Abstract
The tau mutation is a semidominant autosomal allele that dramatically shortens period length of circadian rhythms in Syrian hamsters. We report the molecular identification of the tau locus using genetically directed representational difference analysis to define a region of conserved synteny in hamsters with both the mouse and human genomes. The tau locus is encoded by casein kinase I epsilon (CKIɛ), a homolog of the Drosophila circadian gene double-time. In vitro expression and functional studies of wild-type and tau mutant CKIɛ enzyme reveal that the mutant enzyme has a markedly reduced maximal velocity and autophosphorylation state. In addition, in vitro CKIɛ can interact with mammalian PERIOD proteins, and the mutant enzyme is deficient in its ability to phosphorylate PERIOD. We conclude that tau is an allele of hamster CKIɛ and propose a mechanism by which the mutation leads to the observed aberrant circadian phenotype in mutant animals.
Daily rhythms in biochemical, physiological, and behavioral processes are regulated by biological clocks (1, 2). In natural conditions, the endogenous circadian rhythms that are generated by these clocks are synchronized (entrained) to the 24-hour cycles of the external world by time cues such as the daily light/dark cycle (2). While these clocks are found in organisms as divergent as cyanobacteria, plants, fruit flies, and mammals, there is an extraordinary degree of evolutionary conservation of the underlying generative molecular mechanisms (3–9). The first mammalian circadian gene, Clock, was cloned and characterized in mouse (10–12). Clock encodes a novel member of the basic helix-loop-helix (bHLH) PER-ARNT-SIM (PAS) family of transcription factors. Shortly following the cloning of Clock, three mouse Period orthologs, denoted mPer1 (13, 14), mPer2 (15–17) and mPer3 (18, 19), as well as a Timeless (mTim) ortholog, were cloned (20–24). At the same time, another gene, BMAL1 (25), was found to encode the protein dimerization partner for CLOCK (26), and together the CLOCK/BMAL complex was shown to transactivate mPer1 via conserved E-box elements found in the promoters of Drosophila and mouse period genes (26–28). Finally, in Drosophila and mammals, the negative feedback effects of PER and TIM were shown to act at the level of the CLOCK/BMAL complex (20, 28) in a surprisingly direct manner (29, 30).
In Drosophila, the core elements of the circadian oscillator consist of four proteins: Two stimulatory components, dCLOCK and its partner, dBMAL (CYCLE), and two inhibitory components PERIOD and TIMELESS (31–36). The cycle begins with dCLOCK/dBMAL activation of transcription from the period and timeless genes via E-box motifs in their respective promoters (27, 28, 37). For several hours, the protein products of the per and tim genes, PER and TIM, are synthesized in the cytoplasm, accumulate to a critical level, heterodimerize, and then translocate to the nucleus (34–37). Once in the nucleus, the PER-TIM complex interacts negatively with the dCLOCK/dBMAL complex to inhibit transcription from per and tim (28–30). This inhibition stops the accumulation of PERIOD and TIMELESS, and the two proteins gradually disappear over the next 12 hours. This then allows the cycle to start again with the activation of period and timeless by dCLOCK and dBMAL. In addition to the circadian transcriptional cycle, there are a number of points of post-transcriptional control (6); the cytoplasmic accumulation of PER and TIM, their heterodimerization, and their subsequent nuclear translocation. PER is gradually phosphorylated over time (38) and this occurs at least in part as a consequence of the product of the double-time (dbt) gene, a casein kinase I epsilon (CKIε ) homolog (39, 40). DBT interacts with and phosphorylates PER, which is thought to be critical for PER turnover.
In mammals, a transcription-translation autoregulatory feedback loop also appears to form the core mechanism of the circadian clock system (9). Similar to that seen in Drosophila, CLOCK and BMAL are positive elements that act upstream of the mPer1 gene to activate its transcription (26). While the core four elements (PER, TIM, CLOCK, and BMAL) are conserved, there are a number of differences. All three mammalian Period genes express circadian rhythms of gene expression in the suprachiasmatic nucleus (SCN) of the hypothalamus (41), the site of the mammalian master circadian pacemaker, but are differentially regulated by light (15–19, 42). Regulation of mammalian Timeless also differs from that seen in Drosophila (20, 21, 43). Perhaps the most interesting difference is the role of the mammalian Cryptochrome genes, mCry1 and mCry2, which are essential for the expression of circadian behavioral rhythms (44, 45) and which play a major role in the negative feedback regulation of mPer1 and mPer2 (45–48). Thus, in mammals there are at least eight different genes that are thought to play a role in the circadian clock system of mice (Clock, Bmal1, mPer1, mPer2, mPer3, mTim, mCry1, and mCry2). The positive elements (CLOCK and BMAL) are highly conserved, while the negative elements (PERs, TIM, CRYs) are more diverse, leading to a more complex feedback loop than that seen in Drosophila (9, 49).
The hamster tau mutation
In 1988, Ralph and Menaker (50) discovered a circadian period mutation in a routine laboratory shipment of Syrian hamsters (Mesocricetus auratus). Genetic analysis revealed a semidominant mode of transmission for this spontaneous mutation, named tau. As assayed by wheel-running locomotor activity in constant darkness, the free-running rhythm is about 24 hours in wild-type animals, 22 hours in tau heterozygotes, and just 20 hours in homozygous mutant animals. Over the next decade, critical experiments utilizing the tau mutation established that the suprachiasmatic nucleus (SCN) determines circadian period at the organismal level in mammals (51); demonstrated that a diffusable signal from the SCN can drive circadian rhythms in the animal (52); and demonstrated the existence of SCN-independent circadian oscillators in the retina (53).
Unfortunately, the paucity of genetic resources available in the Syrian hamster has presented a formidable barrier to the identification and molecular analysis of mutants in this organism. The development, however, of new techniques in genome analysis provides promise for the study of mutations in such map-poor species and suggested to us that a molecular genetic analysis of tau might be feasible (54, 55).
In genetically directed representational difference analysis (GDRDA) (56, 57), polymorphic markers tightly linked to a monogenic trait can be identified without prior knowledge of the chromosomal location of that trait or the availability of genetic maps for the organism. GDRDA begins by separating progeny from an experimental cross into two groups based on phenotype, such that the groups differ on average only at the locus under study. For example, homozygous wild type and homozygous mutant animals could be identified and their DNA samples pooled into “tester” and “driver” groups, respectively. Next, genomic representations or “amplicons” are prepared for both pools by a whole genome polymerase chain reaction (PCR) technique. By applying the subtractive hybridization and kinetic enrichment properties of representational difference analysis, of which GDRDA is a modification, it is possible to isolate small DNA fragments that are enriched in the pooled tester samples and absent in the driver pools. Thus, polymorphic DNA markers linked to the trait of interest can be identified directly.
Genetic mapping of tau
The strategy in our molecular genetic analysis of the tau mutation was first to construct a genetic cross with which we could identify markers linked to the mutation by applying GDRDA. Because finding markers linked to a mutation is, by itself, of limited value in searching for a gene in an organism lacking a genetic map, we next sought to identify regions of conserved synteny between hamster and both mouse and human. We could then focus on any obvious positional candidate loci in hamster by referring to the dense genetic maps available in mouse and human around the region of conserved synteny. Candidate genes could then be narrowed by meiotic mapping and identification of mutations.
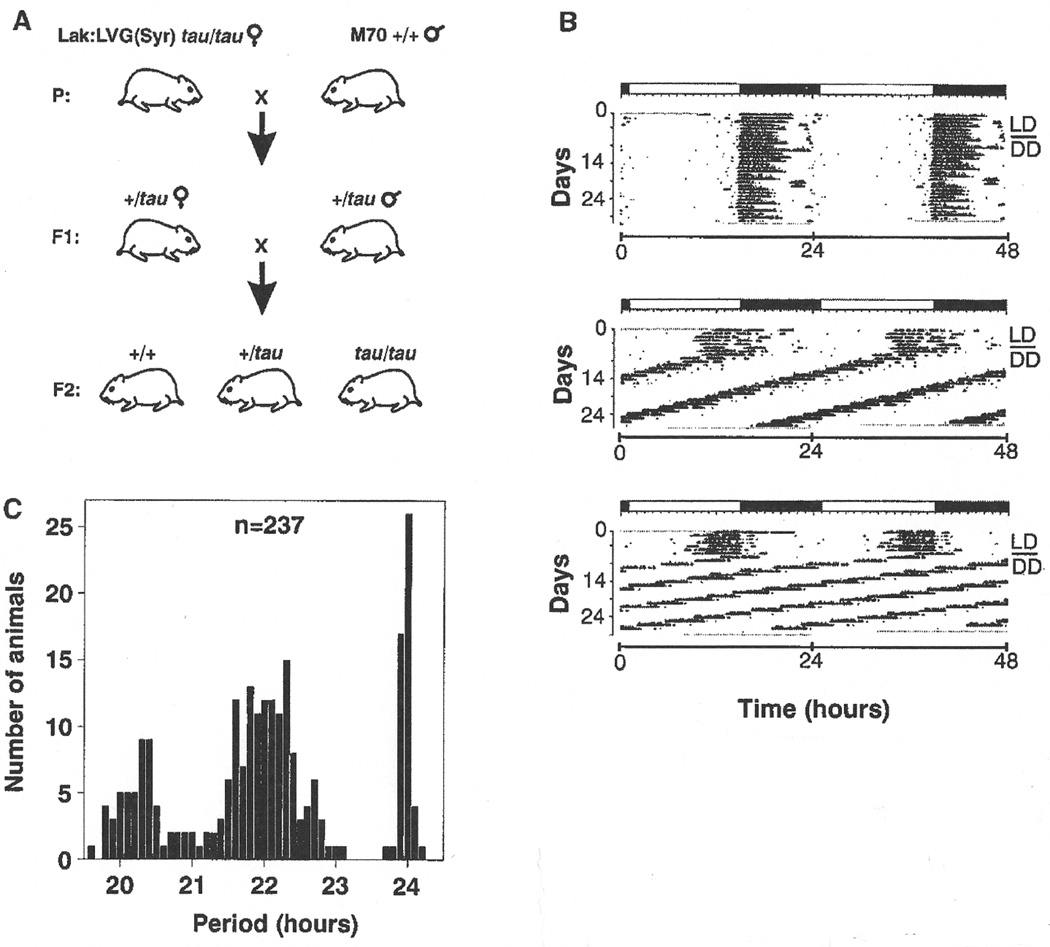
All common hamster strains originate from three littermates born to a female captured in the wild in 1930 (58, 59) and, consequently, their level of heterozygosity is quite low. To create an appropriate Syrian hamster cross that would both segregate the tau mutation and provide the necessary heterozygosity for mapping purposes, we sought a strain derived from a different founder population. The M70 outbred strain maintained at the National Institutes of Health originates from a colony of wild-caught hamsters returned to the U.S. from Syria in 1971 (59). We crossed wild-type males of this strain to Lak:LVG:(Syr) homozygous tau female animals to produce the F1 generation. F1 siblings were then interbred to produce an F2 intercross consisting of 237 progeny (Fig. 1A) (60). Circadian phenotype of all animals was assessed by monitoring wheel-running locomotor activity in constant conditions (Fig. 1B) (61). As expected, the distribution of the period phenotype among the F2 progeny exhibits the classical Mendelian ratio of 1:2:1 (+/+,n = 50 : +/tau,n = 131 : tau/tau,n = 56), indicative of semidominance (Fig. 1C).
Fig. 1.
(A) The Syrian hamster cross used to map the tau mutation (60). Wild-type males of the outbred M70 strain were mated with outbred female Lak:LVG(Syr) hamsters homozygous for the tau mutation. Siblings of the F1 generation were intercrossed to produce 237 F2 progeny. (B) Representative locomotor activity records of the three circadian phenotypes in the F2 hamster progeny (61). Records are double-plotted so that 48 hours are shown for each horizontal trace. Dark regions represent activity. For the first 7 days, animals were housed in a cycle of 14 hours light, 10 hours dark (LD14 :10), noted by the bar above each record. Animals were transferred to constant darkness (DD) for a total of 3 weeks beginning on day 8 (indicated by a line in the right margin). Top record, wild-type animal with a period of 24 hours; middle record, animal heterozygous for the tau mutation with a period of 22 hours; bottom record, homozygous tau animal with a period of 20 hours. (C) Distribution of period among 237 F2 offspring tested for circadian phenotype.
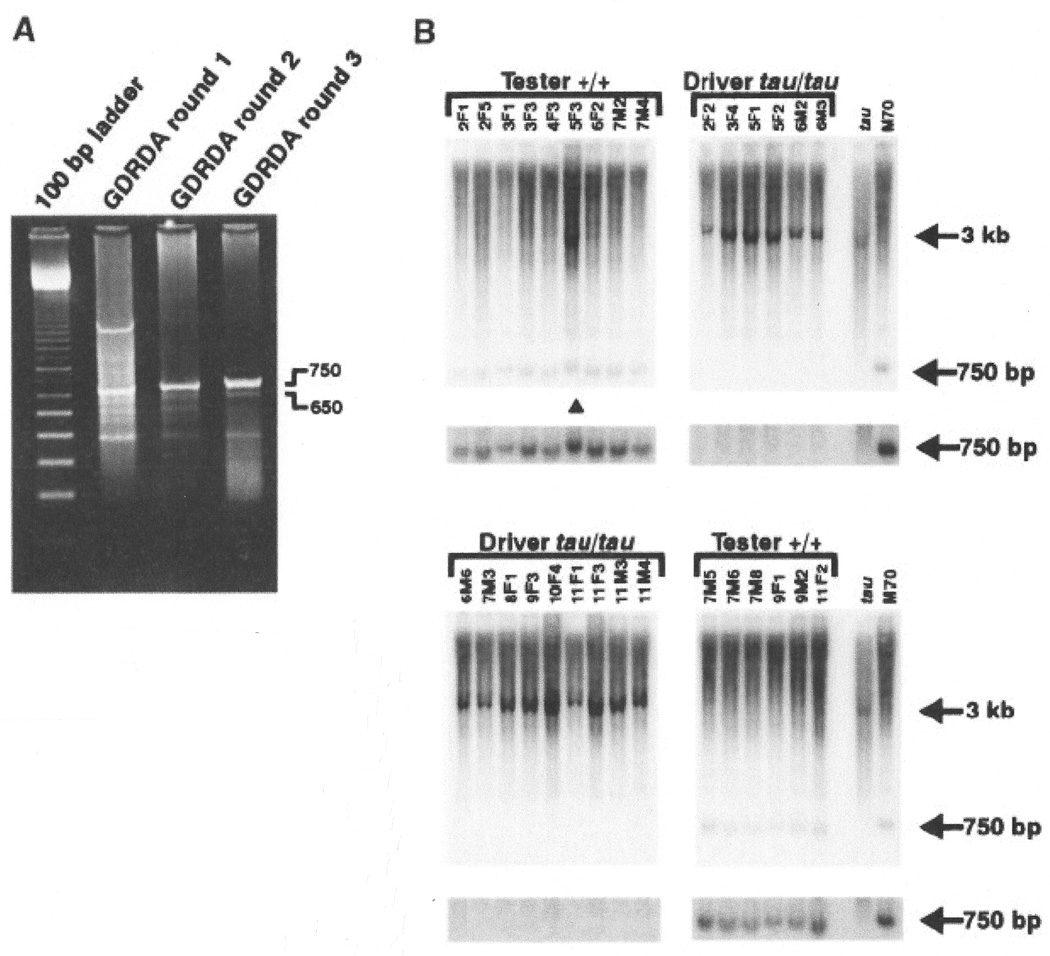
To identify markers linked to the tau mutation via GDRDA, we selected 15 homozygous wild type and 15 homozygous tau F2 animals from a single three-generation pedigree. DNA was extracted from each F2 animal, and an equal amount of genomic DNA from each was pooled into a tester (wild-type) fraction and a driver (homozygous tau) fraction (61). GDRDA was performed essentially as described (56, 61–63). Results of three rounds of subtraction, extension, and amplification yielded several distinct products by ethidium bromide staining (Fig. 2A). Five RDA products from the third round were cloned in a plasmid vector and tested for linkage to the mutation by hybridizing each to Southern blots of the individual F2 DNA samples digested with BamHI (61). Two products, RDA-650 and RDA-750, were polymorphic for the hamster cross. The RDA-750 probe revealed a 750 base pair (bp) product in the tester DNA samples and ∼3 kilobase (kb) fragment in the driver samples (Fig. 2B). Similarly, the RDA-650 probe detected a 650-bp product in the tester DNA samples and a ∼7 kb fragment in the driver samples (not shown). The larger fragments present in the driver samples were not amplified in the original PCR reactions producing the genomic representations. Because the 15 tester and 15 driver animals are the progeny of an F2 intercross, the 30 DNA samples represent a total of 60 meiotic events. Both probes revealed a recombination event in the same animal (Fig. 2B, triangle). As a result, the recombination fraction for both markers with respect to the tester and driver animals is 1/60, which is equivalent to a genetic distance of approximately 1.7 centimorgans (cM) from the tau mutation (64).
Fig. 2.
(A) Ethidium bromide staining of products from three rounds of GDRDA. Two products (650 and 750 bp) from round three exhibited linkage to the tau mutation (61). (B) Southern hybridization analysis of DNA from the 30 F2 animals comprising the tester and driver pools for GDRDA analysis and the parental animals (61). Œ, the single animal in which a recombination event occurred. The regions of the blot containing the 750-bp signal are shown again below each section with an adjustment in brightness to aid visualization of the signal. Numbers identify individual progeny.
To obtain genomic clones containing the GDRDA products, we screened a hamster A genomic DNA library using the RDA-650 and 750 products as probes (65). The resulting clones were then scanned by direct DNA sequencing. In the course of sequencing the RDA-750 A genomic clone, two microsatellite repeats were found (61). PCR reactions with primers flanking the repeats were performed to test for polymorphism in our hamster cross. One of the microsatellites, termed SSR2, was polymorphic and thus simplified the genotyping of the RDA-750 product because it was no longer necessary to type the cross by Southern blotting (66). Typing of SSR2 in the entire F2 cross placed this marker 5.6 cM from the tau locus. We also tested the hamster SSR2 primer pair for amplification of DNA from various mouse strains. We identified the presence of a single amplification product only in the SPRET/Ei strain. This facilitated the mapping of SSR2 in mouse with the Jackson Laboratory BSB [(C57BL/6J x M. spretus) x C57BL/6J)] panel (67, 68). We mapped SSR2 to a region of mouse chromosome 15 (68).
The sequence data from the RDA-650 A genomic clone were analyzed by BLAST and revealed a highly significant sequence similarity in the GenBank database to the mouse gene Celsr1 that, like SSR2, maps to mouse chromosome 15. Two regions of sequence aligned to Celsr1 cDNA for a total of 325 bp with a nucleotide identity of 92%. Given these findings, we reasoned that in mouse, the tau locus could reside in a region with conserved synteny to mouse chromosome 15. To test this hypothesis, we initiated a search for polymorphisms in hamster orthologs of genes that, in mouse, map to chromosome 15. After analyzing the mouse genetic map around the Celsr1 locus, we chose several genes distal and proximal to Celsr1 for which mouse genomic sequence exists in the GenBank database. This facilitated the design of primers to 5'-and 3'-untranslated sequences and to sequences flanking introns of PCR-amplifiable size (69).
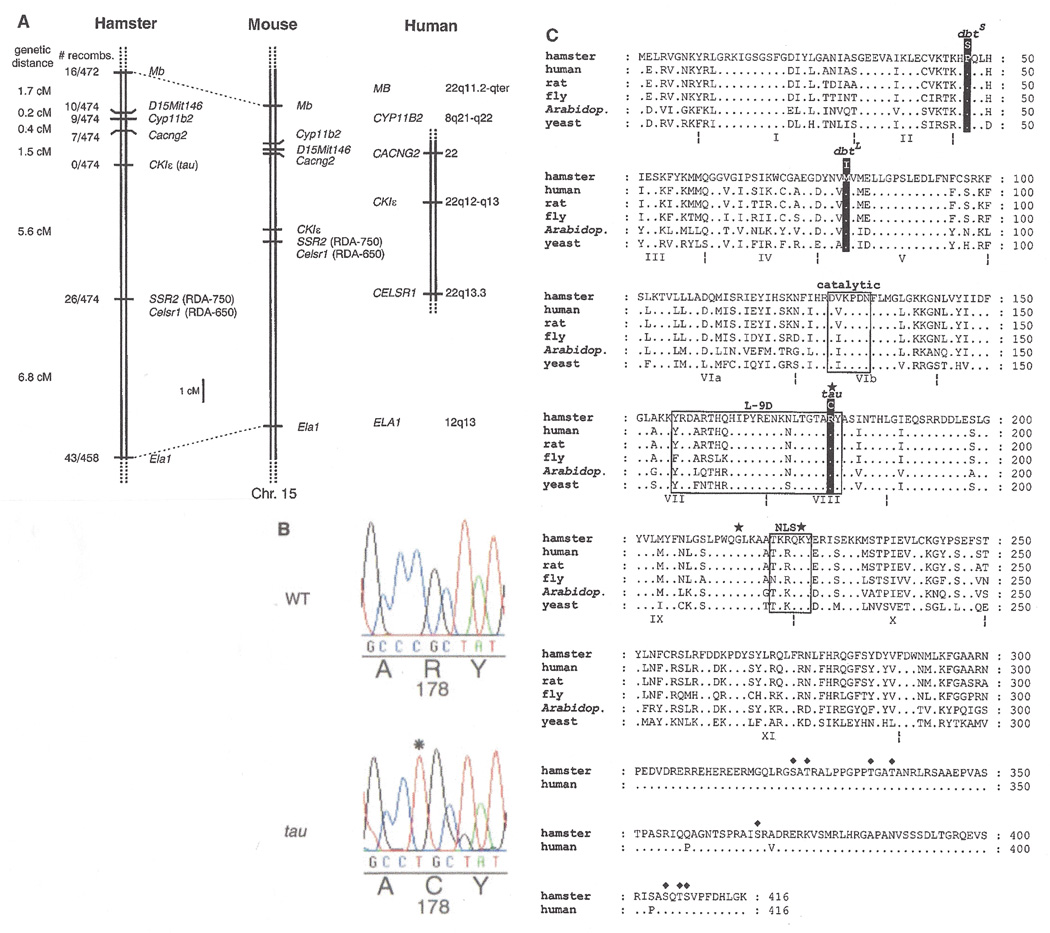
We limited our polymorphism search to untranslated and intronic sequences because the likelihood of discovering differences in those regions is greater. Using this approach, we identified polymorphisms in the following genes, all of which were linked to tau in hamster and thus define a segment of conserved synteny between the two species: Myoglobin (Mb)(70), calcium channel, voltage-dependent, γ subunit 2 (Cacng2, stargazer)(71, 72), aldosterone synthase (Cyp11b2)(73), and elastase (Ela1)(74). We also tested several mouse simple sequence length polymorphism (SSLP) markers from chromosome 15 to determine whether any would amplify hamster DNA. Mouse primer pairs that produced discrete products in hamster were then tested further for their ability to reveal polymorphisms and linkage to the tau locus. One SSLP marker, D15Mit146, out of more than 50 markers tested, was both polymorphic in our hamster cross and cosegregated with tau (75). Hence, using reciprocal mapping techniques between hamster and mouse genomes, we were able to establish a region of conserved synteny between the two species spanning a genetic distance of approximately 15 cM (Fig. 3A) (76). Based on mouse and human comparative maps, the region of conserved synteny spans segments of human chromosomes 8, 12, and 22 (Fig. 3A).
Fig. 3.
(A) Genetic map of the Syrian hamster tau locus, showing regions of conserved synteny in mouse and human (69, 76). Genes are as follows: Myoglobin, Mb; aldosterone synthase, Cyp11b2; calcium channel, voltage-dependent, γ subunit 2, Cacng2, stargazer; casein kinase Iɛ, CKIɛ (tau); cadherin EGF LAG seven-pass G-type receptor, Celsr1; elastase, Ela1. A single mouse SSLP marker, D15Mit146, was mapped in hamster (75). In mouse, the locus amplified by the SSR2 hamster primers has been designated D15Nwu1 in the Mouse Genome Database. (B) Electropherogram of sequence around tau mutation region from wild-type and tau homozygous mutant CKIɛ cDNA. An asterisk denotes the point mutation (C→T transition) in the tau cDNA sequence. Amino acid residues are listed under their corresponding nucleotide codons. (C) Alignment of casein kinase I isoforms from various species (100). Dots indicate identities. Kinase subdomains are bounded by the dotted-lines symbol and are numbered with Roman numerals (101). Boxed residues indicate the catalytic loop, the L-9D loop, and the putative nuclear localization signal (NLS). The star symbol marks each of the three residues (Arg178, Gly215, Lys224) that form the anion binding motif. Putative autophosphorylation sites on the carboxyl-terminal tail are indicated by ♦ (82). Drosophiladouble-time short and long-period mutation sites and the hamster tau mutation site are labeled (39, 40). The amino acid sequences of all proteins except hamster and human CKIɛ are omitted following residue 300 because of reduced homology thereafter. Names and GenBank accession numbers are as follows: Mesocricetus auratus CKIɛ (AF242536); Homo sapiens CKIɛ (L37043); Rattus norvegicus CKI8 (L07578); Drosophila melanogaster double-time (AF055583); Arabidopsis thaliana dual specificity kinase (U48779); Saccharomyces cerevisiae HRR25 (M68605).
Identification of a candidate locus
Mapping to human chromosome 22 in the region of conserved synteny is the gene casein kinase I epsilon (CKIε ) (Fig. 3A), a strong candidate for the tau locus given its homology to the Drosophila circadian gene dbt (40, 77). Because CKIε had not yet been mapped in mouse, we designed primers based on the human cDNA sequence to amplify the corresponding hamster sequence by RT-PCR (61, 77). Separate reactions were performed with wild type and tau total RNA samples isolated from pooled SCN tissue. A single product of approximately 1.3 kb was amplified from each pool, then sequenced and identified as the coding region of the hamster CKIε ortholog.
Comparison of the sequence between wild type and tau animals revealed a single base-pair substitution (Fig. 3B). This C→T transition results in the creation of a recognition sequence for the restriction enzyme BstAPI in the tau product only. This allowed us to PCR-amplify a smaller region around the mutation in all animals from the hamster cross and then to test each product for digestion with BstAPI (78). In doing so, we found no recombinants out of 474 meioses (Fig. 3A). Furthermore, conceptual translation of the tau CKIε cDNA revealed that the mutation causes an arginine-to-cysteine amino acid substitution at residue 178 (Fig. 3C), a highly conserved residue.
To confirm that CKIε maps to chromosome 15 in mouse, we amplified a portion of the coding region of the mouse ortholog and identified a polymorphism between the C57BL/6J and BALB/cJ mouse strains. As a result, we were able to map CKIε to chromosome 15 between SSLP markers D15Mit146 (proximal) and D15Mit35 (distal) using a (BALB/ cJ x C57BL/6J)F2 intercross (79). This finding validated our comparative mapping results and established that the ortholog of our hamster candidate locus mapped to chromosome 15 in mouse.
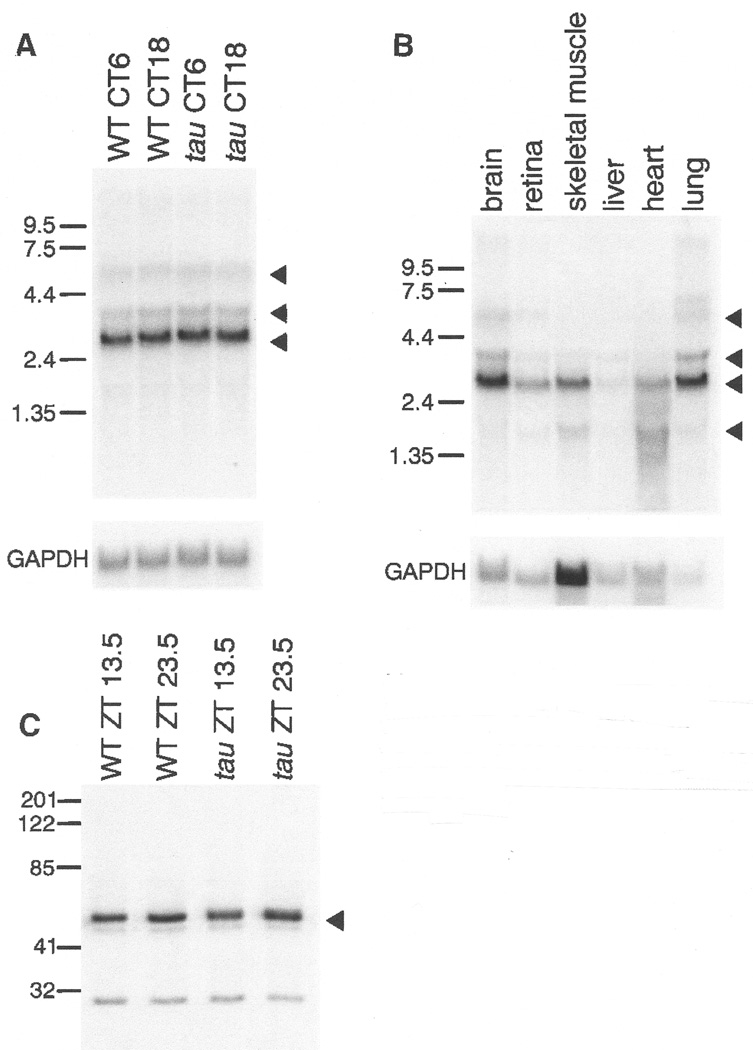
Northern analysis of CKIε from total RNA of hamster brains collected at two circadian times (subjective day and subjective night; CT6 and CT18) reveals no obvious differences in size or abundance between the wild-type and tau transcripts (Fig. 4A) (61). The tissue distribution of hamster CKIε is similar to that reported in human with highest mRNA levels in brain, retina, skeletal muscle, and lung (Fig. 4B) (77). As in human CKIε, there is a major transcript of 2.7 kb, as well as minor transcripts at 1.6, 3.7, and 5.9 kb. Analysis of CKIε protein expression in tissue extracts from brain shows a 48-kD protein in wild type and tau mutant hamsters (Fig. 4C) (61). Finally, preliminary experiments using in situ hybridization with a probe to the COOH-terminal region of hamster CKIε cDNA shows a clear hybridization signal in the SCN and brain when compared to controls with a sense probe (61).
Fig. 4.
(A) Northern analysis (61) of total RNA prepared from brains of wild-type and homozygous tau hamsters collected at two circadian times, CT6 and CT18. A major transcript of 2.7 kb and minor transcripts of 3.5 and 5.9 kb are indicated by triangles. The blot was normalized by hybridization with probe to mouse GAPDH. (B) Northern analysis (61) of total RNA from wild-type hamster brain, retina, skeletal muscle, liver, heart, and lung tissue. A major transcript of 2.7 kb and minor transcripts of 1.6, 3.7, and 5.9 kb are indicated by triangles. The blot was normalized by hybridization with a probe to mouse GAPDH. (C) Immuno-blot of wild-type and tau hamster hypothalamic tissue extracts (61). Tissues were collected at two zeitgeber (ZT) times (13.5 and 23.5). Blots were probed with antibody to human CKIɛ. A strong signal is evident at ∼48 kD (triangle), the reported size of human CKIɛ (77).
Taken together, the genetic linkage analysis and the amino acid substitution with attendant protein structural implications indicate that tau represents a mutant allele of the hamster CKIε gene.
Protein expression and functional analysis
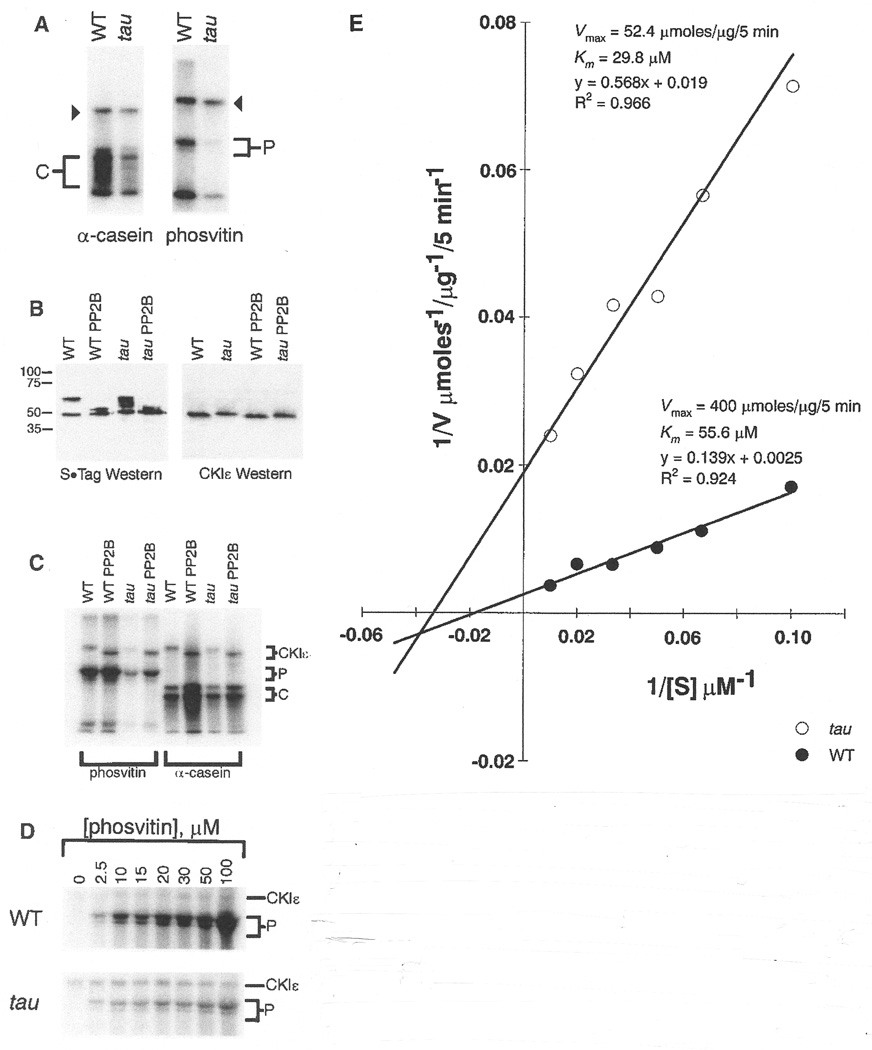
To perform a functional analysis, we cloned hamster wild type and tau full-length CKIε cDNAs in a T7 bacterial expression vector containing sequence producing S•Tag and His-Tag fusions at the amino-terminus of the recombinant protein (80). Like human CKIε, the hamster recombinant protein has a molecular mass of approximately 48 kD. An in vitro casein kinase assay with the expressed proteins and the exogenous CKI substrates phosvitin and a-casein (81) revealed differences in phosphate transfer efficiency, with the tau product exhibiting lower activity than the wild-type enzyme. In addition, autophosphorylation of tau and wild-type enzyme occurred (Fig. 5A).
Fig. 5.
In vitro kinase assays with recombinant homozygous wild type and tau mutant CKIɛ protein. Assays were performed as described (80, 81). Triangles, autophosphorylation products. C and P denote a-casein and phosvitin phosphorylation products, respectively. These results are representative of five independent experiments. (B) Western blot analysis of homozygous wild-type and tau mutant recombinant CKIɛ protein (61). To examine autophosphorylation, kinase assays were performed in the absence of substrate. Following the assay, half of the product was treated with protein phosphatase 2B (PP2B). Blots were incubated with antibody to the S•Tag sequence at the NH2-terminus of the recombinant protein (left) and antibody to the COOH-terminus of human CKIɛ (right). These results are representative of three independent experiments. (C) In vitro kinase assays with homozygous wild type and tau mutant recombinant CKIɛ (84). Enzymes were incubated alone in a kinase reaction prior to addition of substrates to allow autophosphorylation. Half of each reaction was then treated with PP2B. a-casein and phosvitin were used as substrates. C and P denote a-casein and phosvitin phosphorylation products, respectively. The results here are representative of three independent experiments. (D) Kinetic analyses of the kinase activity of homozygous wild-type and tau mutant CKIɛ recombinant protein (61), performed by varying the phosvitin (P) concentration. These results are representative of two independent experiments, each performed in duplicate. (E) The apparent Km and V max values for both enzymes were determined by nonlinear least-squares regression of the untransformed kinetic data (61). The data are represented here graphically on a double-reciprocal plot. Each point represents the average of duplicate samples.
Immunoblot analysis of the recombinant tau and wild-type hamster CKIε with an antibody to the S•Tag fusion sequence revealed a consistent difference in the mobility of the expressed proteins in denaturing gel electrophoresis, suggesting a difference in autophosphorylation state (Fig. 5B) (61). There are two major bands present for wild-type CKIε, likely completely phosphorylated (∼60 kD) and unphosphorylated (∼48 kD) forms of the enzyme. In contrast, the tau mutant enzyme exhibits at least three phosphorylation states (the third form shows intermediate mobility). Treatment with protein phosphatase 2B (PP2B; calcineurin) changes the mobility of the slower migrating species, and the wild-type and tau enzymes comigrate as a doublet of about 48 and 50 kD. When probed with an antibody to part of the COOH-terminus of human CKIε, a single band is evident for both tau and wild-type products, identifying only the unphosphorylated species. PP2B treatment had no effect on the bands identified by the antibody. The recognition of only unphosphorylated CKIε with this COOH-terminal antibody is consistent with the predominance of autophosphorylation sites in the COOH-terminal region of CKIε (82) (Fig. 3C).
Because bacterially expressed CKIε is highly phosphorylated (83), we treated the recombinant wild type and tau enzymes with PP2B prior to assessing the kinase activity (Fig. 5C) (84). Phosphatase treatment enhanced wild-type activity, and to a lesser degree, tau enzyme activity with either phosvitin or a-casein as the substrate, consistent with the reported inhibition of CKI activity by autophosphorylation (82, 85, 86). Nevertheless, autophosphorylation does not account for the attenuated activity in the tau mutant; after dephosphorylation by PP2B, the diminished activity of the tau enzyme relative to the wild-type enzyme persists.
To elucidate the mechanism of attenuation in kinase activity of the tau CKIε enzyme, we performed kinetic analyses of wild-type and tau enzyme using phosvitin as substrate (61). Enzyme activity was linear with time under the conditions of our in vitro kinase assay. Next, using a range of substrate concentrations (0 to 100 [M), we obtained estimates of the Michaelis constant (Km) and maximal enzyme velocity (Vmax) for both enzymes (Fig. 5D) (61). The data are represented graphically as a double-reciprocal plot (Fig. 5E). There is a modest difference in the apparent Km between tau and wild-type enzymes, but a significantly lower Vmax value for the tau enzyme, which provides a kinetic validation of the SDS-PAGE analysis. That the tau kinase binds substrate with a greater affinity than the wild-type enzyme is suggested by the slightly lower Km value obtained for the mutant.
Interaction of hamster CKIɛ with mammalian PERIOD proteins
To determine whether a detectable interaction is possible between the mutant and wild-type hamster CKIε forms and the mammalian PERIOD proteins, we performed in vitro binding assays with the expressed proteins (61). CKIε interacted with the PERIOD proteins; however, there was no consistent difference between mutant and wild-type CKIε forms in this assay: both bound mPER1 and mPER2. This is similar to the finding of others that, in Drosophila, DBT interacts with PER (40).
To determine whether the hamster CKIε forms could phosphorylate mPER1 and mPER2, we developed an affinity kinase assay for in vitro-translated protein substrates (Fig. 6) (87). Because both mutant and wild-type CKIε could interact with PER1 and PER2, we used CKIε as an affinity reagent to bind PER substrate. An affinity kinase assay then demonstrated that tau mutant CKIε phosphorylates mPER1 and mPER2 less well than does wild-type CKIε. Thus, tau and wild-type hamster CKIε can both bind mPER1 and mPER2, but the tau mutant enzyme phosphorylates PER less efficiently.
Fig. 6. Figure 6.
Mutant CKIε is deficient in PERIOD phosphorylation (87). S-agarose beads containing either recombinant wild-type or tau mutant CKIε enzyme were incubated with in vitro– translated, 35S-methionine-labeled substrate and washed extensively. Binding and kinase reactions are indicated for both substrates. Following exposure to detect both the 35S and 32P signals, dried gels were covered with two layers of exposed X-OMAT film to block the 35S signal. PER signal is indicated by filled triangles.
In vivo effects of the tau mutation
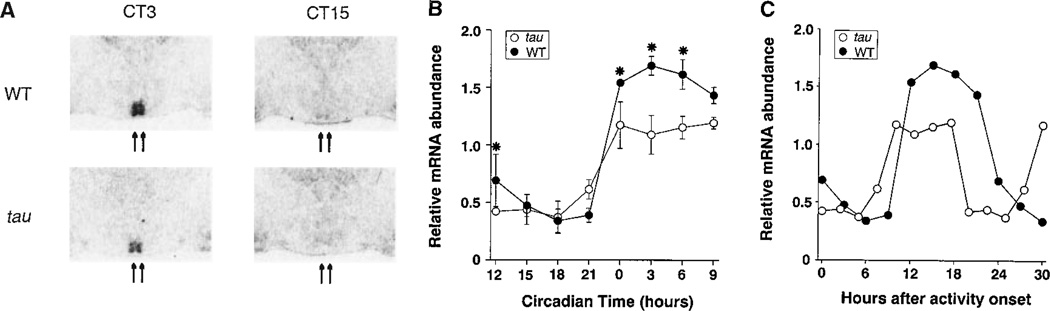
We next examined the effect of the tau mutation in vivo on the expression of Per1 mRNA in the SCN (Fig. 7A) (61). Antisense hamster Per1 riboprobe was hybridized to coronal sections of brains taken every three circadian hours, for a total of eight time points, in both tau and wild-type animals maintained in constant darkness. When normalized for circadian time, there was a reduction in peak values of Per1 mRNA abundance in tau animals as compared to wild type (Fig. 7, A and B). When these results are displayed in real time, Per1 mRNA levels rise approximately 2 hours earlier and decline about 4 hours earlier in homozygous tau hamsters than in wild-type hamsters (Fig. 7C). The decline of Per1 mRNA levels is an indicator of the negative feedback phase of the circadian cycle, and this phase of the cycle occurs about 4 hours earlier in tau mutants, consistent with the shortened circadian period expressed by these animals.
Fig. 7.
(A) Effect of the tau mutation on hamster Per1 gene expression in the suprachiasmatic nucleus (61, 102). Example sections showing hamster Per1 in situ hybridization signals in the SCN region. Arrows indicate the position of the SCN. (B) Circadian rhythm of hamster Per1 mRNA expression in the SCN. Samples were collected every 3 circadian hours following 7 days in constant darkness. Onset of locomotor activity for each hamster (CT12; by convention) was used to determine the circadian phase of the sample. Thus, for the tau mutant hamsters, which have an average period length of 20 hours, samples were collected approximately every 2.5 hours. In situ hybridization Per1 signal was quantified as described in (20). Per1 values are plotted relative to the 24-hour average for the wild-type animals. Two-way analysis of variance was significant for circadian time (F-ratio =30.5, P<10-6,df =7), genotype (F-ratio =12.2, P=0.0015, df =1) and their interaction (F-ratio =2.52, P=0.036, df =7). *Asterisks indicate differences between wild type and tau mutant values at each time point (Tukey’s post hoc, P<0.05). (C) Hamster Per1 mRNA results from (B) plotted in real time. Some points are double-plotted for clarity.
Discussion
The casein kinase I family is a unique group of serine/threonine protein kinases sharing more than 50% identity to one another in the catalytic domain (85). In mammals, seven different isoforms have been identified. Unlike most other protein kinases, CKI members occur as bilobal monomeric proteins, the smaller amino-terminal lobe containing the nucleotide-binding domain and the larger carboxyl-terminal lobe containing the phosphate transfer components. A cleft between the two lobes participates in substrate binding. All members share a near-consensus SV40 T antigen putative nuclear localization signal (NLS). An anion-binding site, which is formed by three residues conserved throughout the CKI family (88–90), recognizes the phosphate ester of the substrate. The CKI family has broad substrate specificity and ubiquitous subcellular localization (85). The reported consensus phosphorylation sequence is S/T/Y(P)X1–2S/T, where S/T/Y(P) is any phosphorylated serine, threonine, or tyrosine residue and X is any amino acid (85, 91–94). It is likely that substrate specificity differs among the many isoforms of CKI.
Although members of the casein kinase I family were among the first of the serine/threonine kinases to be identified, it has been difficult to demonstrate specific biological roles for the various isoforms (85). Recently, the Drosophila gene, double-time (dbt) was identified in a mutagenesis screen for aberrant circadian phenotype. It is regarded as the fly ortholog of mammalian CKIε, owing to its high degree of similarity (92%) to this gene throughout the kinase domain (40, 77). In addition, studies of dbt provided the first in vivo demonstration of a specific CKIε function in a biological system. In this case, DBT most likely functions to phosphorylate the Drosophila circadian clock component, PER. Evidence from the dbtP allele, which is a strong hypomorph, suggests that this phosphorylation event is essential for PER turnover. PER levels are constitutively high in dbtP mutants, and circadian rhythms of behavior and per/tim gene expression are abolished. Two additional mutations in dbt, both missense, give rise to short (dbtS) and long (dbtL) circadian period phenotypes (39, 40), although it is not clear how these mutations lead to these phenotypes.
How might the Arg178Cys tau mutation exert it effects on CKIε? The amino acid substitution occurs within a phosphate recognition site formed by three residues, Arg178, Gly215, and Lys224.In S. pombe Cki1, the corresponding amino acids are: Arg183, Gly220, and Lys229 (Fig. 8, A and B) (61). In the wild-type protein, the basic side groups of arginine and lysine, which are positively charged at physiological pH, are necessary for the formation of the anion binding characteristics of this site. Replacing Arg178 with cysteine and its small hydrophobic sulfhydryl group might modify the ability of the site to bind the negatively charged phosphate ester of the substrate.
Fig. 8.
(A) Cki1 ribbon diagram showing the site of the tau mutation (R183 = hR178) (61). (B) Model of the CKIɛ catalytic surface (61). Coordinates from S. pombe Cki1 are used here to model the analogous region in hamster CKIɛ. ATP (blue) is shown bound in the catalytic cleft. The three residues (yellow) forming the anion binding site are shown binding a sulfate ion (green). In red is the catalytic loop corresponding to residues 128 to 133 (DVKPDN) in hamster CKIɛ (Fig. 3C).
In addition, Arg178 is also located on the L-9D loop of the yeast Cki1 protein (Fig. 3C), which is thought to be involved in activation of catalytic activity in other protein kinases (88, 95, 96). The proximity of Arg183 to the catalytic region of Cki1 and its location on the L-9D loop strongly suggest that the Arg178Cys tau mutation could disrupt catalytic activity (Fig. 8B).
The tau mutation could also affect nuclear localization. In addition to its role in forming the anion binding site, Lys224 is also one of six residues forming a putative nuclear localization signal (TKRQKY). Should a conformational change or interaction occur at this site owing to the Arg178Cys mutation, nuclear localization of the protein could be modified.
How does the attenuation in hamster CKIε activity observed in tau homozygotes give rise to a shortened circadian period? The basis of the circadian mechanism is a transcription translation-based negative autoregulatory feedback loop (Fig. 9) (9). We propose that the shortened period arises as a result of the repression of the CLOCK-BMAL1 complex occurring earlier in tau animals relative to wild-type animals. This explains both the observed reduction in, and earlier appearance of, Per1 mRNA in tau homozygotes in vivo as revealed by in situ hybridization. If the mutant CKIε in homozygous tau animals is less active, and if PER is one of its major substrates in circadian pacemaker cells, then the reduced activity of mutant CKIε likely results in hypophosphorylated PER. Evidence from Drosophila suggests that phosphorylation of PER by the casein kinase DBT is necessary for its turnover (39). Assuming that this is also true of PER in mammalian cells, then hypophosphorylated PER would be expected to accumulate more rapidly in tau animals. This faster rate of PER accumulation in homozygous tau animals relative to wild-type animals could then lead to an earlier transport of PER into the nucleus, causing repression of CLOCK-BMAL1.
Fig. 9.
Model of proposed CKIε role in the mammalian circadian clock, as described in the text. Normal CKIε activity is proposed to cause a delay in the negative feedback signal. LRE, light-responsive element. Blank boxes indicate presumed E-box elements.
Even though the negative feedback signal in tau mutants occurs earlier than that seen in wild type (Fig. 7C), the duration of this negative feedback is not prolonged as one might expect if PER turnover was slower. Indeed, it appears that the duration of negative feedback (as inferred from the trough of the Per1 mRNA in the SCN) is comparable in wild type and tau mutant hamsters. This suggests that the turnover of the entire negative feedback complex (Fig. 9), which may composed of six proteins (PER1, PER2, PER3, TIM, CRY1, CRY2), may not be strongly influenced by the tau mutant CKIε. Recent in vitro and in vivo evidence strongly suggests that multiple interactions occur among these negative feedback components (46, 47, 49). The mCRY proteins can heterodimerize with the mPERs and with mTIM. The mPERs can also dimerize among themselves in various combinations. It is likely that, within the nucleus, a multimeric complex of these negative effectors forms (Fig. 9) (46, 47, 49). It remains to be determined what effects, if any, CKIε has on the CRYs or TIM.
While the time course of the decline of the PER proteins is a major determinant of the derepression of CLOCK-BMAL1 transactivation, this might not be the case in tau homozygotes. A plausible scenario in tau mutants is that, because of the greater availability of PER, derepression of CLOCK-BMAL1 activity might now depend more on the time course of degradation of the PERs’ dimerization partners. With the turnover of their partners, monomeric forms of PER would remain until they are eventually phosphorylated and targeted for degradation by the less active mutant CKIε or some other mechanism, or until they could again dimerize with a partner. This would not be the case in wild-type animals, where a significant delay is imposed by translation both of PER and its partners in addition to the time required for potential posttranslational modifications, such as phosphorylation of PER by CKIε. In tau animals, the hypophosphorylated PER would be available for immediate nuclear translocation upon dimerization and would, therefore, result in an earlier inactivation of CLOCK-BMAL1 relative to wild-type animals. While it is clear from our results that some PER is phosphorylated by the mutant CKIε, these levels should be lower with respect to wild-type animals. This limited amount of phosphorylated PER would be targeted for degradation, yet the synthesis of Per message during CLOCK-BMAL1-mediated transcription would make up for any loss and effectively maintain a pool of monomeric PER. Thus, we propose that a delicate balance must be achieved between the enhancement of the negative feedback signal leading to an earlier repression of CLOCK-BMAL1 on the one hand, and the maintenance of a normal turnover time of the inhibitory complex on the other hand, in order to shorten the period length of the circadian cycle in tau mutants. In contrast, the period-lengthening effect of the dbtL allele can be explained by defective nuclear degradation of the inhibitory complex (39, 40). Finally, none of these interpretations considers the possibility of CKIε acting as a regulator of nuclear entry of PER. Because CKIε and the PER proteins interact, CKIε (or its mutant alleles) could exert an additional level of control by either retaining PER in the cytoplasm or promoting its nuclear localization. We cannot rule out such a mechanism.
We conclude that CKIε is a critical component of the circadian clock in mammals. Using a positional syntenic cloning approach, we have shown that the hamster tau mutation is an allele of CKIε, which establishes that mammalian CKIε is an ortholog of Drosophila double-time, a critical component of the fly circadian system. Biochemical analysis of the mutant CKIε enzyme reveals striking differences in maximal enzyme velocity and autophosphorylation state. We also show that CKIε can interact with and phosphorylate the mammalian PERIOD proteins in vitro. Because Per1 gene expression is altered in tau mutants, we propose that CKIε plays a significant role in delaying the negative feedback signal within the transcription-translation-based autoregulatory loop that composes the core of the circadian mechanism (Fig. 9). Of the nine genes now identified with circadian function in mammals, CKIε is the only enzyme. As such, CKIε makes an ideal target for pharmaceutical compounds influencing circadian rhythms, sleep, and jet lag, as well as other physiological and metabolic processes under circadian regulation.
Supplementary Material
References and Notes
- 1.Aschoff J. Handbook of Behavioral Neurobiology: Biological Rhythms. New York: Plenum; 1981. [Google Scholar]
- 2.Pittendrigh CS. Annu. Rev. Physiol. 1993;55:16. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 3.Kondo T, Ishiura M. Bioessays. 2000;22:10. doi: 10.1002/(SICI)1521-1878(200001)22:1<10::AID-BIES4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 4.Johnson CH, Golden SS. Annu. Rev. Microbiol. 1999;53:389. doi: 10.1146/annurev.micro.53.1.389. [DOI] [PubMed] [Google Scholar]
- 5.Anderson SL, Kay SA. Adv. Genet. 1997;35:1. doi: 10.1016/s0065-2660(08)60446-0. [DOI] [PubMed] [Google Scholar]
- 6.Young MW. Annu. Rev. Biochem. 1998;67:135. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap JC. Cell. 1999;96:271. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 8.Wilsbacher LD, Takahashi JS. Curr. Opin. Genet. Dev. 1998;8:595. doi: 10.1016/s0959-437x(98)80017-8. [DOI] [PubMed] [Google Scholar]
- 9.King DP, Takahashi JS. Annu. Rev. Neurosci. 2000;23:713. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 10.Vitaterna MH, et al. Science. 1994;264:719. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King D, et al. Cell. 1997;89:641. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoch MP, et al. Cell. 1997;89:655. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun ZS, et al. Cell. 1997;90:1003. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 14.Tei H, et al. Nature. 1997;389:512. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 15.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr., Reppert SM. Neuron. 1997;19:1261. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 16.Albrecht U, Sun ZS, Eichele G, Lee CC. Cell. 1997;91:1055. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 17.Takumi T, et al. Genes Cells. 1998;3:167. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 18.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Neuron. 1998;20:1103. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 19.Takumi T, et al. EMBO J. 1998;17:4753. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sangoram A, et al. Neuron. 1998;21:1101. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 21.Zylka MJ, et al. Neuron. 1998;21:1115. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]
- 22.Koike N, et al. FEBS Lett. 1998;441:427. doi: 10.1016/s0014-5793(98)01597-x. [DOI] [PubMed] [Google Scholar]
- 23.Takumi T, et al. Genes Cells. 1999;4:67. doi: 10.1046/j.1365-2443.1999.00238.x. [DOI] [PubMed] [Google Scholar]
- 24.Tischkau SA, et al. J. Neurosci. 1999;19:RC15. doi: 10.1523/JNEUROSCI.19-12-j0005.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda M, Nomura M. Biochem. Biophys. Res. Comm. 1997;233:258. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 26.Gekakis N, et al. Science. 1998;280:1564. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 27.Hao H, Allen DL, Hardin PE. Mol. Cell. Biol. 1997;17:3687. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darlington TK, et al. Science. 1998;280:1599. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 29.Lee C, Bae K, Edery I. Mol. Cell. Biol. 1999;19:5316. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae K, Lee C, Hardin PE, Edery I. J. Neurosci. 2000;20:1746. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardin PE, Hall JC, Rosbash M. Nature. 1990;343:536. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 32.Allada R, White NE, So WV, Hall JC, Rosbash M. Cell. 1998;93:791. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 33.Rutila JE, et al. Cell. 1998;93:805. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 34.Sehgal A, et al. Science. 1995;270:808. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 35.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Science. 1996;271:1736. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 36.Saez L, Young MW. Neuron. 1996;17:911. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 37.So WV, Rosbash M. EMBO J. 1997;16:7146. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2260. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price JL, et al. Cell. 1998;94:83. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 40.Kloss B, et al. Cell. 1998;94:97. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 41.Klein DC, Moore RY, Reppert SM, editors. Supra-chiasmatic Nucleus: The Mind’s Clock. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 42.Shigeyoshi Y, et al. Cell. 1997;91:1043. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 43.Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. J. Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Horst GT, et al. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 45.Vitaterna MH, et al. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12114. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffin EA, Jr., Staknis D, Weitz CJ. Science. 1999;286:768. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 47.Kume K, et al. Cell. 1999;98:193. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 48.Okamura H, et al. Science. 1999;286:2531. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 49.Field MD, et al. Neuron. 2000;25:437. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 50.Ralph MR, Menaker M. Science. 1988;241:1225. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- 51.Ralph MR, Foster RG, Davis FC, Menaker M. Science. 1990;247:975. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 52.Silver R, LeSauter J, Tresco PA, Lehman MN. Nature. 1996;382:810. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 53.Tosini G, Menaker M. Science. 1996;272:419. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 54.Lander ES. Nature Genet. 1993;4:5. doi: 10.1038/ng0593-5. [DOI] [PubMed] [Google Scholar]
- 55.Okuizumi H, et al. Mamm. Genome. 1997;8:121. doi: 10.1007/s003359900370. [DOI] [PubMed] [Google Scholar]
- 56.Lisitsyn NA, et al. Nature Genet. 1994;6:57. doi: 10.1038/ng0194-57. [DOI] [PubMed] [Google Scholar]
- 57.Lisitsyn NA. Trends Genet. 1995;11:303. doi: 10.1016/s0168-9525(00)89087-3. [DOI] [PubMed] [Google Scholar]
- 58.Adler S. Nature. 1948;162:256. doi: 10.1038/162256b0. [DOI] [PubMed] [Google Scholar]
- 59.Murphy M. In: The Hamster: Reproduction and behavior. Siegel HI, editor. New York: Plenum; 1985. pp. 3–20. [Google Scholar]
- 60.Wild-type M70 outbred hamsters were obtained from the National Institutes of Health (National Center for Research Resources, Genetic Resource Section, Bethesda, MD). Hamsters of the outbred strain Lak:LVG(Syr), homozygous for the tau mutation, were obtained from a colony maintained at the University of Virginia (Charlottesville, VA). Animals were bred at Northwestern University (Evanston, IL) by crossing female homozygous tau Lak:LVG(Syr) hamsters with wild-type male M70 animals to produce the F1 generation. Six pairs of parental animals gave rise to separate three-generation pedigrees. F1 siblings were intercrossed to produce the F2 generation consisting of 237 progeny.
- 61.Lisitsyn N, Wigler M. Science. 1993;259:946. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 62.Lisitsyn N, Wigler M. Methods Enzymol. 1995;254:291. doi: 10.1016/0076-6879(95)54021-0. [DOI] [PubMed] [Google Scholar]
- 63.Ott J. Analysis of Human Genetic Linkage. revised edition. Baltimore, MD: Johns Hopkins Univ. Press; 1991. [Google Scholar]
- 64.Large-insert A clones of approximately 12 kb and 20 kb containing the RDA-650 and RDA-750 products, respectively, were obtained by screening a commercially-available Syrian hamster genomic library (A DASH II genomic library, Stratagene #945900). Standard library screening protocols were used (97). CsCl-purified clones were prepared and partially sequenced from both ends by primer walking from T7 and T3 bacteriophage promoter sites.
- 65.A microsatellite sequence, termed SSR2, consisting of the mixed repeat (CA)19(CG)10 was identified in the RDA-750 A genomic clone by sequencing. PCR primers flanking the microsatellite were designed to test for polymorphism: F 5'-GTGCAGT TGAGGAAGATATCTG-3',R5'-CAGTACTAGCAGTTGAATCAAGG-3'. Analysis of products on 7% denaturing polyacrylamide gels using methods modified from (98) revealed a product of approximately 230 bp in tau homozygous animals and of about 225 bp in homozygous wild-type animals.
- 66.Rowe LB, et al. Mamm. Genome. 1994;5:253. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- 67.The locus amplified in mouse by the hamster SSR2 primers has been assigned the mouse locus symbol, D15Nwu1, in the Mouse Genome Database (MGD).
- 68.To identify polymorphisms in hamster orthologs of genes that, in mouse or human, map to the region of conserved synteny, we searched for genes for which 5'-and 3'-untranslated and intronic sequences exist in the GenBank database. Primers were designed to amplify products of 2 kb or less using a standard 50-[l PCR reaction as follows: 1X GeneAmp PCR buffer (Perkin Elmer), 1.5 mM MgCl2, 50 pmol of each primer, 200 [M dNTPs, 100 ng genomic DNA (M70 and tau parental samples), 1 M betaine (Sigma #B-2629), and 2.5 units AmpliTaq DNA polymerase (Perkin Elmer). A “hot start” reaction profile of 30 cycles of 95°C for 1 min, 53°C for 1 min, and 72°C for 2.5 min was used followed by a final extension at 72°C for 10 min. Products were resolved on 1% agarose gels. Those reactions resulting in products in the expected size range were analyzed further by agarose gel electrophoresis, temperature modulated heteroduplex HPLC (WAVE DNA Fragment Analysis System, Trans-genomic) or denaturing polyacrylamide gel electrophoresis as described herein.
- 69.A single nucleotide polymorphism was identified in the Mb gene (myoglobin) by sequencing an intronic PCR product. New primers were designed to amplify a product of approximately 350 bp containing the polymorphism F 5'-CAGAGGGTGCCCT TGACT TCCACC-3',R5'-TGGAGGTAGGTGGCCGCCGCTAAA-3'. Products were analyzed by temperature-modulated heteroduplex HPLC according to manufacturer’s protocols (Transgenomic).
- 70.A polymorphism in the Cacng2 gene (calcium channel, voltage-dependent, gamma subunit 2, stargazer) was identified using the following primers: F' 5'-CCCTGGCGCGCTGTGGAAGCCATC-3' and R5'- AT TCCACTACTAATATAATGGATATATGT-3'. An allele of about 310 bp was amplified in tau homozygous animals and an allele of about 320 bp was amplified in homozygous wild-type animals. These products were resolved on 7% denaturing PAGE gels using protocols modified from (98).
- 71.Letts VA, et al. Nature Genet. 1998;19:340. doi: 10.1038/1228. [DOI] [PubMed] [Google Scholar]
- 72.A polymorphism in the gene Cyp11b2 (aldosterone synthase) was identified using the following primers: F 5'-GCATGCATGCACCAGTGTACAT TC-3' and R 5'-GACCACT TACAACATGTACAAACCACAGCC-3'. Agarose gel analysis revealed an allele of approximately 1 kb in tau homozygous animals and an allele of 1.7 kb in homozygous wild-type animals.
- 73.A polymorphism in the Ela1 gene (elastase) was identified using the following primers: F 5'-GGAAAGTCCTGGGTACTGTGTC-3' and R5'-GT TCGCT TCCCTGGTCCTGTAT-3'. Agarose gel analysis revealed an allele of approximately 730 bp in the tau homozygous animals and an allele of about 760 bp in the homozygous wild-type animals.
- 75.The mouse SSLP marker D15Mit146 was used to amplify a hamster polymorphism of approximately 200 bp in tau homozygous animals and approximately 205 bp in wild-type homozygotes. Products were separated on 7% denaturing PAGE gels using protocols modified from (98).
- 76.Linkage analysis was performed with the program Map Manager QTb2868K (99).
- 77.Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. J. Biol. Chem. 1995;270:14875. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- 78.PCR primers were designed around the CKIɛ point mutation in tau homozygous animals such that a product of approximately 160 bp was amplified: F 5'-GATAACT T TCTCATGGGCT TGG-3' andR5'GGGTGT TGATGGAGGCATAG-3'. PCR products were digested for 2 hours at 60°C directly in PCR reaction buffer without purification, using the restriction enzyme Bst API (New England Biolabs). The point mutation in tau animals results in the creation of a Bst API restriction site. Agarose gel analysis revealed an undigested product of approximately 160 bp in wild-type homozygous animals and a digested product of about 137 bp in tau homozygous animals.
- 79.To map CKIɛ in mouse, we PCR-amplified a 160-bp segment of the mouse ortholog in the strains, C57BL/6J and BALB/cJ, using the primers described (78). By sequencing, we identified a restriction fragment length polymorphism for the enzyme, Hpa II between the two mouse strains. The C57BL/6J PCR product contains two restriction sites for this enzyme, resulting in fragments of 108, 32 and 20 bp upon digestion. The BALB/cJ product contains only one Hpa II site and, following digestion, results in fragments of 108 and 52 bp. We mapped CKIɛ relative to the following mouse SSLP markers using 46 animals from a (BALB/cJ x C57BL/6J)F2 intercross: D15Mit28 (4/88), D15Mit146 (3/90), and D15Mit35 (14/78). The number of recombinations from CKIɛ out of the total number of meioses tested is given in parentheses following the marker name (76). All reactions were separated on 7% PAGE gels using protocols modified from (98).
- 80.Homozygous tau and wild-type full-length cDNA products were cloned into the pET-30 LIC T7 expression vector according to the manufacturer’s protocols (Novagen). Expression of recombinant hamster tau and wild-type CKIɛ was achieved by growing 250 ml cultures of BL21(DE3)pLysS E. coli hosts containing expression constructs for 16 to 18 hours at 23°C in LB medium supplemented with 34 [g/ml chloramphenicol and 30 [g/ml kanamycin. During incubation, cultures were shaken at 150 rpm. Expression of recombinant protein in the soluble phase was higher in the absence of IPTG; the addition of IPTG decreased the yield of soluble recombinant protein. Expressed protein from the soluble phase was purified by means of a 15–amino acid amino-terminal S•Tag fusion using S-protein agarose according to the manufacturer’s protocols (Novagen). S-protein agarose bound with expressed protein was resuspended in 30 mM HEPES, pH 7.5 and stored at 4°C for up to one week. Protein concentrations were determined by taking several 5 [l aliquots of bound S-protein agarose, boiling each for 3 min to release protein, cooling on ice, spinning briefly in a microcentrifuge and then removing supernatants for analysis using a colorimetric protein assay according to the manufacturer’s instructions (DC Protein Assay, Bio-Rad).
- 81.Kinase assays were incubated for 5 min at 37°C in 20 [l of reaction mix [30 mM HEPES (pH 7.5), 7 mM MgCl2, 0.5 mM DT T, 200 µM ATP, 50 µg/ml BSA, 0.5 mg/ml substrate, 5 µCi of [γ-32P]ATP, 3000 Ci/mmol (NEN Research Products)] and approximately 200 to 300 ng of recombinant CKIɛ. Either dephosphorylated a-casein (Sigma #C-8032) or phosvitin (Sigma #P1253) was used as substrate. Initiation of the reaction occurred upon addition of the enzyme. Reactions were terminated by the addition of 2X sample buffer [100 mM DT T, 2% SDS, 80 mM Tris-HCl (pH 6.8), 0.0006% bromophenol blue, 15% glycerol] followed by boiling for 2 min. Products were resolved on 12% SDS-polyacrylamide gels. 32P incorporation was detected in dried gels with a Storm Phosphorimager (Molecular Dynamics) and by exposure of X-OMAT AR (Kodak) film at −80°C.
- 82.Gietzen KF, Virshup DM. J. Biol. Chem. 1999;274:32063. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 83.Cegielska A, Gietzen KF, Rivers A, Virshup DM. J. Biol. Chem. 1998;273:1357. doi: 10.1074/jbc.273.3.1357. [DOI] [PubMed] [Google Scholar]
- 84.Protein phosphatase 2B (PP2B; calcineurin) treatment of recombinant enzyme was performed as follows. Approximately 160 ng of each recombinant enzyme was added to a reaction mixture consisting of [30 mM HEPES (pH 7.5), 7 mM MgCl2, 0.5 mM DT T, 200 µM ATP, 50 µg/ml BSA, 4 units calmodulin (Sigma #P2277), 100 µM CaCl2, 6 units PP2B (Sigma #C-1907)]. Control reactions without PP2B were also performed. Dephosphorylation reactions were incubated at 37°C for 15 min. To stop the reactions, 500 µM cypermethrin (BIOMOL #PR-100) and 1 mM EGTA were added. Kinase reactions were then initiated by adding 0.5 mg/ml substrate (dephosphorylated a-casein or phosvitin) and 5 µCi of [γ-32P]ATP, 3000 Ci/mmol. Reactions were incubated at 37°C for 5 min. To stop reactions, 2X sample buffer was added, followed by boiling for 2 min. Products were separated on 12% SDS-PAGE gels, and dried gels were exposed to phosphor screens and to X-OMAT AR film at −80°C.
- 85.Gross SD, Anderson RA. Cell. Signal. 1998;10:699. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- 86.Graves PR, Roach PJ. J. Biol. Chem. 1995;270:21689. doi: 10.1074/jbc.270.37.21689. [DOI] [PubMed] [Google Scholar]
- 87.Affinity kinase assays were performed by mixing 20 µl of an in vitro translation reaction (TNT T7 Coupled Reticulocyte Lysate System, Promega) containing 35S-methionine-labeled mPER1 or mPER2 with 20 µl S-protein agarose-bound recombinant CKIɛ (either tau or wild-type). Unlabeled amino acid concentrations were increased by using 1X solutions of both methionine and cysteine free amino acid solutions. This resulted in 1 mM methionine, 1 mM cysteine, and 2 mM for all other amino acids in the TNT reaction. Affinity reactions were washed at least 4 times with 1X S-protein agarose bind/wash buffer (Novagen). Half of the bound beads were then added to a kinase reaction as described (81), but without a-casein or phosvitin, and incubated at 37°C for 5 min. 2X loading buffer was added to the binding and kinase reactions which were then boiled for 2 min prior to gel loading. Reactions were separated on 12% SDS-PAGE gels, dried and exposed to phosphor screens. Following exposure to detect both the 35S and 32P signals, dried gels were covered with two layers of exposed XOMAT AR film to block the 35S signal. Covered gels were exposed to phosphor screens for 36 hours.
- 88.Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X. EMBO J. 1995;14:1015. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Longenecker KL, Roach PJ, Hurley TD. J. Mol. Biol. 1996;257:618. doi: 10.1006/jmbi.1996.0189. [DOI] [PubMed] [Google Scholar]
- 90.Longenecker KL, Roach PJ, Hurley TD. Acta Crystallogr. D. 1998;54:473. doi: 10.1107/s0907444997011724. [DOI] [PubMed] [Google Scholar]
- 91.Flotow H, et al. J. Biol. Chem. 1990;265:14264. [PubMed] [Google Scholar]
- 92.Flotow H, Roach PJ. J. Biol. Chem. 1991;266:3724. [PubMed] [Google Scholar]
- 93.Meggio F, Perich JW, Reynolds EC, Pinna LA. FEBS Lett. 1991;283:303. doi: 10.1016/0014-5793(91)80614-9. [DOI] [PubMed] [Google Scholar]
- 94.Songyang Z, et al. Mol. Cell. Biol. 1996;16:6486. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morgan DO, De Bondt HL. Curr. Opin. Cell Biol. 1994;6:239. doi: 10.1016/0955-0674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 96.Taylor SS, Radzio-Andzelm E. Structure. 1994;2:345. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 97.Ausubel FM, et al., editors. Current Protocols in Molecular Biology. New York: Wiley; 1993. [Google Scholar]
- 98.Dietrich W, et al. Genetics. 1992;131:423. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Manly KF, Olson JM. Mamm. Genome. 1999;10:327. doi: 10.1007/s003359900997. [DOI] [PubMed] [Google Scholar]
- 100.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 101.Hanks SK, Hunter T. FASEB J. 1995;9:576. [PubMed] [Google Scholar]
- 102.The Mesocricetus auratus DNA sequences reported here have been assigned the following GenBank accession numbers: casein kinase Iɛ (AF242536) and Period 1 (AF249882).
- 103.Supported by NIMH Grant R37MH39592 ( J.S.T.), an Unrestricted Grant in Neuroscience from Bristol-Myers Squibb ( J.S.T.) NIH Grant R01MH56647 (M.M.) and the NSF Center for Biological Timing. We thank T. Jardetzky for creating the three-dimensional images of Cki1 used here; S. M. Reppert for the expression clones of mouse Per1 and Per2; A. Lin for assistance with genotyping; M. Goto, E. Freuchte, D. Kolker, L. Radcliffe, and V. Alones for technical assistance; and M. H. Vitaterna, S.-H. Yoo, K. J. Seidenman, L. D. Wilsbacher, and A. M. Sangoram for helpful discussions and advice. J. S. T. is an Investigator of the Howard Hughes Medical Institute.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.