Abstract
Lineage tracing involves labeling cells to track their subsequent behavior within the normal tissue environment. The advent of genetic lineage tracing and cell proliferation assays, together with high resolution three-dimensional (3D) imaging and quantitative methods to infer cell behavior from lineage-tracing data, has transformed our understanding of murine epidermal stem and progenitor cells. Here, we review recent insights that reveal how a progenitor cell population maintains interfollicular epidermis, whereas stem cells, quiescent under homeostatic conditions, are mobilized in response to wounding. We discuss progress in understanding how the various stem cell populations of the hair follicle sustain this complex and highly dynamic structure, and recent analysis of stem cells in sweat and sebaceous glands. The extent to which insights from mouse studies can be applied to human epidermis is also considered.
In the mammalian skin, epidermal cells that have been sloughed or injured must be replaced. Lineage-tracing experiments have provided insight into cellular behaviors that underpin epidermal maintenance and repair.
Mammalian epidermis is both highly dynamic and adaptable. There is constant turnover with cells being shed at the epidermal surface and replaced by proliferation in the basal layer (Leblond 1964). In addition, as the epidermis is the frontier with the external environment, it is frequently injured and must rapidly repair any damage (Gurtner et al. 2008). Here we review the recent insights into the cellular behaviors that underpin adult epidermal maintenance and repair provided by lineage tracing. We also consider the challenge of lineage tracing in the hair follicle and the extent to which findings from transgenic mouse studies may be extrapolated to humans.
The simple organization of the epidermis lends itself to studying cell behavior. The organ comprises sheets of keratinocytes that form the interfollicular epidermis (IFE) punctuated by hair follicles and sweat glands. The appearance of the skin varies markedly between different parts of the body with marked variations in the morphology of differentiated keratinocytes, and the number and distribution of epidermal appendages. For example, in the mouse, “typical” epidermis with a high density of hair follicles is found over most of the body. In contrast, tail epidermis is covered in scales and is sparse in hair, whereas the forepaws are covered in thick skin devoid of hair but with numerous sweat glands (Potten 1974; Spearman and Hardy 1977; Braun et al. 2003; Lu et al. 2012). However, all body sites share some common features. Proliferation is confined to the basal cell layer. In adult mice, basal cells divide in parallel with the underlying basement membrane to produce two basal cell daughters (Sherman et al. 1961; Smart 1970; Clayton et al. 2007; Doupé et al. 2010). On commitment to terminal differentiation, basal cells exit the cell cycle and subsequently migrate into the first suprabasal cell layer. From here they progress through a series of differentiating cell layers, culminating in their being shed from the tissue surface.
It has long been argued that both the lifelong production of epidermal cells and the ability of the epidermis to regenerate after injury depend on stem cells within the basal layer (Adami 1901; Potten and Morris 1988). Two models of self-renewal were proposed. The first, based on short-term analysis of the behavior of cells labeled with H3 thymidine and allowed to divide generating cell pairs, argued that all proliferating cells were equivalent and that after division there was a 50:50 chance of every cell differentiating or going on to divide (Leblond 1964; Marques-Pereira and Leblond 1965). The second hypothesis, derived from cell kinetic observations and the histological structure of mouse epidermis, argued that the tissue was split into regularly sized clonal units (Mackenzie 1970; Potten 1974, 1981). Each “epidermal proliferative unit” (EPU) was sustained by a single, slow-cycling, self-renewing stem cell, which divided asymmetrically to produce a stem cell and a transit-amplifying (TA) cell daughter. The TA cell underwent a limited number of divisions after which all of its progeny differentiated, ensuring that 8–10 differentiated keratinocytes resulted from each stem cell division (Potten 1974). It was the second “stem TA” hypothesis that won out and became profoundly influential, being used to interpret numerous experiments in epidermal biology (Jones et al. 2007).
Despite its popularity, there was a body of data inconsistent with the stem/TA model (Jones et al. 2007; Jones and Simons 2008; Doupé and Jones 2012). These inconsistencies were the motivation for lineage-tracing studies to resolve the behavior of the proliferating cells and explain how homeostasis was achieved. The results argue that IFE is maintained by a population of cells termed progenitors. Individual progenitor cell fate is random, but generates progenitor and differentiating daughters with equal probability, ensuring that homeostasis is achieved across the progenitor cell population. There is also evidence that there are rare, slow-cycling cells in parts of the IFE that generate progenitor and stem cell daughters with equal likelihood. We refer to these cells as stem cells. Stem cells make minimal contribution to epidermal maintenance, but are mobilized following epidermal wounding. In summary, progenitors maintain the epidermis, whereas stem cells remain in reserve in case of injury.
LINEAGE TRACING
Labeling an individual cell or population of cells to trace the fate of its descendants is an approach long used in developmental biology that has only been applied to the epidermis comparatively recently (Kretzschmar and Watt 2012). Modern lineage tracing uses genetic labeling to track the fate of keratinocytes and their progeny (Fig. 1) (Snippert and Clevers 2011; Kretzschmar and Watt 2012; Alcolea and Jones 2013). The decisive advantage of this approach over other methods is that cell behavior is studied in a physiological context rather than after gross environmental alterations such as those in tissue culture or transplantation assays.
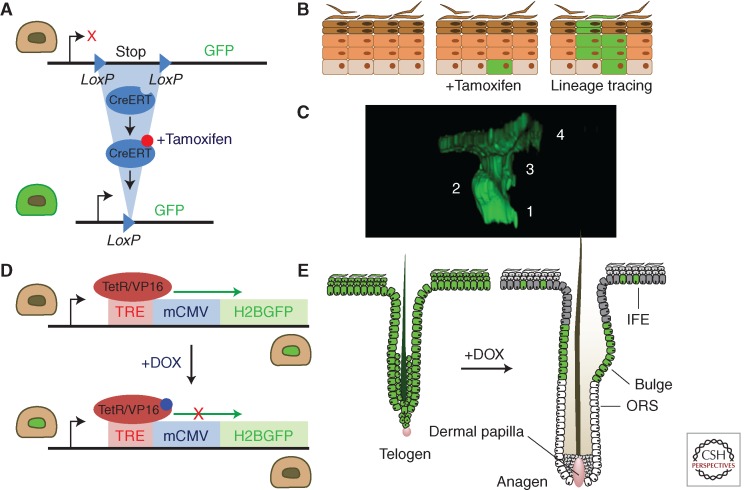
Figure 1.
Lineage tracing and transgenic proliferation assay. (A–C) Genetic lineage tracing with cre recombinase. This is performed in mice expressing two transgenes (A). The first is cre, expressed either from a transgenic promoter or targeted to a specific gene. Cre may be fused to a mutant estrogen receptor (CreERT) so it is only active following treatment with tamoxifen. The second transgene is a reporter, such as green fluorescent protein, (GFP), which often targeted to the ubiquitous Rosa26 locus. The reporter is only expressed following the excision of a loxP flanked “stop” cassette by cre. The progeny of the labeled cell also express the reporter. If cells are labeled at low-frequency, single-cell-derived clones result (B). (C) Rendered confocal z stack showing a typical four-cell clone in AhcreERTR26flEYFP/wt mouse epidermis (numbers indicate individual cells in the clone). (D,E) Transgenic label-retaining assays based on the Histone 2B GFP fusion protein (H2BGFP) system. (D) The expression of the stable H2BGFP protein is under the control of doxycycline (DOX), which activates the TetR/VP16 transcription factor to drive expression of H2BGFP from the tetracycline-responsive regulatory element (TRE) regulatory element. TetR/VP16 is typically expressed from a cell-type-specific promoter. DOX added to the animal’s diet inactivates the H2BGFP expression allowing the fluorescent content to be diluted through cell division. (E) In this example, TetR/VP16 is driven by Keratin 5 promoter, which homogenously labels the epidermal basal layer until DOX treatment. Following cessation of H2BGFP synthesis, the protein is diluted in proliferating cells but retained in more slow-cycling cells (label-retaining cells or LRCs) that remain fluorescent green. Interfollicular LRCs lie adjacent to the hair follicles, whereas in hair follicle, LRCs are found in the bulge region. Sebaceous glands are omitted for clarity. ORS, outer root sheath.
Early lineage-tracing experiments exploited modified deoxyribonucleotides such as tritiated thymidine or bromodeoxyuridine (BrdU), which are taken up by proliferating cells during S phase (Marques-Pereira and Leblond 1965; Taylor et al. 2000; Braun et al. 2003). When the labeled cells divide, the progeny of further divisions inherit the label, enabling them to be visualized until the label is diluted below the detection limit. In adult IFE, both daughters of a cell division are initially found in the basal layer, but over time one or both daughter cells stratify or go on to divide (Doupé et al. 2010).
A second application of labeled nucleotides is the label-retaining cell assay, which is used to detect slow-cycling cells. Historically, it was assumed that LRCs were slow-cycling stem cells based on the prediction of the stem/TA model (Doupé and Jones 2012). In a typical protocol, neonatal mice are administered a course of BrdU to label all the proliferating cells in the epidermis (Bickenbach 1981; Bickenbach and Chism 1998; Braun et al. 2003). Over the succeeding weeks, the label is diluted in dividing cells, but retained in those that divide more slowly. Such assays have shown the presence of LRC in the IFE and the bulge region of the hair follicle (Cotsarelis et al. 1990; Braun et al. 2003). However, these approaches are limited as LRC identified by nucleotide labeling cannot be isolated for molecular characterization.
Nucleotide-based assays remain useful, but have been complemented by transgenic approaches to both track the fate of cycling cells and identify slow-cycling cells. Transgenic lineage tracing exploits genetic labeling to heritably mark a cell and its progeny. This is normally achieved by expressing the enzyme cre recombinase in the cell type under study. Cre activity leads to the expression of a reporter gene, typically β-galactosidase (β-gal) or a fluorescent protein such as GFP, by removing a stop cassette that blocks transcription of the reporter (Fig. 1) (Soriano 1999; Nagy 2000; Srinivas et al. 2001). More recently, multicolor reporters have been developed that randomly label cells with one of three or more colors (Livet et al. 2007; Snippert et al. 2010b; Rinkevich et al. 2011). Many studies exploit drug-regulated forms of cre to control when labeling takes place, most commonly by fusing cre to a mutant estrogen receptor (ERT) (Littlewood et al. 1995). The creERT fusion protein is sequestered in the cytoplasm until the animal is treated with tamoxifen, which frees the protein to enter the nucleus and mediate recombination. To achieve specific labeling of subsets of epidermal cells, many groups have expressed creERT from short transgenic promoters such as keratin 14 (Krt14), which targets basal cells (Vasioukhin et al. 1999). Unfortunately, the random integration of short transgenic promoters into the genome may result in expression patterns that differ significantly from the native gene (Heffner et al. 2012; Murray et al. 2012). For example, a widely used keratin15 (Krt15) inducible cre strain labels the hair follicle bulge as expected, but is also active in IFE (Ito et al. 2005). Similarly, a transgenic creERT expressed from the involucrin (Ivl) promoter is active in the basal epidermis where Ivl is not normally expressed (Mascré et al. 2012). To overcome this problem, gene targeting is increasingly being used to express cre from the endogenous gene (Barker et al. 2007; Snippert et al. 2010a; Wong et al. 2012).
Once cre has been induced, it may label a population of cells, or, if doses of inducing drugs are titrated down, scattered single cells. Single-cell labeling is a powerful technique because the groups of labeled cells that develop as the cell proliferates are clones. To ensure labeling is truly clonal and that the labeled cell clusters do not arise from the fusion of two or more clones, only about 1% of basal cells should be labeled at the start of the experiment for a neutral reporter gene that does not alter cell behavior (Clayton et al. 2007). If clones have a mutation that gives them a proliferative advantage, the labeling efficiency may need to be much lower to resolve individual clones. By combining clonal labeling with confocal imaging and three-dimensional (3D) reconstruction, clones can be visualized at single-cell resolution (Clayton et al. 2007; Doupé et al. 2010). The tail has emerged as a preferred site for study, as preparing whole mounts of tail epidermis is straightforward and the wide spacing of hair follicles facilitates resolving whether clones originate in IFE or the hair follicle (Braun et al. 2003; Clayton et al. 2007). Because there is no detectable apoptosis in the basal layer of normal epidermis (Gandarillas et al. 1999; Clayton et al. 2007), clones capture the entire history of the original cell and its progeny since labeling, recording the number of rounds of cell division, and whether daughter cells have differentiated.
A second important advance has been the development of transgenic proliferation and LRC assays. These exploit DOX-regulated promoters driving expression of a very stable H2BGFP (Fig. 1) (Kanda et al. 1998; Tumbar et al. 2004). These have a substantial advantage over earlier nucleotide experiments as all cells can be labeled, including those that are quiescent, and the potential toxicity of modified nucleotides, which may alter cell behavior, is avoided (Doupé and Jones 2012). A typical experiment is to express high levels of H2BGFP in basal keratinocytes and then shut off transcription of the protein; H2BGFP is then diluted by proliferation, being partitioned equally between daughter cells (Kanda et al. 1998). The rate of cell proliferation can be inferred from measuring levels of H2BGFP fluorescence either by flow cytometry or quantitative confocal microscopy (Doupé et al. 2012; Mascré et al. 2012). Additionally, slow-cycling H2BGFP LRCs may be readily identified and sorted for molecular and functional characterization (Tumbar et al. 2004; Mascré et al. 2012).
IFE IS MAINTAINED BY PROGENITORS AND REPAIRED BY STEM CELLS
So, what have these new transgenic tools revealed about cell behavior in IFE? An early but significant result was the demonstration that IFE is not dependent on hair follicle stem cells but is self-sustaining under normal conditions (Ito et al. 2005; Levy et al. 2005). The progeny of genetically labeled bulge stem cells populate the hair follicles, but do not cross into IFE; also, transgenic deletion of this population leads to loss of hair, but has no effect on IFE (Ito et al. 2005).
The next key insight into IFE homeostasis was achieved by lineage tracing of a representative sample of proliferating cells in tail epidermis using the AhcreERT strain crossed onto a yellow fluorescent protein (YFP) reporter (Kemp et al. 2004; Clayton et al. 2007). One in 500 basal cells was inducibly labeled in a cohort of adult animals, generating single-cell-derived clones. Whole mounts were prepared at multiple time points over the next year, and clones were imaged using confocal microscopy and 3D reconstruction. Counting each cell in more than 4000 clones yielded a large-scale data set that was used to resolve cell behavior (Clayton et al. 2007). The surprising conclusion of this analysis was that epidermis was not maintained by slow-cycling stem cells and TA cells, but by a single population of functionally equivalent cells, termed progenitors (Fig. 2). These cells divide once a week, generating equal numbers of progenitor and differentiating cells, which subsequently leave the basal layer without dividing again. However, rather than every cell dividing asymmetrically, as was long assumed to be the case for stem cells, cycling epidermal cell divisions have one of three potential outcomes, producing two differentiating daughters, two progenitors, or one cell of each type. Further, although the fate of individual progenitors is random, the probabilities of each type of division are tuned so as to achieve homeostasis across the large number of divisions in the entire progenitor cell population (Jones et al. 2007; Jones and Simons 2008). The molecular basis of the balanced probabilities in the three-way cell fate decision of progenitors remains an intriguing mystery. Subsequent studies have confirmed these findings using a different inducible cre line (ivlcreERT) in tail epidermis (Mascré et al. 2012).
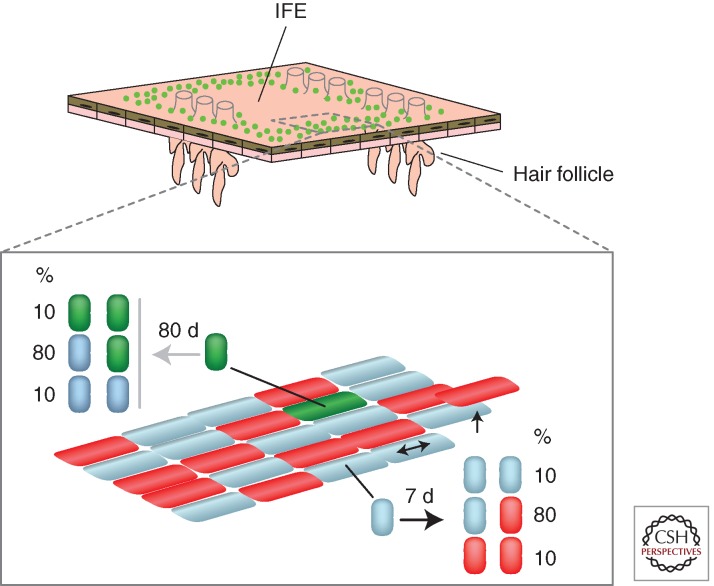
Figure 2.
Cell behavior in IFE. Analysis of large clonal lineage-tracing studies reveals mouse tail epidermis containing two populations of proliferating cells, stem cells (green), and progenitor cells (blue), along with differentiated basal cells (red) that have exited the cell cycle and are waiting to stratify. Epidermal maintenance is achieved by the self-renewing population of progenitors. As a differentiating cell leaves the basal layer through stratification, a neighboring progenitor divides. Progenitor division occurs once a week on average, and results in two progenitors, two differentiated cells, or one cell of each type as shown. The outcome of a given progenitor division is random, but the probabilities are balanced, so homeostasis is achieved across the progenitor population. Stem cells (green) are clustered around the hair follicles and under the edges of the overlying scales, and only divide every 3 months, generating stem or progenitor daughters with the probabilities shown. Stem cells make a negligible contribution to tissue maintenance, but proliferate following wounding.
In addition, both ear epidermis and the stratified squamous epithelium of the esophagus are maintained in a similar manner to tail IFE (Doupé et al. 2010, 2012). Indeed, the principle of tissue maintenance by “population self-renewal,” first shown in epidermis, now appears to extend to multiple tissues and be highly conserved in evolution from Drosophila to mouse (Klein et al. 2010b; Lopez-Garcia et al. 2010; Snippert et al. 2010b; Sheng and Mantunis 2011; de Navascues et al. 2012).
If progenitors maintain epidermis, how is it repaired? BrdU assays showed the presence of LRC in tail epidermis, a finding confirmed by a recent study using a transgenic H2BGFP approach (Braun et al. 2003; Mascré et al. 2012). The LRC lie adjacent to the hair follicles and in the basal layer beneath the margins of the scales that cover the outer surface of the tail, but there are very few LRC cells over the rest of the IFE (Fig. 2) (Mascré et al. 2012). the fortuitous discovery that treating transgenic Krt14creERT mice with very low doses of tamoxifen preferentially labels slow-cycling cells with a similar distribution to the LRC enables the fate of this population to be resolved (Mascré et al. 2012). A plausible quantitative fit to the clonal lineage-tracing data is obtained with a cell division time of 10–12 weeks and a similar model of population self-renewal to that seen in progenitors, but with the slow-cycling cells generating progenitors or slow-cycling daughters with equal probability (Mascré et al. 2012). A recent study used Krt14creERT mice induced with topical low-dose tamoxifen to track clones from 2 days of age, when scale formation begins (Schweizer and Marks 1977; Gomez et al. 2013). At 2 days of age, clones appeared to be randomly induced across the whole IFE; a week later the proportion of clones crossing the scale–interscale boundary was only 4%, although this increased to 9% by 3 months. These observations argue that, during the rapid postnatal expansion of tail IFE, the scale and interscale regions are mostly sustained by different cell populations (Gomez et al. 2013). Larger quantitative studies in adult mice are required to determine whether the scale and interscale regions are discrete compartments, possibly with differing stem and progenitor cell dynamics, in homeostatic IFE.
This picture changes dramatically when tail epidermis is wounded, however. The slow-cycling cells “wake up” and contribute numerous cells to repair the defect and generate clones that persist in the healed epidermis (Mascré et al. 2012). The slow-cycling cells thus have the potential to regenerate the epidermis with a localized distribution suggestive of a niche and show population self-renewal, so may reasonably be termed stem cells (Mascré et al. 2012). Together with stem cells in epidermal appendages, IFE stem cells function as “reserve cells,” mobilized to repair IFE after injury, but only making a negligible contribution to epidermal maintenance (Ito et al. 2005; Levy et al. 2005; Snippert et al. 2010a; Lu et al. 2012; Mascré et al. 2012).
Although they are backed up by the mobilization of slow-cycling stem cells, is there any evidence that progenitors also play any role in injury repair? Early studies revealed widespread cell synchronization and acceleration of cell division following abrasion of the epidermal surface, arguing that progenitors have the ability to respond to injury (Morris and Argyris 1983). This is confirmed by recent lineage-tracing studies that show that progenitor-derived clones are substantially expanded in the vicinity of an excisional wound. However, by the time the wound heals, progenitor clones are lost, presumably by differentiation (Mascré et al. 2012).
The apparent dependence of epidermis on reserve stem cells contrasts with murine esophageal epithelium (EE), a stratified squamous epithelium, which resembles IFE, but lacks any appendages such as glands (Doupé et al. 2012). Transgenic H2BGFP assays show that there are no LRCs in the basal layer of EE, and lineage tracing reveals a single progenitor population with balanced stochastic behavior similar to that seen in IFE (Clayton et al. 2007; Doupé et al. 2012). When EE is wounded, progenitors close to the injury switch to producing an excess of progenitor cell daughters until the epithelium is repaired, when they revert to homeostatic behavior. This shows that in EE both maintenance and wound repair are achieved by a single-cell population without any support from slow-cycling stem cells (Doupé et al. 2012). Further studies to resolve the dynamics of the epidermal progenitor response to injury are required.
FROM MOUSE TO HUMAN?
Human IFE has some obvious differences from the mouse. There are many more layers of keratinocytes and in most body sites thickness of the epidermis undulates, projecting into the dermis in the form of rête ridges (RR), separated by dermal papillae (DP) (Chacko and Vaidya 1968; Lavker and Sun 1982). However, as in the mouse, the dynamic turnover of keratinocytes is supported by the proliferation of basal cells, although this is sustained over a 50-fold longer lifespan (Epstein and Maibach 1965). We now consider whether the cellular dynamics in mouse epidermis are conserved in humans. The challenge addressing this issue is the indirect nature of the evidence. Cell culture and transplantation studies perturb the environment so that extrapolation of the results back into homeostatic tissue may be unreliable, whereas lineage-tracing data is very limited.
As in the mouse, there appears to be heterogeneity in the proliferative potential of basal cells in human IFE. Early evidence of this came from studies of established cultures of human keratinocytes. When single-cell-derived colonies are subcloned, three distinct types of colony-initiating cells are revealed. Holoclones have a very high proliferative potential, whereas paraclones generate small, differentiated colonies. Cells with intermediate properties, termed meroclones, are also observed (Barrandon and Green 1987). There is a hierarchy of colony-forming potential. Holoclones and meroclones generate paraclones, but paraclones cannot generate larger colonies.
Subsequent studies showed that cultured human keratinocytes could be fractionated on the basis of the expression and function of β1 integrin extracellular matrix receptors (Jones and Watt 1993). Cells expressing high levels of active β1 integrins isolated directly from neonatal skin form large growing colonies and regenerate human epidermis when xenografted, a finding that parallels the high expression of β1 integrins in mouse tail IFE stem cells (Jones et al. 1995; Mascré et al. 2012). In contrast, keratinocytes with lower levels of functional integrin form small colonies in which all cells eventually undergo terminal differentiation and are unable to regenerate epidermis following xenografting. Additional markers of highly clonogenic keratinocytes, which are coexpressed in cells high in β1 integrins, have also been defined. These are a high-level expression of the Notch ligand Delta1, MCSP, and LRIG1, as well as a low expression of DSG3 (Lowell and Watt 2001; Legg et al. 2003; Wan et al. 2003, 2007; Jensen and Watt 2006; Estrach et al. 2007). Analysis of expression of these markers in human IFE reveals that cells expressing high levels of clonogenic markers lie in irregular clusters localized around the tips of DP (Jones et al. 1995; Legg et al. 2003; Jensen and Watt 2006; Wan et al. 2007). Strikingly, the great majority of cells within the clusters are quiescent, whereas proliferating cells lie between the clusters (Jensen et al. 1999; Legg et al. 2003).
The study closest to in vivo lineage tracing took normal neonatal human epidermis, grafted it onto immune compromised mice, waited for 6 weeks to let the tissue stabilize and then injected lentiviral vectors carrying red and green fluorescent protein reporters (Ghazizadeh and Taichman 2005). This resulted in confluent reporter adjacent to the injection site, but low-frequency labeling more distant from it. Sections of grafted epidermis were imaged after 6 months, revealing groups of labeled cells that ranged widely in size and shape, and arose throughout the basal layer. The interpretation of the data requires caution as it is unclear whether labeled areas are single-cell clones. However, it is clear that the capacity to generate long-lived clones is found throughout the basal layer, as it is in the mouse. Further, no evidence of regular-sized EPU is seen, but rather irregularly shaped clusters consistent with the stochastic cell behavior seen in mouse epidermis (Ghazizadeh and Taichman 2005; Doupé et al. 2010). More recently, it has been proposed that human IFE may actually be maintained by progenitor cells behaving in the same manner as those in the mouse, allowing the clusters of stem cells to remain quiescent (Klein et al. 2011). Although the data are insufficient to conclusively resolve human epidermal cell dynamics, these observations suggest that the basic maintenance mechanism might be relatively similar in human and mouse epidermis.
Another set of informative results comes from studies of aging, sun-exposed skin. The basal layer of heavily sun-exposed human epidermis flattens, losing the undulating pattern of RR and DP, and the clustered staining of the putative stem cell markers β1 integrin and MCSP, arguing that the clusters of marker positive cells are not essential for epidermal maintenance (Giangreco et al. 2010). Analysis of the sizes of p53 mutant clones in sun-exposed human epidermis indicates that they are formed by cells with stochastic fate, but instead of this being in homeostatic balance, the odds of producing proliferating daughter cells are increased resulting in exponential clone growth (Jonason et al. 1996; Jensen et al. 1999; Klein et al. 2010a; Roshan and Jones 2012). The quantitative signature of the human clone size distribution was the same as that seen in UV-exposed mouse epidermis (Zhang et al. 2001; Klein et al. 2010a). This argues that in human and mouse epidermis, the effect of p53 mutation and UV exposure results in a small tilt in stochastic cell fate toward proliferation.
To summarize, the evidence available for human epidermis is consistent with the hypothesis of slow-cycling stem cells with a minimal role in tissue maintenance and progenitor cells that support homeostasis, as is found in the mouse. However, the evidence is far from conclusive, and innovative approaches are required to fully resolve human IFE stem and progenitor cell dynamics. As culture methods improve, however, human keratinocytes will increasingly provide a platform for investigating gene function far more rapidly than is feasible in the mouse (Kretz et al. 2013). Seeding a low proportion of cells carrying a fluorescent reporter and a short hairpin RNA in 3D culture allows the effects of gene knockdown in clones to be studied in the context of wild-type cells (Mulder et al. 2012).
HAIR FOLLICLES
Hair follicles are complex organs that have been a focus of intense research, including several lineage-tracing studies (Lee and Tumbar 2012). Lineage tracing in hair follicles is challenging because of their complex 3D structure, the multiple cell lineages involved, and the alternate apoptosis and regeneration that characterize the cycle of the lower hair follicle (Fig. 3). Nevertheless significant progress is being made and we will highlight some key advances here.
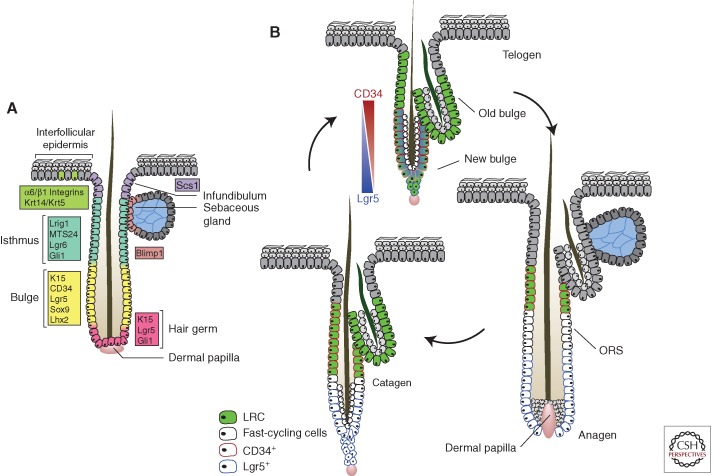
Figure 3.
Hair follicle stem cell dynamics. (A) Summary of the expression of proposed stem cell markers in the hair follicle. The extent to which these markers identify functionally distinct populations has yet to be fully resolved. (B) Stem cell proliferation and migration in the hair cycle. During anagen, cells from the lower bulge region start proliferating, contributing to the formation of the outer root sheath (ORS). Genetic label retaining shows that some bulge cells remain quiescent (green), whereas others migrate into the lower follicle, losing their stem cell properties, and proliferate (white). In catagen, the proliferating cells return to the bulge where they provide a niche sustaining the quiescent stem cells through telogen and into the next hair cycle. Marker expression changes dynamically through the cycle. CD34 is expressed in the telogen bulge and retained by quiescent stem cells throughout the cycle. Lgr5 expression overlaps with CD34 in telogen, but is localized to the proliferating cells in the lower follicle in anagen. The functional significance of the changes in Lgr5 expression is unclear. (Figure based on data from Jaks et al. 2008 and Hsu and Fuchs 2012.)
Labeled nucleotide studies identified the hair follicle bulge as a region that contained slow-cycling cells in mouse (Cotsarelis et al. 1990; Braun et al. 2003). This was later confirmed using transgenic label-retaining assays, showing that cells were able to generate all epidermal lineages when transplanted (Tumbar et al. 2004). The first lineage-tracing experiment on bulge cells used a transgenic inducible cre driven by a Krt15 promoter to label bulge cells and their progeny (Morris et al. 2004). This showed that the progeny of bulge cells labeled in the resting (telogen) phase of the hair cycle contributed extensively to the expansion of the lower follicle below the bulge in the next growth (anagen) phase of the hair cycle. Subsequent studies showed that the progeny of bulge cells labeled with Krt15 or sonic hedgehog-driven cre make no contribution to the IFE in homeostasis (Ito et al. 2005; Levy et al. 2005). Combining transgenic label-retaining assays and lineage tracing from a Krt14-driven cre line suggests that although bulge stem cells cycle slowly, they may undergo both symmetric and asymmetric cell divisions to achieve population self-renewal (Waghmare et al. 2008; Zhang et al. 2009). More recently, H2BGFP and nucleotide labeling were used in combination to show that some bulge stem cells migrate from the bulge into the lower follicle, proliferating to contribute to cells to the ORS in anagen. These proliferating cells return to the bulge to contribute to the stem cell niche in telogen (Fig. 3) (Hsu et al. 2011).
A series of studies has used ubiquitously expressed CreERT lines to analyze clone size and distribution in the proliferating cells in synchronized follicles (Legue and Nicolas 2005; Legue et al. 2010; Sequeira and Nicolas 2012). The results reveal a wide range of size and appearance in clones in the hair matrix and inner root sheath and ORS cells. These observations may reflect distinct cell populations with divergent behavior, stochastic differentiation, and proliferation or a combination of both. Live imaging will be key to resolving the dynamics of the hair follicle cells (Rompolas et al. 2012).
Another set of lineage-tracing studies exploited mouse lines in which cre was targeted to candidate stem cell marker genes, avoiding the potential misexpression that can occur with short randomly inserted promoters. Lgr5 is a wnt target gene that is expressed in stem cells in the intestinal and gastric epithelium of mice (Barker et al. 2007, 2010). Using a transgenic strain in which both CreERT and EGFP are targeted to the Lgr5 locus revealed that the gene is expressed in “cycling” cells in the bulge and secondary germ of telogen follicles, but that the zone of Lgr5 expression expands to encompass the ORS below the bulge in anagen follicles (Fig. 3) (Jaks et al. 2008). There is partial overlap between Lgr5 expression and that of another bulge stem cell marker, CD34, in telogen, but not in anagen (Trempus et al. 2003; Jaks et al. 2008). This changing expression pattern is indicative of a general problem with hair follicle stem cell markers, that gene expression is driven by multiple regulators, and a marker may label stem cells in telogen, but is a functionally distinct population(s) in anagen, when stem cells migrate into the lower follicle from the bulge (Hsu et al. 2011). Lineage tracing revealed that inducing labeling in telogen leads to widespread labeling of the lower follicle that persists for at least 6 months, but not to labeling of the sebaceous gland and upper follicle. The proportion of hair follicles that were labeled remained constant over time, arguing that Lgr5+ cells are a self-maintaining population (Jaks et al. 2008). In summary, lgr5 labels stem cells in the bulge region of telogen follicles that maintain the lower follicle through successive hair cycles.
A similar strategy has been applied to lineage trace the progeny of cells expressing a related gene, Lgr6 (Snippert et al. 2010a). In telogen, Lgr6 is expressed in a distinct population of nondividing cells lying directly above the CD34 and Krt15-expressing cells in the bulge and below other proliferative populations in the “junctional zone” of the upper follicle expressing the markers MTS24 and Lrig1 (Nijhof et al. 2006; Jensen et al. 2009). Lineage tracing in juvenile mice indicated that Lgr6 cells contributed labeled progeny to the upper follicle and sebaceous gland, which persisted for up to a year. Labeling was also present in IFE, a finding at variance with previous studies, and may result from the presence of Lgr6+ cells in IFE rather than a flux of cells from the hair follicle into the epidermis (Snippert et al. 2010a). Analysis of labeled cells in uninduced Krt14creER mice suggests that junctional zone cells may also contribute to the infundibulum, sebaceous gland, and IFE (Jensen et al. 2009).
How do hair follicle stem cells respond to epidermal wounding? In back skin, lineage tracing indicates that progeny of hair follicle stem cells in the upper follicle and bulge migrate out of the follicle to contribute to IFE repair, and that some of these cells persist long term in the healed epidermis (Ito et al. 2005; Levy et al. 2005, 2007; Jaks et al. 2008; Snippert et al. 2010a; Arwert et al. 2012). In the tail, where hair follicles are sparse, both IFE and hair follicle stem cells are mobilized, although the observation that wounds heal in Edar mutant mice, which lack hair follicles in tail skin, argues that the hair follicle contribution is nonessential (Langton et al. 2008; Mascré et al. 2012). The relative contribution of IFE and hair follicle stem cells to wound healing in typical mouse epidermis remains to be determined.
Can the findings in the mouse be extrapolated to human hair follicles? The lineage-tracing studies of mouse hair follicle stem cells highlight how misleading the culture and transplantation assays used in human studies may be. For example, mouse lgr5+ stem cells that only contribute to the lower hair follicle in vivo generate epidermis and sebaceous gland when transplanted (Jaks et al. 2008). This said, there is evidence for proliferative heterogeneity in human hair follicles. Microdissection studies indicate that different regions of human hair follicles vary in their ability to generate keratinocyte colonies in culture, with cells in the lower follicle just below the bulge region having the highest proliferative potential, perhaps reflecting the migration of stem cells out of the bulge (Rochat et al. 1994; Hsu et al. 2011). Markers for the clonogenic cells have been identified with the highest colony-forming efficiency residing in cells positive for CD200, but negative for the mouse bulge marker CD34 (Ohyama et al. 2006). Studies of patients with androgenic alopecia show that CD200+ cells are lost in bald follicles, although expression of the bulge marker KRT15 persists (Garza et al. 2011). The recent development of a combined culture and transplantation protocol that generates hair follicles from human bulge cells will significantly enhance the analysis of the potential of human hair follicle stem cells (Toyoshima et al. 2012).
SEBACEOUS AND SWEAT GLANDS
Hair follicles do not just comprise hair, but also contain apocrine sebaceous glands located just above the bulge. Lineage tracing in homeostatic adult epidermis indicates that different follicle stem cell populations may contribute progeny to the sebaceous gland including Krt15 positive bulge cells, the Lgr6 positive population, and junctional zone stem cells (Jensen et al. 2009; Snippert et al. 2010a; Petersson et al. 2011). However, other studies support the existence of Blimp1-expressing sebaceous progenitors on the margins of the gland and a self-maintaining population within the gland itself (Ghazizadeh and Taichman 2001; Horsley et al. 2006).
Compared with the hair follicle and sebaceous glands, the sweat glands (SG) and the ducts that connect them to the epidermal surface have received scant attention in lineage-tracing studies until recently (Lu et al. 2012). In the mouse, SG are confined to the paws. Adult SG are quiescent, but the ducts are continually turned over (Fig. 4). Transgenic lineage-tracing studies within the glands suggest the existence of unipotent myoepithelial and luminal progenitor cell populations that are able to regenerate their own lineage after specific cells have been deleted by transgenic expression of diphtheria toxin receptor (Fig. 4) (Lu et al. 2012).
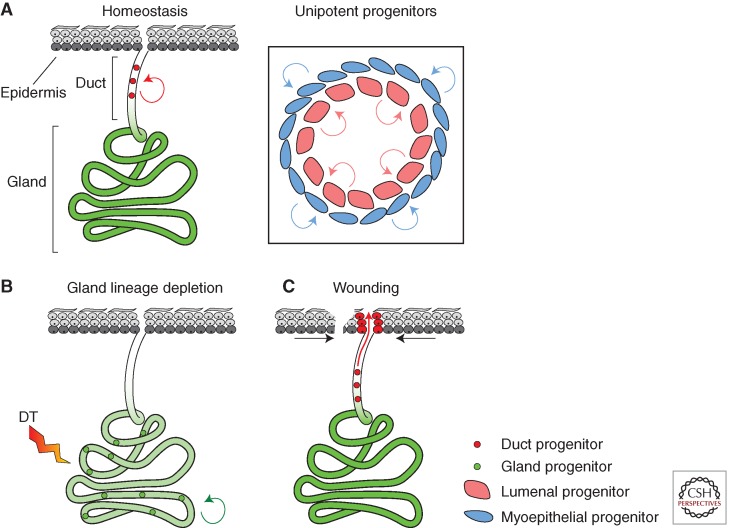
Figure 4.
Mouse sweat-gland maintenance and wound repair. (A) In adult skin, SG are quiescent, whereas the myoepithelial and luminal cells that form the sweat duct are continually turned over, being replaced by lineage committed progenitor cells (inset). (B) Transgenic deletion of glandular cells using diphtheria toxin (DT) receptor mobilizes glandular progenitors to replenish the lost cells. (C) Wounding of the epidermis adjacent to the sweat gland triggers proliferation of the duct progenitor cells, but not the sweat gland, to repair the connection of the duct to the epidermal surface. Green shading indicates nondividing cells as determined H2BGFP proliferation assay (see Fig. 1).
Following injury of mouse forepaw epidermis, the cells of the sweat duct, but not the gland, contribute to wound repair (Lu et al. 2012). The role of sweat duct cells may be much more significant in other species however (Miller et al. 1998). In humans, eccrine SG outnumber hair follicles several-fold. A recent study used 3D reconstruction of sections of human epidermis regenerating after laser ablation to show that the ducts of SG make a substantial contribution to the regeneration of human IFE, at least matching that from the hair follicles (Rittie et al. 2013). Further studies of sweat ducts are needed to determine which cells participate in wound healing and if they can be manipulated to promote epidermal regeneration.
CONCLUDING REMARKS
Lineage tracing has revealed normal cell behavior and produced startling insights into stem cell behavior. Rather than every stem cell behaving in the same manner, the maintenance of IFE and possibly the hair follicle depends on balancing the probabilities of differentiation and self-renewal over the stem/progenitor cell population. Although the task of maintaining IFE is achieved by progenitors, they are underpinned by stem cells, possibly confined to a niche, that are activated in response to injury.
Despite significant progress, resolving the stem cell populations within the hair follicle remains a challenge. Further lineage-tracing studies are required to resolve the behavior of upper follicle stem cells, such as those expressing Lrig1, and address whether the multiplicity of stem cell markers reveals multiple functionally distinct stem cell populations or reflects alterations in gene expression as a consequence of the varying microenvironment of the follicle (Jensen et al. 2009; Watt and Jensen 2009). The relationships between multilineage and lineage-restricted progenitors such as for the sebaceous gland also need further definition, as does the role of nonkeratinocyte lineages in regulating stem cell behavior (Horsley et al. 2006; Festa et al. 2011; Rikiishi 2012). An exciting new development is the live imaging of hair follicle cells expressing fluorescent proteins enabling the highly dynamic process of hair regeneration to be tracked directly for up to 8 hours (Rompolas et al. 2012). This will allow short-term cell dynamics at key stages of the hair cycle to be linked with long-term cellular outputs assayed by genetic lineage tracing.
This review has focused on epidermal homeostasis and wound repair, but lineage tracing is also beginning to make inroads into resolving cell behavior in disease. Recent studies have focused on cancer, but in principle the technique can be applied to any disease for which there is a mouse model, and promises to transform our understanding of the role of stem cells in epidermal pathology (Driessens et al. 2012).
ACKNOWLEDGMENTS
We thank David Doupé, Allon Klein, Kim Jensen, Kasumi Murai, Amit Roshan, Ben Simons, and Doug Winton for stimulating discussions.
Footnotes
Editors: Anthony E. Oro and Fiona M. Watt
Additional Perspectives on The Skin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Adami JG 1901. An address on the causation of cancerous and other new growths: Delivered before the Yale University Medical Alumni Association, New Haven, Conn. Br Med J 1: 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcolea MP, Jones PH 2013. Tracking cells in their native habitat: Lineage tracing in epithelial neoplasia. Nat Rev Cancer 13: 161–171 [DOI] [PubMed] [Google Scholar]
- Arwert EN, Hoste E, Watt FM 2012. Epithelial stem cells, wound healing and cancer. Nat Rev Cancer 12: 170–180 [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, et al. 2010. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6: 25–36 [DOI] [PubMed] [Google Scholar]
- Barrandon Y, Green H 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci 84: 2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickenbach JR 1981. Identification and behavior of label-retaining cells in oral mucosa and skin. J Dent Res 60: 1611–1620 [DOI] [PubMed] [Google Scholar]
- Bickenbach JR, Chism E 1998. Selection and extended growth of murine epidermal stem cells in culture. Exp Cell Res 244: 184–195 [DOI] [PubMed] [Google Scholar]
- Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM 2003. Manipulation of stem cell proliferation and lineage commitment: Visualisation of label-retaining cells in whole mounts of mouse epidermis. Development 130: 5241–5255 [DOI] [PubMed] [Google Scholar]
- Chacko LM, Vaidya MC 1968. The dermal papillae and ridge patterns in human volar skin. Acta Anat (Basel) 70: 99–108 [DOI] [PubMed] [Google Scholar]
- Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH 2007. A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189 [DOI] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, Lavker RM 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61: 1329–1337 [DOI] [PubMed] [Google Scholar]
- De Navascues J, Perdigoto CN, Bian Y, Schneider MH, Bardin AJ, Martinez-Arias A, Simons BD 2012. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J 31: 2473–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupé D, Jones PH 2012. Interfollicular homeostasis: Dicing with differentiation. Exp Dermatol 21: 249–253 [DOI] [PubMed] [Google Scholar]
- Doupé DP, Klein AM, Simons BD, Jones PH 2010. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell 18: 317–323 [DOI] [PubMed] [Google Scholar]
- Doupé DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, Jones PH 2012. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337: 1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C 2012. Defining the mode of tumour growth by clonal analysis. Nature 488: 527–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein WL, Maibach HI 1965. Cell renewal in human epidermis. Arch Dermatol 92: 462–468 [PubMed] [Google Scholar]
- Estrach S, Legg J, Watt FM 2007. Syntenin mediates Delta1-induced cohesiveness of epidermal stem cells in culture. J Cell Sci 120: 2944–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa E, Fretz J, Berry R, Schmidt B, Rodeheffer M, Horowitz M, Horsley V 2011. Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell 146: 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandarillas A, Goldsmith LA, Gschmeissner S, Leigh IM, Watt FM 1999. Evidence that apoptosis and terminal differentiation of epidermal keratinocytes are distinct processes. Exp Dermatol 8: 71–79 [DOI] [PubMed] [Google Scholar]
- Garza LA, Yang CC, Zhao T, Blatt HB, Lee M, He H, Stanton DC, Carrasco L, Spiegel JH, Tobias JW, et al. 2011. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest 121: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh S, Taichman LB 2001. Multiple classes of stem cells in cutaneous epithelium: A lineage analysis of adult mouse skin. EMBO J 20: 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazizadeh S, Taichman LB 2005. Organization of stem cells and their progeny in human epidermis. J Invest Dermatol 124: 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Goldie SJ, Failla V, Saintigny G, Watt FM 2010. Human skin aging is associated with reduced expression of the stem cell markers beta1 integrin and MCSP. J Invest Dermatol 130: 604–608 [DOI] [PubMed] [Google Scholar]
- Gomez C, Chua W, Miremadi A, Quist S, Headon Denis J, Watt FM 2013. The interfollicular epidermis of adult mouse tail comprises two distinct cell lineages that are differentially regulated by Wnt, Edaradd, and Lrig1. Stem Cell Rep 1: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT 2008. Wound repair and regeneration. Nature 453: 314–321 [DOI] [PubMed] [Google Scholar]
- Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, Eppig JT, Murray SA 2012. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun 3: 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, O’Carroll D, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E 2006. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell 126: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Fuchs E 2012. A family business: Stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol 13: 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E 2011. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R 2008. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40: 1291–1299 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Watt FM 2006. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci 103: 11958–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen UB, Lowell S, Watt FM 1999. The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: A new view based on whole-mount labelling and lineage analysis. Development 126: 2409–2418 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM 2009. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA, Leffell DJ, Tarone RE, Brash DE 1996. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci 93: 14025–14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Simons BD 2008. Epidermal homeostasis: Do committed progenitors work while stem cells sleep? Nat Rev Mol Cell Biol 9: 82–88 [DOI] [PubMed] [Google Scholar]
- Jones PH, Watt FM 1993. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73: 713–724 [DOI] [PubMed] [Google Scholar]
- Jones PH, Harper S, Watt FM 1995. Stem cell patterning and fate in human epidermis. Cell 80: 83–93 [DOI] [PubMed] [Google Scholar]
- Jones PH, Simons BD, Watt FM 2007. Sic transit gloria: Farewell to the epidermal transit amplifying cell? Cell Stem Cell 1: 371–381 [DOI] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 8: 377–385 [DOI] [PubMed] [Google Scholar]
- Kemp R, Ireland H, Clayton E, Houghton C, Howard L, Winton DJ 2004. Elimination of background recombination: Somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res 32: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Brash DE, Jones PH, Simons BD 2010a. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc Natl Acad Sci 107: 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD 2010b. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell 7: 214–224 [DOI] [PubMed] [Google Scholar]
- Klein AM, Nikolaidou-Neokosmidou V, Doupé DP, Jones PH, Simons BD 2011. Patterning as a signature of human epidermal stem cell regulation. J R Soc Interface 8: 1815–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM 2012. Lineage tracing. Cell 148: 33–45 [DOI] [PubMed] [Google Scholar]
- Langton AK, Herrick SE, Headon DJ 2008. An extended epidermal response heals cutaneous wounds in the absence of a hair follicle stem cell contribution. J Invest Dermatol 128: 1311–1318 [DOI] [PubMed] [Google Scholar]
- Lavker RM, Sun TT 1982. Heterogeneity in epidermal basal keratinocytes: Morphological and functional correlations. Science 215: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Leblond CP 1964. Classification of cell populations on the basis of their proliferative behavior. Natl Cancer Inst Monogr 14: 119–150 [PubMed] [Google Scholar]
- Lee J, Tumbar T 2012. Hairy tale of signaling in hair follicle development and cycling. Semin Cell Dev Biol 23: 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg J, Jensen UB, Broad S, Leigh I, Watt FM 2003. Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development 130: 6049–6063 [DOI] [PubMed] [Google Scholar]
- Legue E, Nicolas JF 2005. Hair follicle renewal: Organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development 132: 4143–4154 [DOI] [PubMed] [Google Scholar]
- Legue E, Sequeira I, Nicolas JF 2010. Hair follicle renewal: Authentic morphogenesis that depends on a complex progression of stem cell lineages. Development 137: 569–577 [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA 2005. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell 9: 855–861 [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA 2007. Epidermal stem cells arise from the hair follicle after wounding. FASEB J 21: 1358–1366 [DOI] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23: 1686–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62 [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ 2010. Intestinal stem cell replacement follows a neutral drift. Science 330: 822–825 [DOI] [PubMed] [Google Scholar]
- Lowell S, Watt FM 2001. Delta regulates keratinocyte spreading and motility independently of differentiation. Mech Dev 107: 133–140 [DOI] [PubMed] [Google Scholar]
- Lu CP, Polak L, Rocha AS, Pasolli HA, Chen SC, Sharma N, Blanpain C, Fuchs E 2012. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 150: 136–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IC 1970. Relationship between mitosis and the ordered structure of the stratum corneum in mouse epidermis. Nature 226: 653–655 [DOI] [PubMed] [Google Scholar]
- Marques-Pereira JP, Leblond CP 1965. Mitosis and differentiation in the stratified squamous epithelium of the rat esophagus. Am J Anat 117: 73–87 [DOI] [PubMed] [Google Scholar]
- Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohee S, Sotiropoulou PA, Simons BD, Blanpain C 2012. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489: 257–262 [DOI] [PubMed] [Google Scholar]
- Miller SJ, Burke EM, Rader MD, Coulombe PA, Lavker RM 1998. Re-epithelialization of porcine skin by the sweat apparatus. J Invest Dermatol 110: 13–19 [DOI] [PubMed] [Google Scholar]
- Morris R, Argyris TS 1983. Epidermal cell cycle and transit times during hyperplastic growth induced by abrasion or treatment with 12-O-tetradecanoylphorbol-13-acetate. Cancer Res 43: 4935–4942 [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G 2004. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol 22: 411–417 [DOI] [PubMed] [Google Scholar]
- Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmany G, Donati G, Uribe-Lewis S, Pavlidis P, et al. 2012. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol 14: 753–763 [DOI] [PubMed] [Google Scholar]
- Murray SA, Eppig JT, Smedley D, Simpson EM, Rosenthal N 2012. Beyond knockouts: cre Resources for conditional mutagenesis. Mamm Genome 23: 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A 2000. Cre recombinase: The universal reagent for genome tailoring. Genesis 26: 99–109 [PubMed] [Google Scholar]
- Nijhof JG, Braun KM, Giangreco A, van Pelt C, Kawamoto H, Boyd RL, Willemze R, Mullenders LH, Watt FM, de Gruijl FR, et al. 2006. The cell-surface marker MTS24 identifies a novel population of follicular keratinocytes with characteristics of progenitor cells. Development 133: 3027–3037 [DOI] [PubMed] [Google Scholar]
- Ohyama M, Terunuma A, Tock CL, Radonovich MF, Pise-Masison CA, Hopping SB, Brady JN, Udey MC, Vogel JC 2006. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J Clin Invest 116: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson M, Brylka H, Kraus A, John S, Rappl G, Schettina P, Niemann C 2011. TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J 30: 3004–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS 1974. The epidermal proliferative unit: The possible role of the central basal cell. Cell Tissue Kinet 7: 77–88 [DOI] [PubMed] [Google Scholar]
- Potten CS 1981. Cell replacement in epidermis (keratopoiesis) via discrete units of proliferation. Int Rev Cytol 69: 271–318 [DOI] [PubMed] [Google Scholar]
- Potten CS, Morris RJ 1988. Epithelial stem cells in vivo. J Cell Sci Suppl 10: 45–62 [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL 2011. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature 476: 409–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittie L, Sachs DL, Orringer JS, Voorhees JJ, Fisher GJ 2013. Eccrine sweat glands are major contributors to reepithelialization of human wounds. Am J Pathol 182: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat A, Kobayashi K, Barrandon Y 1994. Location of stem cells of human hair follicles by clonal analysis. Cell 76: 1063–1073 [DOI] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, Greco V 2012. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487: 496–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshan A, Jones PH 2012. Chronic low dose UV exposure and p53 mutation: Tilting the odds in early epidermal preneoplasia? Int J Radiat Biol 88: 682–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer J, Marks F 1977. A developmental study of the distribution and frequency of Langerhans cells in relation to formation of patterning in mouse tail epidermis. J Invest Dermatol 69: 198–204 [DOI] [PubMed] [Google Scholar]
- Sequeira I, Nicolas JF 2012. Redefining the structure of the hair follicle by 3D clonal analysis. Development 139: 3741–3751 [DOI] [PubMed] [Google Scholar]
- Sheng XR, Matunis E 2011. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138: 3367–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman FG, Quastler H, Wimber DR 1961. Cell population kinetics in the ear epidermis of mice. Exp Cell Res 25: 114–119 [DOI] [PubMed] [Google Scholar]
- Smart IH 1970. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. Br J Dermatol 82: 276–282 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Clevers H 2011. Tracking adult stem cells. EMBO Rep 12: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. 2010a. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385–1389 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. 2010b. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144 [DOI] [PubMed] [Google Scholar]
- Soriano P 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21: 70–71 [DOI] [PubMed] [Google Scholar]
- Spearman RI, Hardy JA 1977. Ultrastructure of the contrasting types of keratinization seen in the tail epidermis of the laboratory mouse Mus musculus. Arch Dermatol Res 258: 33–40 [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM 2000. Involvement of follicular stem cells in forming not only the follicle but also the epidermis. Cell 102: 451–461 [DOI] [PubMed] [Google Scholar]
- Toyoshima KE, Asakawa K, Ishibashi N, Toki H, Ogawa M, Hasegawa T, Irie T, Tachikawa T, Sato A, Takeda A, et al. 2012. Fully functional hair follicle regeneration through the rearrangement of stem cells and their niches. Nat Commun 36: 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, Tennant RW 2003. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol 120: 501–511 [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E 2004. Defining the epithelial stem cell niche in skin. Science 303: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E 1999. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci 96: 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmare SK, Bansal R, Lee J, Zhang YV, McDermitt DJ, Tumbar T 2008. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J 27: 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Stone MG, Simpson C, Reynolds LE, Marshall JF, Hart IR, Hodivala-Dilke KM, Eady RA 2003. Desmosomal proteins, including desmoglein 3, serve as novel negative markers for epidermal stem cell-containing population of keratinocytes. J Cell Sci 116: 4239–4248 [DOI] [PubMed] [Google Scholar]
- Wan H, Yuan M, Simpson C, Allen K, Gavins FN, Ikram MS, Basu S, Baksh N, O’Toole EA, Hart IR 2007. Stem/progenitor cell-like properties of desmoglein 3dim cells in primary and immortalized keratinocyte lines. Stem Cells 25: 1286–1297 [DOI] [PubMed] [Google Scholar]
- Watt FM, Jensen KB 2009. Epidermal stem cell diversity and quiescence. EMBO Mol Med 1: 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MW, et al. 2012. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 14: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Remenyik E, Zelterman D, Brash DE, Wikonkal NM 2001. Escaping the stem cell compartment: Sustained UVB exposure allows p53-mutant keratinocytes to colonize adjacent epidermal proliferating units without incurring additional mutations. Proc Natl Acad Sci 98: 13948–13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T 2009. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell 5: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]