Abstract
As the interactions of phage with mammalian innate and adaptive immune systems are better delineated and with our ability to recognize and eliminate toxins and other potentially harmful phage gene products, the potential of phage therapies is now being realized. Early efforts to use phage therapeutically were hampered by inadequate phage purification and limited knowledge of phage–bacterial and phage–human relations. However, although use of phage as an antibacterial therapy in countries that require controlled clinical studies has been hampered by the high costs of patient trials, their use as vaccines and the use of phage components such as lysolytic enzymes or lysozymes has progressed to the point of commercial applications. Recent studies concerning the intimate associations between mammalian hosts and bacterial and phage microbiomes should hasten this progress.
Our knowledge of phage–bacterial and phage–human interactions has grown, as has our ability to manipulate phage genomes. This should result in the rapid development of phage-based therapies (e.g., vaccines and antibiotics).
The human superorganism is made up of eukaryotic and prokaryotic cells, in which the prokaryotic cells far outnumber the eukaryotic cells. Given our current limited understanding of ourselves as super organisms it is remarkable that we have achieved successes, albeit limited, in developing therapies for infectious diseases. This was especially true in the 19th and 20th centuries when specific strains of bacteria were first being discovered and in some cases associated with animal or human disease states.
The therapeutic advances realized at that time were based on limited observations and heroic empirical efforts, such as the discovery of the antisyphilitic compound Salvarsan or 606 (in reference to the hundreds of compounds screened) by S. Hata in Paul Ehrlich’s laboratory in the early 1900s (Sneade 2005).
Given the state of knowledge at the beginning of the 20th century concerning microbiology and the immune defense mechanisms, the discovery of bacteriophage (phage) viruses that kill specific strains of bacteria was welcomed. However, soon after their discovery a number of issues arose, including a controversy as to whether the bacteriophages were self-replicating particles or a lytic enzymatic activity activated in the bacteria. One of the pioneers associated with their discovery, Félix d’Herelle, consulted with Albert Einstein who reassured d’Herelle that his experiments designed to prove that they were self-replicating particles were valid (Summers 1999). Perhaps more serious were the problems that arose owing to a lack of appreciation of the narrow host range of most phage strains, so that a phage isolated from an infected patient did not appear to be effective in treating another patient with a seemingly similar infection. In addition, there was a lack of knowledge of bacterial toxins and methods to purify phage preparations so that they were not contaminated with such toxins. These problems coupled with inadequately designed animal and clinical experiments were some of the factors that led, in the United States, to the development of a government agency to oversee therapeutic developments and claims (Merril et al. 2003, 2006).
In addition to these early problems, the discovery of the broad host range antibiotics—beginning with penicillin—more than 85 years ago, resulted in the virtual abandonment of efforts to continue in the development of the phage as a therapeutic antibacterial agent in the United States. However, the increasing incidence of antibiotic-resistant bacterial strains, particularly the bacterial strains commonly found in hospital settings, has prompted a reexamination of the potentials of phage antibacterial therapies. In addition, there is now an increased awareness of clinical problems associated with the extended bacterial host range of most commonly used antibiotics. Although an extended bacterial host range was one of the criteria used in searching for clinically relevant antibiotics, as this property partially alleviated the need to accurately identify infecting bacterial species before therapy was initiated, it is now recognized that such extended host range antibiotics often result in collateral damage to the natural human microbiome (Jernberg et al. 2010). Even before the medical community recognized the extent of this problem, it was clear that individuals treated successfully for one species of infectious agent would often succumb to an infection by another secondary foreign bacterial strain.
As most phage strains have a narrow bacterial host range, they are expected to cause minimal perturbations of the normal human microbiome when they are used to treat bacterial infections. Also, as their mechanisms of antibacterial activity of phage are not related to the mode of action of the antibiotic therapies, it should be possible to find effective phage therapies for the antibiotic-resistant disease-associated bacterial strains.
In addition our ability to develop molecular tools that permit manipulations of phage genomes and our knowledge of the microbiology and physiology of phage along with some knowledge of their interactions with the human immune system, they are beginning to be adapted to serve as vaccines. The phage strains are also being used to produce antibacterial proteins that are encoded in their genomes. By manipulating the promoters for these genes, it is possible to produce antibacterial proteins such as the phage-based lysozymes in quantity. The clinical applications of phage, including the development of phage-based vaccines and phage products are now beginning to enter commercial development.
PHAGE ANTIBACTERIAL THERAPY
One of the discoverers of phage, Félix d'Herelle, clearly recognized the clinical potential of phage as an antibacterial agent (Summers 1999). The ability of phage to kill bacteria and in addition to replicate exponentially suggests some of the potential advantages of phage antibacterial therapy. Although d’Herelle’s reported successes in his first clinical applications of antibacterial phage therapies, others had difficulties when they tried to scale up the therapies to larger and/or different patient populations (Merril et al. 2003, 2006). Despite these problems, numerous phage strains were isolated and used clinically in Europe, the Middle East, and Asia (Alisky et al. 1998; Sulakvelidze et al. 2001). Unfortunately, most of the phage data in these studies are presented in a qualitative manner with inadequate clinical details and controls.
Potential Problems for Use of Phage as Antibacterial Therapy
Oral and topical antibacterial applications of phage should present few problems other than an occasional inflammation owing to an immune response or a reaction to bacterial by-products that have not been sufficiently reduced in concentration during the phage preparation. However, system applications require greater care in reducing bacterial contaminations including endo and exotoxins. In addition, phage pharmacokinetic data is rudimentary at best, as many phage strains have not been administered systemically to animals and almost none have been studied carefully in humans. Animal studies have clearly shown that most phage are taken up by the liver with some trapping by the spleen (Merril et al. 2003). In some cases, it has been possible to select phage strains that can remain in the circulation, and remain available for interaction with possible systemic bacterial infectious agents by genetic selection methods (Merril et al. 1996; Vitiello et al. 2005). In a similar manner it should be possible to select phage strains that have a reduced immunogenicity. Reduced immunogenicity, to minimize adverse reactions to phage therapy, may be also accomplished by developing phage strains displaying epitopes that match surface proteins on normal human blood cells. In this regard it might be useful to develop a “platform” phage that is free of toxin and antibiotic resistance genes, but with the ability to stay in the circulation longer, and with reduced immunogenicity. Such a platform phage could be engineered so that it could be easily modified for enhancement of its host range for specific bacterial strains. For example, this could be accomplished with a phage in which the tail proteins could be specified in a manner similar to that found in coliphage K1-5. This phage is a “dual” specificity phage that encodes two different tail proteins allowing it to attack and replicate on both K1 and K5 strains of Escherichia coli (Scholl et al. 2001). Alternatively one could use a site-specific recombination system that permits the phage to switch between alternative tail fiber proteins, or by using a reverse transcriptase controlled tropism switch to generate alternate tail fibers (Sandmeier 1994; Liu et al. 2002). Phage strains considered for therapeutic purposes should be screened for toxins, antibiotic resistance, and genes that increase bacterial pathogenicity (Merril et al. 2003). With the growth of the genomic data banks and the development of bioinformatics, tools such as screens will be become more effective (Pauwels et al. 2009; Verheust et al. 2010).
Animal Models for Treatment of Infectious Diseases
At the beginning of international hostilities in the 1940s, the U.S. Department of Defense sponsored carefully designed phage therapy experiments for animal models of infectious diseases. Despite the encouraging results obtained in these experiments this approach was virtually abandoned in the United States following the discovery and development of broad-spectrum antibiotics (Merril et al. 2003, 2006). However, with the advent of antibiotic-resistant pathogenic bacterial strains, research on the potential of phage therapy has been rejuvenated. In addition, with the expanded knowledge of phage molecular biology and interactions with mammalian immune systems it is possible to genetically engineer phage that might be more efficacious than the wild types found in nature. This potential was shown in the development of long circulating phage strains that could stay in the circulatory system longer than the laboratory strains from which they were derived. There have been a number of successful demonstrations of the effectiveness of phage in animal models of bacterial infectious diseases (e.g., see Dubos et al. 1943; Smith and Huggins 1982; Merril et al. 1996; Biswas et al. 2002; Bull et al. 2002; Gu et al. 2012) including a recent success in the treatment of rats using an animal model system for sepsis and meningitis caused by S242, a fatal neonatal meningitis E. coli strain (Pouillot et al. 2012).
Clinical Studies
One of the first problems encountered by the infectious disease physician is to determine what agent would offer an efficacious treatment for the patient’s disease. Although the narrow host range of phage strains would appear to offer an impediment for the clinical adaptation of phage therapy, it should be possible to address this problem by developing libraries of clinically relevant phage strains carrying marker genes. The use of such strains could provide both an identification of the infecting bacterial strains along with information as to which strains of phage might offer an effective therapy for the patient’s infection (Merril et al. 2003, 2006). One of the major problems confronting phage therapy is the need and cost of adequate clinical trials. This is of course a problem for any new antibacterial therapy. The early clinical applications often lacked the new standard double-blind statistically valid experimental designs (Carlton 1999). The years of experience devoted to developing an understanding of the molecular biology, physiology, and methods for the purification of phage should be an asset in helping to design clinical applications and evaluations. Clinical trials needed to meet the full requirements of the U.S. Food and Drug Administration (FDA) for therapeutic phage have not yet been fulfilled for any of the phage strains that have been shown to be effective in animal models of infectious diseases. Such trials are expensive; however, given the need for such agents, for the treatment of antibiotic-resistant bacterial infectious organisms, approval should be gained for some in the not-too-distant future.
USE OF PHAGE COMPONENTS
As Prophylactic Agents
Despite reasonable success of phage therapy in diagnosis and treatment of bacterial infections by bacteriophages as discussed above, parallel research has been progressing in the use of bacteriophage components mostly as prophylaxis. Lysozymes, originally discovered by Alexander Fleming (Fleming 1922), are ubiquitous in nature, being associated with phage in mammals. Lysozymes have been used for decades as a prophylactic agent to kill Gram-positive bacteria because of their ability to degrade cell walls from outside by hydrolysis of one of the four major bonds in the peptidoglycan, which results in hypotonic bursting of the inner membrane and leaking of intracellular components and thereby causing cell death. Lysozymes from different sources including phage have been used as an adduct in the brewing industry and in yogurt and other milk products. As discussed below, phage-encoded lysozymes are of two kinds: (1) endolysin, which is made by phage during phage lytic growth after infection of bacterial host, and which lyses the cell wall from inside to help release phage progeny particles; and (2) phage tail-associated murein lytic enzymes (TAME), which can hydrolyze cell wall bonds from outside after phage adsorption to the host.
Brewing industries face problems with their fermentation systems, as airborne bacteria frequently contaminate them. Bacterial contamination can make a beer turbid, aromatic, odious, or ropy. The most common contaminations are lactic-acid-producing Gram-positive bacteria (Lactobacillus and Pediococcus) and acetic-acid-producing Gram-negative bacteria (Acetobacter). The spoilage comes from the fact that acetic acid and lactic acids, among other by-products, are made by bacteria from sugar. Some measured amount of specific strains of lactic-acid-producing bacteria are allowed during fermentation to keep the pH low, thus facilitating beer production as well as lowering acidity by “malolactic acid” production. In wine making, unwanted contaminations are routinely taken care of by sanitation practices. In fact, direct lysozyme addition acts as a preservative for storage of foods like yogurt, tofu, cheese, and sake (Larson 2005).
More recently, the addition of phage endolysins has been shown to protect against a broad spectrum of Gram-positive bacteria. If microbial contamination is envisioned, lysozyme is used to kill the lactic acid bacteria. In fact, lysozyme can be added to the final product. Depending on factors like temperature, pH, etc., lysozyme starts working immediately after addition and kills bacteria at once. Lysozyme in beer remains 50% active for at least a 6-month period. Lysozyme present in beer has minimal effects on the physical and sensory properties of the beer (Feeney and Nagy 1952; Daeschel et al. 1991; Landschoot 2005; Waite and Daeschel 2007).
As Curative Agents
More recently, however, lysozymes are being developed for treatment of mammalian infections. Endolysin, used by phage to release progeny particles from bacterial hosts, acts by breaking up the cell from inside. This occurs after another phage product, called holin, creates pores in the inner membrane through which lysozymes can pass through the membrane and reach the cell wall (Wang et al. 2003). However, peptidoglycan-degrading enzymes have the ability to digest bacterial cell walls. The phage-encoded endolysins precisely do that except in a species- or genus-specific manner (Nelson et al. 2001; Fischetti 2010). Phage-encoded endolysin usually can lyse from outside only the corresponding host. Recently, cell wall lysis-mediated killing activity of phage-encoded pure lysozymes has been successfully used to its maximal level by, among others, Fischetti and colleagues, and Loessner and colleagues. Like the current surge in bacteriophage therapy, development of endolysin-mediated therapy of infection was motivated by the emergence of antibiotic-resistant bacteria in clinics. Endolysins create holes in the cell from outside by peptidoglycan digestion and expansion of the inner cytoplasmic membrane and subsequent hypotonic lysis.
Most human infections begin at the mucosal membranes followed by their colonization, which are usually a reservoir of many pathogens. Very few anti-infective agents are known that prevent mucosal colonization. Nonetheless, if one can reduce the bacterial load of the mucous membranes in the community, in hospitals, and in nursing homes, the incidence of the diseases would very likely reduce. Precisely, endolysins are expected to be very effective in such cases. Specific endolysins have now been identified to very effectively kill Gram-positive bacteria (Nelson et al. 2001; Loeffler et al. 2003). Interestingly, the endolysins bind very tightly to their cell wall substrates and thus and do not have a turnover number requiring multiple molecules to bind and hydrolyze several cell wall bonds to make effective cell wall lysis (Loessner et al. 2002; Jervis et al. 2005). An oral colonization animal model has been developed with Streptococcus pyogenes, a nasal model with pneumococci, and a vaginal model with group B streptococci. These studies showed, for example, that nanogram quantities of endolysin kill S. pyogenes 106-fold in 2–4 h after lysozyme treatments. All current success in dealing with endolysin-mediated cell killing are only with Gram-positive bacteria simply because the enzyme when added from outside can access the cell wall peptidoglycan. Gram-negative bacteria, however, have an outer membrane that sterically interferes with lysozyme action thus making lysozyme-mediated cell killing generally ineffective. However, recently, a structurally engineered phage lysozyme containing the FyuA-binding domain of pesticin fused to the amino terminus of T4 lysozyme has given encouraging results against Gram-negative pathogens (Lukacik et al. 2012).
Immunity and Resistance
Both in bacteriophage therapy and in direct lysozyme (of phage or any other source) therapy, it has always been feared that such foreign objects or proteins when used systematically would develop neutralizing antibodies and thus hinder their antibacterial action in the future. However, it has been found that highly immune serum slows down but does not block bacteriolytic activities of pneumococcal-specific endolysins (Loeffler et al. 2001). Another concern of endolysin treatment was its short circulation time after administration (Loeffler et al. 2001). But this issue did not affect the treatment because of the very rapid action time of the lysozyme molecules. Both in antibiotic and phage therapy, another major concern is the phenomenon of the increase of resistant pathogenic strains. Interestingly, every attempt to generate resistant mutants against endolysins in the laboratory setup has so far failed (Loeffler et al. 2001). Although this gives more credence to the potential of lysozyme therapy, we note that most antibiotic-resistance elements that have infiltrated into antibiotic therapy are because of bacteria-acquiring resistance elements from natural environments by lateral gene transfer.
Use of Lysozyme for Treating Bacterial Contamination in Plant Cell Cultures
Even egg white lysozyme has been successfully used to reduce Bacillus circulans and Sphingomonas paucimobilis infection of in vitro shoot cultures quince and hybrid rootstock (Marino et al. 2003). Although lysozyme did not have a negative effect on shoot growth, under optimal conditions it was effective in eliminating B. circulans in quince shoot and hybrid rootstock cultures, and has a bacteriostatic effect on S. paucimobilis. These results suggest that lysozyme may be able to replace antibiotic treatments of in vitro shoot cultures although it may not be effective against every plant bacterial infection.
Use of Bacteriophage Tail-Associated Lytic Enzymes as Antibacterial Therapeutic Agents
It has been known for a while that peptidoglycan-degrading murein hydrolases cleave bacterial cell walls very efficiently in a species-specific manner. Thus, they have the potential of being therapeutic agents against specific pathogens. Recently, this idea has been successfully exploited. These hydrolases are different from the phage-encoded endolysins, which are made and used by phage to come out of host cells by cell wall degradation from inside as discussed above. Unlike the endolysins the murein hydrolases are present at the tip of tails of phage virions. It is believed that the tail-attached hydrolases help DNA injection after phage adsorption. If a bacterial cell is infected by phage particles at very high multiplicities, then the cell lyses before phage DNA starts replication. It has been called “lysis from without” (Delbruck 1940). These tail hydrolases are more or less ubiquitous among tailed phages. Motivated by the emergence of drug-resistant human pathogen Staphylococcus aureus, Paul et al. (2011) identified the muralytic activity of the tail of the broad host range staphylococcal-specific phage, called phage K. These investigators showed the efficacy of the K-encoded purified hydrolase as a bacteriocidal agent in cell culture. The efficacy of this protein was further enhanced by making a hybrid protein comprising the catalytic domain of the hydrolase and the staphylococcal cell wall binding domain of a bacteriocin called lysostaphin (Baba and Schneewind 1998) to generate a protein called P128. The bacteriolytic activity of P128 was more than two orders of magnitude higher than the catalytic domain of the hydrolase alone because of increased substrate specificity. The hybrid has already been tested in experimental nasal colonization of methicillin-resistant S. aureus (MRSA) in experimental rats; the protein has been shown extremely effective in decolonizing the animals. Thus it is an excellent prospective therapeutic against infection.
PHAGE-MEDIATED VACCINE
Prevention of infectious diseases by direct inoculation of a minute quantity of pathogens was shown in China during the 15th century, where healthy children were inoculated with a minute quantity of exudates collected from the pustule of smallpox-infected persons. It was observed that the process of this crude inoculation technique protected the children from future epidemics of smallpox (Tizard 1984; Needham 1999). Despite the fact that the science behind the treatment of infectious diseases through vaccination was first shown by Edward Jenner more than three centuries ago (Riedel 2005) and that use of vaccine in treatment of diseases is widespread now, there are still problems and limitations in preparing vaccines by traditional means. This set the stage to explore other approaches for producing better vaccines (Burdin et al. 2004; O’Hagan and Rappuoli 2004). The advancement of recombinant DNA technology led us to produce vaccine candidates that are recombinant protein molecules displaying subunits of pathogens. However, such vaccine candidates lack many immunogenic features of the original pathogens (Petrovsky and Aguilar 2004). Although many of these recombinant vaccines contain succession of immunodominant epitopes, they fail to stimulate the production of neutralizing antibodies. To overcome this situation, currently many of these recombinant vaccines are mixed with adjuvants to enhance immunostimulatory effects. Unfortunately, the adjuvants that have been tried are either questionable or the process is time consuming and thus not an ideal method of developing vaccines for rapidly evolving diseases like swine flu (Barrett and Stanberry 2008).
Previously immunostimulatory effects of synthetic-immunodominant peptides (epitopes) of antigen and recombinant viruses or nucleotide acid-based (DNA) vaccines were evaluated through various clinical trials. Results of these trials indicated that these vaccines were lacking an effective delivery system for proper activation of the immune system (Petrovsky and Aguilar 2004). These activation deficiencies were reflected in the inability of the vaccines to elicit both humoral and cell-mediated immunity against targeted pathogens. Thus, development of a proper delivery system for any type of subunit or DNA vaccine is of paramount importance (Burdin et al. 2004).
Vaccinologists are now evaluating bacteriophages (phages) as vaccine delivery agents, and the idea is gaining popularity owing to the inherited character of phages to stimulate both arms of the immune system (humoral and cell-mediated immunity).
The ability of phage to deliver gene(s) to mammalian cells was discovered in studies in which lambda (λ) phage carrying a galactose transferase gene was shown capable of inducing galactose transferase enzyme activity in human fibroblast cells from a child lacking the capacity to make the Gal-transferase enzyme (Merril et al. 1971). This ability of phage to deliver genes to human cells was later independently confirmed with a phage carrying the β-galactosidase gene in human cells deficient in that enzyme activity (Horst et al. 1975).
A concern that was expressed concerning these experiments is that the DNA used to correct the genetic defects in the eukaryotic cells is bacterial or prokaryotic. However, it has now been clearly shown that prokaryotic genes, such as the genes in a segment of the Ti plasmid of the bacteria Agrobacterium tumefaciens can function in eukaryotes. These genes have been shown to be capable of transforming eukaryotic cells in plants and they are the cause of the commonly observed plant crown gall tumors. In addition, the bacterial Ti plasmid genes have also been shown to be capable of affecting yeast, filamentous fungi, cultivated mushrooms, and human cells in culture (Lacroix et al. 2006).
Currently, two separate approaches of antigen delivery systems are attempted with phage vaccines. These systems are designated as “phage display vaccines” and “phage DNA vaccines” (Clark and March 2004a). Broadly, phage display vaccines are mainly referred for producing immunogenic phage particles that display foreign antigen on phage surface. This generally is achieved by expressing antigens as fusion products of one of the major surface proteins of phage virion; phage DNA vaccines, on the other hand, are produced by incorporation of foreign antigen genes in phage genome under the control of strong eukaryotic promoters. In the latter case, phage acts as a passive carrier to transfer the foreign DNA in mammalian cells where the antigen gene is expressed. These two methods are often combined to produce phages that carry a foreign antigen gene along with a display protein or peptide on the surface. The display proteins or peptides are generally selected for their specific binding affinity of antigen-presenting cells. Thus, such phage constructs pose the additional ability of delivering the antigen gene directly to the immunoreactive cells like dendritic cells, Kupffer cells, etc. (Zanghi et al. 2007).
Phage Display Vaccine
A filamentous phage M13 display system was first used for expression of antibody fragments and their affinity maturation (Benhar 2001). Although M13 is successful in several cases in displaying melanoma-specific tumor antigen to produce cancer vaccines effective in reducing tumor growth in animal models (Fang et al. 2005), the M13 display system has limitations (Gupta et al. 2003). The system is not efficient in displaying larger proteins when using the high copy coat protein gp VIII. The other coat protein, gpIII, which can effectively display foreign proteins or peptides, has a low copy number in the M13 virion (Smith 1985).
Soc and Hoc, two of the virion proteins of phage T4, are more effective major display platforms for foreign antigens. Unlike M13, T4 is able to display peptides of various sizes and sequences without much problem. Hoc and Soc are also highly immunogenic and therefore act as good adjuvants. Thus, T4 phage is considered as a better display platform than filamentous phages (Li et al. 2006a). Currently, several vaccines for infectious diseases are prepared by using the T4 phage display system, which has shown promising results in animal models (Jiang et al. 1997).
T7 is another phage capable of displaying proteins and peptides including antigens. Fusion products are generated by cloning the coding sequences for the peptides and proteins at the carboxyl end of the capsid protein, 10B (Lindner et al. 2011). Lewis lung cancer vaccine prepared by T7 phage display of vascular endothelial growth factor (VEGF) has been successfully used to break immunologic tolerance. When injected in mice, T7-VEGF display produced strong immunogenic response against cancer cells and inhibited cancer growth (Li et al. 2006b). The comparative aspects of different phages for antigen display are shown in Table 1.
Table 1.
List of phages and their virion proteins, which are used for display
| Vector phage | Phage virion proteins for fusion display | Copy number of display | Size of display shown |
|---|---|---|---|
| M13 | gpVIII | ≈2700 | Small peptide (6–8 amino acids) |
| gpIII | ≈5 | Large protein with reduced viability | |
| T4 | Soc | ≈960 | Large protein (up to 837 amino acids) |
| Hoc | ≈160 | Protein (up to 183 amino acids) | |
| T7 | 10A | ≈415 | Protein (40–50 amino acids) |
| 10B | ≈1 | Protein (up to 1200 amino acids) | |
| Lambda (λ) | gpD | ≈420 | Large protein (1024 amino acids) |
Data were taken from Smith (1985), Markland et al. (1991), Mikawa et al. (1996), Ren et al. (1996, 1997), Ren and Black (1998), Iwasaki et al. (2000), Savinov and Austin (2001), Sche et al. (2001), Gupta et al. (2003), Pacheco et al. (2006), and Li et al. (2007).
The above phages are used for presentation of displayed antigens to mammalian cells; the λ system was also developed as a DNA delivery platform to antigen-presenting cells (APC).
Recently, the common cloning vector phage lambda (λ) is also gaining popularity as a phage display platform. The complex cloning method of λ phage display is considerably simplified by using a Cre-loxP site-specific recombination process to develop a λ phage display system that facilitates rapid high efficiency cloning of foreign DNA in λ genome (Gupta et al. 2003). The method allows displaying foreign genes fused to λ capsid protein gene D; nearly about 450 copies of foreign antigen can display as D fusion on λ surface (Gupta et al. 2003). The ability of λ to display multiple copies of the same antigen on the surface of a single phage particle is more effective in eliciting a strong immune response than low copy display (Clark and March 2004a). Production of successful λ display antigen to produce neutralizing antibodies against infectious porcine circovirus infection has been shown by Hayes et al. (2010), using gpD fusion antigen on phage λ. Activation of immune system by phage display vaccine is described in Figure 1.
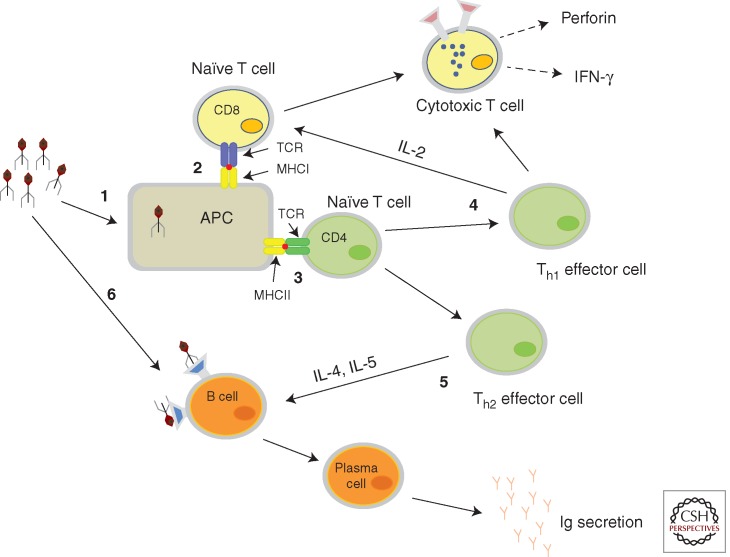
Figure 1.
Immune activation of mammalian system with phage display antigens. (1) Particulate nature of phage activates the antigen-presenting cells (APC), which process the antigens for immune presentation. (2) Presentation of processed antigens by major histocompatibility complexes (MHC) class I molecules to CD8 T cells, which leads to T-cell activation. (3) Antigens are presented by MHC class II molecules to CD4 T cell, which in turn activates Th1 and Th2 effector cells. (4) Th1 cells generate cytotoxic T-cell (CTL) responses and help produce interferon γ (IFN-γ). (5) Th2 cells activate B cells to make antibodies. (6) Direct activation of B cells by phage vaccines also leads to massive antibody response. T-cell receptors are denoted by TCR.
Phage DNA Vaccine
Successful delivery and expression of foreign genes in eukaryotic cells by phage-mediated gene transfer is also in common use now. The use of strong eukaryotic promoters has extended the capacity of phage to deliver foreign antigen genes to mammalian cells. Generally for production of vaccine, the antigen genes under the control of a eukaryotic promoter are cloned inside of a nonessential region of phage genome. When injected in a mammalian system, this phage vaccine acting as a DNA vaccine can induce potent immune response by expressing foreign antigen inside of APCs (Fig. 2) (Clark and March 2006). Several λ-based DNA vaccines for infectious diseases have been prepared that have shown promising results in animal models (Clark and March 2004b; March et al. 2004).
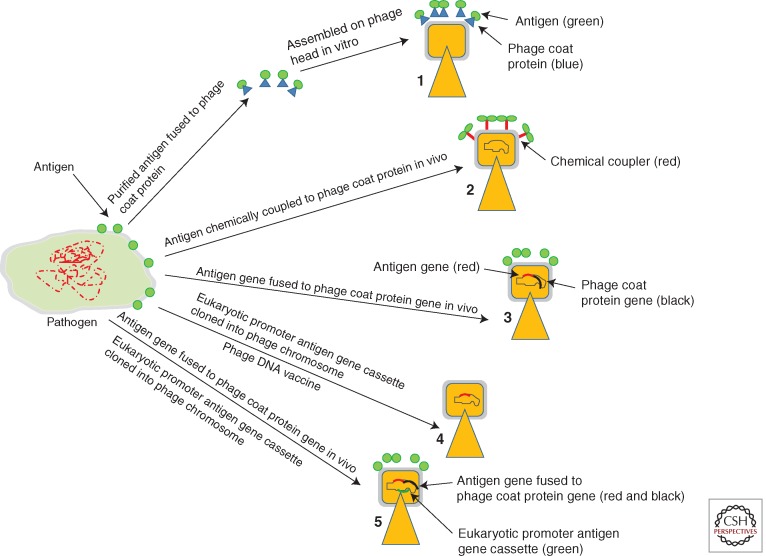
Figure 2.
Schematic representations of several phage-mediated vaccine delivery systems. (1) Production of virus-like particles (VLPs) by adding purified phage capsid proteins fused to antigens during assembly of phage particles in vitro. (2) Antigens are attached by chemical conjugation on preassembled phage head. (3) Antigens are displayed on phage surface by fusion of antigen genes with phage capsid protein genes. (4) Phage as a DNA delivery vehicle where antigen genes are cloned in phage genome under the control of eukaryotic promoters. (5) Phage for DNA vaccine, where phage carries antigen genes under the control of eukaryotic promoters. The phage also displays foreign proteins on its surface as fusion of phage capsid proteins. This protein targets the phage to antigen-presenting cells (APC).
Advantages of Phage-Mediated Vaccine Delivery
Recent global surveillance indicates that infectious microorganisms are emerging at an alarming rate (Barrett and Stanberry 2008). There is also a significant concern about the potential of generating highly virulent microorganisms using genetic engineering and synthetic biology techniques. The outbreak caused by these types of natural or man-made variants can spread rapidly and has to be stopped before diseases become epidemic. The quarantine and rapid vaccination against targeted pathogens is the only way to prevent epidemics during massive disease outbreaks. One of the best advantages of phage vaccine is that it can be produced inexpensively and deployed rapidly during disease outbreaks.
Utilizing advanced knowledge in bioinformatics, gene sequencing, and phage genetics, it is possible to create and manufacture vaccines in record time. Genomic and proteomic data of any old or newly emerging pathogenic bacteria or virus can be scanned using existing algorithms and database management systems to identify novel antigenic motifs that can be cloned into phage genome to produce phage vaccine. The resulting phage vaccine can propagate rapidly using bacteriological media and does not need cell culture or a chicken-egg-based system for manufacturing sufficient quantities of vaccine to protect against a new pathogen; such preparations can be made in weeks rather than months. The phage particles are nonpathogenic to humans and the resultant vaccines do not require adjuvants. By using phage vaccine, all relevant information can be delivered to the immune system to allow for an immune response that includes antibody production and the development of immunity to a pathogen (Fig. 1). The particulate nature of phage vaccine attracts APCs that engulf, process, and present phage-mediated antigens through MHCs (class I and class II pathways) (Manoutcharian et al. 1999; Gaubin et al. 2003) to evoke both cell-mediated and humoral immunities (Fig. 1). It was expected that phage as an extracellular antigen will only activate antibody production through MHC class II bias pathway, but it has been shown that phage as a particulate antigen can access the MHC I pathway through cross priming, which activates a cellular immune response (Gaubin et al. 2003). In addition, the immunostimulatory unmethylated CpG motifs of the phage genome are recognized by the innate immune cells through Toll-like receptors (TLRs) (Krieg et al. 1995; Mason et al. 2005), which further enhance immunity. Immunostimulatory effect of phage vaccination through oral route has been evaluated by Clark et al. They have succeeded in making a phage DNA vaccine against Yersina pestis by cloning V-antigen in phage λ genome. Upon oral administration, it produced a significant immune response in mouse (Clark and March 2004b). Others have also explored the oral delivery route of phage vaccination, and showed antigen-specific response (Delmastro et al. 1997; Zuercher et al. 2000). These results indicate the possibility of oral delivery of phage vaccine in developing countries.
In summary, phage particles pose several intrinsic characters that make them lucrative for developing vaccine-delivery platforms. The natural stability of phage particles provides easy processing and cheaper manufacturing capability for large-scale production of vaccines. The particulate nature of phage makes them easy to purify by simple centrifugation steps, which are far easier and cheaper than the methods applied for purifying soluble recombinant protein vaccines. Additionally, phage itself acts as a strong adjuvant and thus enhances excellent immune response against any antigens present along with phage (Frenkel et al. 2000; Manoutcharian et al. 2004). In fact, the response against phage-displayed antigens can break immunological tolerance against self-proteins and thus is an ideal vehicle for delivering cancer antigens (Fang et al. 2005).
PERSPECTIVES
The ever-increasing list of antibiotic-resistant bacterial strains and newly emerging viruses is forcing a reexamination of alternate modes of therapies for infectious diseases. Of these, the phage and their molecular components and products offer a number of new therapeutic alternatives. Although the development of phage-based applications will require new directions, there are advantages over the prior art of treating infectious disease. A useful aspect is the ability to develop therapies for bacterial infection that minimally perturb the human microbiome. As recent research of the microbiome has shown, even small alterations can have effects that range from alterations in the perception of hunger to increased susceptibility to lethal infections by foreign bacterial strains. These advances in our ability to work with phage extend from the treatment of human infections to increasing the efficiency of industrial processes, particularly those that rely on fermentation. The development of phage-based delivery of antigens and genes will result in new vaccines for protection from numerous diseases and new therapeutic approaches to both infectious and noninfectious diseases, such as some forms of cancer. The newly developed genomic sequencing methods along with other high-throughput technologies including cell sorters and multiwell metabolic monitoring robots should result in the rapid development of the needed information for phage-based therapies.
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Alisky J, Iczkowski K, Rapoport A, Troitsky N 1998. Bacteriophages show promise as antimicrobial agents. J Infect 36: 5–15 [DOI] [PubMed] [Google Scholar]
- Baba T, Schneewind O 1998. Instruments of microbial warfare: Bacteriocin synthesis, toxicity and immunity. Trends Microbiol 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Barrett ADT, Stanberry LR 2008. Vaccines for biodefence and emerging and neglected diseases. Academic, Waltham, MA [Google Scholar]
- Benhar I 2001. Biotechnological applications of phage and cell display. Biotechnol Adv 19: 1–33 [DOI] [PubMed] [Google Scholar]
- Biswas B, Adhya S, Washart P, Paul B, Trostel AN, Powell B, Carlton R, Merril CR 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun 70: 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Levin BR, DeRouin T, Walker N, Bloch CA 2002. Dynamics of success and failure in phage and antibiotic therapy in experimental infections. BMC Microbiol 2: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdin N, Guy B, Moingeon P 2004. Immunological foundations to the quest for new vaccine adjuvants. BioDrugs 18: 79–93 [DOI] [PubMed] [Google Scholar]
- Carlton RM 1999. Phage therapy: Past history and future prospects. Arch Immunol Ther Exp 47: 267–274 [PubMed] [Google Scholar]
- Clark JR, March JB 2004a. Bacterial viruses as human vaccines? Expert Rev Vaccines 3: 463–476 [DOI] [PubMed] [Google Scholar]
- Clark JR, March JB 2004b. Bacteriophage-mediated nucleic acid immunisation. FEMS Immunol Med Microbiol 40: 21–26 [DOI] [PubMed] [Google Scholar]
- Clark JR, March JB 2006. Bacteriophages and biotechnology: Vaccines, gene therapy and antibacterials. Trends Biotechnol 24: 212–218 [DOI] [PubMed] [Google Scholar]
- Daeschel MA, Jung DS, Watson BT 1991. Controlling wine malolactic fermentation with nisin and nisin-resistant strains of Leuconostoc oenos. Appl Environ Microbiol 57: 601–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbruck M 1940. The growth of bacteriophage and lysis of the host. J Gen Physiol 23: 643–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmastro P, Meola A, Monaci P, Cortese R, Galfre G 1997. Immunogenicity of filamentous phage displaying peptide mimotopes after oral administration. Vaccine 15: 1276–1285 [DOI] [PubMed] [Google Scholar]
- Dubos RJ, Straus JH, Pierce C 1943. The multiplication of bacteriophage in vivo and its protective effect against an experimental infection with Shigella dysenteriae. J Exp Med 78: 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Wang G, Yang Q, Song J, Wang Y, Wang L 2005. The potential of phage display virions expressing malignant tumor specific antigen MAGE-A1 epitope in murine model. Vaccine 23: 4860–4866 [DOI] [PubMed] [Google Scholar]
- Feeney RE, Nagy DA 1952. The antibacterial activity of the egg white protein conalbumin. J Bacteriol 64: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA 2010. Bacteriophage endolysins: A novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol 300: 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A 1922. On a remarkable bacteriolytic element found in tissues and secretions. P Roy Soc London B 93: 306–317 [Google Scholar]
- Frenkel D, Katz O, Solomon B 2000. Immunization against Alzheimer’s β-amyloid plaques via EFRH phage administration. Proc Natl Acad Sci 97: 11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubin M, Fanutti C, Mishal Z, Durrbach A, De Berardinis P, Sartorius R, Del Pozzo G, Guardiola J, Perham RN, Piatier-Tonneau D 2003. Processing of filamentous bacteriophage virions in antigen-presenting cells targets both HLA class I and class II peptide loading compartments. DNA Cell Biol 22: 11–18 [DOI] [PubMed] [Google Scholar]
- Gu J, Liu X, Li Y, Han W, Lei L, Yang Y, Zhao H, Gao Y, Song J, Lu R, et al. 2012. A method for generation phage cocktail with great therapeutic potential. PloS ONE 7: e31698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Onda M, Pastan I, Adhya S, Chaudhary VK 2003. High-density functional display of proteins on bacteriophage λ. J Mol Biol 334: 241–254 [DOI] [PubMed] [Google Scholar]
- Hayes S, Gamage LN, Hayes C 2010. Dual expression system for assembling phage λ display particle (LDP) vaccine to porcine Circovirus 2 (PCV2). Vaccine 28: 6789–6799 [DOI] [PubMed] [Google Scholar]
- Horst J, Kluge JF, Beyreuther K, Gerok W 1975. Gene transfer to human cells: Transducing phage λ plac gene expression in GM1-gangliosidosis fibroblasts. Proc Natl Acad Sci 72: 3531–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, Trus BL, Wingfield PT, Cheng N, Campusano G, Rao VB, Steven AC 2000. Molecular architecture of bacteriophage T4 capsid: Vertex structure and bimodal binding of the stabilizing accessory protein, Soc. Virology 271: 321–333 [DOI] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK 2010. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156: 3216–3223 [DOI] [PubMed] [Google Scholar]
- Jervis EJ, Guarna MM, Doheny JG, Haynes CA, Kilburn DG 2005. Dynamic localization and persistent stimulation of factor-dependent cells by a stem cell factor/cellulose binding domain fusion protein. Biotechnol Bioeng 91: 314–324 [DOI] [PubMed] [Google Scholar]
- Jiang J, AbuShilbayeh L, Rao VB 1997. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect Immun 65: 4770–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM 1995. Cpg motifs in bacterial-DNA trigger direct B-cell activation. Nature 374: 546–549 [DOI] [PubMed] [Google Scholar]
- Lacroix B, Tzfira T, Vainstein A, Citovsky V 2006. A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet 22: 29–37 [DOI] [PubMed] [Google Scholar]
- Landschoot AV 2005. Antibacterial properties of hen egg white lysozyme against beer spoilage bacteria and effect of lysozyme on yeast fermentation. Cerevisia 32: 219–225 [Google Scholar]
- Larson AE 2005. Lysozyme in antimicrobial in food. Taylor and Francis, London [Google Scholar]
- Li Q, Shivachandra SB, Leppla SH, Rao VB 2006a. Bacteriophage T4 capsid: A unique platform for efficient surface assembly of macromolecular complexes. J Mol Biol 363: 577–588 [DOI] [PubMed] [Google Scholar]
- Li XH, Tang L, Liu D, Sun HM, Zhou CC, Tan LS, Wang LP, Zhang PD, Zhang SQ 2006b. Antitumor effect of recombinant T7 phage vaccine expressing xenogenic vascular endothelial growth factor on Lewis lung cancer in mice. Ai Zheng 25: 1221–1226 [PubMed] [Google Scholar]
- Li Q, Shivachandra SB, Zhang Z, Rao VB 2007. Assembly of the small outer capsid protein, Soc, on bacteriophage T4: A novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid. J Mol Biol 370: 1006–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner T, Kolmar H, Haberkorn U, Mier W 2011. DNA libraries for the construction of phage libraries: Statistical and structural requirements and synthetic methods. Molecules 16: 1625–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, et al. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295: 2091–2094 [DOI] [PubMed] [Google Scholar]
- Loeffler JM, Nelson D, Fischetti VA 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294: 2170–2172 [DOI] [PubMed] [Google Scholar]
- Loeffler JM, Djurkovic S, Fischetti VA 2003. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect Immun 71: 6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessner MJ, Kramer K, Ebel F, Scherer S 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol Microbiol 44: 335–349 [DOI] [PubMed] [Google Scholar]
- Lukacik P, Barnard TJ, Keller PW, Chaturvedi KS, Seddiki N, Fairman JW, Noinaj N, Kirby TL, Henderson JP, Steven AC, et al. 2012. Proc Natl Acad Sci 109: 9857–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoutcharian K, Terrazas LI, Gevorkian G, Acero G, Petrossian P, Rodriguez M, Govezensky T 1999. Phage-displayed T-cell epitope grafted into immunoglobulin heavy-chain complementarity-determining regions: An effective vaccine design tested in murine cysticercosis. Infect Immun 67: 4764–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoutcharian K, Diaz-Orea A, Gevorkian G, Fragoso G, Acero G, Gonzalez E, De Aluja A, Villalobos N, Gomez-Conde E, Sciutto E 2004. Recombinant bacteriophage-based multiepitope vaccine against Taenia solium pig cysticercosis. Vet Immunol Immunopathol 99: 11–24 [DOI] [PubMed] [Google Scholar]
- March JB, Clark JR, Jepson CD 2004. Genetic immunisation against hepatitis B using whole bacteriophage λ particles. Vaccine 22: 1666–1671 [DOI] [PubMed] [Google Scholar]
- Marino G, Ferrarini V, Giardini S, Biavati B 2003. Use of lysozyme for treatment of bacterial contamination in in vitro shoot cultures of fruit plants. In Vitro Cell Dev Biol 39: 327–331 [Google Scholar]
- Markland W, Roberts BL, Saxena MJ, Guterman SK, Ladner RC 1991. Design, construction and function of a multicopy display vector using fusions to the major coat protein of bacteriophage M13. Gene 109: 13–19 [DOI] [PubMed] [Google Scholar]
- Mason KA, Ariga H, Neal R, Valdecanas D, Hunter N, Krieg AM, Whisnant JK, Milas L 2005. Targeting toll-like receptor 9 with CpG oligodeoxynucleotides enhances tumor response to fractionated radiotherapy. Clin Cancer Res 11: 361–369 [PubMed] [Google Scholar]
- Merril CR, Geier MR, Petricciani JC 1971. Bacterial virus gene expression in human cells. Nature 233: 398–400 [DOI] [PubMed] [Google Scholar]
- Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S 1996. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci 93: 3188–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril CR, Scholl D, Adhya SL 2003. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2: 489–497 [DOI] [PubMed] [Google Scholar]
- Merril CR, Scholl D, Adhya S 2006. The bacteriophage. Oxford University Press, London [Google Scholar]
- Mikawa YG, Maruyama IN, Brenner S 1996. Surface display of proteins on bacteriophage λ heads. J Mol Biol 262: 21–30 [DOI] [PubMed] [Google Scholar]
- Needham J 1999. Science and civilisation in China. Cambridge University Press, Cambridge [Google Scholar]
- Nelson D, Loomis L, Fischetti VA 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci 98: 4107–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan DT, Rappuoli R 2004. Novel approaches to vaccine delivery. Pharm Res 21: 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco S, Gomez I, Sato R, Bravo A, Soberon M 2006. Functional display of Bacillus thuringiensis Cry1Ac toxin on T7 phage. J Invertebr Pathol 92: 45–49 [DOI] [PubMed] [Google Scholar]
- Paul VD, Sundarrajan S, Rajagopalan SS, Hariharan S, Kempashanaiah N, Padmanabhan S, Sriram B, Ramachandran J 2011. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol 11: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels K, Gijsbers R, Toelen J, Schambach A, Willard-Gallo K, Verheust C, Debyser Z, Herman P 2009. State-of-the-art lentiviral vectors for research use: Risk assessment and biosafety recommendations. Curr Gene Ther 9: 459–474 [DOI] [PubMed] [Google Scholar]
- Petrovsky N, Aguilar JC 2004. Vaccine adjuvants: Current state and future trends. Immunol Cell Biol 82: 488–496 [DOI] [PubMed] [Google Scholar]
- Pouillot F, Chomton M, Blois H, Courroux C, Noelig J, Bidet P, Bingen E, Bonacorsi S 2012. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b:H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob Agents Chemother 56: 3568–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Black LW 1998. Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene 215: 439–444 [DOI] [PubMed] [Google Scholar]
- Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW 1996. Phage display of intact domains at high copy number: A system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci 5: 1833–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZJ, Baumann RG, Black LW 1997. Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: Construction of a T4 phage hoc gene display vector. Gene 195: 303–311 [DOI] [PubMed] [Google Scholar]
- Riedel S 2005. Edward Jenner and the history of smallpox and vaccination. Proc (Bayl Univ Med Cent) 18: 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol Microbiol 12: 343–350 [DOI] [PubMed] [Google Scholar]
- Savinov SN, Austin DJ 2001. The cloning of human genes using cDNA phage display and small-molecule chemical probes. Comb Chem High T Scr 4: 593–597 [DOI] [PubMed] [Google Scholar]
- Sche PP, McKenzie KM, White JD, Austin DJ 2001. Corrigendum to: “Display cloning: Functional identification of natural product receptors using cDNA-phage display” [Chemistry & Biology 6 (1999) 707–716]. Chem Biol 8: 399–400 [DOI] [PubMed] [Google Scholar]
- Scholl D, Rogers S, Adhya S, Merril CR 2001. Bacteriophage K1–5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J Virol 75: 2509–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP 1985. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228: 1315–1317 [DOI] [PubMed] [Google Scholar]
- Smith HW, Huggins MB 1982. Successful treatment of experimental Escherichia coli infections in mice using phage: Its general superiority over antibiotics. J Gen Microbiol 128: 307–318 [DOI] [PubMed] [Google Scholar]
- Sneade W 2005. Drug discovery: A history. Wiley-Interscience, Hoboken, NJ [Google Scholar]
- Sulakvelidze A, Alavidze Z, Morris JG Jr 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers WC 1999. Félix ďHerelle and the origins of molecular biology. Yale University Press, New Haven and London [Google Scholar]
- Tizard IR 1984. Immunology: An introduction. Cengage Learning, Stamford, CT [Google Scholar]
- Verheust C, Pauwels K, Mahillon JD, Helinski R, Herman P 2010. Contained use of bacteriophages: Risk assessment and biosafety recommendations. Appl Biosafety 15: 32–44 [Google Scholar]
- Vitiello CL, Merril CR, Adhya S 2005. An amino acid substitution in a capsid protein enhances phage survival in mouse circulatory system more than a 1000-fold. Virus Res 114: 101–103 [DOI] [PubMed] [Google Scholar]
- Waite JG, Daeschel MA 2007. Contribution of wine components to inactivation of food-borne pathogens. J Food Sci 72: M286–M291 [DOI] [PubMed] [Google Scholar]
- Wang IN, Deaton J, Young R 2003. Sizing the holin lesion with an endolysin-β-galactosidase fusion. J Bacteriol 185: 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanghi CN, Sapinoro R, Bradel-Tretheway B, Dewhurst S 2007. A tractable method for simultaneous modifications to the head and tail of bacteriophage λ and its application to enhancing phage-mediated gene delivery. Nucleic Acids Res 35: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuercher AW, Miescher SM, Vogel M, Rudolf MP, Stadler MB, Stadler BM 2000. Oral anti-IgE immunization with epitope-displaying phage. Eur J Immunol 30: 128–135 [DOI] [PubMed] [Google Scholar]