cells. The importance of bacterial amyloids as PAMPs is further supported by work in animal models, where secretion of curli fibrils contributes significantly to host responses against E. coli or S. Typhimurium (Bian et al., 2001, Tukel et al., 2005, Tukel et al., 2009). Although expression of curli fibrils is induced in E. coli at ambient temperature (26°C) in rich medium, expression at body temperature (37°C) can be observed after culturing bacteria under conditions of iron starvation, which is encountered in the mammalian host (Romling et al., 1998). Serum from mice infected with S. Typhimurium or patients recovering from E. coli sepsis contain anti—CsgA antibodies, which provides indirect evidence for in vivo expression of curli fibrils (Humphries et al., 2005, Bian et al., 2000). Collectively, these studies identify bacterial amyloid from biofilm material as a significant PAMP recognized by the innate immune system. Amyloid fibrils are not only produced by members of the Enterobacteriaceae, but are also commonly present in biofilm material from bacteria belonging to the Firmicutes, Bacteroides and Actinobacteria phyla (Larsen et al., 2007, Jordal et al., 2009), whose representatives are dominant constituents of the intestinal microbiota. Thus recognition of amyloids is of broad significance for detection of bacterial cells by the innate immune system.
Material and Methods
Bacterial strains and plasmids
Curli producing wild type E. coli strain MC4100, an isogenic curli mutant LSR13 (csgBA mutant) and a csgA::TN105 mutant (MHR204) were kindly provided by Dr. Scott Hultgren and Dr. Matthew Chapman (Wang & Chapman, 2008)(Hammar et al., 1995). To complement curli production in strain LSR13, the csgBA genes were amplified from S. Typhimurium by using primers 5′ GGGATCCGGGTGACAGCATGAAAAACAAATTGTTA 3′ and 5′ GGAATTCTTTATTAGCGCAGACGCTAAATTAATACTGGTTAGCCGTGGC 3′. The resulting PCR product was digested with EcoRI and BamHI and ligated into low copy plasmid pWSK29 giving rise to plasmid pCsgBA.
Growth conditions for biofilm formation and purification of curli
Extracellular matrix formation was induced on T-medium plates at 28°C for 48 h (Collinson et al., 1991). Curli production was monitored by the addition of congo red to this medium to a final concentration of 20 mg/L (Hammar et al., 1995). E. coli cells were recovered from the plates in phosphate buffered-saline and O.D. 600 was adjusted to 0.5. Bacteria were either killed by the addition of 1% sodium azide for 5 min or lyzed by heat treatment at 100°C for 5 min. Na-Azide was removed by washing in PBS three times.
Curli was purified from the surface of the S. Typhimurium msbB mutant (RPW3) according to an established protocol (Collinson et al., 1991). Briefly, cells were removed from T-medium plates and lyzed by sonication followed by enzymatic digestion and preparative sodium dodecyl sulphate-gel electrophoresis (SDS-PAGE). Insoluble material (curli fibrils) retained in the well of the SDS-PAGE gel was collected after the electrophoresis.
Western Blot
Bacteria were recovered from the T-medium plates and curli was depolymerized by 90% formic acid treatment as described previously (Collinson et al., 1991). The sample was resuspended in SDS-PAGE sample buffer, boiled for 10 min and analysed by SDS-PAGE on a 15% gel. Following electrophoresis, proteins were transferred to Immobilon-P membrane (Millipore) using a Trans-Blot SD semi-dry electrophoretic transfer cell (Biorad) according to the manufacturer’s instructions. CsgA antiserum (Humphries et al., 2003) and a horse radish preoxidase conjugated goat anti-rabbit secondary antibody (Biorad) were used to detect curli expression.
Purification of GST fusion proteins
Plasmid pSW5-50, containing the csgA gene cloned in the gluthathione S transferase (GST) fusion protein vector pGEX-4T-2 was described previously (Humphries et al., 2003). Fusion proteins GST and GST-CsgA were purified from E. coli strains DH5α(pGEX-4T-2) and DH5α(pSW5-50), respectively, using glutathione sepharose (Amersham Pharmacia) columns as described previously(Humphries et al., 2003). The protein concentration in each sample was determined by Bradford assay (Ausubel et al., 1994).
Tissue culture cells and reagents
The HeLa 57A cell line stably transfected with a NF-kB luciferase reporter was kindly provided by Dr. R.T. Hay (the Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, UK). HeLa 57A cells were maintained in DMEM containing 10% FBS at 37°C in 5% CO2 atmosphere. Human monocytic cell line THP-1 was kindly provided by Dr. Vernon Tesh. THP-1 cells were maintained in RPMI containing 10% FBS and glutamine. The synthetic diacylated lipopeptide (Pam2CSK4) and synthetic triacylated lipopeptide (Pam3CSK4) were purchased from InvivoGen.
Transfection of HeLa cells
HeLa 57A cells were seeded at a density of 1×104 cells per well in 96-well tissue culture plates containing DMEM+10% FBS 24 hours prior to transfection. Then cells were transiently transfected using ExGen 500 reagent (Fermentas) according to the manufacturer’s instructions. Vectors carrying mTLR4, mMD2, mCD14, hTLR2, hTLR1, hTLR6 and LacZ (Keestra et al., 2007, Keestra & van Putten, 2008) were added in various combinations to a total amount of 150 ng transfected plasmid DNA. Human dominant negative MyD88 (DNMyD88) vector was purchased from Invivogen and added to the transfection reaction. In all transfections, the LacZ vector was used to normalize the transfection efficiency. Luciferase assays were performed at 48h post-transfection.
Luciferase Assay
The Luciferase Assay System (containing luciferase reagent and reporter lysis buffer) and β-galactosidase Enzyme Assay System were purchased from Promega. TLR signaling was assessed using the NF-kB luciferase reporter system. For luciferase assays, cells were stimulated with purified curli (2.5 μg/well), GST (2.5 μg/well), GST-CsgA (2.5 μg/well), Pam3CSK4 (0.05 μg/well), or Pam2CSK4 (0.05 μg/well) for 6 hours prior to determining luciferase activity. For luciferase assays with whole bacterial cells, HeLa cells were stimulated with 4×105 colony forming units (CFU)/well of the indicated bacterial strains for 4 hours prior to determining luciferase activity. To determine luciferase activity, cells were washed three times with PBS, and lysed with 60μl of reporter lysis buffer. Luciferase activity was measured using a multimode plate reader (Analyst GT, Molecular Devices). Luciferase values were adjusted to β-galactosidase values to normalize the efficiency of transfection. Results were expressed in relative luciferase units (RLU).
Stimulation of THP-1 cells
To differentiate monocytes into macrophages, THP-1 cells were stimulated with 50ng/ml phorbol myristic acid (PMA) (Sigma) and seeded at a density of 5x105 cells per well in 24 well plates. Following a 48 hour incubation at 37°C in 5% CO2 atmosphere, PMA containing media was removed. Macrophages were washed twice with PBS, and replaced with PMA free medium. Fresh media was added daily (500μl), and cell assays were performed after 4 days of incubation.
Blocking anti-human TLR2 (T2.5) and TLR1 (GD2.F4), antibodies were purchased from eBioscience. Macrophages were treated with 10μg per well of antibody or medium (control) 1 hour prior to stimulation. Purified curli fibrils, synthetic CsgA111-151 (5μM) or Pam3CSK4 (0.1μg/ml) were added to the wells for 4 hours. After stimulation, RNA was extracted to determine Il-8 mRNA levels by Real Time PCR.
Real Time PCR
RNA samples were prepared using TRIzol reagent (Molecular Research center). Real time PCR was performed using the SYBR Green method (Applied Biosystems, CA) according to the manufacturer’s instructions. Reverse transcription of total RNA (1μg) was performed at 48°C for 30 minutes. Real time PCR was performed for each cDNA sample (5μl/reaction) in duplicate using gene specific primers (human GAPDH and human IL8) (Stylianou et al., 2002) and an ABI Prism 77000 thermocycler (95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute). Real time PCR amplification of GAPDH transcripts was used to normalize the cDNA concentrations of the other gene transcripts. Results were given as 2−ΔCT±S, where S is the standard deviation.
Animal Experiments
For mouse experiments, 4 to 6 week old female C57BL/6 mice or congenic TLR2-deficient mice (Takeuchi et al., 1999) were purchased from Jackson Laboratories. Groups of 4 mice were intraperitoneally infected with 1×108 CFU in PBS or sterile PBS (mock infection). At 8 hours after infection, mice where sacrificed a sample of the liver was collected from each mouse, immediately snap-frozen in liquid nitrogen and stored at −80°C. RNA was extracted from snap-frozen tissue with TriReagent (Molecular Research Center) according to the instruction of the manufacturer. 1000 ng of RNA from each sample was reverse transcribed in 0.05 ml volume (Taqman reverse transcription reagent, Applied Biosystems). 0.005 ml of cDNA was used for each Real-Time reaction. Real-time PCR was performed using SYBR Green (Applied Biosystems) and the 7900HT Fast Real-Time PCR System. The data were analyzed using the comparative Ct method (Applied Biosystems). Fold-increases in cytokine expression in infected mice were calculated relative to the average level of the respective cytokine in four mock-infected mice. The primers for Gapdh and Nos2 have been described previously (Wilson et al., 2008, Roux et al., 2007).
Acknowledgements
Work in AJB’s laboratory is supported by Public Health Service grants AI040124, AI044170, AI073120, AI076246, AI079173 and AI088122. CT is supported by Scientist Development Grant 0835248N from the American Heart Association.
Literature
- Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WS, Hampel H, Hull M, Landreth G, Lue L, Mrak R, Mackenzie IR, McGeer PL, O’Banion MK, Pachter J, Pasinetti G, Plata-Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss-Coray T. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. J. Wiley & Sons; 1994. [Google Scholar]
- Bian Z, Brauner A, Li Y, Normark S. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- Bian Z, Yan ZQ, Hansson GK, Thoren P, Normark S. Activation of inducible nitric oxide synthase/nitric oxide by curli fibers leads to a fall in blood pressure during systemic Escherichia coli infection in mice. J Infect Dis. 2001;183:612–619. doi: 10.1086/318528. [DOI] [PubMed] [Google Scholar]
- Brokx SJ, Ellison M, Locke T, Bottorff D, Frost L, Weiner JH. Genome-wide analysis of lipoprotein expression in Escherichia coli MG1655. Journal of bacteriology. 2004;186:3254–3258. doi: 10.1128/JB.186.10.3254-3258.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buwitt-Beckmann U, Heine H, Wiesmuller KH, Jung G, Brock R, Akira S, Ulmer AJ. TLR1- and TLR6-independent recognition of bacterial lipopeptides. The Journal of biological chemistry. 2006;281:9049–9057. doi: 10.1074/jbc.M512525200. [DOI] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Song Z, Willett MH, Heine S, Yung RC, Liu MC, Groshong SD, Zhang Y, Tuder RM, Moller DR. Serum Amyloid A Regulates Granulomatous Inflammation in Sarcoidosis through Toll-like Receptor-2. American journal of respiratory and critical care medicine. 181:360–373. doi: 10.1164/rccm.200905-0696OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, He R, Tian J, Ye PP, Ye RD. Cutting edge: TLR2 is a functional receptor for acute-phase serum amyloid A. J Immunol. 2008;181:22–26. doi: 10.4049/jimmunol.181.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. Journal of bacteriology. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Tapping RI, Martin MH, Nawar H, Lyle EA, Russell MW, Connell TD. Toll-like receptor 2 mediates cellular activation by the B subunits of type II heat-labile enterotoxins. Infection and immunity. 2005;73:1343–1349. doi: 10.1128/IAI.73.3.1343-1349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Molecular microbiology. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hantke K, Braun V. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. European journal of biochemistry/FEBS. 1973;34:284–296. doi: 10.1111/j.1432-1033.1973.tb02757.x. [DOI] [PubMed] [Google Scholar]
- He RL, Zhou J, Hanson CZ, Chen J, Cheng N, Ye RD. Serum amyloid A induces G-CSF expression and neutrophilia via Toll-like receptor 2. Blood. 2009;113:429–437. doi: 10.1182/blood-2008-03-139923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AD, DeRidder S, Bäumler AJ. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect. Immun. 2005;73:5329–5338. doi: 10.1128/IAI.73.9.5329-5338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S, Figueiredo J, Khare S, Nunes J, Adams LG, Tsolis RM, Bäumler AJ. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Molecular microbiology. 2003;48:1357–1376. doi: 10.1046/j.1365-2958.2003.03507.x. [DOI] [PubMed] [Google Scholar]
- Jana M, Palencia CA, Pahan K. Fibrillar amyloid-beta peptides activate microglia via TLR2: implications for Alzheimer’s disease. J Immunol. 2008;181:7254–7262. doi: 10.4049/jimmunol.181.10.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordal PB, Dueholm MS, Larsen P, Petersen SV, Enghild JJ, Christiansen G, Hojrup P, Nielsen PH, Otzen DE. Widespread abundance of functional bacterial amyloid in mycolata and other gram-positive bacteria. Applied and environmental microbiology. 2009;75:4101–4110. doi: 10.1128/AEM.02107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keestra AM, de Zoete MR, van Aubel RA, van Putten JP. The central leucine-rich repeat region of chicken TLR16 dictates unique ligand specificity and species-specific interaction with TLR2. J Immunol. 2007;178:7110–7119. doi: 10.4049/jimmunol.178.11.7110. [DOI] [PubMed] [Google Scholar]
- Keestra AM, van Putten JP. Unique properties of the chicken TLR4/MD-2 complex: selective lipopolysaccharide activation of the MyD88-dependent pathway. J Immunol. 2008;181:4354–4362. doi: 10.4049/jimmunol.181.6.4354. [DOI] [PubMed] [Google Scholar]
- Kurt-Jones EA, Sandor F, Ortiz Y, Bowen GN, Counter SL, Wang TC, Finberg RW. Use of murine embryonic fibroblasts to define Toll-like receptor activation and specificity. Journal of endotoxin research. 2004;10:419–424. doi: 10.1179/096805104225006516. [DOI] [PubMed] [Google Scholar]
- Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environmental microbiology. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- Liang S, M Wang,, Tapping RI, Stepensky V, Nawar HF, Triantafilou M, Triantafilou K, Connell TD, Hajishengallis G. Ganglioside GD1a is an essential coreceptor for Toll-like receptor 2 signaling in response to the B subunit of type IIb enterotoxin. The Journal of biological chemistry. 2007;282:7532–7542. doi: 10.1074/jbc.M611722200. [DOI] [PubMed] [Google Scholar]
- Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Olsen A, Wick MJ, Morgelin M, Bjorck L. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infection and immunity. 1998;66:944–949. doi: 10.1128/iai.66.3.944-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environmental microbiology. 2000;2:450–464. doi: 10.1046/j.1462-2920.2000.00128.x. [DOI] [PubMed] [Google Scholar]
- Reed-Geaghan EG, Savage JC, Hise AG, Landreth GE. CD14 and toll-like receptors 2 and 4 are required for fibrillar A{beta}-stimulated microglial activation. J Neurosci. 2009;29:11982–11992. doi: 10.1523/JNEUROSCI.3158-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard KL, Filali M, Prefontaine P, Rivest S. Toll-like receptor 2 acts as a natural innate immune receptor to clear amyloid beta 1-42 and delay the cognitive decline in a mouse model of Alzheimer’s disease. J Neurosci. 2008;28:5784–5793. doi: 10.1523/JNEUROSCI.1146-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Z Bian,, M Hammar,, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. Journal of bacteriology. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschape H. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int J Med Microbiol. 2003;293:273–285. doi: 10.1078/1438-4221-00268. [DOI] [PubMed] [Google Scholar]
- Roux CM, Rolan HG, Santos RL, Beremand PD, Thomas TL, Adams LG, Tsolis RM. Brucella requires a functional Type IV secretion system to elicit innate immune responses in mice. Cellular microbiology. 2007;9:1851–1869. doi: 10.1111/j.1462-5822.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. The Journal of biological chemistry. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- Shibata K, Hasebe A, Into T, Yamada M, Watanabe T. The N-terminal lipopeptide of a 44-kDa membrane-bound lipoprotein of Mycoplasma salivarium is responsible for the expression of intercellular adhesion molecule-1 on the cell surface of normal human gingival fibroblasts. J Immunol. 2000;165:6538–6544. doi: 10.4049/jimmunol.165.11.6538. [DOI] [PubMed] [Google Scholar]
- Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, Lacy-Hulbert A, Khoury JE, Golenbock DT, Moore KJ. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nature immunology. 11:155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou E, Yndestad A, Sikkeland LI, Bjerkeli V, Damas JK, Haug T, Eiken HG, Aukrust P, Froland SS. Effects of interferon-alpha on gene expression of chemokines and members of the tumour necrosis factor superfamily in HIV-infected patients. Clin Exp Immunol. 2002;130:279–285. doi: 10.1046/j.1365-2249.2002.01980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. International immunology. 2001;13:933–940. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- Tang SC, Lathia JD, Selvaraj PK, Jo DG, Mughal MR, Cheng A, Siler DA, Markesbery WR, Arumugam TV, Mattson MP. Toll-like receptor-4 mediates neuronal apoptosis induced by amyloid beta-peptide and the membrane lipid peroxidation product 4-hydroxynonenal. Experimental neurology. 2008;213:114–121. doi: 10.1016/j.expneurol.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Girardin SE, Philpott DJ, Blanot D, Nahori MA, Werts C, Boneca IG. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO reports. 2004;5:1000–1006. doi: 10.1038/sj.embor.7400248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tükel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Bäumler AJ. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Molecular microbiology. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- Tükel C, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Bäumler AJ. Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell host & microbe. 2009;6:45–53. doi: 10.1016/j.chom.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan ML, Ajit D, Crouse NR, Nichols MR. Toll-like receptors 2 and 4 mediate Abeta(1-42) activation of the innate immune response in a human monocytic cell line. J Neurochem. 2008;104:524–533. doi: 10.1111/j.1471-4159.2007.05001.x. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- Vidal O, Longin R, Prigent-Combaret C, C Dorel, M. Hooreman, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. Journal of bacteriology. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Dziarski R, Kirschning CJ, Muzio M, Gupta D. Micrococci and peptidoglycan activate TLR2-->MyD88-->IRAK-->TRAF-->NIK-->IKK-->NF-kappaB signal transduction pathway that induces transcription of interleukin-8. Infection and immunity. 2001;69:2270–2276. doi: 10.1128/IAI.69.4.2270-2276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. Journal of molecular biology. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Bäumler AJ. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cellular microbiology. 2008;10:876–890. doi: 10.1111/j.1462-5822.2007.01090.x. [DOI] [PubMed] [Google Scholar]