Abstract
The function of microbial interactions is to enable microorganisms to survive by establishing a homeostasis between microbial neighbors and local environments. A microorganism can respond to environmental stimuli using metabolic exchange—the transfer of molecular factors, including small molecules and proteins. Microbial interactions not only influence the survival of the microbes but also have roles in morphological and developmental processes of the organisms themselves and their neighbors. This, in turn, shapes the entire habitat of these organisms. Here we highlight our current understanding of metabolic exchange as well as the emergence of new technologies that are allowing us to eavesdrop on microbial conversations comprising dozens to hundreds of secreted metabolites that control the behavior, survival and differentiation of members of the community. The goal of the rapidly advancing field studying multifactorial metabolic exchange is to devise a microbial ‘Rosetta stone’ in order to understand the language by which microbial interactions are negotiated and, ultimately, to control the outcome of these conversations.
Microbial interactions (Fig. 1) exist in nearly every niche on this planet, ranging from the oral cavity, intestine and skin of humans, to the cocoons of wasps and down to grains of sand. When at equilibrium, many microorganisms coexist in stable mixed communities. When these communities are perturbed, our ecosystems can be considerably affected, resulting in catastrophic events that have an impact on our society, such as loss of food supplies, destruction of concrete buildings, deadly animal diseases and pandemics. Additionally, modern health care, agriculture and other commercial processes have been shaped by biologically active metabolites produced by fungi and bacteria1–8. For instance, the antibiotics penicillin and vancomycin facilitate the control of microbial infections, the immunosuppressant rapamycin allows routine organ transplantation and paclitaxel (Taxol) is a critical treatment for many cancers. Similarly, microbially produced molecules protect our food supplies from microbial and insect invasions; enhance growth of plants, poultry and cattle; and are also used in many consumer products, including soap, toothpaste and paints. Thus, when considering microbially produced metabolites, we often think in terms of how these metabolites influence our quality of life but frequently overlook their impact on complex microbial interactions and as initiators of multicellular behavior in microbial communities—the purposes for which these metabolites are primarily produced. For microbes themselves, microbial interactions provide access to nutrients and protection from external communities and allow adaptation to changing ecological niches.
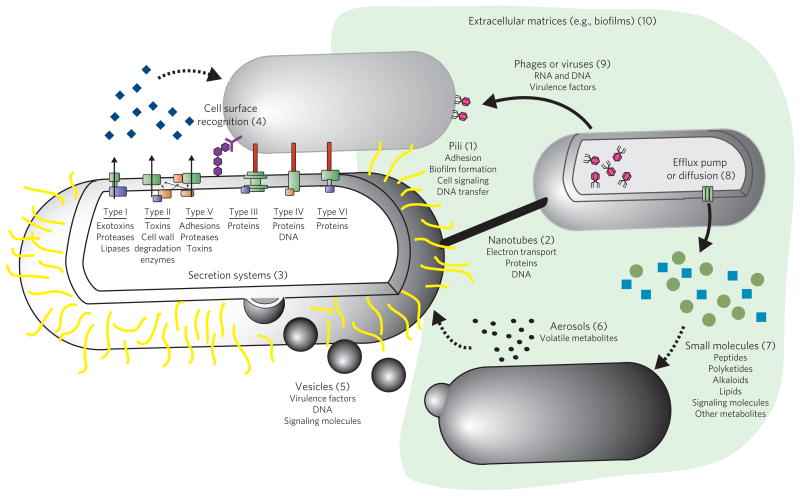
Figure 1. Microbial interactions.
Microbial interactions may be parasitic, such that one organism benefits at the cost of another; mutualistic, such that both organisms benefit; or commensal, such that one organism benefits at no cost or benefit to the other. all of these interactions, regardless of the outcome, occur through a diverse set of mechanisms by which genetic and molecular information is transferred. The most widely studied mechanisms of microbial interaction, some of which remain controversial, are shown. These include pili (1)69,70, nanotubes (2)71–73, secretion systems (3)74–77, cell surface recognition (4)78,79, vesicles (5)80–82, aerosols (6)83–85, small molecules (7)86–91 transported via efflux pumps or diffusion (8)92, phages or viruses (9)93–95 and biofilms (10)96. Each of these types of interaction plays a vital part in microbial metabolic exchange and provides the basis for microbial survival. Although some of these interactions are dependent on cell-to-cell contact, many do not occur through physical contact. Contact-independent metabolic exchange is advantageous because the signals are dispersed, enabling them to reach many neighboring cells and communities as opposed to only one cell at a time. The dispersion of metabolic exchange factors allows them to serve as nutrients or cues to neighboring microbes, thereby controlling the behavior of the larger microbial community and, in effect, leading to behavior as a multicellular entity.
Microbes dedicate enormous resources to microbial interactions. The percentages of microbial genomes that are dedicated to the production of secondary metabolites, a subset of metabolic exchange factors, have been defined by several studies as approximately 5–15%. Amazingly, however, the total number of open reading frames (ORFs) dedicated to microbial interactions has not been determined despite the importance of microbial interactions to the survival and fitness of the individual microbe and the larger microbial community as a whole9–13. To estimate the proportion of the bacterial proteome involved in microbial interactions, the genomes of Staphylococcus aureus subsp. aureus USA300_FPR3757, Pseudomonas aeruginosa str. PAO1 and Bacillus subtilis subsp. subtilis str. 168 were obtained from the Pathosystems Resource Integration Center (PATRIC) database14 and were manually curated using the National Center for Biotechnology Information (NCBI) basic local alignment search tool (BLAST) in an attempt to assign a function to every predicted ORF.
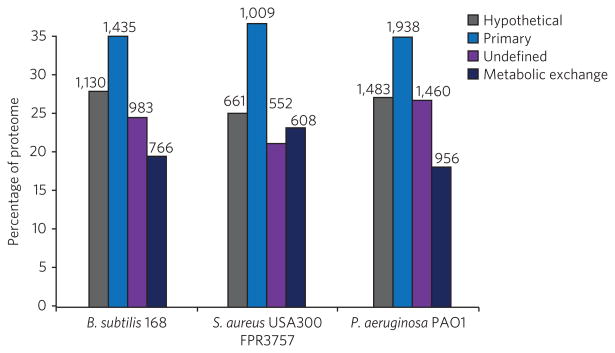
According to our analysis, 17–42% of the predicted ORFs are dedicated to microbial interactions (Fig. 2 and Supplementary Data Set 1). The ability to assign functions to several ORFs was limited by misannotations within the NCBI database, which includes annotations that do not have a basis in the biology of a given organism. For instance, several ORFs of P. aeruginosa strains have been assigned to be involved in sporulation. As P. aeruginosa does not sporulate, these annotations are unfounded and were therefore corrected in our analysis. Additionally, after manual BLAST analysis, many ORFs did not seem to encode the conserved domains required for the previously assigned functions. This analysis, albeit rudimentary, indicates that a much larger portion of the proteomic capacity of these microbes is involved in establishing their interactome than is appreciated at present. By and large, the interdependencies and diversity of interactions in the interactome are severely underappreciated and poorly understood.
Figure 2. Percentages of the predicted ORFs used in microbial interactions.
On the basis of BLAST analysis, the predicted ORFs of S. aureus subsp. aureus USA300 FPR3757, P. aeruginosa str. PAO1 and B. subtilis subsp. subtilis str. 168 were categorized by function into four groups: hypothetical or unassigned ORFs (gray), ORFs involved in primary metabolism (light blue), ORFs for which homologs exist but whose role in metabolic exchange is unclear (purple), and ORFs involved in microbial interactions and metabolic exchange (dark blue). The number above each group corresponds to the number of ORFs in that category. The roles of these ORFs were putatively assigned on the basis of BLAST analysis or inferred from clustering within the genome. For example, hypothetical or primary genes that clustered within gene clusters involved in the production of secondary metabolites were assigned to metabolic exchange, even though their roles in biosynthesis remain unknown. It should be noted that nutrition-based sensing and signaling were not included in this assessment.
In this Perspective, we discuss one of the major aspects of microbial interactions, microbial metabolic exchange, highlighting the chemical and functional diversity of the metabolites involved, the important roles they have in cellular differentiation within microbial colonies and in the maintenance of ecosystems, the emerging tools that allow us to study microbial interactions in a spatially organized and systematic approach, and how we anticipate this knowledge will shape research and biotechnological applications.
Metabolic exchange factors
Quorum-sensing factors are the most widely recognized and most thoroughly studied metabolic exchange factors produced by microbial populations15–18. The secretion and detection of quorum-sensing factors is used to gauge cell density and to sense the presence of neighboring species by eavesdropping on the quorum-sensing factors they produce, resulting in the initiation of appropriate group behaviors. The structural diversity of quorum-sensing factors is broad (Fig. 3). Furthermore, depending on the molecules involved, quorum-sensing responses of microbial communities can vary substantially and may be affected by neighboring microbes and by abiotic factors such as pH, temperature and nutrient availability. Quorum sensing controls developmental processes such as cell differentiation and, in turn, influences many microbial systems of interest, including symbiotic interactions, virulence, competence, conjugation, antibiotic production, motility, sporulation and biofilm formation. Although quorum-sensing inhibition has been speculated to be a good therapeutic approach for targeting many pathogens, there are no molecules in this category with FDA approval at present.
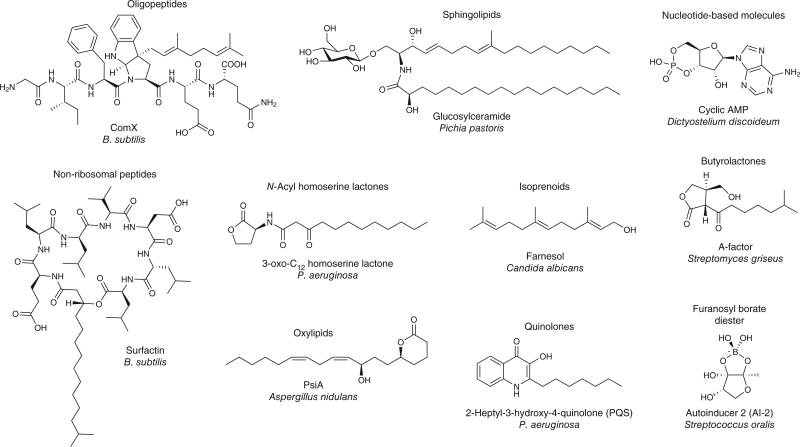
Figure 3. Chemical diversity of quorum-sensing molecules.
The diversity of quorum-sensing molecules described in the literature is shown. The chemical scaffolds of quorum-sensing factors range in structural complexity from simple isoprenoids and cyclic nucleotides to quinolones to complex peptide scaffolds.
Quorum-sensing responses are often studied as responses to independent molecules rather than in the context of a multifactorial metabolic exchange. For instance, quorum sensing in the human pathogen S. aureus involves the autoinducer system locus agr (accessory gene regulator), which encodes the quorum sensor autoinducing peptide. This quorum-sensing system is thought to regulate at least 23 secreted factors, including δ-toxin, α-hemolysin, and other toxins and proteases that affect interactions with neighboring microbes as well as with hosts19. Thus, in this case and, we suspect, in many others, quorum sensing is not responsible for only one specific phenotype but rather is the first instruction in a negotiation involving multiple metabolic exchange factors.
Quorum-sensing factors are just one well-studied portion of an even larger collection of molecules that develops and maintains distinct cell populations within communities. Microbes produce a large repertoire of structurally varied metabolic exchange factors that is critical for establishing microbial communities composed of one or several species (Fig. 4). The largest force shaping microbial communities is nutrition. Mutualistic metabolic dependence (or syntrophism) has led to the evolution of sophisticated mechanisms for nutrient sensing and signaling, allowing mixed communities to thrive by efficiently using even marginal nutritional strategies20,21. Akin to quorum-sensing factors, many syntrophic molecules are directives to neighboring organisms. Recent reports highlighting the importance of syntrophic interactions have shown that diffusible metabolic exchange factors from one species can enable cultivation of another species. Specifically, it was shown that isolated marine bacteria would only grow in the presence of a shared ‘helper’ molecule produced by other organisms. The helper molecule was identified as a siderophore required for iron uptake22,23. Such helper molecules have also been observed in interactions between microbes on iron-rich media, and other molecules, such as the cell wall component N-acetylglucosamines (GlcNAcs), have been shown to regulate the production of these helper molecules24,25. Therefore, the proposal that multimolecular signal response systems exist and many secreted molecules have a specific story to tell, especially when viewed in combination, is an attractive one. What has been largely ignored in research is that multiple signals, loosely defined here as secreted molecules that influence community behavior, alter the multicellular and/or social behavior of microorganisms.
Figure 4. Chemical diversity of metabolic exchange factors.
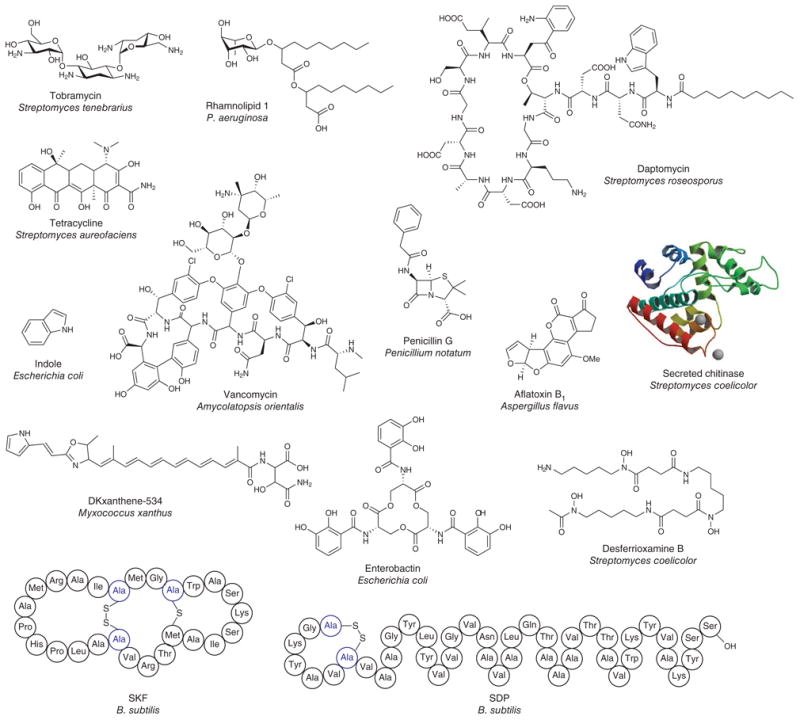
The metabolites involved in metabolic exchange have diverse structural scaffolds, ranging from small molecules and peptides to proteins (hydrolases, chitinases, protease and so on).
The boundary between metabolites involved in quorum sensing and those involved in other signaling roles often remains unclear. For example, in B. subtilis, two quorum-sensing peptides, competence pheromone (ComX) and surfactin, are responsible for paracrine signaling, in which some cells within the population produce a signal targeted to a specific subpopulation26. ComX triggers the production of surfactin, which in turn causes a subpopulation of B. subtilis cells within the colony to produce the extracellular matrix that holds the biofilm together. In addition to its role in intraspecies signaling, surfactin is an antibiotic and an interspecies signal that controls the production of aerial hyphae in Streptomyces species27. It is the most effective biological surfactant identified thus far, and this surfactant property is a trait that B. subtilis cells harness to move over solid surfaces, including those of plants28. Clearly, surfactin is a critical molecule for B. subtilis, but even in this case one suspects that we are only beginning to piece together the many key roles it has in the natural history of this species.
Overall, it is striking how little we know about the roles of metabolic exchange factors, including antibiotics, in natural communities. Although traditionally antibiotics were thought of as agents of microbial warfare, these molecules have also been proposed to have roles as quorum-sensing signals or to have other functions that help establish and stabilize microbial communities29,30. In the vicinity of the producer, an antibiotic concentration may be high; however, it remains unclear whether antibiotic concentrations in the natural microbial environment are sufficient to kill competing organisms31. For example, although high concentrations of the antibiotics tobramycin, ciprofloxacin and tetracycline lead to decreased growth or replication and, eventually, cell death in P. aeruginosa, subinhibitory concentrations of these antibiotics increase transcription of the genes involved in biofilm formation, in effect serving as a signal. Other documented microbial processes affected by sublethal concentrations of antibiotics include motility, hypha formation and sporulation, and it is likely that social interactions and multicellular behavior are also affected26–28.
Metabolic exchange in multicellular behavior
Understanding how multicellular behavior of microbial colonies is influenced by metabolic exchange factors is crucial for the development of a microbial Rosetta stone. This multicellular behavior is visible in the metabolic output of microbial colonies at both the colony level (Fig. 5a) and the cellular level (Fig. 5b), as seen in B. subtilis26. Although the distribution and localization of metabolic exchange factors are important, temporally regulated production of different classes of factor is essential for mediating cellular differentiation within a microbial colony. An array of secreted quorum-sensing factors and metabolites controls differentiation, but it remains unclear exactly how the combination of signals is processed and integrated to allow the elegant spatial organization of B. subtilis colonies to develop32. We speculate that, as we are beginning to see in B. subtilis, the primary function of metabolic exchange may be to spatially and temporally govern cell differentiation within the colony and to modulate this differentiation as necessitated by neighboring species and environmental conditions.
Figure 5. Cell differentiation of Bacillus subtilis at the colony and cellular levels.
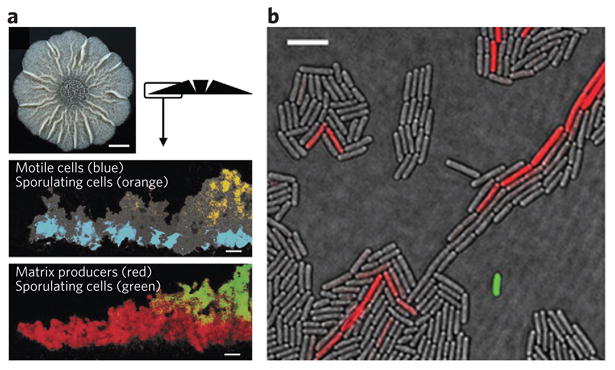
Monospecies bacterial communities are in fact multicellular communities with various subpopulations. (a) using an overlay of fluorescence and transmitted light micrographs, distinct populations of a B. subtilis biofilm can be observed where motile cells, sporulating cells and matrix-producing cells are false-colored. Scale bars, 50 μm. (b) using an overlay of fluorescence and transmitted light microscopy, distinct populations of surfactin-producing cells (expressing surfactin synthase subunit 1 (SrfAA)-YFP; artificially colored green) and matrix-producing cells (expressing YqxM-CFP; YqxM is a protein involved in anchoring cells together in B. subtilis biofilms; artificially colored red) can be observed. Although just 1 in 1,000–3,000 cells in a colony produces surfactin, this allows production of up to 1 g l−1 in liquid culture (4b)97,98. Scale bar, 3 μm. Figure adapted from ref.26 with permission.
We anticipate that an understanding of the role of metabolic exchange in bacterial colony differentiation and the formation of mixed communities will lead to an improved ability to cure or prevent disease. This knowledge could potentially lead to an increased understanding of biofilm formation or to ways of decreasing the number of latent cells in infections such as tuberculosis33. Furthermore, we anticipate that through understanding and manipulating the multicellular behavior of microorganisms, we could improve the commercial production of microbial compounds either by increasing the number of cells actively producing an antibiotic or by inducing the expression of orphan genes that are involved in antibiotic biosynthesis but whose expression is not observed in pure culture34. Simply increasing the number of antibiotic- or biofuel-producing cells from 1 in 1,000 to 4 in 1,000 may increase the yield enough to make a product commercially viable. Thus, systematic approaches that allow the identification of conditions that induce metabolite production represent an important opportunity for biotechnology applications. To fully exploit this opportunity, we will have to understand the nature of the forces driving multicellularity, and in particular how multiple metabolic exchange factors converge on a cell and how this drives cellular differentiation. Ultimately, an appreciation of the roles of metabolic exchange in multicellular microbial communities in the context of biotechnological applications could be exploited for modern multibillion-dollar, biology-based economies (bioeconomies).
Ecological effects of metabolic exchange
In addition to being a potential driver of future bioeconomies, metabolic exchange drives ecology. One elegant early example of the critical part played by microbial metabolic exchange in marine ecology was described in the late 1980s (Fig. 6a)35. It was demonstrated that the small molecule istatin, produced by an Alteromonas sp. bacterial symbiont of brine shrimp (Palaemon macrodactylus), controls the growth of the pathogenic fungus Lagenidium callinectes, a recognized infective agent of many crustaceans. When embryos of the brine shrimp were treated with antibiotics to remove the Alteromonas strain, the fungal pathogen killed the embryos within 6 h. It is now clear that there is a rich diversity of microbial symbionts that use metabolic exchange to maintain the balance within the marine biome36,37.
Figure 6. Ecological roles of microbial metabolic exchange.
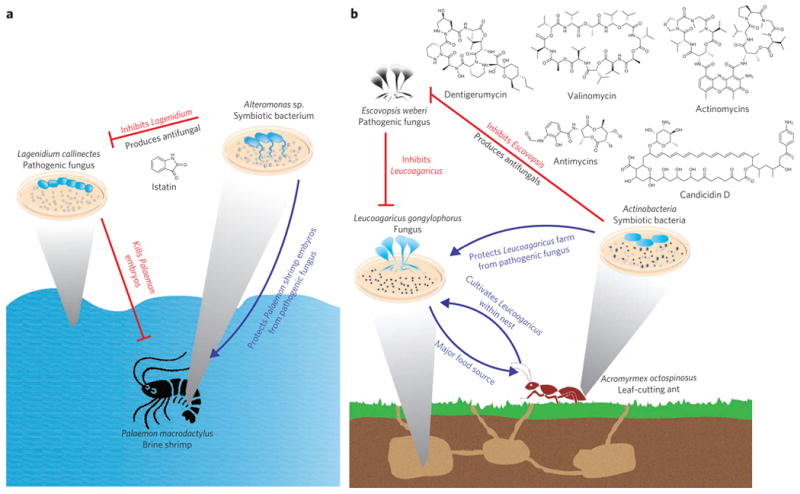
Microbial metabolic exchange has important roles in ecology and the survival of higher organisms. (a) Symbiotic bacteria of the brine shrimp produce the antifungal compound istatin, thereby protecting shrimp embryos from pathogenic fungi. (b)Actinomyces spp. symbionts of leaf-cutting ants produce metabolites that protect the fungus farmed by the ants from a pathogenic fungus.
This rich diversity of metabolic exchange and symbiosis in the marine environment is mirrored in terrestrial ecosystems. A recently uncovered example is the microbial community associated with the life cycle of the leaf-cutting ant (Fig. 6b). Leaf-cutting ants, such as Acromyrmex octospinosus, maintain a mutualistic relationship with the fungus Leucoagaricus gongylophorus in which the ants provide the fungus with nutrition by supplying it with harvested leaf material, and in turn the fungus serves as a major food source for the ants38,39. Microbial symbionts of leaf-cutting ants, mainly Pseudonocardia spp. and Streptomyces spp., protect the fungal garden against the microbial pathogen of the genus Escovopsis by producing antifungal compounds such as actinomycins, valinomycin, antimycins, candi-cidin macrolides and the cyclodepsipeptide dentigerumycin, thereby protecting the ants' food supply and colony from destruction40–43.
In addition to maintaining the external environment, microbial metabolic exchange plays an important part in human microecology. In the human body, it is estimated that mammalian cells are outnumbered by microbial cells ten to one. These microbes protect us from infection, degrade unused substrates, train the immune system and produce vitamins such as biotin or vitamin K44–46. The diversity of microbes and their metabolic exchange factors in human communities is staggering. For example, the human mouth provides a habitat for ∼500 different species of naturally occurring and transient bacteria. Initial colonization is undertaken by several Streptococcus species that bind to salivary receptors and then act as a platform for additional bacteria to aggregate together. In an artificial dental plaque, initial colonization was dominated by Streptococcus, Prevotella, Actinomyces and Veillonella species47. Cells of the genera Prevotella and Actinomyces showed the most interspecies associations, and it is therefore proposed that the role of these genera is to establish and maintain biofilm complexity47. One example of the role of metabolic exchange factors in the human oral microbiome comes from the quorum-sensing molecules of the autoinducer 2 (AI-2) family, which are essential for mutualistic and abundant biofilm growth of two bacteria: Actinomyces naeslundii str. T14V and Streptococcus oralis str. 34 (ref.48). In this case, direct contact between A. naeslundii and S. oralis through coaggregation and coadhesion leads to the upregulation of AI-2 by S. oralis. The locally high concentration of AI-2 subsequently allows the formation of a mutualistic mixed biofilm. However, this example is just a sentence in the conversation between the 500 species that compose a mature dental film49. To understand these communities, we must identify the molecules that dictate behavior and then understand their meaning and the hierarchy with which they are listened to and decoded.
It is clear that metabolic exchange plays a critical part in environmental and human microecology, but it is evident from sequencing of environmental DNA that these communities are even more complex than is illustrated by these examples50. Microbial metabolic exchange has a vital role in maintaining larger multicellular systems, and disruptions of the community may lead to the proliferation of pathogens. However, the system-wide effect of perturbing these interactions has not been established for any system. To truly examine the complexities of microbial metabolic exchange in ecological terms, new tools must be developed and applied.
Challenges in studying microbial metabolic exchange
The discovery of penicillin, the result of an interaction between a fungus and S. aureus, by Alexander Fleming ignited interest in studying single microbial metabolic exchange factors for medical applications. However, few studies have investigated the effects of the multitude of factors in multicellular communities. Daunting challenges remain, including the thorough identification of all microbes and metabolites involved in microbial metabolic exchange. Complex microbial communities, such as those found in the human intestine, dental plaque, the rhizosphere and biofouling communities, are estimated to contain several hundred to a few thousand different organisms. In the human intestine alone, as many as 1,000 different species of naturally occurring and transient microbes may participate in metabolic exchange51. On the basis of the available microbial genome sequences, it is reasonable to estimate that each of the microbes involved in these complex communities has the genetic capacity to produce approximately ten molecules capable of influencing the behavior of neighboring cell populations52. Therefore, it is likely that metabolic exchange in these communities involves thousands of molecules, posing a substantial challenge for understanding how they affect behavior. Furthermore, single molecules can participate in several aspects of the community, as exemplified by the roles of surfactin in motility and in intraspecies signaling and as an interspecies antibiotic26. It is possible that other metabolic exchange factors also have multiple roles in microbial interactions, but our ability to identify and verify these roles, or to identify alternative responsible factors, is hampered by the lack of tools available to study these complex interactions in context.
Emerging methods for studying metabolic exchange
The challenges of unraveling microbial interactions, combined with the ecological and medicinal importance of microbial metabolic exchange, have motivated the recent adaptation of a variety of powerful scientific techniques to the study of microbe-microbe and microbe-host interactions. Successful investigations of microbial metabolic exchange require traditional methods in microbiology, such as molecular biology, genetics, systems biology or biochemistry, to be combined with emerging and newly developed methods (Table 1), such as multiplexed fluorescence microscopy, NMR imaging, imaging mass spectrometry (IMS), microfluidics or approaches designed to investigate global genomic, proteomic, peptidomic or metabolic profiles. Ultimately, to truly understand the complexity of metabolic exchange in microbial communities, it is necessary to develop tools to identify the microbial players, understand their overall physical and metabolic states, characterize the metabolic outputs associated with those states and the surrounding environment and completely integrate results from diverse experiments.
Table 1. Investigational methods for microbial interactions and communities.
| Method | Advantages | Disadvantages |
|---|---|---|
| IMS | Gathers mass information in both a chemical and a spatial manner | Each sample can be measured only at a single time point |
| Correlates specific phenotypes with metabolites | Not all media are compatible with current tools | |
| NMR imaging | Gathers mass information in both a spatial and a temporal manner | Requires specialized equipment |
| Direct, non-invasive monitoring of metabolite concentrations | Deconvolution is complex | |
| ‘Omics’ approaches | Provides genetic, proteomic and metabolomic information for single species | Requires biochemical verification |
| Correlates genotypes to chemotypes | Reliable, user-friendly databases are unavailable | |
| Metagenomics | Generates a catalogue of microorganisms in a microbiome | Limited to identifying the microbes involved |
| Allows comparison of microbiomes in healthy versus diseased individuals | Data analysis is complex | |
| Microfluidics | Can precisely control the microenvironment | Requires expert knowledge for use |
| Allows the confinement and quantification of cells | Device materials may be toxic to cells |
The study of microbial interactions has led to the convergence of many traditional techniques, as well as the development of new methods. Highlighted are several approaches now being used to investigate microbial metabolic exchange and microbial communities.
The first step in studying microbial metabolic exchange is the identification of the members of a microbial community. This might include traditional methods such as biochemical assays, microscopy and 16S and 18S ribosomal RNA sequencing that are robust but low throughput, still high in cost and not feasible in all cases (not all organisms are amenable to facile DNA extractions). There is a need for the development of tools to efficiently and effectively identify all microbes in a community. To facilitate the quick identification of microbes, fluorescence in situ hybridization (FISH) methods are continuously being developed53. In FISH, specifically designed fluorescent nucleotide sequences are used as probes to label and identify targeted strains. This was recently accomplished for an artificial dental plaque in which 15 different phylotypes were simultaneously visualized and differentiated.
Another path toward understanding microbial interactions is to identify and characterize the metabolic exchange factors involved in a specific interaction. Typically, a single species is grown in liquid culture, and the resulting cell broth is chemically extracted by organic solvents, solid-phase resins or methods that allow identification of small polar metabolites. These crude extracts undergo bioactivity-guided fractionation against a panel of microbial or cell lines. Using analytical techniques such as HPLC, MS, NMR or X-ray crystallography, individual compounds are purified and structurally characterized. Complementing traditional approaches, recent advances in genome sequencing and protein structure prediction have greatly facilitated identification of biosynthetic genes and prediction of the biochemical activities of their encoded products. A number of tools are available for in silico identification of gene clusters involved in the production of potential metabolic exchange factors54–56. The most recent addition to these tools is antiSMASH (antibiotics and secondary metabolite analysis shell), a web-based and stand-alone software pipeline for the identification, annotation and analysis of secondary metabolite gene clusters57. However, important challenges remain in this area. Specifically, although there are many public databases for the analysis of proteomic, RNA sequencing, lipidomic and metabolomic data, our ability to analyze the vast amounts of data generated by ‘omic’ tools and to integrate findings from diverse approaches is limited by a lack of centralization or maintenance and by unfriendly user interfaces.
The integration of biochemical and analytical techniques, as in IMS, also provides a new dimension to investigating the physical and metabolic states of interacting microbes58. MALDI-IMS, for example, has been adapted by our laboratory to agar-based microbial cultures, enabling the investigation of the chemical identity and spatial distribution of metabolic exchange factors that ionize by MS. The MALDI-IMS approaches in our laboratory (outlined in Fig. 7) require the sample to be covered with matrix and then dehydrated for analysis. We used MALDI-IMS to investigate B. subtilis cannibalism59, as well as the bacterium's single-neighbor interactions with Streptomyces coelicolor and S. aureus60,61 and its interactions within marine microbial assemblages62. Using MALDI-IMS, we were able to identify several metabolites produced by these organisms, including the cannibalistic factors sporulation delaying protein (Sdp) and sporulation killing factor (Skf) of B. subtilis. IMS allowed direct observation of these cannibalistic metabolites, which eased their isolation and subsequent structural analysis by MS and targeted NMR. Although MALDI-IMS has provided us with a new way to investigate microbial interactions at the chemical level, it is limited by the required application of matrix and the necessary dehydration before sample analysis. The process of matrix application and dehydration kills the microbes and therefore restricts analysis to only a single colony at one point in time. In addition, in using our current MALDI-IMS methods, not all types of agar medium are compatible; therefore, each medium must be evaluated before an experiment is initiated. Furthermore, not all molecules ionize or are stable under the conditions in use at present60. To address these limitations of MALDI-IMS of microbial cultures, we are currently developing other MS-based techniques to complement MALDI-IMS to enable metabolic analysis of live microbial colonies.
Figure 7. MALDI-IMS links chemistry to bacterial phenotypes.
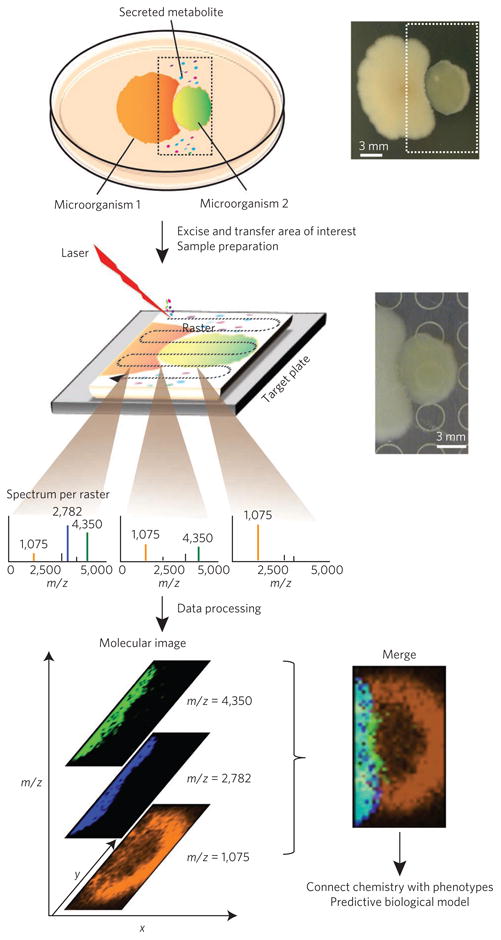
MALDI-IMS can be used to visualize the spatial distribution of metabolic exchange factors and to aid in their identification. To prepare the sample for MALDI-IMS, microbial colonies are cultured on an agar plate, excised, transferred to a MALDI target surface, covered with matrix, dried and subjected to rastering MALDI-MS. A MALDI-MS image is generated by directing a laser at different x,y positions of a sample in a predefined manner, creating a two-dimensional molecular profile of the molecules that are present in the top layer of the sample. Any ion observed in these spectra can be spatially visualized with a false-color that reflects the intensity of the MS signal60.
Concurrent with the development of live MS analysis of microorganisms, NMR imaging is being developed as a tool to investigate live biofilms63,64. These tools have been used to interrogate the structure and the dynamic metabolic processes of Shewanella oneidensis str. MR-1 and Streptococcus mutans str. UA159 by integrating NMR with confocal laser scanning microscopy63,64. NMR provides direct, time-resolved, noninvasive monitoring of metabolite concentration, metabolic pathways and flux rates, while a confocal laser scanning microscope within the NMR magnet can monitor fluorescent tags to follow gene expression or an individual strain. The combination of these techniques provides an exciting opportunity to produce a full three-dimensional image of a live biofilm that includes NMR data showing the spatial distribution of small metabolites.
Recently, microfluidics has been presented as a tool that can be used to study metabolic exchange65. For a cell to differentiate into a specific cell type within a microbial colony, several signals may need to converge at specific concentrations. Microfluidics may provide us with a method to achieve those concentrations in vitro. This ability will enable researchers to investigate the impact of gradients of multiple metabolic exchange factors on the behaviors of single cells or biofilm communities.
The methods mentioned above are a small subset of the advanced tools that have been developed to facilitate the study of microbial metabolic exchange. Although there have been many advances in the tools and methods available for investigating microbial interactions, research still suffers from the limitations of these tools, even when used in combination. We can visualize single cells with microscopy. We can visualize metabolites involved in metabolic exchange with IMS. However, the current tools available are not capable—or have not yet been demonstrated to be capable—of the resolution required to differentiate single cells and simultaneously detect the metabolites produced and their distribution in communities on a surface of choice. Ideally, we need to identify each microbe and visualize each individual cell and the metabolites it produces on materials such as mucosal epithelium, plant stems, flower petals or soil and to connect this information to genomic signatures. This will require substantial technological advances.
Although many current studies of microbial metabolic interactions are simplified to focus on just a single molecule or one interaction at a time, we envision a future in which multiplexed interactions can be studied in their natural context and the outcome of interspecies interactions can be controlled to benefit human health and agriculture. Thus, the next frontier is the development of tools that connect genotypes, chemotypes and phenotypes in complex environments and communities. To accomplish this, we need to continue to push the boundaries of what is possible, often with creative adaptations of existing instrumentation and the fusion of heretofore separate approaches and fields of microbiology. Our laboratories use IMS, in addition to other MS and cell biology approaches, as a tool to connect chemotypes to phenotypes, providing insight into the largely invisible molecular sphere and an opportunity to observe the multiplexed nature of spatial systems microbiology66. However, to continue to push this field forward, it is critical to develop new methods that correlate the observed chemistry with changes in cell architecture, behavior and development to elucidate the function of newly discovered molecules. Connecting the chemistry to the genes responsible for metabolite biosynthesis and cellular responses and determining the developmental state of individual cells within those regions of the colonies that are producing specific metabolites will be critical for our understanding of microbial interactions in nature.
We already know that metabolic exchange factors serve as cues or signals in microbial communities; they are required for morphological and developmental processes, the survival of individual microbes and the fitness of the microbial community as a whole67,68. Typically, the language of microbes is studied one signal at a time, with entire subfields focusing on just a few molecules. In reality, there are many different classes of molecules necessary for the social behavior of microbes. Describing just one molecule at a time, typically out of context, is akin to us using one sentence instead of a full dialogue when we speak49. Fortunately, tools are emerging that enable us to listen in on microbial conversations consisting of several molecules, allowing microbial metabolic exchange to be documented in outstanding detail and in context. It is our opinion that these approaches provide the only way to develop a Rosetta stone for microbial communication as they will allow us to establish functional translations of the molecules that microbes secrete and the signaling networks that these molecules affect. In our quest to generate a Rosetta stone, we should not underestimate the complexity of microbial interactions in the context of multicellular and community behavior. If every organism in a community of 500 microbes differentiates into 5 cell types, each producing 3 molecules that provide specific instructions to neighboring cells, there would be 7,500 molecules present and actively providing instruction in this community. Perhaps other disciplines, such as those concerning human population dynamics and interactions, can provide some critical insight. Typically, attention has been given to single factors and not to the multifactorial nature of metabolic exchange. To develop a Rosetta stone, we must understand what types of molecule are produced and how they act in concert, and we must continue to develop tools to study metabolic exchange.
There is little doubt that the next two decades will see the development of user-friendly tools to study the metabolic exchange of microbes both at the cellular level and in real time. Accomplishing this goal will require continued collaborative contributions from biologists, chemists, chemical engineers, informaticians, mathematicians and other investigators to design and build tools and methods for observing genetic and molecular changes spatially and temporally. It is these tools that will allow us to develop the Rosetta stone for microbial conversations, enabling us to understand, in molecular detail, how microbial communities are established and maintained in nature and in human hosts.
Supplementary Material
Acknowledgments
The authors would like to acknowledge J. Yang, W. Moree and C. Rath (University of California, San Diego) for providing critical reviews of the manuscript and E. Shank (Harvard Medical School) for insightful discussions. The P.C.D. laboratory is supported by US National Institutes of Health grants GM094802, GM086283 and AI095125 and by the Beckman Foundation. The K.P. laboratory is supported by US National Institutes of Health grants GM057045 and AI095125.
Footnotes
Competing financial interests: The authors declare no competing financial interests.
Supplementary information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Banat IM, Makkar RS, Cameotra SS. Potential commercial applications of microbial surfactants. Appl Microbiol Biotechnol. 2000;53:495–508. doi: 10.1007/s002530051648. [DOI] [PubMed] [Google Scholar]
- 3.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 4.Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006;39:235–251. [Google Scholar]
- 5.Fujinami S, Fujisawa M. Industrial applications of alkaliphiles and their enzymes–past, present and future. Environ Technol. 2010;31:845–856. doi: 10.1080/09593331003762807. [DOI] [PubMed] [Google Scholar]
- 6.Fusetani N. Antifouling marine natural products. Nat Prod Rep. 2011;28:400–410. doi: 10.1039/c0np00034e. [DOI] [PubMed] [Google Scholar]
- 7.Dayan FE, Cantrell CL, Duke SO. Natural products in crop protection. Bioorg Med Chem. 2009;17:4022–4034. doi: 10.1016/j.bmc.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Babalola OO. Benefcial bacteria of agricultural importance. Biotechnol Lett. 2010;32:1559–1570. doi: 10.1007/s10529-010-0347-0. [DOI] [PubMed] [Google Scholar]
- 9.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda H, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 11.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 12.Ohnishi Y, et al. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol. 2008;190:4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder EE, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowery CA, Dickerson TJ, Janda KD. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem Soc Rev. 2008;37:1337–1346. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 16.Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu Rev Microbiol. 2009;63:99–118. doi: 10.1146/annurev.micro.091208.073248. This review discusses the influence that a bacterium has within a community through participating in metabolic exchange. [DOI] [PubMed] [Google Scholar]
- 17.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. This paper discusses the quorum-sensing networks in Vibrio spp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. This work discusses quorum-sensing regulation in P aeruginosa. [DOI] [PubMed] [Google Scholar]
- 19.Antunes LC, Ferreira RB, Buckner MM, Finlay BB. Quorum sensing in bacterial virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 20.Little AE, Robinson CJ, Peterson SB, Rafa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 21.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 22.D'Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis K, Epstein S, D'Onofrio A, Ling LL. Uncultured microorganisms as a source of secondary metabolites. J Antibiot (Tokyo) 2010;63:468–476. doi: 10.1038/ja.2010.87. [DOI] [PubMed] [Google Scholar]
- 24.Rigali S, et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat Prod Rep. 2011;28:1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- 26.López D, Kolter R. Extracellular signals that defne distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. This review discusses the cascades of cell differentiation pathways that are triggered by metabolic exchange. [DOI] [PubMed] [Google Scholar]
- 27.Straight PD, Willey JM, Kolter R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol. 2006;188:4918–4925. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angelini TE, Roper M, Kolter R, Weitz DA, Brenner MP. Bacillus subtilis spreads by surfing on waves of surfactant. Proc Natl Acad Sci USA. 2009;106:18109–18113. doi: 10.1073/pnas.0905890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies J. Everything depends on everything else. Clin Microbiol Infect. 2009;15(suppl 1):1–4. doi: 10.1111/j.1469-0691.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- 30.Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Phil Trans R Soc Lond B. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. This review is one of the first to discuss bacterial populations as multidimensional organisms. [DOI] [PubMed] [Google Scholar]
- 33.Strauss E. Grand challenge commentary: Exploiting single-cell variation for new antibiotics. Nat Chem Biol. 2010;6:873–875. doi: 10.1038/nchembio.483. [DOI] [PubMed] [Google Scholar]
- 34.Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 35.Gil-Turnes MS, Hay ME, Fenical W. Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science. 1989;246:116–118. doi: 10.1126/science.2781297. This provides an excellent example of the roles of metabolic exchange factors produced by symbiotic bacteria in host survival. [DOI] [PubMed] [Google Scholar]
- 36.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 37.Hentschel U, et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68:4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–704. [Google Scholar]
- 39.Currie CR, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 40.Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci USA. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci USA. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoenian I, et al. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci USA. 2011;108:1955–1960. doi: 10.1073/pnas.1008441108. This work used new methodologies to characterize the metabolic exchange between microbial communities within the nests of leaf-cutting ants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 45.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci USA. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. This paper represents an example of the use of labeling and imaging approaches to visualize and diferentiate phylotypes of microbial communities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rickard AH, et al. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60:1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- 49.Schloss PD, Handelsman J. The last word: books as a statistical metaphor for microbial communities. Annu Rev Microbiol. 2007;61:23–34. doi: 10.1146/annurev.micro.61.011507.151712. [DOI] [PubMed] [Google Scholar]
- 50.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 51.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baltz RH. Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol. 2008;8:557–563. doi: 10.1016/j.coph.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Moter A, Gobel UB. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods. 2000;41:85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 54.de Jong A, van Heel AJ, Kok J, Kuipers OP. BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res. 2010;38:W647–W651. doi: 10.1093/nar/gkq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32:W405–W413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Medema MH, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. This paper describes the development of the most comprehensive software pipeline available at present that is capable of identifying potential biosynthetic gene clusters for the whole range of known secondary metabolite classes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watrous JD, Dorrestein PC. Imaging mass spectrometry in microbiology. Nat Rev Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. This thorough review covers IMS techniques and their applicability in microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu WT, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci USA. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. This is the first work to apply IMS to discover specific factors that are active in microbial intraspecies interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. This is the original report on the development of IMS to study microbial metabolic exchange. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez D, et al. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology published online. 2011 Jun 30; doi: 10.1099/mic.0.048736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang YL, et al. Connecting chemotypes and phenotypes of cultured marine microbial assemblages by imaging mass spectrometry. Angew Chem Int Edn Engl. 2011;50:5839–5842. doi: 10.1002/anie.201101225. This paper highlights the utility of applying IMS to the investigation of metabolic exchange in microbial assemblages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McLean JS, et al. Investigations of structure and metabolism within Shewanella oneidensis MR-1 biofilms. J Microbiol Methods. 2008;74:47–56. doi: 10.1016/j.mimet.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 64.McLean JS, Ona ON, Majors PD. Correlated biofilm imaging, transport and metabolism measurements via combined nuclear magnetic resonance and confocal microscopy. ISME J. 2008;2:121–131. doi: 10.1038/ismej.2007.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boedicker JQ, Vincent ME, Ismagilov RF. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Edn Engl. 2009;48:5908–5911. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kersten RD, et al. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Sordi L, Muhlschlegel FA. Quorum sensing and fungal-bacterial interactions in Candida albicans: a communicative network regulating microbial coexistence and virulence. FEMS Yeast Res. 2009;9:990–999. doi: 10.1111/j.1567-1364.2009.00573.x. [DOI] [PubMed] [Google Scholar]
- 68.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. This review focuses on how bacterial small molecules modulate interspecies interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 70.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria -structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 72.Gorby YA, et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci USA. 2006;103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reguera G, et al. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- 74.DiGiuseppe Champion PA, Cox JS. Protein secretion systems in Mycobacteria. Cell Microbiol. 2007;9:1376–1384. doi: 10.1111/j.1462-5822.2007.00943.x. [DOI] [PubMed] [Google Scholar]
- 75.Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet. 2010;44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. This comprehensive review discusses the contact-dependent systems that microbes use to interact with each other and their environment. [DOI] [PubMed] [Google Scholar]
- 76.Holland IB. The extraordinary diversity of bacterial protein secretion mechanisms. Methods Mol Biol. 2010;619:1–20. doi: 10.1007/978-1-60327-412-8_1. [DOI] [PubMed] [Google Scholar]
- 77.Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 78.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 79.Ubbink J, Schar-Zammaretti P. Probing bacterial interactions: integrated approaches combining atomic force microscopy, electron microscopy and biophysical techniques. Micron. 2005;36:293–320. doi: 10.1016/j.micron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Haurat MF, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286:1269–1276. doi: 10.1074/jbc.M110.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 82.Roze LV, Chanda A, Linz JE. Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet Biol. 2011;48:35–48. doi: 10.1016/j.fgb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kai M, et al. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81:1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- 84.Minerdi D, Bossi S, Gullino ML, Garibaldi A. Volatile organic compounds: a potential direct long-distance mechanism for antagonistic action of Fusarium oxysporum strain MSA 35. Environ Microbiol. 2009;11:844–854. doi: 10.1111/j.1462-2920.2008.01805.x. [DOI] [PubMed] [Google Scholar]
- 85.Schulz S, Dickschat JS. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 2007;24:814–842. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 86.Borges-Walmsley MI, Walmsley AR. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 2000;8:133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- 87.Boyer M, Wisniewski-Dye F. Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol Ecol. 2009;70:1–19. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 88.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 89.Ratcliff WC, Denison RF. Microbiology. Alternative actions for antibiotics. Science. 2011;332:547–548. doi: 10.1126/science.1205970. This article discusses the possibility that there may be several roles for antibiotics in the native environment of microbes. [DOI] [PubMed] [Google Scholar]
- 90.Singh A, Del Poeta M. Lipid signalling in pathogenic fungi. Cell Microbiol. 2011;13:177–185. doi: 10.1111/j.1462-5822.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 92.Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64:672–693. doi: 10.1128/mmbr.64.4.672-693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boyd EF, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–529. doi: 10.1016/s0966-842x(02)02459-9. [DOI] [PubMed] [Google Scholar]
- 94.Coleman DC, et al. Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Gen Microbiol. 1989;135:1679–1697. doi: 10.1099/00221287-135-6-1679. [DOI] [PubMed] [Google Scholar]
- 95.Kuroda M, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 96.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 97.Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 98.López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.