Abstract
Previously, we reported strong influences of genetic variants on metabolic phenotypes, some of them with clinical relevance. Here, we hypothesize that DNA methylation may have an important and potentially independent effect on human metabolism. To test this hypothesis, we conducted what is to the best of our knowledge the first epigenome-wide association study (EWAS) between DNA methylation and metabolic traits (metabotypes) in human blood. We assess 649 blood metabolic traits from 1814 participants of the Kooperative Gesundheitsforschung in der Region Augsburg (KORA) population study for association with methylation of 457 004 CpG sites, determined on the Infinium HumanMethylation450 BeadChip platform. Using the EWAS approach, we identified two types of methylome–metabotype associations. One type is driven by an underlying genetic effect; the other type is independent of genetic variation and potentially driven by common environmental and life-style-dependent factors. We report eight CpG loci at genome-wide significance that have a genetic variant as confounder (P = 3.9 × 10−20 to 2.0 × 10−108, r2 = 0.036 to 0.221). Seven loci display CpG site-specific associations to metabotypes, but do not exhibit any underlying genetic signals (P = 9.2 × 10−14 to 2.7 × 10−27, r2 = 0.008 to 0.107). We further identify several groups of CpG loci that associate with a same metabotype, such as 4-vinylphenol sulfate and 4-androsten-3-beta,17-beta-diol disulfate. In these cases, the association between CpG-methylation and metabotype is likely the result of a common external environmental factor, including smoking. Our study shows that analysis of EWAS with large numbers of metabolic traits in large population cohorts are, in principle, feasible. Taken together, our data suggest that DNA methylation plays an important role in regulating human metabolism.
INTRODUCTION
Metabolomics aims at the holistic measurement of ideally all small molecules (metabolites) in a biological sample. It has been shown in many studies that the metabolic phenotype (metabotype), as it can be determined in a sample of human blood, carries information on important biological processes and that some metabolic traits represent intermediate phenotypes linking genetic and environmental factors to endpoints of complex disorders (1). However, in order to translate this knowledge into actionable therapeutic evidence, the precise nature of the biological processes that lead from genetic variances and environmental factors to disease outcomes requires further elucidation. Epigenetic regulation of metabolic processes via DNA methylation and gene expression may play a major role in this system. The recent availability of array-based whole-genome DNA methylation measurements now allows for epigenome-wide association studies (EWAS) with disease-relevant phenotypes, which address such questions in a basically non-biased manner.
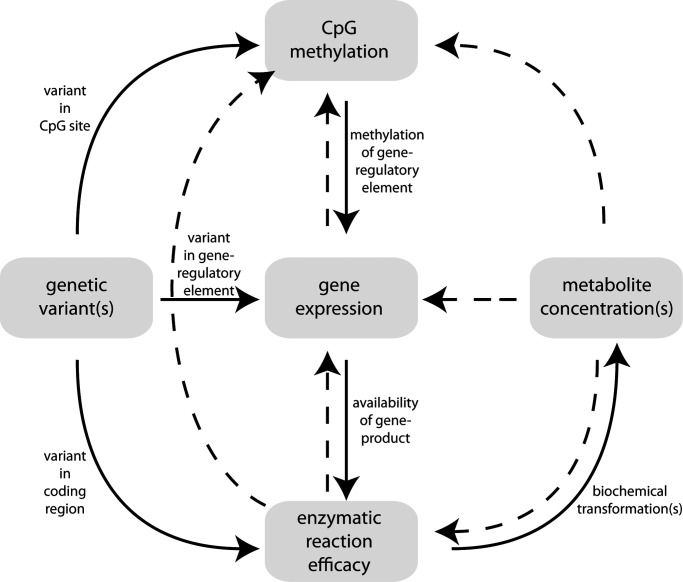
Many blood serum metabolic traits represent intermediate phenotypes that link genetic and environmental factors to disease and often represent indicators of complex disorders (2). Identification of key factors that determine disease-relevant human metabolic traits is essential to our understanding of their role in the etiology of complex disorders (3). In previous genome-wide association studies (GWAS) with metabolic traits, we identified many instances of single nucleotide polymorphism (SNP)–metabotype associations with large effect sizes, the so-called genetically influenced metabotypes (GIMs) (1). Some of these GIMs have already been shown to play a role in complex disorders; others are presently under investigation. DNA methylation is an important gene-regulatory mechanism (4,5) and is therefore expected to also play a role in determining disease-relevant metabotypes (6). For instance, Menni et al. reported three associations between DNA methylation and C-glycosyl tryptophan levels. This metabolite also associated with bone mineral density and birth weight in their study (7). DNA methylation is influenced by genetic and environmental factors, and many feedback mechanisms between DNA methylation, gene expression and other biological processes are known or suspected (5). However, in contrast to genetic variation, where it is clear that a genetic variant is causal for an association between a SNP and a phenotype, this is not so for associations between variance in DNA methylation and metabolic traits (see Fig. 1). The availability of whole-genome methylation assays makes EWAS now possible and thereby allows for a basically bias-free approach to these questions (8,9). Here, we present the first EWAS with metabolic traits in human blood (the study design is sketched in Fig. 2).
Figure 1.
Schematic view of processes that link genetic variance and CpG–methylation to metabolic phenotypes. Possible feedback reactions are depicted by dashed lines, such as transcription activity leaving traces on the DNA by CpG–methylation, alosteric inactivation of enzymatic reactions or transcription regulation by metabolite-mRNA binding. Other potential regulatory and feedback mechanisms, involving for instance microRNA silencing and histone modifications, may exist but are not depicted here.
Figure 2.
Study design.
RESULTS
All data used here were obtained from the Kooperative Gesundheitsforschung in der Region Augsburg (KORA) F4 population study and have been described previously in the publications referenced below. Briefly, the Illumina Infinium HumanMethylation450 BeadChip platform was used to determine DNA methylation (10). This platform quantifies relative methylation of CpG sites using the Illumina DNA bead array technology and DNA bi-sulfite conversion (8). The percentage of methylation of a given cytosine is reported as a beta-value (b-value), which is a continuous variable between 0 and 1, corresponding to the ratio of the methylated signal over the sum of the methylated and unmethylated signals. After quality control, data on b-values for a total of 457 004 CpG sites observed for 1805 individuals entered the analysis.
The metabolite data set has been described previously in a number of GWAS with metabolic traits. It consists of measurements obtained on three different metabolomics platforms: (i) platform ‘Biocrates’ implements a kit-based-targeted quantitative FIA-MS/MS method (151 traits), (ii) platform ‘Metabolon’ uses non-targeted, semi-quantitative liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and GC-MS methods (483 traits) and (iii) platform ‘Lipofit’ derives lipid-related parameters from 1H NMR measurements (15 traits). Detailed descriptions of these data sets can be found in the following papers: Jourdan et al. and Illig et al. (11,12) for metabolites form the Biocrates platform, Suhre et al. (13) for known metabolites from the Metabolon platform, Petersen et al. (14) for lipoprotein classes from the Lipofit platform and Krumsiek et al. (15) for non-annotated (unknown) metabolites from the Metabolon platform. In total, 649 metabolic traits were used in this study (see Supplementary Material, Table S1 for a full list). In the metabolomics data, depending on technology and metabolic trait, some values are missing. However, in most cases, data for >1700 subjects are available. The exact number of observations used in any specific analysis is reported in the tables. Based on prior experience, we tested log-transformed metabolite concentrations (Biocrates and Lipofit platforms) or ion counts (Metabolon platform) for association with DNA methylation (b-values), using age, gender, body mass index (BMI) and white blood cell count (WBC) as covariates in a linear model. The genome-wide level of significance at an alpha level of 0.05 after the Bonferroni correction for 649 × 457 004 tests is pgw = 1.69 × 10−10. Note that due to biochemical interactions, the concentrations of some metabolic traits correlate with each other. Bonferroni correction may, therefore, be overly conservative. We report association data below this level as Supplementary Material.
Genetic variance near or in CpG sites may influence DNA methylation and thus may be an unidentified confounding factor behind some of the associations. To explicitly account for the effect of genetic variance and also to identify associations that are driven by non-genetic factors, we conducted a second association analysis where we included three SNPs from the vicinity of the CpG site into the model. These three SNPs were selected iteratively as follows: first we tested the association of each b-value for linear additive dependence on every individual genotyped SNP within a window of ±5 Mb around the CpG site, using age, gender, BMI and WBC as covariates. We then selected the SNP that showed the strongest association with CpG methylation (called SNP1). We then selected a second SNP (SNP2) and then a third (SNP3) following the same procedure, including the already selected SNP(s) as covariates.
Strong CpG–metabotype associations were found at loci that harbor previously reported SNP–metabotype associations
Manhattan plots for the associations of CpG–methylation with metabolite concentrations (CpG–metabotype associations) are presented in Figure 3. We identified 621 CpG–metabotype associations at Bonferroni threshold (Supplementary Material, Table S2). Manual inspection of the top ranking associations revealed the presence of eight loci (ACADS, PYROXD2, NAT8, ACADM, OPLAH, FADS1, UGT1A and SULT2A1) that have been found previously in association between a genetic variant and a same metabolic phenotype as the one identified here (12,13,15) (Table 1, Supplementary Material, Fig. S1). At this stage of the analysis, we initially suspected artificial associations: Microarray-based methylation chips are susceptible to interference with SNPs in the CpG region. Such SNPs may interfere with the oligo-based determination of CpG methylation (16). To exclude such experimental artifacts, we attempted the validation of the methylation measurements using the Sequenom EpiTYPER system, which is an array-independent method based on mass spectrometry (17). DNA samples from a subset of 30–40 samples were analyzed using the EpiTYPER system for five of the eight loci (see Section ‘Methods’ and Supplementary Material, Table S3). Validation of the three remaining sites was not attempted for lack of suitable amplicons. For all successfully analyzed loci (ACADS, PYROXD2, ACADM, OPLAH and FADS1) we observed strong correlation between DNA methylation determined using the EpiTYPER system and the Infinium HumanMethylation450 BeadChip (see r2 in Table 1). However, the EpiTYPER system is also susceptible to the presence of SNPs in the DNA sequence that may potentially induce a false methylation signal. The MassArray software (18) allows for the detection of the presence of such SNPs. Using this software, we did not observe a significant number of SNPs within the analyzed fragments (the number of samples with a SNP in the fragment is reported in Table 1, output from MassArray is provided as Supplementary Material, Fig. S4). However, using the UCSC Genome Browser, we further checked for the presence of SNPs within the analyzed CpG sites. In the case of ACADS, PYROXD2, NAT8 and SULT2A1 a frequent SNP were reported in the C or G nucleotide of the CpG site. No SNPs were present in the vicinity of the CpG sites of the ACADM, OPLAH, FADS1 and UGT1A cases. In the first four cases, genetic variance in the CpG site may thus be at the origin of the observed genotype-dependant methylation, whereas in the latter four cases such an effect can be clearly ruled out.
Figure 3.

Manhattan plots of CpG–metabotype associations without (top) and including (bottom) three SNPs into the model to account confounding genetic factors. Associations with P-values < 10−13 are indicated by vertical lines. Associations with P-values < 10−25 are indicated by red dots. Manhattan plots comparing these CpG–metabotype associations to previously published SNP–metabotype associations are provided as Supplementary Material, Figure S1.
Table 1.
CpG–metabotype associations limited to loci that also show a strong association with a genetic variant
| Locus name | CpG | Chr | Pos | Metabolic trait | Beta′ | r2 | P-value | N | r2 (NEpi) | Fragment | # Samples with SNP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACADS | cg24768164 | 12 | 121 163 261 | Butyrylcarnitinea | −0.998 | 0.221 | 2.0 × 10−108 | 1744 | 0.907 (35) | CpG_9 | 0 |
| PYROXD2 | cg26690318 | 10 | 100 167 465 | X-12092b | 2.171 | 0.138 | 2.2 × 10−60 | 1725 | 0.904 (31) | CpG_14 | 0 |
| NAT8 | cg13584399 | 2 | 73 907 327 | N-acetylornithinea | −0.950 | 0.120 | 8.9 × 10−52 | 1731 | Not analyzed | — | |
| ACADM | cg10523679 | 1 | 76 189 770 | Hexanoylcarnitinea | −0.456 | 0.065 | 1.8 × 10−30 | 1749 | 0.954 (31) | CpG_4 | 2 |
| OPLAH | cg06239191 | 8 | 145 163 136 | 5-oxoprolinea | 0.813 | 0.056 | 8.0 × 10−25 | 1737 | 0.872 (32) | CpG_1 | 0 |
| FADS1 | cg11250194 | 11 | 61 601 937 | PC aa C38:4c | 11.41 | 0.054 | 1.0 × 10−24 | 1781 | 0.653 (35) | CpG_5 | 0 |
| UGT1A | cg26799339 | 2 | 234 664 336 | bilirubin (Z,Z)a | −0.973 | 0.054 | 2.9 × 10−24 | 1706 | Not analyzed | — | |
| SULT2A1 | cg00365481 | 19 | 48 362 237 | X-11440b | 1.358 | 0.0363 | 3.9 × 10−20 | 1742 | Not analyzed | — | |
CpG id (cg-numbers), chromosome (Chr) and chromosome position (Pos, human genome build 37), metabolic trait, the effect size (beta’), variance of metabolic trait explained by CpG methylation (r2), P-value of the linear model, and number of samples (N); for the EpiTYPER validation, the correlation coefficient (Pearson r2) between the Infinium HumanMethylation450 BeadChip derived b-values and the EpiTYPER methylation is reported for the EpiTYPER fragment corresponding to associated CpG site; NEpi is the number of samples used in the EpiTYPER replication; the number (#) of samples that contain a SNP in the quantified amplicon [identified using MassArray (18)] are given; scatterplots between Infinium HumanMethylation450 BeadChip derived b-values and the EpiTYPER-based methylation levels are provided in Supplementary Material, Figure S3; graphical output from the SNP detection analysis software for the individual amplicons is shown in Supplementary Material, Figure S4. We initiated the EpiTYPER replication early-on in the project. Eventually, after adjusting for the covariates described in the methods part, in two cases CpG sites that differ from those selected for replication (cg14631276 at OPLAH and cg19610905 at FADS1) exhibited stronger signals of association. As these sites display only slightly stronger signals of association, we did not repeat the replication on these CpG sites.
aA genetic association at this locus with this metabolic trait was reported in Suhre et al., 2011 (13)
bKrumsiek et al., 2012 (15).
cIllig et al., 2010 (12).
After elimination of potentially underlying genetic signals, the majority of CpG–metabotype associations were driven by a common, but yet unidentified external factor
In an effort to eliminate all underlying genetic signals from the CpG–metabotype associations, we conducted a second association study, explicitly accounting for a potential genetic effect by including three SNPs from the vicinity of the CpG site into the model. Using this approach, we expected to obtain an association signal that can be clearly attributed to processes of DNA methylation that are not driven by a genetic variance near the CpG site. This study identified 562 CpG–metabotype associations at the Bonferroni threshold of significance, most of which were already present in the initial study (Supplementary Material, Table S4). As expected, all associations reported in Table 1 disappeared. To group neighboring CpG sites that exhibit identical association signals, we grouped CpG sites together that were no more than 1 000 000 bases apart. In this grouping process, we included all CpG sites with association P-values < 10−9 (1260 sites). This process resulted in a total of 459 distinct loci. When inspecting the associating metabolic traits at these 459 loci, we observed that some metabolic traits were strongly over-represented. We, therefore, grouped multiple loci that exhibit a common pattern of CpG–metabotype associations into larger loci groups. This annotation procedure led to the definition of six loci groups (named VINYLPHENOL, STEROIDS, ‘PC ae C4 … ’, ‘PC aa … ’, ‘tyr/trp’, ‘Lipofit traits’ by the predominant associating metabolic traits; Table 2 and Supplementary Material, Table S4). We further annotated seven distinct loci, where a metabolic trait specifically associates with only a single locus (named UGT2B15, TXNIP, DHCR24, MYO5C, ABCG1, SLC25A22, CPT1A by neighboring genes). All but 14 of the 459 loci were thus annotated. The remaining 14 loci, which had P-values > 2 × 10−12, were not further investigated. The association at the seven loci and six loci groups that we discuss below all had P-values < 10−13 for at least one of their CpG–metabotype associations.
Table 2.
CpG–metabotype associations after correction for genetic effects and exclusion of inflated loci
| Locus name | CpG | Chr | Pos | Metabolic trait | Beta’ | r2 | P-value | N | r2 (NEpi) | Fragment | # samples with SNP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UGT2B15 | cg09189601 | 4 | 69 514 031 | X-11491 | −0.865 | 0.087 | 2.69 × 10−27 | 1283 | Not analyzed | – | |
| TXNIP | cg19693031 | 1 | 145 441 552 | Chylo-A | −0.996 | 0.038 | 1.11 × 10−21 | 1771 | 0.842 (41) | CpG_5 | 0 |
| DHCR24 | cg17901584 | 1 | 55 353 706 | PC ae C36:5 | 4.001 | 0.036 | 3.65 × 10−18 | 1780 | 0.744 (41) | CpG_5 | 0 |
| MYO5C | cg06192883 | 15 | 52 554 171 | Glycine | −0.659 | 0.030 | 1.61 × 10−15 | 1744 | 0.257 (41, n.s.) | CpG_4 | 31 |
| ABCG1 | cg06500161 | 21 | 43 656 587 | SM C16:0 | −0.817 | 0.008 | 1.04 × 10−14 | 1781 | 0.507 (33) | CpG_2.3 | 21 |
| SLC25A22 | cg09441501 | 11 | 798 350 | Arg | −1.000 | 0.035 | 1.66 × 10−14 | 1780 | Not analyzed | – | |
| CPT1A | cg00574958 | 11 | 68 607 622 | VLDL-A | −1.000 | 0.025 | 9.23 × 10−14 | 1773 | 0.332 (41) | CpG_5 | 1 |
| SLC7A11 (STEROIDS) | cg06690548 | 4 | 139 162 808 | A-diol | −0.980 | 0.071 | 6.83 × 10−39 | 1746 | 0.123 (40, n.s.) | CpG_2 | 0 |
| PHGDH (STEROIDS) | cg14476101 | 1 | 120 255 992 | A-diol | −0.929 | 0.035 | 6.50 × 10−21 | 1742 | 0.205 (41, n.s.) | CpG_2 | 4 |
| LOC100132354 (STEROIDS) | cg18120259 | 6 | 43 894 639 | A-diol | −0.932 | 0.023 | 1.10 × 10−14 | 1747 | 0.667 (41) | CpG_3 | 0 |
| SLC1A5 (STEROIDS) | cg22304262 | 19 | 47 287 778 | A-diol | −0.954 | 0.022 | 6.49 × 10−14 | 1744 | 0.608 (41) | CpG_11 | 13 |
| cg13526915 (STEROIDS) | cg13526915 | 14 | 24 164 078 | A-diol | −0.924 | 0.020 | 3.15 × 10−13 | 1746 | 0.181 (31, n.s.) | CpG_3 | 15 |
| AHRR (VINYLPHENOL) | cg05575921* | 5 | 373 378 | 4-vs | −0.953 | 0.107 | 3.52 × 10−49 | 1709 | 0.977 (41) | CpG_3 | 0 |
| ALPPL2 (VINYLPHENOL) | cg21566642* | 2 | 233 284 661 | 4-vs | −0.945 | 0.079 | 7.03 × 10−37 | 1706 | Not analyzed | – | |
| F2RL3 (VINYLPHENOL) | cg03636183* | 19 | 17 000 585 | 4-vs | −0.977 | 0.063 | 5.63 × 10−30 | 1708 | Not analyzed | – | |
| cg06126421 (VINYLPHENOL) | cg06126421* | 6 | 30 720 080 | 4-vs | −0.952 | 0.048 | 4.12 × 10−25 | 1709 | 0.951 (41) | CpG_4 | 0 |
| RARA (VINYLPHENOL) | cg19572487* | 17 | 38 476 024 | 4-vs | −0.887 | 0.034 | 6.12 × 10−16 | 1707 | 0.822 (40) | CpG_2 | 0 |
| GFI1 (VINYLPHENOL) | cg09935388* | 1 | 92 947 588 | 4-vs | −0.899 | 0.030 | 3.01 × 10−15 | 1709 | 0.816 (42) | CpG_4.5.6 | 0 |
| TPM1 (VINYLPHENOL) | cg10403394 | 15 | 63 349 192 | 4-vs | 6.198 | 0.013 | 4.63 × 10−13 | 1709 | 0.934 (41) | CpG_5.6 | 0 |
| cg23079012 (VINYLPHENOL) | cg23079012 | 2 | 8 343 710 | 4-vs | −0.996 | 0.026 | 9.07 × 10−13 | 1709 | 0.870 (35) | CpG_5.6 | 14 |
Legend as in Table 1; 4-androsten-3beta, 17beta-diol disulfate (A-diol); 4-vinylphenol sulfate (4-vs); cases where no statistically significant correlation between the methylation readouts from the Infinium HumanMethylation450 BeadChip and the EpiTYPER system was observed are marked as ‘n.s.’. Loci in the VINYLPHENOL group that are marked by a ‘*’ were reported by Zeilinger et al. (10) in association with smoking.
We then noted that the majority of all loci (391) was covered by only three of the loci groups (‘PC ae C4 … ’, ‘trp/tyr’ and ‘Lipofit’). Manual inspection of the associations related to these groups revealed that the metabolic traits that associated with the CpG sites at these loci had very similar patterns and should thus be considered as one single loci group. In particular, the phosphocholine PC ae C44:5 was found as a common leading trait in 374 out of the 391 loci. Other metabolic traits that associated with many CpG sites in these loci groups are the phospholipids PC ae C42:4, PC ae C42:5, PC ae C44:4, PC aa C26:0, the lipid traits Chylo-A, VLDL-A from the Lipofit platform and the amino acids tyrosine and tryptophan (measured both on the Biocrates and on the Metabolon platform). The P-value distributions of the individual associations for these traits were highly inflated (see QQ-plots in Supplementary Material, Fig. S3). This inflation indicates a correlation between these metabolic traits and non-loci-specific DNA-methylation, which is likely driven by a common external factor. However, after testing the numerous phenotypes available in KORA (i.e. age, gender, BMI, blood lipid parameters, smoking, alcohol consumption, diabetes, and hypertension, and data from our previous metabolome-wide association studies for association with these traits), we were unable to identify a potential common external factor. Further research is needed to elucidate this question.
Only few highly significant loci- and two loci group-specific CpG–metabotype associations remain after removal of the confounding genetic effects
After exclusion of the inflated association signals and confounding genetic effects, only seven distinct loci (UGT2B15, TXNIP, DHCR24, MYO5C, ABCG1, SLC25A22 and CPT1A) and two loci groups (named STEROIDS—comprising five loci—and VINYLPHENOL—comprising eight loci—by reference to the associated traits) remain (Table 2). Validation of CpG methylation was attempted for all loci using the EpiTYPER system and is reported in Table 2. Although this validation was generally successful, some of the methylation measurements could not be fully validated. These cases should thus be interpreted with care.
DISCUSSION
This is the first EWAS with metabolic traits. We identified two types of methylome–metabotype associations. One type is driven by an underlying genetic effect (eight loci, Table 1); the other type is independent of genetic variation and potentially driven by common environmental and life-style-dependent factors (seven loci and two loci groups, Table 2). Given the early state of this field of research and the associated computational complexity, some choices had to be made early-on in the study, which may be improved in future work. In this light, we discuss here the main results of our study, mentioning potential caveats as we go. First of all, it needs to be noted that the Infinium HumanMethylation450 BeadChip as an array-based technique only queries a subset of all potentially methylated sites in the genome. Future studies using full sequencing may thus alter some of the results presented here. It has further been suggested that the standard pipeline for Infinium HumanMethylation450 BeadChip data processing using the GenomeStudio software is suboptimal (19). The Infinium HumanMethylation450 BeadChip combines two distinct probe types, Infinium I and II, which may cause a bias in the analysis if all signals are merged as a unique source of methylation measurement. Infinium I considers two bead types (methylated and unmethylated) for the same CpG locus, both sharing the same color channel, whereas Infinium II utilizes a single bead type and two color channels (19). There have been several efforts to develop new methodologies and pipelines to overcome the shift between Infinium I and II, such as subset-quantile within array normalization (20) and beta mixture quantile dilation (21). Although those methods have been compared using different criteria, the method of choice is still subject of discussion (22). These alternative methods are potentially preferable in future studies and may lead to higher statistical power, however, we believe that these caveats do not put our observed associations into question. To further test this point, we obtained preliminary methylation data for the CpG sites reported in Tables 1 and 2, which have been normalized using the pipeline described in Touleimat and Tost (19). We find that all associations reported in this paper are robust with respect to these changes in the normalization of the HumanMethylation450 BeadChip data (data not shown).
Another limitation of this study is the fact that we determine DNA methylation in cells obtained from whole blood. However, blood metabolite levels are largely determined by metabolic transformations that occur in the liver, kidney, muscle and adipose tissue. Furthermore, in this study, we only have DNA available from unsorted cells that were extracted from the whole blood. The DNA methylation reported here is, therefore, a readout from a mixture of different types of blood cells (23). The CpG–metabotype associations we report here are thus likely limited to processes of DNA methylation that are not cell-type specific. This is in particular reflected in the CpG–metabotype associations that have an underlying genetic variant, since all cells carry the same genetic variants. It also suggests that most (if not all) associations without an underlying genetic effect are driven by an external environmental factor that affects the organism as a whole.
For instance, using the same population study and DNA methylation data that is used in this study, Zeilinger et al. (10) show that tobacco smoking leads to extensive genome-wide changes in DNA methylation, identifying the CpG sites of the VINYLPHENOL loci group we report here. Here, we find an association of the VINYLPHENOL loci group with 4-vinylphenol sulfate (4-vs) . Interestingly, Manini et al. (24) showed that 4-vinylphenol associates with smoking, and these authors provide a biochemical explanation for this association: they found urinary 4-vinylphenol to be significantly correlated with airborne styrene, but also found a measurable background excretion of 4-vinylphenol in all urine samples from controls not occupationally exposed to styrene. This background appeared to be highly correlated to smoking (P < 0.001), which can be explained by the fact that styrene is one of many chemicals found in cigarettes. Therefore, it is likely that the association between CpG–methylation and 4-vinylphenol sulfate for the sites of the VINYLPHENOL loci group is driven by the common environmental factor smoking.
Similarly, we suspect a common external environmental factor that may be driving the associations observed in the STEROIDS case. We note that there is a mutual theme between the biological function of some of the genes at these loci. The steroid 4-androsten-3-beta,17-beta-diol (A-diol) belongs to the class of androgenic anabolic steroids (androstenedione) and is an intermediate in the biochemical pathway that produces the androgen testosterone and the estrogens estrone and estradiol. The gene product of the SLC7A11 (solute carrier family 7) gene is involved in metabolism and transport systems induced by estrogen and therefore an estrogen-responsive gene (25). PHGDH (phosphoglycerate dehydrogenase) catalyses the first and rate-limiting step in the phosphorylated pathway of serine biosynthesis (note that genetic variance in PHGDH also associates with serine (12,13), but that the CpG–serine association is robust to the inclusion of the relevant SNPs into the model). PHGDH is part of a key metabolic pathway that is essential in estrogen receptor (ER)-negative breast cancer (26). SLC1A5 [solute carrier family 1 (neutral amino acid transporter), member 5] also known as ASCT2 (ASC stands for ‘alanine-serine-cysteine–preferring’) is a Na+ (and K+)-dependent glutamate transporter, accepting as substrates all neutral amino acids, including glutamine, asparagine and branched-chain and aromatic amino acids, and excludes methylated amino acids, anionic amino acids and cationic amino acids. Tumor cells are known for their high requirement of glutamine that serves multiple functions within the cells, including nutritional and energy source and ASCT2 mediates net uptake of glutamine (6,7). Tamoxifen and raloxifene, which are selective ER modulators, suppress the proliferation of ER-negative cells through inhibition of glutamine uptake in a dose-dependent manner through inhibition of ASCT2 (27). The common theme of these three associations is thus related to steroid metabolism. One may speculate that differences in steroid metabolism may impact on the specific methylation of the genes of the STEROID locus-group. However, the external factor that leads to differences in steroid metabolism remains to be identified.
A detailed discussion of the potential biological background of the seven associations that involve only single loci is beyond the scope of the present paper. These loci require further in-depth analysis before we can get a clearer picture of their biological importance. We only briefly highlight the TXNIP case as an example: this case is well validated using EpiTYPER (r2 = 0.842). In addition to the association of cg19693031 with chylomicrons (size class A of the Lipofit platform, P = 3.65 × 10−18), this site also associates with known metabolic markers of diabetes, such as a number of other lipid parameters, hexose (P = 4.35 × 10−12), and alpha-hydroxybutyrate (P = 7.24 × 10−8) (see Supplementary Material, Table S5 for a full list). TXNIP is functionally involved in glucose regulation. In a recent study with 4450 individuals, TXNIP expression was consistently elevated in the muscle of pre-diabetic and diabetic participants. However, the authors found no evidence for association between common genetic variation in the TXNIP gene and type 2 diabetes (28). Our data suggest that DNA methylation may play a regulatory role in this case.
Regarding the influence of genetic variance on methylation, the situation is quite complex. We identified eight loci at which the association of CpG–methylation with metabotype is confounded by a frequent SNP in the gene region. The CpG–metabotype association disappears when genetic variance is included into the model. Since array-based methylation assays are susceptible to artifacts that may be induced by SNPs within the probe region, we attempted to validate the methylation measurements using the mass spectrometry-based EpiTYPER system. Although we observed strong correlation between methylation measured on both systems in the five cases that were successfully analyzed, due to the presence of SNPs in the CpG sites, we cannot totally rule out interference of frequent SNPs with the Infinium HumanMethylation450 BeadChip in two of the cases (ACADS and PYROXD2). However, genetic variants in the CpG sites themselves can also explain the data (see below).
Nevertheless, some cases are clear: For instance in the case of ACADM (Fig. 4), no SNPs have been reported in any database in the vicinity of the cg10523679 site, and DNA methylation has been well validated using the EpiTYPER system (r2 = 0.954). Moreover, two other assayed CpG sites in close vicinity of cg10523679 also show a strong association signal (cg22875332, P = 1.1 × 10−29; cg03433033, P = 3.3 × 10−30; Supplementary Material, Table S2). This suggests that DNA methylation at this locus is not limited to a single CpG-pair. A genetic variant in the observed CpG site itself can be ruled out. Alternative scenarios are that genetic variants in a gene regulatory element at the ACADM locus influence gene expression or that a variant in a coding region modifies the enzymatic reaction efficacy of ACADM. Both scenarios can explain the observed changes in the metabolite concentrations. A feedback mechanism would then be required in order to explain the changes in DNA methylation (see possibilities suggested in Fig. 1). Potentially, epistasis may also play a role. However, it is also possible that the genetic variant is merely a confounding factor, and that no functional relation exists between variance in DNA methylation and variation in metabolic traits.
Figure 4.
Association between genotype, CpG–methylation and metabolic phenotype at the ACADM locus. (A) Scatterplot of b-values at cg10523679 and hexanoylcarnitine, colored by the genotype of SNP rs12134854; (B) correlation between methylation of cg10523679 determined by EpiTYPER and by the Infinium HumanMethylation450 BeadChip for selected samples (r2 = 0.954); (C) as in (A), but for cg10523679 methylation determined on a subset of samples using the EpiTYPER system (fragment 4, which contains cg10523679); (D) boxplots of hexanoylcarnitine concentrations as a function of rs12134854 genotype; (E) methylation of cg10523679 determined using the Infinium HumanMethylation450 BeadChip as a function of the rs12134854 genotype. This figure shows that there is a strong three-way association between genotype, CpG methylation, and hexanoylcarnitine concentrations at the ACADM locus. Note that hexanoylcarnitine is essentially a substrate of the ACADM enzyme, rs12134854 is in linkage equilibrium of the ACADM gene, and cg10523679 is located in the promoter region of the ACADM gene.
Taken together, our data suggest that some of the here observed associations between CpG methylation and metabolite concentrations may be explained by genetic confounders or by non-genetic external factors. Four possible scenarios are sketched in Figure 5: First, a genetic variant in linkage disequilibrium (LD) with a SNP that influences metabolite concentrations may result in the loss of a CpG site that is observed by the Illumina platform. For four of the eight cases with a possible genetic confounding factor (ACADS, PYROXD2, NAT8 and SULT2A1) frequent variants in the CpG site are reported in dbSNP and are thus likely candidates for such a confounding factor (Fig. 5a). Second, a genetic variant may result in the loss of a CpG site that is not queried by the Illumina platform. Maintenance of DNA methylation (e.g. during mitosis) may then lead to a spill-over of methylation of that site to neighboring CpG sites, including the one observed on the Infinium HumanMethylation450 BeadChip platform. No frequent SNP was found in dbSNP and data from the 1000 Genomes Project for the other four cases with a genetic background (ACADM, FADS1, UGT1A and OPLAH; Fig. 5b). Potentially, a SNP within the CpG probe, but not resulting in the loss of a CpG site may result in an artificial association signal (16) (Fig. 5c). However, for the eight cases with a potential genetic effect, we exclude such an effect since no other frequent SNPs were detected in the vicinity of these CpGs using the EpiTYPER platform. By accounting for three SNPs in our second model, we also ruled out this possibility for the associations reported in Table 2 and Supplementary Material, Table S4. In these cases, the scenario depicted in Figure 5d appears to be the most parsimonious. For the VINYLPHENOL, case we have shown that smoking is the common external factor (10). For the STEROID case, we have shown that there is a common theme between some of the differentially methylated gene loci, but we were not able to identify the nature of the external factor. Regarding the ‘inflated loci’, we also suspect a common environmental factor, which still needs to be identified. This could be a factor that is presently not captured in the KORA phenotype data set, such as sleep deprivation, general fatigue or exposure to a common environmental agent.
Figure 5.
Possible scenarios that may result in an observed CpG–metabotype association induced by a confounding genetic variant or by an external environmental factor.
Regarding future EWAS, a number of lessons can be learned from our study. Concerning the numerical treatment of inflation of some traits in the association, methods based on PCA have been recently suggested to control for potential confounders in data sets with large numbers of CpGs (29). Such techniques may be used in the future to increase the statistical power of EWAS. In terms of coping with potential artifacts due to interaction of the assay oligo-probes with genetic variants in the region, we developed an approach that may help alleviate this problem: By including three SNPs from the CpG region into the model, we account for variance in the metabolic traits that can be explained by genetic variance in the vicinity of the CpG site, regardless of whether this is a true genetic effect or an artifact induced by interaction between a SNP and the oligo-probe on the methylation array. This approach may also be applied in future EWAS with other phenotypes.
Cell-type mixtures may also be a problem. For blood samples, it is possible to use correlation structures in the methylation data to determine the cell-type composition of the analyzed blood cells (30). For instance, Liu et al. (31) successfully used this approach to adjust for cell-type proportions in an EWAS with rheumatoid arthritis. Studies with DNA methylation using DNA from human tissue biopsies (e.g. muscle and adipose tissue) may reveal more specific and potentially stronger CpG–metabotype associations. However, such samples are generally not available on an epidemiological scale, so that such studies may in turn be limited by their statistical power. In this context, it should be noted that a study using tissue samples from six different tissues and individuals has been published (16), while other studies showed that DNA methylation patterns were substantially different between lymphocyte cell lines and whole blood (9) and also across a large spectrum of samples, tissues and diseases (32).
In summary, we have shown that EWAS with large numbers of metabolic traits in big population cohorts are in principal feasible and can shed new light on the role of DNA methylation in human metabolism. However, we did not observe similarly strong effects of DNA methylation on metabotypes compared to what we previously reported for associations of genotype with metabotypes. We also found that results of associations from EWAS with metabolic traits may be difficult to interpret in terms of causality, and that it is hard to distinguish between true functional association and mere correlation that is driven by an unidentified common factor. Our data may now be used in future studies where a role of DNA methylation in the etiology of complex disorders is suspected.
MATERIAL AND METHODS
Study population and blood sampling
The KORA study is an independent population-based sample from the general population living in the region of Augsburg, southern Germany. KORA has been described in detail (33 and references therein). Here, we use data from the KORA F4 survey, which was conducted between 2006 and 2008. A total of 3080 subjects participated in the examination, comprising individuals who, at that time, were aged 32–81 years. Blood samples for metabolic analysis and DNA extraction were collected at the time of the KORA F4 visit. To avoid variation due to circadian rhythm, blood was drawn in the morning between 08:00 and 10:30 after a period of at least 10 h overnight fasting. Material was drawn into serum gel tubes, gently inverted twice and then allowed to rest for 30 min at room temperature (18–25°C) to obtain complete coagulation. The material was then centrifuged for 10 min (2750 g at 15°C). Serum was divided into aliquots and kept for a maximum of 6 h at 4°C, after which it was frozen at −80°C until analysis. Written informed consent has been given by all participants and the studies have been approved by the local ethics committee (Bayerische Landesärztekammer).
Metabolomics data set
The metabolite data used here consist of 649 measurements of metabolic traits that were obtained on three different metabolomics platforms: platform ‘Biocrates’ implements a kit-based-targeted quantitative FIA-MS/MS method (151 traits), platform ‘Metabolon’ uses non-targeted, semi-quantitative LC-MS/MS and GC-MS methods (483 traits), and platform ‘Lipofit’ derives lipid-related parameters from 1H NMR measurements (15 traits). The full metabolite set is provided as Supplementary Material, Table S1. These data sets have been extensively described previously. Quality control and platform-related details can be found in Jourdan et al. and Illig et al. (11,12) (Biocrates platform), Suhre et al. (13) (known metabolites from the Metabolon platform), Petersen et al. (14) (lipoprotein classes from the Lipofit platform) and Krumsiek et al. (15) (non-identified metabolic traits from the Metabolon platform).
Array-based DNA methylation analysis
DNA methylation was determined for 1814 samples using the Infinium HumanMethylation450 BeadChip platform (8). A total of 1000 ng genomic DNA from each sample was bisulfate-converted using the EZ-96 DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's procedure, with the alternative incubation conditions recommended when using the Infinium Methylation Assay. Genome-wide DNA methylation was assessed using the Infinium HumanMethylation450 BeadChip, following the Infinium HD Methylation protocol. This consists of a whole-genome amplification step using 4 µl of each bisulfite-converted sample, followed by enzymatic fragmentation and application of the samples to BeadChips (Illumina). The arrays were fluorescently stained and scanned with the Illumina HiScan SQ scanner. The percentage of methylation of a given cytosine is reported as a beta-value, which is a continuous variable between 0 and 1, corresponding to the ratio of the methylated signal over the sum of the methylated and unmethylated signals. GenomeStudio (version 2010.3) with methylation module (version 1.8.5) was used to process the raw image data generated by BeadArray Reader. Initial quality assessment of assay performance was conducted using the GenomeStudio software integrated controls dashboard and included assessment of DNP and Biotin staining, extension, hybridization, target removal, bisulfite conversion, specificity, negative and non-polymorphic controls. Nine samples had to be excluded because of deviations from optimal performance that also remained when the complete Infinium HD Methylation protocol was repeated, suggesting insufficient DNA quality. All 1805 approved samples were preprocessed with Genome Studio (background subtraction and control normalization) and the corrected beta-values were then extracted with the same software. Since the average success rate was larger than 99% for all samples, we did not exclude any samples. The beta-values ranged from 4 × 10−5 to 0.99881. After exclusion of non-autosomal sites, a total of 457 004 CpG sites were used for analysis. To control for reproducibility of methylation data, a positive control was included per Illumina run, summing up to a total of six replicates. As additional quality check, we calculated a CV (coefficient of variance) for all CpG site using these six replicates. The maximum CV was 2.5%. Furthermore, using control samples which were designed with 0, 20, 60 and 100% methylation, we checked for variation in the mean of the beta-values over different categories of CpG sites (Exon, UTR3, UTR5, Body, TSS1500, TSS200, Island, N_Shelf, N_Shore, S_Shelf, S_Shore). The mean for each control sample was comparable across the different categories. For all of these individuals, genome-wide SNP data were already available. These data have been used and described extensively in the past in the context of several GWAS [e.g. (12,13)].
Experimental validation using EpiTYPER
We attempted experimental validation of most of the loci reported in Tables 1 and 2 using the EpiTYPER system. EZ-96 DNA methylation kits (Zymo Research, CA, USA) were used for bisulfite treatment of 500 ng of genomic DNA. Amplicons were designed for each gene. The target regions were then amplified to allow further in vitro transcription using the primer pairs and annealing temperatures (Ta) described in Supplementary Material, Table S3. PCR products were treated according to the standard protocol (Sequenom EpiTyper Assay) including SAP treatment and T-cleavage reaction as described previously (17). Resin-cleaned samples were dispensed to a 384 SpectroCHIP preloaded with matrix (SEQUENOM, Inc., San Diego, CA, USA) by Nanodispenser. Mass spectra were then collected using a Sequenom MALDI-TOF MS Compact Unit mass spectrometer and analyzed using proprietary peak picking and signal-to-noise calculations (Sequenom Epityper v1.2).
SNPs can hamper correct quantification of methylation status at one or more CpG sites and thus may complicate interpretation of results. Each novel peak among the MassArray spectrum can be explained by any number of potential SNPs. By using an exhaustive string substitution approach as implemented in the R package ‘MassArray’ (18), putative SNPs can be identified by comparing expected and observed data. The putative nucleotide sequences underlying novel peaks are identified by the EpiTYPER software and can be used for comparison with the original input sequence. The applied algorithm substitutes each base pair at a time in the original input sequence with the other three remaining bases or a gap, i.e. a deletion, and then assesses the ability of the altered input sequence to explain the observed fragmentation pattern due to new peaks. This is done by fragmentation of the altered input sequence and finding base compositional matches to the putative base pair composition of the new peak. Once these new peaks are mapped to the appropriate fragments, the expected peaks corresponding to these fragments are analyzed in order to determine whether they are missing or if there is a diminished signal-to-noise ratio (SNR), which is done by comparison of the expected peak SNR to the average SNR of the sample. The SNR of the novel peak is also compared with the average SNR of the sample and granted more reliability if it exceeds this average. Finally, the SNP's quality is then calculated as function of new peak SNR and expected peak SNR. For samples showing a putative high-confidence SNP that maps to a fragment containing one or more CpGs, methylation data from that site should be interpreted with caution. The numbers of SNPs that were detected within the relevant fragments are reported in Tables 1 and 2.
Statistical analysis
A linear model with covariates age, gender, BMI and white WBC was used to test for association between DNA methylation (b-values) and log-transformed metabolite concentrations (Biocrates and Lipofit platforms) or ion counts (Metabolon platform). The genome-wide level of significance at an alpha level of 0.05 after correction for the number of metabolic traits (649) and DNA methylation sites (457 004) is pgw = 1.69 × 10−10. To account for the effect of genetic variance, we conducted a second association analysis where we included three SNPs from the vicinity of the CpG site into the model. These three SNPs were selected iteratively as follows: first we tested the association of each b-value for linear additive dependence on every individual genotyped SNP within a window of ±5 Mb around the CpG site, using age, gender, BMI and WBC as covariates. We then selected the SNP that showed the strongest association with CpG methylation (called SNP1). We then selected a second SNP (SNP2) and then a third (SNP3) following the same procedure, including the already selected SNP(s) as covariates. The lm subroutine from the R stats package (version 2.15; R Foundation for Statistical Computation) was used for statistical analysis and SPSS (version 20; IBM) for graphical visualization.
SUPPLEMENTARY MATERIAL
FUNDING
The KORA study was initiated and financed by the Helmholtz Zentrum München—German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. This work was supported by the DFG/Tr22-Z03. The research leading to these results has also received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 261433 and by the DFG/Tr22-Z03. Part of the metabolomics measurements were funded by BMBF grant 0315494A (project SysMBo). A.K.P. was supported by the ENGAGE Exchange and Mobility Program (HEALTH-F4-2007-201413) and W.R.M. is supported by BMBF grant 03IS2061B (project Gani_Med). This work was also supported in part by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V.). C.G. has received funding from the European Union's Seventh Framework Programme (FP7-Health-F5-2012) under grant agreement no. 305280 (MIMOmics). K.S. is supported by ‘Biomedical Research Program’ funds at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation. The statements made herein are solely the responsibility of the authors. Funding to pay the Open Access publication charges for this article was provided by Weill Cornell Medical College in Qatar.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Othmane Bouhali for providing us with high-performance computing resources at Texas A&M in Qatar. We further thank Dr Cornelia Prehn, Julia Scarpa, Katharina Sckell and Arsin Sabunchi for metabolomics measurements performed at the Helmholtz Zentrum München, Genome Analysis Center, Metabolomics Core Facility. We thank all KORA study participants and all members of the field staff in Augsburg who planned and conducted the study and Nadine Lindemann and Franziska Scharl for excellent technical support.
Conflict of interest statement. F.H. is employed by numares Health GmbH. He contributed only to logistics and optimization of NMR spectroscopy and to NMR data interpretation. R.P.M. is employed by Metabolon, Inc. He contributed only to the logistics and optimization of MS spectroscopy and to MS data interpretation. numares health GmbH and Metabolon, Inc. were not involved in the design of the study, statistical analyses or interpretation of the results. All other authors declare that they have no competing interests.
REFERENCES
- 1.Suhre K., Gieger C. Genetic variation in metabolic phenotypes: study designs and applications. Nat. Rev. Genet. 2012;13:759–769. doi: 10.1038/nrg3314. doi:10.1038/nrg3314. [DOI] [PubMed] [Google Scholar]
- 2.Gieger C., Geistlinger L., Altmaier E., Hrabe de Angelis M., Kronenberg F., Meitinger T., Mewes H.W., Wichmann H.E., Weinberger K.M., Adamski J., et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. doi:10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mootha V.K., Hirschhorn J.N. Inborn variation in metabolism. Nat. Genet. 2010;42:97–98. doi: 10.1038/ng0210-97. doi:10.1038/ng0210-97. [DOI] [PubMed] [Google Scholar]
- 4.Portela A., Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. doi:10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 5.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. doi:10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 6.Rakyan V.K., Down T.A., Balding D.J., Beck S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 2011;12:529–541. doi: 10.1038/nrg3000. doi:10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menni C., Kastenmuller G., Petersen A.K., Bell J.T., Psatha M., Tsai P.C., Gieger C., Schulz H., Erte I., John S., et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013 doi: 10.1093/ije/dyt094. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. doi:10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Aberg K., Khachane A.N., Rudolf G., Nerella S., Fugman D.A., Tischfield J.A., van den Oord E.J. Methylome-wide comparison of human genomic DNA extracted from whole blood and from EBV-transformed lymphocyte cell lines. Eur. J. Hum. Genet. 2012;20:953–955. doi: 10.1038/ejhg.2012.33. doi:10.1038/ejhg.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeilinger S., Kuhnel B., Klopp N., Baurecht H., Kleinschmidt A., Gieger C., Weidinger S., Lattka E., Adamski J., Peters A., et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. doi:10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jourdan C., Petersen A.K., Gieger C., Doring A., Illig T., Wang-Sattler R., Meisinger C., Peters A., Adamski J., Prehn C., et al. Body fat free mass is associated with the serum metabolite profile in a population-based study. PLoS ONE. 2012;7:e40009. doi: 10.1371/journal.pone.0040009. doi:10.1371/journal.pone.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Illig T., Gieger C., Zhai G., Romisch-Margl W., Wang-Sattler R., Prehn C., Altmaier E., Kastenmuller G., Kato B.S., Mewes H.W., et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010;42:137–141. doi: 10.1038/ng.507. doi:10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B., Altmaier E., Deloukas P., Erdmann J., Grundberg E., et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. doi:10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen A.K., Stark K., Musameh M.D., Nelson C.P., Romisch-Margl W., Kremer W., Raffler J., Krug S., Skurk T., Rist M.J., et al. Genetic associations with lipoprotein subfractions provide information on their biological nature. Hum. Mol. Genet. 2012;21:1433–1443. doi: 10.1093/hmg/ddr580. doi:10.1093/hmg/ddr580. [DOI] [PubMed] [Google Scholar]
- 15.Krumsiek J., Suhre K., Evans A.M., Mitchell M.W., Mohney R.P., Milburn M.V., Wagele B., Romisch-Margl W., Illig T., Adamski J., et al. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012;8:e1003005. doi: 10.1371/journal.pgen.1003005. doi:10.1371/journal.pgen.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byun H.M., Siegmund K.D., Pan F., Weisenberger D.J., Kanel G., Laird P.W., Yang A.S. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum. Mol. Genet. 2009;18:4808–4817. doi: 10.1093/hmg/ddp445. doi:10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrich M., Nelson M.R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C.R., Field J.K., van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. doi:10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson R.F., Suzuki M., Lau K.W., Greally J.M. A pipeline for the quantitative analysis of CG dinucleotide methylation using mass spectrometry. Bioinformatics. 2009;25:2164–2170. doi: 10.1093/bioinformatics/btp382. doi:10.1093/bioinformatics/btp382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Touleimat N., Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. doi:10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 20.Maksimovic J., Gordon L., Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. doi:10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. doi:10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marabita F., Almgren M., Lindholm M.E., Ruhrmann S., Fagerstrom-Billai F., Jagodic M., Sundberg C.J., Ekstrom T.J., Teschendorff A.E., Tegner J., et al. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics. 2013;8:333–346. doi: 10.4161/epi.24008. doi:10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinius L.E., Acevedo N., Joerink M., Pershagen G., Dahlen S.E., Greco D., Soderhall C., Scheynius A., Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. doi:10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manini P., De Palma G., Andreoli R., Goldoni M., Poli D., Lasagni G., Mutti A. [Urinary excretion of 4-vinyl phenol after experimental and occupational exposure to styrene] G. Ital. Med. Lav. Ergon. 2003;25(Suppl):61–62. [PubMed] [Google Scholar]
- 25.Buterin T., Koch C., Naegeli H. Convergent transcriptional profiles induced by endogenous estrogen and distinct xenoestrogens in breast cancer cells. Carcinogenesis. 2006;27:1567–1578. doi: 10.1093/carcin/bgi339. doi:10.1093/carcin/bgi339. [DOI] [PubMed] [Google Scholar]
- 26.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.K., Jang H.G., Jha A.K., et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. doi:10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todorova V.K., Kaufmann Y., Luo S., Klimberg V.S. Tamoxifen and raloxifene suppress the proliferation of estrogen receptor-negative cells through inhibition of glutamine uptake. Cancer Chemother. Pharmacol. 2011;67:285–291. doi: 10.1007/s00280-010-1316-y. doi:10.1007/s00280-010-1316-y. [DOI] [PubMed] [Google Scholar]
- 28.Parikh H., Carlsson E., Chutkow W.A., Johansson L.E., Storgaard H., Poulsen P., Saxena R., Ladd C., Schulze P.C., Mazzini M.J., et al. TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. doi:10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W., Gao G., Nerella S., Hultman C.M., Magnusson P.K., Sullivan P.F., Aberg K.A., van den Oord E.J. MethylPCA: a toolkit to control for confounders in methylome-wide association studies. BMC Bioinfor. 2013;14:74. doi: 10.1186/1471-2105-14-74. doi:10.1186/1471-2105-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koestler D.C., Christensen B.C., Marsit C.J., Kelsey K.T., Houseman E.A. Recursively partitioned mixture model clustering of DNA methylation data using biologically informed correlation structures. Stat. Appl. Genet. Mol. Biol. 2013;12:225–240. doi: 10.1515/sagmb-2012-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Aryee M.J., Padyukov L., Fallin M.D., Hesselberg E., Runarsson A., Reinius L., Acevedo N., Taub M., Ronninger M., et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. doi:10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez A.F., Assenov Y., Martin-Subero J.I., Balint B., Siebert R., Taniguchi H., Yamamoto H., Hidalgo M., Tan A.C., Galm O., et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–419. doi: 10.1101/gr.119867.110. doi:10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wichmann H.E., Gieger C., Illig T. KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):S26–S30. doi: 10.1055/s-2005-858226. doi:10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.