Abstract
Incidence and mortality associated with hepatocellular carcinoma (HCC) is rising throughout the world. Accurate, noninvasive biomarkers for the early detection of HCC are urgently needed to reduce worldwide morbidity and mortality related to HCC. MicroRNAs (miRNAs), 17- to 25-nucleotide noncoding RNAs that are frequently dysregulated in HCC, have shown great promise as tissue-based markers for HCC diagnosis and prognosis. Moreover, they are stably expressed in serum and urine, and these circulating microRNAs (cmiRNAs) are emerging as novel noninvasive biomarkers for the early detection and prognosis of HCC. This article summarizes the latest findings on the role of circulating miRNAs as potential minimally invasive diagnostic and prognostic biomarkers for HCC.
Keywords: circulating microRNAs (cmiRNAs), biomarkers, HCC, diagnosis and prognosis
Introduction
Primary liver cancer mainly includes hepatocellular carcinoma (HCC), cholangiocarcinoma, and hepatic angiosarcoma. Hepatocellular carcinoma (HCC) is the most common and most highly malignant hepatoma, the fifth most common cancer, and the third most common cause of cancer-related death in the world [1]. In the US, approximately 6000 new cases are diagnosed for HCC each year and HCC is not a chemosensitive tumor with development of resistance to many anticancer drugs [2]. The major risk factor for HCC is chronic viral hepatitis B or C, which accounts for 80-90% of all cases of HCC worldwide [3, 4]. Other risk factors include alcohol abuse, aflatoxin B1 or vinyl chloride exposure, primary biliary cirrhosis, diabetes, non-alcoholic fatty liver disease, and genetic disorders [5, 6]. The risk factors vary widely from country to country: in countries where hepatitis B is endemic, such as China, hepatitis B is the predominant cause of HCC [7], whereas in the United States, where hepatitis B is rare because of high vaccination rates, the major cause of HCC is cirrhosis. HCC has an overall 5-year survival rate of 5-9% from the time of clinical diagnosis, and the dismal prognosis is largely caused by late detection of the tumors [8, 9]. However, the 5-year survival rate is 69% for patients who undergo hepatectomy if the tumor is detected early, particularly when the tumor is a single nodule and smaller than 2 cm [10, 11]. Thus, early detection of HCC at a surgically resectable stage offers the best chance of survival for patients.
Sensitive and specific cancer biomarkers are essential for early detection and diagnosis of HCC, as well as for developing preventive screening and therapeutic trials. However, current diagnostic methods are insufficient for detecting HCC at early stages. MicroRNAs (miRNAs) have shown great promise as tissue-based diagnostic and prognostic markers for HCC. Moreover, they are stably expressed in serum and urine, and these circulating miRNAs (cmiRNAs) are emerging as novel non-invasive biomarkers for the early detection and prognosis of HCC. This article summarizes the latest findings on the role of circulating miRNAs as potential minimally invasive diagnostic and prognostic biomarkers for HCC.
Current Diagnostic Methods for HCC
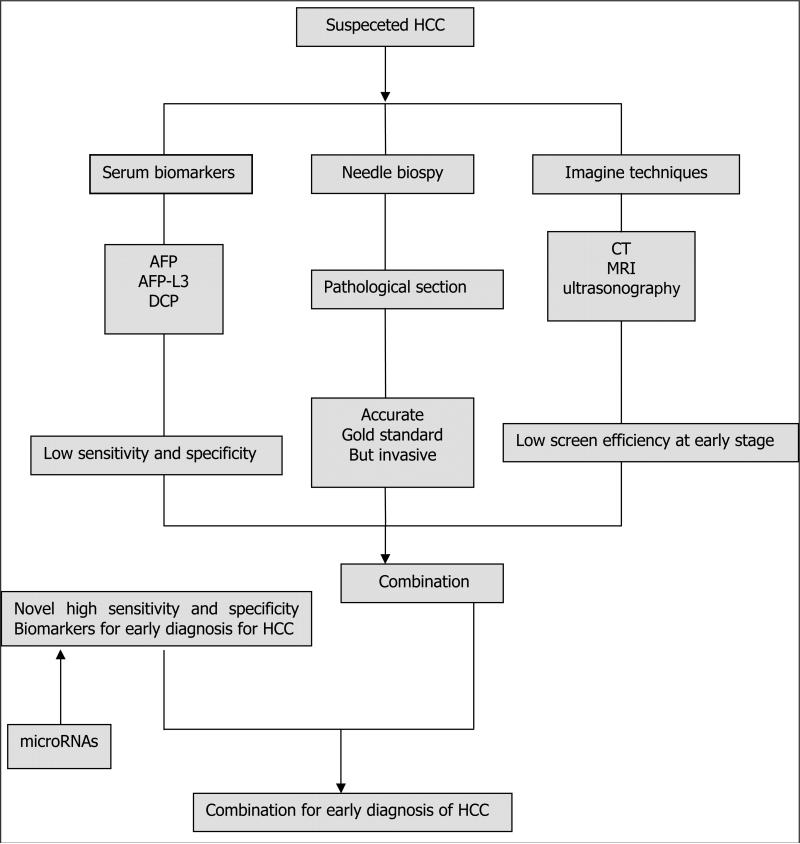
Diagnosis of HCC is usually based on imaging [abdominal ultrasonography, magnetic resonance imaging (MRI), and contrast-enhanced computed tomography (CT)] and laboratory analysis [serum α-fetoprotein (AFP) and Des-gamma carboxyprothrombin (DCP) levels], sometimes verified by biopsy results. Advances in MRI and CT have greatly improved imaging of focal hypervascular masses consistent with HCC, but few radiologists are skilled at finding tumors on MRI studies used for screening, and these procedures are costly and not readily available in developing countries. Ultrasonography can detect large lesions but fails to detect small tumors, and because this procedure is operator-dependent, the diagnostic accuracy varies.
Most doctors and investigators still depend on clinical laboratory analyses to diagnose HCC. Serum AFP and DCP levels have long been used as tumor biomarkers. Serum AFP <20 ng/mL is considered normal and AFP >400 ng/mL is generally considered positive for HCC; European Society for Medical Oncology guidelines recommend that serum AFP >400 ng/mL can be used instead of fine-needle cytologic analysis to diagnose HCC, especially in patients with liver cirrhosis [12]. However, the accuracy of AFP is modest (sensitivity: 39-65%; specificity: 76-94%). One-third of cases of early-stage HCC (tumors <3 cm) are missed using AFP analysis [13], and serum AFP levels are also elevated in patients with benign liver diseases, such as hepatitis and cirrhosis [14, 15]. Serum DCP has been identified as an effective serologic tool in early diagnosis of HCC. Circulating DCP is primarily produced by HCC tissues and is elevated in proportion to tumor size [16]. Choi et al have evaluated 2 new autoanalyzers (μTAS and Lumipulse) for DCP assay [17]. The sensitivity and specificity for μTAS were 68.3% (95% CI = 59.2-76.5%) and 95.0% (95% CI = 89.3-98.1%), respectively, and for Lumipulse, the sensitivity was 76.7% (95% CI = 68.1-83.9%) and the specificity was 92.2% (95% CI = 85.8-96.4%). The 97.5-percentile upper reference limit for μTAS in healthy individuals was 29.5 mAU/mL, and for Lumipulse, 35.0 mAU/mL [17]. However, elevated DCP activity is present in only 44-47% of HCC patients with tumors <3 cm [15].
Despite major efforts, a large proportion of individuals are still diagnosed at a stage when effective treatments are lacking [18]. Given the limitations of imaging studies and the limited sensitivity of the laboratory analyses available, the development of non-invasive biomarkers with high sensitivity and specificity that can be used for large-scale clinical investigations would be highly beneficial. Figure 1 displays the flow chart of current diagnosis for HCC.
Figure 1.
The flow chart of current diagnosis
Role of miRNAs in cancer
miRNAs are 17- to 25-nucleotide non-coding RNAs that can bind to complementary sequences in 3-untranslated regions (3’-UTR) of target mRNAs to induce degradation or translational repression. miRNAs have emerged as critical components of the complex functional pathway networks controlling important cellular processes, such as proliferation, development, differentiation, stress response, and apoptosis. Abnormal expression of miRNAs has been implicated in the oncogenic process; miRNAs have been found to function both as oncogenes and as tumor suppressor genes [19]. These deregulated miRNAs contribute to tumor initiation and progression by favoring uncontrolled proliferation and survival, promoting invasive behavior, and by regulating apoptosis, which depends on the cellular context and on their target genes.
miR-21 is unique in that its expression is elevated in most human cancers [20, 21] and promotes cell proliferation and tumor invasiveness in hepatocellular carcinomas by targeting PTEN, PDCD4, and RECK [22]. Oncogenic miR-155 was also significantly up-regulated in primary human HCCs promoted hepatocyte proliferation and tumorigenesis by increasing Wnt signaling [23, 24]. High miR-155 expression was an independent predictor of poor prognosis in HCC [25]. miR-221 and miR-222 are two highly homologous microRNAs and upregulated in several types of human tumors, which act as oncogenes or tumor suppressors, depending on tumor system [26]. Overexpression of miR-221 can accelerate hepatocyte proliferation during liver regeneration and stimulate the growth of tumorigenic murine hepatic progenitor cells [27, 28]. miR-222 was upregulated in a larger series of primary HCC tumors and has a strong relationship between the high expression of miR-222 with tumor progression and patient survival. Overexpression of miR-222 confers cell migratory advantages in HCC through enhancing AKT signaling [29]. miR-181, as an activator of hepatic progenitor cells, maintains “stemness” during hepatocarcinogenesis and promotes hepatocarcinogenesis by targeting CDX2, GATA6, NLK and TIMP3 [30, 31]. The depletion of miR-181b inhibited tumor growth of HCC in nude mice [30]. Functional analysis of these oncomiRs and their targeted genes in liver cancer will help the understanding the role of miRNAs in hepatocarcinogenesis as molecular biomarkers and possible targets for developing oncomiR-targeted therapy of HCC.
Recent studies also demonstrated that many miRNAs such as miR-1, miR-16, miR-27b, miR-30d, miR-122, miR-126, miR-133, miR-143 and the let-7 family were abundantly expressed in adult liver tissue and modulated a diverse spectrum of liver functions [32, 33]. Biological and molecular functions are also affected by miRNA deregulation in HCC: miRNAs have been found to play important roles in HCC progression, cell proliferation and differentiation, avoidance of apoptosis, and metastasis [34-37]. Thus, identification of the deregulated miRNAs and their targets in HCC may provide promising therapeutic opportunities. For example, MET (a tyrosine kinase hepatocyte growth factor receptor), which is overexpressed in 40-70% of HCCs and is involved in cell motility, invasion, and metastasis, is posttranscriptionally regulated by miR-199a/a* and miR-1 in HCC [37, 38]. Silencing miR-1 can both inhibit HCC growth and mediate HCC cell invasion [37]. The liver-specific miR-122 is the most abundant miRNA in the liver, and it plays an important role in regulating hepatocyte development and differentiation [39, 40]. miR-122 is downregulated in HCC tumor tissues and cancer cell lines, and overexpression of miR-122 has been found to induce apoptosis and suppress proliferation in HepG2 and Hep3B cells [36]. Some investigators found that cyclin G1 was a direct target of miR-122 [41] and that miR-122/cyclin G1 interaction modulated p53 activity and affected doxorubicin sensitivity in human HCC cells [42]. miR-101, which is repressed in HCC, has been found to target expression of the FOS oncogene and inhibit HGF-induced cell invasion and migration [43].
Circulating miRNAs (cmiRNAs) in patients with cancer
The use of miRNAs as biomarkers has received increasing attention in recent years [44, 45]. The role of miRNAs in regulating a great variety of targets and, as a consequence, multiple pathways allows for their use as a diagnostic tool for early cancer detection, risk assessment, and prognosis, and for the design of innovative therapeutic strategies. Identifying tumor-specific alterations in circulating nucleic acids of cancer patients could be a novel, noninvasive means of diagnosing the cancer, particularly at early stages [46, 47]. Some studies have clearly shown that circulating miRNAs can originate from cancer tissues and are protected from endogenous RNase activity [47]. Although miRNAs are unstable RNA molecules, they are in fact highly stable and readily detected in serum and plasma. Biochemical analyses have indicated that circulating miRNAs are resistant to RNase activity, as well as to extreme pH and temperature [46, 47]. Further study has demonstrated that miRNAs remain largely intact in routinely collected formalin-fixed, paraffin-embedded clinical tissues [48] and that they are remarkably stable in both formalin-fixed, paraffin-embedded tissues and fresh snap-frozen specimens [49, 50].
miRNA microarrays, real-time polymerase chain reaction (PCR) arrays, and next-generation Solexa sequencing technology have been applied in examinations of circulating miRNA levels. All 3 high-throughput techniques can generate miRNA signatures in body fluids. In one study, analysis of the combined expression levels of 4 circulating miRNAs (miR-21, miR-126, miR-210, and miR-486-5p) distinguished patients with stage I non-small cell lung cancer from healthy controls with a sensitivity of 73.33% and a specificity of 96.55% [51]. Quantitative reverse-transcriptase PCR analysis also identified a profile of 7 serum miRNAs (miR-10a, miR-22, miR-100, miR-148b, miR-223, miR-133a, and miR-127-3p) as biomarkers for esophageal squamous cell carcinoma [52]. Analysis of the combined expression of miR-21, miR-210, miR-155, and miR-196a in plasma was also found to discriminate patients with pancreatic adenocarcinoma from controls [53].
Although most studies examining circulating miRNAs assessed their levels in serum and plasma, miRNAs have also been detected in tears, breast milk, bronchial lavage, colostrum, seminal fluid, amniotic fluid, pleural fluid, peritoneal fluid, and cerebrospinal fluid [54, 55]. Urine levels of miR-1236, miR-374a, and miR-767-3p were found to be increased in patients with bladder urothelial cancers compared with healthy controls [54]. miR-125a and miR-200a were also present at significantly lower levels in the saliva of the patients with oral squamous-cell carcinoma than in matched healthy controls [56]. These findings could be useful if associations are found between specific miRNA levels in body fluids and specific diseases, which would allow the expression profiles of these circulating miRNAs to be used as minimally invasive biomarkers for monitoring and especially for diagnosing human cancers.
Circulating miRNAs (cmiRNAs) as biomarkers for HCC
Because many miRNAs are dysregulated in HCC, it is to be expected that circulating miRNA levels are also affected by HCC progression. It is interesting that miR-15b, miR-21, miR-130b and miR-183 were highly expressed in culture supernatants of HCC cell lines [57]. The findings of studies examining altered levels of circulating miRNAs in patients with HCC are summarized in Table 1. For example, circulating miR-21 [58, 60-62], miR-222 [59, 61] and miR-223 [58, 59] were found to be upregulated in the serum/plasma of HCC patients carrying the hepatitis B or C virus by most investigators. Circulating miR-21 level in the patients with HCC was significantly higher than in patients with chronic hepatitis and healthy controls. ROC analysis of miR-21 yielded an AUC of 0.773 with 61.1% sensitivity and 83.3% specificity when differentiating HCC from chronic hepatitis, and an AUC of 0.953 with 87.3% sensitivity and 92.0% specificity when differentiating HCC from healthy control. Both sets of values were superior to AFP as biomarker in HCC (62). At the same times, serum miR-1, miR-25, miR- 92a, miR-206, miR-375, and let-7f were also significantly upregulated in HCC patients [63].
Table 1.
Findings of studies examining circulating miRNA (cmiRNAs) expression in patients with hepatocellular carcinoma (HCC)
| microRNA | Body fluids | Alteration | Etiology of HCC* | Source |
|---|---|---|---|---|
| let-7f | Serum | Up | HBV | (63) |
| miR-1 | Serum | Up | HBV | (63) |
| miR-15b | Serum | Up | HBV | (57) |
| miR-16 | Serum | Down | HBV, HCV, other | (60) |
| miR-21 | Serum/plasma | Up | HBV, HCV | (58, 60, 61,62) |
| miR-21 | Serum | Down | HBV | (59) |
| miR-25 | Serum | Up | HBV | (63) |
| miR-92a | Serum | Up | HBV | (63) |
| miR-130b | Serum | Up | HBV | (57) |
| miR-122 | Serum | Up | HBV | (58, 59) |
| miR-146a | Serum | Up | HBV | (70) |
| miR-183 | Serum | Up | HBV | (57) |
| miR-199a | Serum | Down | HBV, HCV, other | (60) |
| miR-206 | Serum | Up | HBV | (63) |
| miR-215 | Serum | Up | HBV | (70) |
| miR-221 | Serum | Up | HBV | (61) |
| miR-222 | Serum | Up | HBV | (59, 61) |
| miR-223 | Serum | Up | HBV | (58, 59) |
| miR-224 | Serum | Up | HBV | (61,70) |
| miR-375 | Serum | Up | HBV | (63) |
| miR-574-3p | Serum | Up | HBV | (70) |
| miR-885-5p | Serum | Up | HBV | (70) |
| miR-7 | Urine | Up | HCV | (15) |
| miR-92b | Urine | Down | HCV | (15) |
| miR-200a | Urine | Up | HCV | (15) |
| miR-219 | Urine | Down | HCV | (15) |
| miR-323 | Urine | Down | HCV | (15) |
| miR-335 | Urine | Up | HCV | (15) |
| miR-449 | Urine | Down | HCV | (15) |
| miR-453 | Urine | Up | HCV | (15) |
| miR-486 | Urine | Down | HCV | (15) |
| miR-502d | Urine | Down | HCV | (15) |
| miR-516-5p | Urine | Down | HCV | (15) |
| miR-520a* | Urine | Up | HCV | (15) |
| miR-521 | Urine | Up | HCV | (15) |
| miR-532 | Urine | Up | HCV | (15) |
| miR-610 | Urine | Up | HCV | (15) |
| miR-618 | Urine | Up | HCV | (15) |
| miR-625 | Urine | Up | HCV | (15) |
| miR-640 | Urine | Up | HCV | (15) |
| miR-650 | Urine | Down | HCV | (15) |
| miR-765 | Urine | Up | HCV | (15) |
HBV, hepatitis B virus; HCV, hepatitis C virus.
However, circulating miRNA expression levels are not always consistent with miRNA expression levels in tissue. For instance, miR-122 was found to be downregulated in HCC tumor tissues and cancer cell lines [36] but upregulated in the serum of HCC patients carrying the hepatitis B virus [58, 59]. Because miR-122 is a liver disease-specific tumor suppressor [36], the dysregulation of its target genes in patients with liver disease has shown a strong association with tumorigenesis [64]. A reduction of miR-122 expression in HCC cells has been observed in both a rat model for HCC and in a study of human HCC samples compared with pair-matched control tissue samples [65]. Restoration of miR-122 expression in HCC cell lines has been found to impair in vitro migration, anchorage-independent growth, invasion, angiogenesis, and intrahepatic metastasis [66]. Similar findings have been obtained by other research groups [67, 68]. This inverse relationship between tissue miRNA expression levels and circulating miRNA expression levels suggests that secreted miRNAs from cells may be an important component of circulating miRNA expression analysis.
Serum miR-15b and miR-130b levels were also found to be upregulated in HCC [57]. miR-130b had the largest area under the curve (0.913), with a sensitivity of 87.7% and a specificity of 81.4% for detecting HCC, and miR-15b had the highest sensitivity for detecting HCC of the miRNAs examined (98.3%), although its specificity was very low (15.3%). The high sensitivity of circulating miR-15b and miR-130b as biomarkers for HCC holds promise for patients with early-stage HCC, who may have low AFP levels despite the presence of disease. Similarly, serum miR-16 was found to be a more sensitive biomarker for HCC than serum AFP, DCP, and Lens culinaris agglutinin-reactive AFP (AFP-L3) levels [3]. Thus, analysis of the combination of serum miR-16 expression levels and serum AFP, DCP, and AFP-L3 levels allowed detection of 92.4% of HCC cases, with a high specificity (78.5%) [3].
Although most studies assessing circulating miRNAs in HCC focused on serum and plasma miRNA levels, recent studies have shown that urine miRNA levels are also useful biomarkers for HCC at early stages [15]. In one study, 5 miRNAs were examined in patients with HCC; 3 were upregulated (miR-625, miR-532, and miR-618) and 2 were downregulated (miR-516-5P and miR-650) in the urine of HCC patients carrying the hepatitis C virus relative to healthy controls. miR-618 was significantly upregulated (≥3 fold difference of expression; p < 0.05) in the urine of 35 of the 74 hepatitis C virus positive patients , and miR-650 was significantly downregulated in 42 of the 74 hepatitis C virus positive patients [15]. The sensitivity of urine miR-650 levels and the specificity of the combination of miR-618 and miR-650 levels for detecting HCC were greatly improved compared with the sensitivity and specificity of AFP-based detection (sensitivity 68%; specificity 75%) [15]. Urine offers a non-invasive and easily acquired sample source for developing screening tools, and the presence of miRNAs in body fluids such as urine may represent a gold mine of noninvasive biomarkers for cancer.
Advantages and limitations of circulating miRNAs (cmiRNAs) as biomarkers for HCC
Because no optimal blood tumor marker for HCC has been developed so far—the performance of AFP, AFP-L3, and DCP is limited in a surveillance setting and for early detection of HCC [69]—there is a need for novel, noninvasive biomarkers that can serve as a reliable method for early detection of HCC. Microarray, real-time PCR, next generation sequencing, microfluidics and bioinformatics have enabled the discovery of a great number of circulating miRNAs as potential biomarkers for early detection or prognosis in cancer, and some circulating miRNAs have even been found to be related to cancer progression or metastasis.
Even though the prospect of circulating miRNAs as minimally invasive biomarkers for HCC is encouraging, the magnitude of the challenge must be conquered by our clinical and basic researchers in the future. A simple standard assay for the quantification of circulating miRNAs in various body fluids should be established, and the specificity and sensitivity of circulating miRNA profile-based biomarkers should be validated in larger samples. Specific circulating miRNAs associated with HCC should be definitively identified, and their functions and networks must be further examined. Although the sensitivity and stability of miRNAs as biomarkers are suitable for a clinical setting, appropriate controls need to be used in a research setting because HCC is often accompanied by viral hepatitis, cirrhosis, or other underlying liver conditions. When assessing the specificity of a miRNA for detecting HCC, it is critical to ensure that patients and controls are matched not only by age and sex, but also by the etiology and severity of the underlying liver disease.
Despite the miRNA classifier gave a high positive predictive value in some HCC cohorts, there are several limitations. First, the classifier need to be validated in other ethnic populations, such as in Europe and Japan, in which hepatitis C virus is the major etiology of HCC. Next, the postsurgical serum samples were small and obtained at different time points. It is urgent to test more longitudinal samples to verify the specific time or period of time in which the circulating miRNAs return to basal levels [57].
Circulating miRNAs could be used as first-line detection for an early diagnosis of HCC patients, if the sensitivity and specificity outperform those of presently used tumor markers. The discovery of circulating miRNAs offers an attractive clinical perspective but this field is quite new and more work remains to be performed. For instance, it has yet to be established which specific circulating miRNAs can reliably and accurately detect HCC at early stage [3].
Conclusion
Ideal biomarkers for tumors should have a satisfactory specificity and sensitivity. Currently, no biomarkers exist that can accurately identify subjects who are at risk of developing HCC: although AFP, AFP-L3, and DCP are fairly accurate at detecting HCC, their use is limited in a surveillance setting and for early detection of HCC. Thus, circulating miRNAs are attractive potential biomarkers for HCC.
Early studies clearly demonstrated that circulating miRNAs (cmiRNAs) had sufficient specificity and sensitivity to detect HCC, and tremendous efforts have been devoted to identify novel, noninvasive circulating miRNA (cmiRNAs) -based biomarkers for early tumor detection, diagnosis, and prognosis. However, extensive investigation is still needed to validate the potential of circulating miRNAs as biomarkers. Because circulating miRNAs have shown promising results as potential biomarkers for HCC in pre-clinical studies, circulating miRNAs could be used for first-line diagnostic and prognostic testing if they outperform currently used HCC biomarkers in the first-line setting. With the accessibility of large sample sets, a wide range of available technology, and increasing evidence that miRNAs are involved in cancer-driven changes in the blood, circulating miRNAs, perhaps accompanying other markers, are likely to become widely used minimally invasive biomarkers for cancer in the future.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program) (2012CB720600, 2012CB720605), the National Natural Science Foundation of China (81271919) and the United States National Cancer Institute grant UO1CA111302. The University of Texas MD Anderson Cancer Center is supported in part by a Cancer Center Support Grant (CA016672) from the United States National Institutes of Health.
References
- 1.Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Chan JY, Fong CC, Tzang CH, Fung KP, et al. Transcriptional analysis of doxorubicin- induced cytotoxicity and resistance in human hepatocellular carcinoma cell lines. Liver Int. 2009;29:1338–1347. doi: 10.1111/j.1478-3231.2009.02081.x. [DOI] [PubMed] [Google Scholar]
- 3.Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371–1383. doi: 10.1016/j.jhep.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 4.Bosch FX, Ribes J, Cléries R, Díaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191–211. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebouissou S, Bioulac-Sage P, Zucman-Rossi J. Molecular pathogenesis of focal nodular hyperplasia and hepatocellular adenoma. J Hepatol. 2008;48:163–170. doi: 10.1016/j.jhep.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Katayama F, Kato H, Tanaka H, Wang J, et al. Hepatitis B and C Virus Infection and Hepatocellular Carcinoma in China: A Review of Epidemiology and Control Measures. J Epidemiol. 2011;21:401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz M, Roayaie S, Konstadoulakis M. Strategies for the management of hepatocellular carcinoma. Nat Clin Pract Oncol. 2007;4:424–432. doi: 10.1038/ncponc0844. [DOI] [PubMed] [Google Scholar]
- 9.Poon RT, Fan ST. Hepatectomy for hepatocellular carcinoma: patient selection and postoperative outcome. Liver Transpl. 2004;10:S39–45. doi: 10.1002/lt.20040. [DOI] [PubMed] [Google Scholar]
- 10.Yuen MF, Cheng CC, Lauder IJ, Lam SK, Ooi CG, et al. Early detection of hepatocellular carcinoma increases the chance of treatment: Hong Kong experience. Hepatology. 2000;31:330–335. doi: 10.1002/hep.510310211. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda S, Itamoto T, Nakahara H, Kohashi T, Ohdan H, et al. Clinicopathologic features and prognostic factors of resected solitary small-sized hepatocellular carcinoma. Hepatogastroenterology. 2005;52:1163–1167. [PubMed] [Google Scholar]
- 12.Jelic S, Sotiropoulos GC. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21:v59–64. doi: 10.1093/annonc/mdq166. [DOI] [PubMed] [Google Scholar]
- 13.Collier J, Sherman M. Screening for hepatocellular carcinoma. Hepatology. 1998;27:273–278. doi: 10.1002/hep.510270140. [DOI] [PubMed] [Google Scholar]
- 14.Oka H, Tamori A, Kuroki T, Kobayashi K, Yamamoto S. Prospective study of alpha-fetoprotein in cirrhotic patients monitored for development of hepatocellular carcinoma. Hepatology. 1994;19:61–66. [PubMed] [Google Scholar]
- 15.Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19–31. doi: 10.7150/jca.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasahara A, Hayashi N, Fusamoto H, Kawada Y, Imai Y, et al. Clinical valuation of plasma des-gamma-carboxy prothrombin as a marker protein of hepatocellular carcinoma in patients with tumors of various sizes. Dig Dis Sci. 1993;38:2170–2176. doi: 10.1007/BF01299891. [DOI] [PubMed] [Google Scholar]
- 17.Choi J, Park Y, Kim JH, Kim HS. Evaluation of automated serum des-gamma-carboxyprothrombin (DCP) assays for detecting hepatocellular carcinoma. Clin Biochem. 2011;44:1464–1468. doi: 10.1016/j.clinbiochem.2011.08.1144. [DOI] [PubMed] [Google Scholar]
- 18.Bruix J, Llovet JM. Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology. 2002;35:519–524. doi: 10.1053/jhep.2002.32089. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Sen S. MicroRNA functional network in pancreatic cancer: From biology to biomarkers of disease. J Biosci. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- 20.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Yu J, Yu S, Lavker RM, Cai L, et al. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J Hepatol. 2010;53:98–107. doi: 10.1016/j.jhep.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Wei W, Cheng N, Wang K, Li B, et al. Hepatitis C Virus-induced upregulation of miR-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012 doi: 10.1002/hep.25849. doi: 10.1002/hep.25849. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25.Han ZB, Chen HY, Fan JW, Wu JY, Tang HM, et al. Up-regulation of microRNA-155 promotes cancer cell invasion and predicts poor survival of hepatocellular carcinoma following liver transplantation. J Cancer Res Clin Oncol. 2012;138:153–161. doi: 10.1007/s00432-011-1076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12:27–33. doi: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan Q, Loya K, Rani B, Möbus S, Balakrishnan A, et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2012 doi: 10.1002/hep.25984. doi: 10.1002/hep.25984. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Wong QW, Ching AK, Chan AW, Choy KW, To KF, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 30.Wang B, Hsu SH, Majumder S, Kutay H, Huang W, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–1797. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009;15:1665–1672. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim CJ, Park NH, Chung YH. Roads towards a new tailored therapy for hepatocellular carcinoma: diagnostic, therapeutic and prognostic implications of microRNAs. Liver Int. 2012;32:695–697. doi: 10.1111/j.1478-3231.2012.02779.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, et al. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92–97. doi: 10.1016/j.gene.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Augello C, Vaira V, Caruso L, Destro A, Maggioni M, et al. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma. Liver Int. 2012;32:772–782. doi: 10.1111/j.1478-3231.2012.02795.x. [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Zhu X, Wu L, Yang R, Yang Z, et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int. 2012;32:752–760. doi: 10.1111/j.1478-3231.2011.02750.x. [DOI] [PubMed] [Google Scholar]
- 37.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in heatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 39.Morita K, Taketomi A, Shirabe K, Umeda K, Kayashima H, et al. Clinical significance and potential of hepatic microRNA-122 expression in hepatitis C. Liver Int. 2011;31:474–484. doi: 10.1111/j.1478-3231.2010.02433.x. [DOI] [PubMed] [Google Scholar]
- 40.Chang J, Nicolas E, Marks D, Sander C, Lerro A, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 41.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 42.Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- 43.Li S, Fu H, Wang Y, Tie Y, Xing R, et al. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellularcarcinoma. Hepatology. 2009;49:1194–1202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 44.Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastroenterol. 2009;15:5633–5640. doi: 10.3748/wjg.15.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Ba Y, Ma L, Cai X, Yin Y, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 49.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J, Smyth P, Flavin R, Cahill S, Denning K, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang C, Wang C, Chen X, Yang C, Li K, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 53.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, et al. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, et al. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu J, Wu C, Che X, Wang L, Yu D, et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–142. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- 59.Qi P, Cheng SQ, Wang H, Li N, Chen YF, et al. Serum MicroRNAs as Biomarkers for Hepatocellular Carcinoma in Chinese Patients with Chronic Hepatitis B Virus Infection. PLoS One. 2011;6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45:355–360. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;4:406, 70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 62.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167–175. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 63.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, et al. Serum microRNA Profiles Serve as Novel Biomarkers for HBV Infection and Diagnosis of HBV-Positive Hepatocarcinoma. Cancer Res. 2010;70:9798–9807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 64.Boutz DR, Collins PJ, Suresh U, Lu M, Ramírez CM, et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem. 2011;286:18066–18078. doi: 10.1074/jbc.M110.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 67.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, et al. Alphafetoprotein, des- gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110–118. doi: 10.1053/j.gastro.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gui J, Tian Y, Wen X, Zhang W, Zhang P, et al. Serum microRNA characterization identifiesmiR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]