Abstract
Long non-coding RNAs (lncRNAs) have gained massive attention in recent years as a potentially new and crucial layer of gene regulation. LncRNAs are prevalently transcribed in the genome, but their roles in gene regulation and disease development are largely unknown. HOX antisense intergenic RNA (HOTAIR), a lncRNA located in the HOXC locus, has been shown to repress HOXD gene expression and promote breast cancer metastasis. Mechanistically, HOTAIR interacts with and recruits polycomb repressive complex 2 (PRC2) and regulates chromosome occupancy by EZH2 (a subunit of PRC2), which leads to histone H3 lysine 27 trimethylation of the HOXD locus. Moreover, HOTAIR is pervasively overexpressed in most human cancers compared with noncancerous adjacent tissues. This review summarizes the studies on the HOTAIR lncRNA over the past 6 years.
Keywords: lncRNA, HOTAIR, cancer, metastasis, carcinogenesis
Introduction
Only a very small portion of transcripts in the whole human genome are associated with protein-coding genes; the vast majority are non-coding RNAs (ncRNAs) [1]. It is estimated that 98% of the human genome DNA is non-protein coding [2]. Long non-coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides; this length distinguishes lncRNAs from small regulatory RNAs such as microRNAs, short interfering RNAs, piwi-interacting RNAs, small nucleolar RNAs, and other short RNAs [3]. lncRNAs are pervasively transcribed from the human genome, but their involvement in human disease is just beginning to be revealed. The lncRNA HOX antisense intergenic RNA (HOTAIR) was identified in 2007 [4]. Numerous other lncRNAs were subsequently identified by next-generation sequencing and computational prediction, followed by studies to uncover lncRNAs' roles in different aspects of gene expression regulation and disease, such as carcinogenesis and metastasis of various cancers. In this review, we discuss the studies from the past 6 years on the HOTAIR lncRNA, with a focus on HOTAIR's identification, functional characterization, clinical implications, and regulation of gene expression in carcinogenesis and cancer metastasis.
Identification and Characterization of HOTAIR
In 2007, Rinn et al. [4] and Woo and Kingston [5] identified a lncRNA that regulates the HOX genes. HOTAIR, a 2158-nucleotide ncRNA, is located on chromosome 12q13.13, which is a regulatory boundary in the HOXC cluster [4]. Notably, knockdown of HOTAIR did not affect expression of the HOXC cluster but instead led to derepression of a 40-kb region of the HOXD cluster [4].
Many small RNAs, such as microRNAs, are well conserved across diverse species [6]. In contrast, long ncRNAs lack strong conservation. For example, many well-described lncRNAs, such as Xist, are poorly conserved [7], suggesting that lncRNAs may be subject to different selection pressures. In mammals, HOTAIR also evolved more quickly than the nearby HOX genes, as indicated by HOTAIR's poorly conserved sequences and structures [8]. The cognate murine HOTAIR has only 58% sequence identity to human HOTAIR, and deleting the HOXC cluster containing HOTAIR in mice produced no obvious phenotypic manifestation and did not affect HOXD10 expression, which also suggests a rapid evolution of HOTAIR [9].
Deregulation of HOTAIR in Cancers
Breast cancer
Breast cancer was the first report of HOTAIR deregulation in cancers. Gupta et al. [10] found that HOTAIR expression levels are higher in primary breast tumor than in adjacent noncancerous tissue. Using quantitative polymerase chain reaction, they also showed that HOTAIR overexpression ranges from hundreds of times greater to nearly two-thousand times greater in breast cancer metastases than those in noncancerous tissue, and that HOTAIR expression in primary breast tumors is a powerful predictor of metastasis and eventual death. Importantly, overexpression of HOTAIR in epithelial cancer cells promoted tumor invasion and metastasis. Conversely, loss of HOTAIR can inhibit cancer invasiveness [10]. A recent study revealed that estradiol transcriptionally induces HOTAIR; this study also identified multiple functional estrogen response elements in HOTAIR's promoter region. This mechanism may contribute to HOTAIR upregulation and breast cancer progression [11].
However, in another recent study that examined HOTAIR expression and methylation in breast cancer and evaluated their associations with patient outcome, HOTAIR expression varied widely in primary breast cancers and did not have significant associations with clinical or pathologic characteristics [12]. The possible reason for this discrepancy is due to the different approaches used for profiling gene expression, and the cut-off values used to define the HOTAIR expression level. Further studies are thus needed to resolve this discrepancy.
Hepatocellular carcinoma
A previous study reported that HOTAIR expression was significantly higher in hepatocellular carcinoma (HCC) tissue than that in adjacent noncancerous tissues [13]. Patients whose HCC tumors overexpressed HOTAIR had an increased risk of HCC recurrence after hepatectomy, and there was also a correlation between HOTAIR overexpression and increased risk of lymph node metastasis [13]. Overexpression of HOTAIR was an independent prognostic factor for HCC recurrence in liver transplant patients. Furthermore, patients with high expression of HOTAIR in the tumor had significantly shorter recurrence-free survival than those with low expression of HOTAIR [14]. In HCC cells, silencing of HOTAIR by RNA interference led to decreased cell viability and invasiveness, to sensitized tumor necrosis factor alpha-induced apoptosis, and to increased chemotherapeutic sensitivity of cancer cells to cisplatin and doxorubicin [14]. In vitro assays demonstrated that depletion of HOTAIR reduced cell proliferation and decreased expression of matrix metalloproteinase-9 and vascular endothelial growth factor proteins, both of which are important for cell migration and metastasis [13]. A later study performed in HCC also showed that HOTAIR was overexpressed in HCC patients and was associated with a worse prognosis and an increased risk of metastasis in these patients [15].
Pancreatic cancer
Corroborating the findings of HOTAIR in breast cancer and HCC, studies of pancreatic cancer have shown that HOTAIR expression is higher in pancreatic cancers than that in noncancerous pancreatic tissue and is associated with more aggressive tumors. Silencing of HOTAIR in pancreatic cancer cells decreased cell proliferation, inhibited cell-cycle progression, and induced apoptosis [16]. Xenograft assays showed that HOTAIR knockdown in L3.6pL pancreatic cancer cells inhibited tumor growth in mice, which indicates the anti-proliferative activities of HOTAIR in pancreatic cancer [16].
Non-small cell lung cancer
HOTAIR expression has also been associated with carcinogenesis and metastasis in non-small cell lung cancer (NSCLC) patients. A recent study compared HOTAIR expression levels in 77 NSCLC tumors with expression levels in corresponding noncancerous lung tissue. High expression of HOTAIR correlated with advanced lymph node metastasis or lymph-vascular invasion and with a short disease-free interval in patients with NSCLC [17]. Interestingly, tumor-promoting collagen type 1 (Col-1) upregulates the expression of HOTAIR in NSCLC cells, and has been shown to activate a reporter gene under the control of the HOTAIR promoter [18]. These findings suggest the potential roles for HOTAIR in lung carcinogenesis and metastasis [18].
Other cancers
Studies of colorectal cancer (CRC) patients revealed that HOTAIR expression levels were higher in CRC tissue than those in the corresponding noncancerous tissue. High expression levels of HOTAIR correlated with the presence of liver metastasis, and patients whose CRC had a high HOTAIR expression level also had a worse prognosis than did patients whose tumors had a low HOTAIR level [19]. Based on an in situ hybridization assay, 91 of 160 (56.87%) paraffin-embedded nasopharyngeal carcinoma (NPC) biopsy specimens had elevated levels of HOTAIR. NPC patients with high HOTAIR expression levels in the tumor had a worse prognosis for overall survival rate than those with low HOTAIR expression levels [20]. HOTAIR expression levels were also significantly higher in laryngeal squamous cell cancer than in corresponding adjacent noncancerous tissues, and high HOTAIR expression was detected in patients whose tumors had a high histological grade or an advanced clinical stage [21]. Depletion of HOTAIR by short interfering RNA led to decreased invasion and increased apoptosis of HEp-2 cells, and tumor growth was significantly inhibited in mice injected with HOTAIR-deficient cells [21]. Overexpression of HOTAIR was also strongly associated with high-grade tumor and metastasis in gastrointestinal stromal tumor specimens [22]. RNA interference-mediated knockdown of HOTAIR altered the expression of reported HOTAIR target genes and suppressed gastrointestinal stromal tumor cell invasiveness [22]. It is reported that expression of HOTAIR in ovarian tumor tissue was higher than that in normal ovarian tissue [23]. Esophageal squamous cell carcinoma patients with elevated levels of HOTAIR had substantially lower 5-year survival rates than did HOTAIR-negative patients [24]. High levels of HOTAIR in primary sarcoma also correlated with a high probability of metastasis [25]. Table 1 summarizes the expression of HOTAIR in different types of cancers.
Table 1.
HOTAIR expression in various human cancers as reported in published studies
| Cancer types | Expression | Method of analysis | References |
|---|---|---|---|
| Breast cancer | Upregulated in both primary and metastatic tumors | Array, qPCR | [10,12,16] |
| Hepatocellular carcinoma | Upregulated in both primary and metastatic tumors | qPCR | [13–15] |
| Colorectal cancer | Upregulated in both primary and metastatic tumors | qPCR | [19] |
| Sarcoma | Upregulated in both primary and metastatic tumors | qPCR | [25] |
| Gastrointestinal stromal tumor | Upregulated in both primary and metastatic tumors | qPCR | [22] |
| Pancreatic cancer | Upregulated in primary tumor | qPCR | [16] |
| Laryngeal squamous cell cancer | Upregulated in primary tumor | qPCR | [21] |
| Nasopharyngeal carcinoma | Upregulated in primary tumor | ISH | [20] |
| Non-small cell lung cancer | Upregulated in both primary and metastatic tumors | qPCR | [17,18] |
| Ovarian cancer | Upregulated in primary tumor | qPCR | [23] |
| Esophageal squamous cell carcinoma | Upregulated in primary tumor | ISH, qPCR | [24] |
qPCR, quantitative polymerase chain reaction; ISH, in situ hybridization.
Mechanism of Action of HOTAIR
The PRC2 maintains stem cell pluripotency and suppresses cell differentiation by promoting histone H3K27 trimethylation and transcriptional repression of differentiation genes. The HOTAIR study pioneered by Chang's group [4] uncovered a new mechanism whereby HOTAIR leads to transcriptional silencing of a distant chromosomal region through epigenetic regulation. As shown in Fig. 1, HOTAIR interacts with PRC2 and is required for PRC2 occupancy of the HOXD locus, leading to H3K27 trimethylation [4]. Enforced HOTAIR expression in epithelial cancer cells was found to increase invasive and metastatic abilities and reprogram the PRC2 occupancy pattern to resemble embryonic fibroblasts [10]. Conversely, depletion of HOTAIR in highly metastatic, mesenchymal cancer cells inhibited cell invasiveness. Gene expression analysis showed that hundreds of genes were induced or repressed upon HOTAIR overexpression. Depletion of PRC2 in HOTAIR-overexpressing cells reversed the global gene expression pattern to that of cells without HOTAIR overexpression [10]. PRC2 is specifically required for HOTAIR to promote cell invasiveness. Subsequent studies further elucidated that a 5′ domain of HOTAIR binds PRC2, whereas a 3′ domain of HOTAIR binds the LSD1/CoREST/REST complex [26]. HOTAIR not only functions as a molecular scaffold to link PRC2 and LSD1/CoREST/REST protein complexes, but also coordinates the chromatin targeting of these proteins and in turn couples histone H3K27 methylation and H3K4 demethylation for epigenetic gene silencing [27]. Chu et al. [28] used chromatin isolation by RNA purification (ChIRP) to identify the chromatin regions interacted with HOTAIR, and found HOTAIR preferentially occupies a GA-rich DNA motif on chromatin. This finding suggests that HOTAIR can dictate chromatin states and gene transcription, just like sequence-specific transcription factors.
Figure 1.
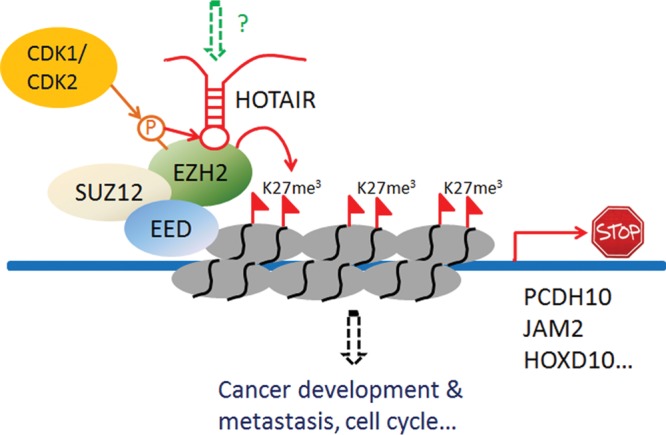
Schematic illustration of HOTAIR associated with PRC2 in the regulation of epigenetic markers and cancer metastasis
Another group reported independently that the BRCA1-binding region in EZH2 overlaps with the HOTAIR-binding domain, and that BRCA1 inhibits the binding of EZH2 to HOTAIR [29]. As a consequence, decreased expression of BRCA1 causes genome-wide EZH2 retargeting and elevates H3K27me3 levels at PRC2 target loci in both murine embryonic stem cells and human breast cancer cells [29]. Both BRCA1 and HOTAIR bind the PRC2 subunit EZH2 and coordinate PRC2-dependent epigenetic regulation on the chromosome, indicating that HOTAIR regulates gene transcription and expression by competing with other proteins. HOTAIR binding to EZH2 also depends on EZH2 protein modification. EZH2 is phosphorylated by cyclin-dependent kinase 1 (CDK1) at threonine residues 345 and 487 in a cell-cycle-dependent manner [30]. A phospho-mimetic mutation at residue T345 increased HOTAIR binding to EZH2, whereas substitution of residue T487 with either alanine or aspartic acid had no effect (Fig. 1). An EZH2 domain composed of threonine 345 was found to be essential for binding to HOTAIR and to the 5′ end of another lncRNA, Xist [30]. Similar to threonine 345 phosphorylation of murine EZH2, human EZH2 was found to be phosphorylated at threonine 350 by CDK1 and CDK2 [31,32]. However, whether threonine 350 phosphorylation regulates HOTAIR binding to EZH2 in humans is not yet known. These findings implied that phosphorylation of EZH2 by CDK1 and CDK2 may enhance EZH2 function during the S and G2 phases of the cell cycle. Enhanced EZH2 function leads to K27 trimethylation of de novo synthesized histone H3, which is incorporated into nascent nucleosomes before cell division. However, whether HOTAIR expression is regulated during cell cycle progression and whether HOTAIR is involved in the regulation of cell cycle remain to be determined. This speculation was supported by a recent gene set enrichment analysis (GSEA) of pancreatic cancer patient gene-profiling data to identify gene set differences between patients with high HOTAIR levels and low levels, which demonstrated that HOTAIR regulates gene sets that are mainly associated with cell proliferation and cell cycle progression [16].
Perspectives
Deletion of the HOXC cluster encompassing HOTAIR in mice has surprisingly little effect on the gene expression pattern, the transcription efficiency, or the level of H3K27me3 coverage of different HOXD genes [9]. Nevertheless, in vivo confirmation of HOTAIR's functions still awaits a specific HOTAIR-deficient mouse model. However, HOTAIR may have a rapid evolvement within mammals and have functions in humans that are not easily revealed in mice, and thus studies using human cells may be highly important. Given the potential importance of HOTAIR in various human cancers, further studies on how exactly HOTAIR contributes to carcinogenesis and metastasis are needed, and it is necessary to discover how HOTAIR is regulated and which pathway is involved in the induction of HOTAIR. DNA methylation may play important roles in regulating HOTAIR expression in the breast cancer [12]. HOTAIR is highly expressed in the metastatic cancers, which is known to have poor outcomes in patients receiving chemo- or radiotherapy; it would be interesting to see whether HOTAIR contribute to resistance to chemo- or radiotherapy in patients and cancer cells. The subcellular localization of HOTAIR is not restricted in nucleus, and it is also expressed in the cytoplasm [33]. It is very likely, besides the regulation on chromatin states by acting on PRC2 and other epigenetical modulators, HOTAIR also plays other roles in the cytoplasm. Moreover, since HOTAIR regulates the expression of hundreds or even thousands of genes [10,16], further investigation is necessary to discover other roles played by HOTAIR.
Funding
The noncoding RNA work in the Ma Lab is supported by the grants from the US National Institutes of Health (R00CA138572 and R01CA166051; to L.M.) and a Cancer Prevention and Research Institute of Texas Scholar Award R1004 (to L.M.).
Acknowledgements
We are grateful for the professional manuscript editing by Diane S. Hackett and Jill R. Delsigne from the Department of Scientific Publications at MD Anderson Cancer Center.
References
- 1.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 2.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–934. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo CJ, Kingston RE. HOTAIR lifts noncoding RNAs to new levels. Cell. 2007;129:1257–1259. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 7.Nesterova TB, Barton SC, Surani MA, Brockdorff N. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol. 2001;235:343–350. doi: 10.1006/dbio.2001.0295. [DOI] [PubMed] [Google Scholar]
- 8.He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. doi: 10.1186/1471-2148-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;pii:S0022-2836(13)00038–7. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, Risch HA, et al. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 13.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 15.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 16.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, Sato I, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 18.Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui X, Fewell C, Flemington EK, et al. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol. 2013;6:35. doi: 10.1186/1756-8722-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 20.Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 22.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 23.Cui L, Xie XY, Wang H, Chen XL, Liu SL, Hu LN. Expression of long non-coding RNA HOTAIR mRNA in ovarian cancer. J Sichuan Univ (medical science edition) 2013;44:57–59. [PubMed] [Google Scholar]
- 24.Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8:e63516. doi: 10.1371/journal.pone.0063516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milhem MM, Knutson T, Yang S, Zhu D, Wang X, Leslie KK, Meng X. Correlation of MTDH/AEG-1 and HOTAIR expression with metastasis and response to treatment in sarcoma patients. J Cancer Sci Ther. 2011;(Suppl 5):004. [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L, Fang J. Writer meets eraser in HOTAIR. Acta Biochem Biophys Sin. 2011;43:1–3. doi: 10.1093/abbs/gmq110. [DOI] [PubMed] [Google Scholar]
- 28.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Zeng X, Chen S, Ding L, Zhong J, Zhao JC, Wang L, et al. BRCA1 is a negative modulator of the PRC2 complex. EMBO J. 2013;32:1584–1597. doi: 10.1038/emboj.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, Bagchi A, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng X, Chen S, Huang H. Phosphorylation of EZH2 by CDK1 and CDK2: a possible regulatory mechanism of transmission of the H3K27me3 epigenetic mark through cell divisions. Cell Cycle. 2011;10:579–583. doi: 10.4161/cc.10.4.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]


