Abstract
The 26S proteasome is the major eukaryotic ATP-dependent protease, yet the detailed mechanisms utilized by the proteasomal heterohexameric AAA+ unfoldase to drive substrate degradation remain poorly understood. To perform systematic mutational analyses of individual ATPase subunits, we heterologously expressed unfoldase subcomplex from Saccharomyces cerevisiae in Escherichia coli and reconstituted the proteasome in vitro. Our studies demonstrate that the six ATPases play distinct roles in degradation, corresponding to their positions in spiral staircases adopted by the AAA+ domains in the absence and presence of substrate. ATP hydrolysis in subunits at the top of the staircases is critical for substrate engagement and translocation. While the unfoldase relies on this vertical asymmetry for substrate processing, interaction with the peptidase exhibits three-fold symmetry with contributions from every other subunit. These diverse functional asymmetries highlight how the 26S proteasome deviates from simpler, homomeric AAA+ proteases.
Introduction
The ubiquitin-proteasome system (UPS) is the major pathway for selective protein degradation in all eukaryotic cells, where it mediates protein quality control and the destruction of critical regulatory proteins1,2. Protein substrates are covalently modified with a poly-ubiquitin chain and targeted to the 26S proteasome for ATP-dependent proteolysis. Despite this crucial role of the UPS for protein degradation, the mechanistic principles of the proteasome still remain largely elusive.
The eukaryotic 26S proteasome is a ∼2.5 MDa molecular machine formed from at least 33 different subunits. It consists of a barrel-shaped 20S core particle that is capped on one or both ends by the 19S regulatory particle. The core particle contains an internal chamber with sequestered proteolytic active sites and gated axial pores to restrict substrate entry3. Access to this chamber is controlled by the regulatory particle, which is also responsible for recognition, deubiquitination, engagement, unfolding, and translocation of substrate4-7. The regulatory particle includes 19 different subunits and can be divided into the base and lid subcomplexes. The base contains a ring of six distinct AAA+ ATPases in the order Rpt1–2–6–3–4–58, which constitute the ATP-dependent motor of the proteasome and dock atop the core particle. Additional integral components of the base are the ubiquitin receptor Rpn13 as well as two large scaffolding subunits, Rpn1 and Rpn29-11. The nine-subunit lid, which includes the deubiquinating enzyme Rpn1112,13, binds asymmetrically to the side of the base-core complex and positions the second ubiquitin receptor, Rpn10, close to the lid-base interface above the entrance of the processing pore9,10,14.
Once a substrate is tethered to a proteasomal ubiquitin receptor, a complex set of enzymatic activities defines the pathway to degradation. The substrate must be deubiquitinated by Rpn11, while the ATPase ring of the base engages an unstructured degradation-initiation region of the protein, mechanically disrupts globular structures, and translocates the unfolded polypeptide into the peptidase chamber. ATP hydrolysis by the Rpt subunits of the base is crucial for substrate degradation, yet it remains unclear how these six distinct subunits work together to drive ATP-dependent unfolding and translocation.
Previous studies of the related homohexameric unfoldase ClpX suggest that ATP hydrolysis occurs in one subunit at a time with a certain degree of coordination, such that subunits may contribute additively and equally to substrate processing15. However, the homomeric nature of ClpX hinders assessment of whether all six subunits indeed sequentially progress through the different stages of the ATP-hydrolysis cycle. The unique heterohexameric architecture of the proteasomal ATPase ring has thus prompted the fundamental question whether the six Rpts are functionally equivalent or play distinct roles in ATP hydrolysis and substrate processing. While Rpt1–6 share highly homologous AAA+ ATPase domains, they differ substantially in their N-terminal coiled-coil domains, which interact with the laterally bound lid, and in their C-terminal unstructured tails, which mediate interaction with the core particle and trigger gate opening to the proteolytic chamber7,16-18. Furthermore, recent EM structures of the apo and substrate-bound 26S proteasome revealed distinct vertical asymmetries within the base ATPase ring9,10,19,20. In the absence of substrate, the large AAA+ subdomains of Rpt1–6 adopt a pronounced spiral-staircase configuration, with Rpt3 at the top and Rpt2 at the bottom position. Strikingly, upon substrate engagement, the base switches to a more planar ring conformation that is characterized by a spiral staircase with Rpt1 at the top and Rpt4 at the bottom20. However, the functional significance of these staircase configurations, potentially manifested as differential subunit contributions to substrate degradation or subcomplex interactions within the holoenzyme, has yet to be determined.
Endogenous 26S proteasome has been used to investigate the role of individual Rpts, revealing functional differences in their contributions to base assembly, 26S holoenzyme formation, ATP hydrolysis, peptidase gate opening, and substrate degradation7,21-28. Despite these results, limitations in working with endogenous proteasome, in part due to in-vivo assembly problems or lethal degradation defects, have largely prevented extensive systematic studies and a quantitative mechanistic understanding of the individual processes involved in substrate degradation.
Here, we investigated the mechanisms underlying ATP-dependent substrate processing by the heterohexameric unfoldase of the 26S proteasome. To define the differential contributions of individual Rpts to ATP hydrolysis, substrate degradation, peptidase binding, and gate opening, we developed systems for the heterologous expression of the base subcomplex and the in-vitro reconstitution of partially recombinant proteasomes, and performed systematic mutational analyses of key catalytic and structural motifs.
Results
Heterologous expression of the base subcomplex
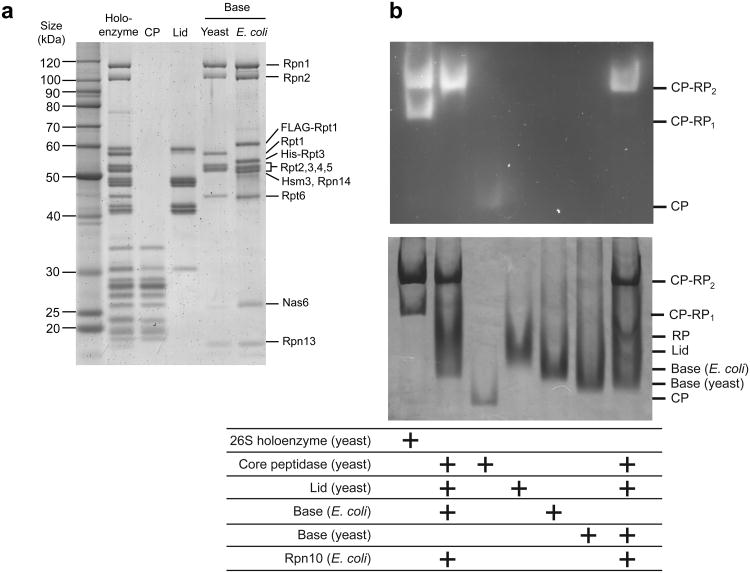
For the present studies, the base subcomplex of the proteasome from Saccharomyces cerevisiae was produced in Escherichia coli by co-expression of thirteen yeast proteins, including nine integral base subunits (Rpt1–6, Rpn1, Rpn2, Rpn13) and four proteasome assembly chaperones (Nas2, Nas6, Rpn14, Hsm329-32). We isolated assembled base by tandem-affinity purification, using tags on two different subunits, followed by gel-filtration chromatography. The purified base exhibited appropriate stoichiometry and no subunit truncations, as revealed by SDS-PAGE (Fig. 1a) and mass spectrometry. We observed Nas6, Hsm3, and Rpn14 stably associated with the recombinant base, whereas these chaperones were not present in the base purified from yeast, as indicated by SDS-PAGE, native PAGE (Fig. 1b), and size-exclusion chromatography (Supplementary Fig. 1). This result is consistent with studies of in-vivo proteasome assembly, indicating that Nas6, Hsm3, and Rpn14 are displaced upon base binding to the core particle and lid, whereas Nas2 dissociates at an earlier stage of base assembly26,29,30,33. One model for in-vivo base assembly proposes that the core particle might act as a template to facilitate the proper arrangement of Rpts in the hexameric ring26. However, our successful constitution of the base subcomplex in E. coli rules out a strict requirement for such templated assembly.
Figure 1. Expression of yeast base subcomplex in E. coli and reconstitution of 26S proteasome.
(a) Sypro-Ruby stained SDS PAGE of the purified proteasomal subcomplexes used in this study. Endogenous complexes were isolated from yeast using FLAG tags on Rpn11 for holoenzyme and lid, on Pre1 for core particle (CP), and on Rpn2 for base preparations. Recombinant base expressed in E. coli was purified using a FLAG tag on Rpt1 and a His6 tag on Rpt3. Proteasome chaperones Nas6, Hsm3 and Rpn14 only co-purify with the base subcomplex produced in E. coli. (b) 26S holoenzyme reconstituted with CP, lid, Rpn10, and either endogenous or recombinant base was analyzed by native gel electrophoresis. Endogenous yeast 26S holoenzyme and individual CP, lid, and base subcomplexes were also analyzed for comparison. Yeast holoenzyme migrated as two bands corresponding to proteasomes singly (CP-RP1) and doubly (CP-RP2) capped with regulatory particles (RP). Excess lid and base was used for reconstituted proteasome samples, which therefore migrated only as doubly capped holoenzyme.
We compared the activities of the recombinant base to endogenous yeast base. Both base subcomplexes hydrolyzed ∼ 51 ATP enz−1 min−1 in the absence of substrate (Table 1). The ability of the ATP-bound base to interact with core particle and induce gate opening was determined by monitoring the fluorescence increase upon peptidase cleavage of the fluorogenic peptide Suc-LLVY-AMC. In the presence of ATP, recombinant base stimulated core-particle activity approximately 20-fold, similar to endogenous yeast base. In agreement with previous reports, we measured about two-fold higher peptide hydrolysis with the non-hydrolysable analog ATPγS compared to ATP (unpublished data, R.B.)4,34, which may be due to potential differences in the ATPase-ring conformation19 or the dynamics of base-core interactions.
Table 1. Biochemical data for base variants with individual ATPase mutations.
Summary of ATP hydrolysis rates (base), peptidase stimulation (CP and base), and degradation rates (reconstituted 26S proteasomes) for all base variants included in this study. Values for each assay are expressed both in absolute terms and as a normalized percentage of wild-type base activity. Base mutants are designated as EQ (Walker-B), YA (pore-1 loop), DN (pore-2 loop), NT (C-terminal tail truncation), and KS (Walker-A), followed by a number indicating which Rpt subunit contained the mutation (1–6). Degradation activities for WT base and EQ mutants are included for both multiple turnover (left) and single turnover (right) conditions. Errors were estimated to be ±10% (s.d.) based on repeat measurements (n ≥ 3 technical replicates).
| Residue Mutated | basal ATPase rate | peptidase stimulation | degradation rate (kdeg) | ||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| min−1 | % WT | fold increase | % WT | (enz−1 min−1) | % WT | ||
| Holoenzyme | - | 107 | - | - | - | 0.32 | - |
|
| |||||||
| WT (E. coli) | - | 51 | 100 | 21 | 100 | 0.30 | 100 |
| WT (yeast) | - | 54 | 106 | 22 | 103 | 0.29 | 97 |
| EQ hexamer | - | 2 | 4 | 24 | 110 | 0.00 | 0.00 |
|
| |||||||
| EQ1 | 310 | 35 | 68 | 15 | 68 | 0.08 / 0.12 | 27 / 40 |
| EQ2 | 284 | 82 | 161 | 29 | 134 | 0.28 / 0.24 | 92 / 79 |
| EQ3 | 273 | 17 | 33 | 19 | 89 | 0.005 / 0.00 | 2 / 0 |
| EQ4 | 282 | 55 | 108 | 20 | 95 | 0.001 / 0.00 | 0.41 / 0 |
| EQ5 | 282 | 34 | 67 | 25 | 118 | 0.13 / 0.21 | 44 / 72 |
| EQ6 | 249 | 83 | 164 | 35 | 161 | 0.02 / 0.01 | 6 / 5 |
|
| |||||||
| YA1 | 284 | 66 | 129 | 18 | 84 | 0.30 | 101 |
| YA2 | 257 | 75 | 148 | 8 | 38 | 0.15 | 52 |
| YA3 | 246 | 44 | 87 | 17 | 81 | 0.19 | 62 |
| YA4 | 255 | 38 | 75 | 14 | 65 | 0.16 | 54 |
| YA5 | 255 | 89 | 174 | 15 | 68 | 0.22 | 74 |
| YA6 | 222 | 38 | 76 | 18 | 82 | 0.26 | 87 |
|
| |||||||
| DN1 | 327 | 39 | 76 | 23 | 106 | 0.21 | 71 |
| EN2 | 300 | 59 | 115 | 24 | 110 | 0.27 | 91 |
| DN3 | 289 | 65 | 127 | 20 | 91 | 0.16 | 52 |
| DN4 | 298 | 141 | 278 | 41 | 192 | 0.22 | 74 |
| DN5 | 298 | 90 | 177 | 32 | 149 | 0.40 | 134 |
| DN6 | 265 | 58 | 113 | 18 | 84 | 0.29 | 96 |
|
| |||||||
| NT1 | 464–468 | 61 | 121 | 20 | 91 | 0.29 | 98 |
| NT2 | 434–438 | 45 | 88 | 2 | 10 | 0.13 | 44 |
| NT3 | 424–428 | 65 | 127 | 1 | 5 | 0.05 | 16 |
| NT4 | 433–437 | 49 | 97 | 23 | 108 | 0.25 | 83 |
| NT5 | 430–434 | 86 | 169 | 1 | 7 | 0.14 | 47 |
| NT6 | 401–405 | 53 | 104 | 5 | 24 | 0.25 | 83 |
|
| |||||||
| KS1 | 257 | 32* | 62* | 15 | 71 | 0.07* | 24* |
| KS2 | 230 | 13* | 25* | 2 | 10 | nd* | nd* |
| KS3 | 219 | 29* | 57* | 12 | 58 | 0.08* | 26* |
| KS4 | 228 | 15* | 29* | 15 | 68 | 0.001* | 0.40* |
| KS5 | 228 | 74* | 147* | 13 | 59 | 0.02* | 6* |
| KS6 | 195 | 22* | 43* | 12 | 54 | 0.04* | 15* |
nd = not determined
ATPase and degradation activities for the WA-KS mutants represent lower bounds due to varying degrees of misassembly observed for these base mutants.
Importantly, we also reconstituted 26S holoenzyme, using either endogenous or recombinant base and the lid and core particle purified from yeast. Successful reconstitution was assessed by native PAGE (Fig. 1b) and in-vitro degradation of a polyubiquitinated model substrate (Supplementary Fig. 2), a green fluorescent protein (GFP)-titinV15P-cyclin-PY fusion, whose degradation could be measured through the decrease of GFP fluorescence (Fig. 2). Proteasomes reconstituted with saturating recombinant or endogenous base degraded substrate at a maximal rate of 0.3 enz−1 min−1, comparable to 0.32 enz−1 min−1 observed for holoenzyme purified from yeast (Table 1). Substrate degradation by reconstituted proteasomes strictly required addition of recombinant Rpn10, an intrinsic ubiquitin-receptor that does not co-purify with isolated lid or base subcomplexes. Consistent with previously described degradation defects in the absence of Rpn1035, we found that omitting Rpn10 or deleting its ubiquitin-interacting motif resulted in 40-fold slower degradation (Fig. 2a, Supplementary Fig. 3), despite the presence of the second ubiquitin receptor, Rpn13. Since proteasome formation did not depend on Rpn10 (data not shown, R.B.) and degradation was not facilitated by Rpn10 lacking its ubiquitin-interacting motif, this result suggests that Rpn10 is either the primary receptor for our model substrates or required in combination with Rpn13 for multivalent ubiquitin-chain binding.
Figure 2. Proteasomes reconstituted with endogenous or heterologously expressed base exhibit similar degradation activities for a polyubiquitinated substrate.
(a) Degradation of a polyubiquitinated GFP fusion substrate by endogenous yeast holoenzyme or 26S proteasomes reconstituted with saturating recombinant base (ebase) or endogenous yeast base (ybase). Substrate degradation was monitored by the loss of GFP fluorescence and strictly required the addition of Rpn10, despite the presence of Rpn13 (see Fig. 1A). (b) Michaelis-Menten analyses of substrate degradation by proteasomes reconstituted with endogenous or recombinant base. Degradation reactions were performed using limiting base and excess core particle, lid and Rpn10 to ensure that reconstituted proteasome particles were singly capped. KM and Vmax values were 0.63 μM and 0.21 enz−1 min−1 for holoenzyme with endogenous base, and 0.35 μM and 0.18 enz−1 min−1 for holoenzyme with recombinant base.
Functional asymmetry of the heterohexameric AAA+ unfoldase
To examine the roles of Rpt1–6 in nucleotide-dependent substrate processing, we individually abolished their ATP hydrolysis by systematically introducing a catalytic mutation in the recombinant base. In the homohexameric bacterial unfoldase ClpX, mutation of the conserved Walker-B glutamate prevents hydrolysis and induces a permanently ATP-bound state in the mutated subunit36, but other AAA+ unfoldases require distinct Walker-B mutations to eliminate ATP-hydrolysis activity37,38. We therefore tested the effects of various substitutions of the conserved Walker-B aspartate and glutamate residues by simultaneously placing them in all six Rpts (Supplementary Note 1). Ultimately, mutation of glutamate to glutamine (EQ) allowed proper assembly of base that exhibited wild-type levels of peptidase-binding and gate-opening activities despite being inactive in ATP hydrolysis (Table 1), indicating that this mutation in fact traps Rpt subunits in a permanently ATP-bound state. We next introduced a single EQ mutation per hexamer to fix individual Rpts in the ATP-bound state and test their contributions to ATP hydrolysis as well as core-gate opening. Depending on which subunit was mutated, we observed considerable differences in activities (Table 1), indicating that the Rpt subunits are functionally non-equivalent.
ATP hydrolysis by the isolated base decreased by more than 60% upon mutation of Rpt3, whereas inactivating other subunits had either minor effects (Rpt1, Rpt4, Rpt5) or notably increased the hydrolysis rate (Rpt2, Rpt6). Peptidase stimulation by these base mutants also varied: mutated Rpt5, Rpt2, and Rpt6 caused 20%, 30%, and 60% stronger gate opening compared to wild-type base, mutated Rpt4 did not lead to noticeable changes, while mutations in Rpt3 or Rpt1 decreased gate-opening activity by 20% or 30% despite proper complex formation with the core particle. It is important to consider that the measured ATPase and gate-opening activities reflect an average of the unmutated five Rpt subunits and are influenced by subunit communication. The increase in ATPase activity resulting from mutation of Rpt2 and Rpt6, for instance, is likely caused by the response of neighboring subunits to a permanently ATP-bound state. Some of these stimulated ATP-hydrolysis events may be non-productive and thus not result in increased substrate-degradation rates. This is supported by the fact that most base variants containing a mutant Rpt showed no stimulation or even slight repression in ATP-hydrolysis activity upon the addition of substrate, whereas the ATPase rate of wild-type base approximately doubled (Supplementary Fig. 4).
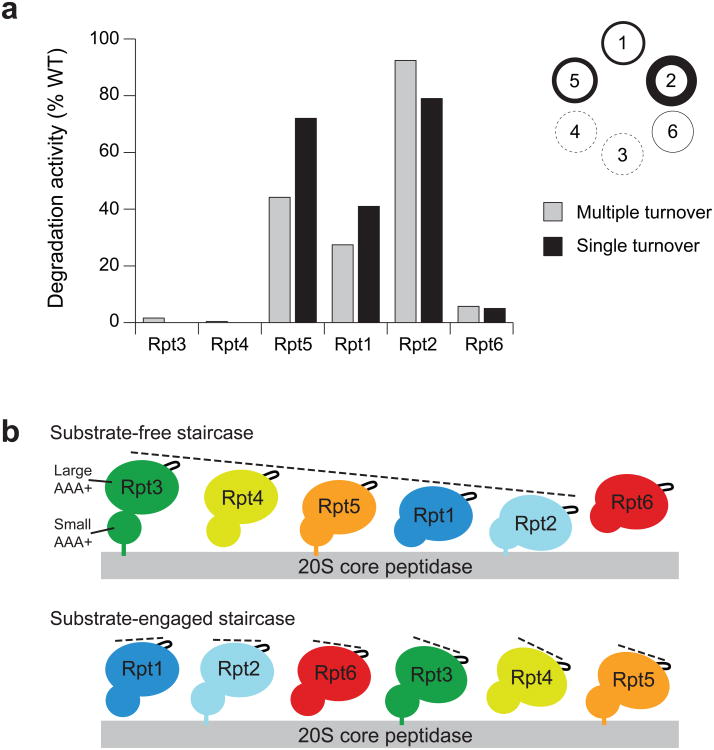
To explore the distinct roles of individual Rpt subunits in substrate processing, we reconstituted proteasomes with base variants containing single-subunit EQ mutations and compared their rates of ubiquitin-mediated substrate degradation under multiple-turnover conditions (Fig. 3a). An EQ mutation in either Rpt3 or Rpt4 completely eliminated substrate degradation, and a mutated Rpt6 resulted in a 90% decrease in degradation rate. Mutations in Rpt1 and Rpt5 lowered the degradation rate by 73% and 56%, respectively, whereas the Rpt2 mutant showed no defect. Importantly, the observed degradation defects were not simply the result of compromised proteasome assembly, as all mutants stimulated peptidase-gate opening (Table 1) and bound lid and core particle (Supplementary Fig. 5a,b). Considering the order of ATPase subunits within the base (Rpt1–2–6–3–4–5), it is strikingly evident that mutants with severe degradation defects (Rpt6, Rpt3, and Rpt4) map to one half of the ring.
Figure 3. Degradation activities for base variants with a Walker-B EQ mutation in individual Rpt subunits correlate with the subunit's position in the spiral staircase arrangement of the base.
(a) In-vitro degradation rates for reconstituted proteasomes containing base variants with single Rpt subunits fixed in a permanent “ATP-bound” state by the EQ mutation. Degradation under multiple-turnover (gray) and single-turnover (black) conditions was monitored by the loss of fluorescence resulting from degradation of a polyubiquitinated GFP fusion substrate. Degradation activities were measured relative to reconstituted proteasome containing wild-type (WT) recombinant base. Errors for multiple-turnover degradation rates were estimated to be ±10% (s.d.) of the mean WT value based on repeat measurements (n = 3 technical replicates). The circular diagram is an alternative representation of the multiple-turnover data, with the line thickness corresponding to the observed degradation activities for a mutation in the respective subunit. (b) The large AAA+ subdomains of Rpt1–6 adopt distinct spiral staircase arrangements in the absence and presence of substrate. Shown are cartoon representations of the pre-engaged (top) and substrate-engaged (bottom) staircases based on cryoEM reconstructions9,20, with the individual Rpt subunits splayed out and their pore-facing side pointing to the right. In the pre-engaged spiral, the small AAA+ subdomains of Rpt1–6 are arranged in a relatively planar fashion, while the large AAA+ subdomains are differentially lifted out of the ring plane, resulting in a pronounced spiral staircase with Rpt3 in the highest and Rpt2 in the lowest position. In the substrate-engaged spiral, the small and large AAA+ subdomains are mostly level, and the staircase orientation of pore loops primarily originates from differential rotations of subdomains in the plane of the ring.
Next we investigated the mechanistic role of individual Rpts at different stages of substrate processing by performing single-turnover degradation experiments (Fig. 3a, Supplementary Fig. 5c). In contrast to steady-state analyses, these measurements discriminate between potential defects in substrate engagement versus translocation and unfolding. The resulting data for GFP-substrate turnover were best fit by a double-exponential decay. Since GFP loses fluorescence in a single unfolding step, this double-exponential behavior was likely due to two types of substrates that probably varied in their ubiquitin tagging and were degraded at different rates. As expected, the two rates averaged to roughly match the multiple-turnover rate, and the base mutants exhibited the same ranking of activities as in multiple-turnover degradation (Fig. 3a, Table 1). Even under these single-turnover conditions, Rpt3 or Rpt4 mutants did not show any measurable degradation, whereas the Rpt6 mutant degraded substrate at 5% of the wild-type rate. This Rpt6 mutant exhibited a short lag preceding the exponential fluorescence decay (Supplementary Fig. 5c), which may indicate delayed substrate translocation and unfolding due to defects in engagement. The 95% decrease in degradation rate may thus originate from slower engagement instead of or in addition to compromised unfolding and translocation. The complete lack of substrate degradation for ATPase-deficient Rpt3 or Rpt4 may similarly be a consequence of severe engagement defects.
Spiral staircase configurations of the base
EM reconstructions of the ATP-bound 26S proteasome in the absence of substrate revealed that the large AAA+ subdomains of the Rpts adopt a pronounced spiral-staircase configuration around the ring9,10 (Fig. 3b). Rpt3 occupies the highest and Rpt2 the lowest position relative to the core particle, with Rpt6 bridging the vertical gap between the two subunits. The degradation defects of EQ mutants and the inferred contributions of individual Rpts to substrate degradation largely correlate with the vertical positions of subunits in this pre-engaged staircase. Conformational changes in subunits at the top of the apo spiral (Rpt3, Rpt4 and Rpt6) thus appear to be critical to engage a substrate and initiate translocation, whereas subunits in lower positions are less important.
Our recent EM reconstruction of the translocating proteasome demonstrated that substrate engagement induces substantial conformational changes in the regulatory particle, leading to a more planar ATPase-ring with a rearranged staircase of pore loops, in which Rpt1 is at the top and Rpt4 at the bottom position (Fig. 3b)20. Based on the static appearance of the spiral, in combination with single-molecule data for the related protease ClpXP, we previously proposed that the substrate-engaged staircase represents a default dwell state that is adopted by the base before or after coordinated ATP-hydrolysis events progress around the ATPase ring20. Interestingly, in the present study we observed an approximately 70% reduction in degradation activity when ATP hydrolysis was eliminated in Rpt1, located at the top of the substrate-engaged staircase. Rpt1 may therefore play a special role in processive substrate translocation, possibly by triggering the coordinated ATP hydrolysis of subunits.
Differential contributions of pore loops
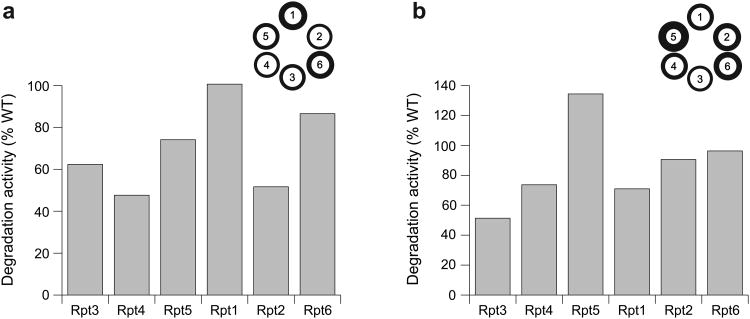
Previous work on various AAA+ unfoldases proposed that a conserved aromatic-hydrophobic (Ar-Φ) loop protrudes from every ATPase subunit into the central channel and undergoes nucleotide-dependent power strokes to drive substrate translocation22,39-46. To study these loop functions in the proteasome, we constructed base variants with tyrosine-to-alanine mutations in the Ar-Φ loop of individual Rpts (Table 1, Fig. 4A). Loop mutations in Rpt3, Rpt4, or Rpt6 decreased the rate of ATP hydrolysis by 15–25%, whereas mutations in Rpt1, Rpt2, and Rpt5 stimulated ATPase activity by 30–75% compared to wild-type base. Ar-Φ-loop mutations have been observed to stimulate ATP hydrolysis in some AAA+ unfoldases, including ClpX and the proteasome22,40, likely due to reduced steric constraints on loop movements within the central pore. In addition, subunit communication may induce changes in neighboring, non-mutated subunits, making it infeasible to specify individual Rpt contributions to overall ATP-hydrolysis activity using only the Ar-Φ-loop data.
Figure 4. Degradation activities for base variants containing single-subunit pore-loop mutations.
Degradation of a polyubiquitinated GFP fusion substrate was monitored by the loss of fluorescence upon addition of proteasomes reconstituted with base variants containing (a) YA mutations in the Ar-Φ loop of single Rpt subunits, and (b) DN mutations in the pore-2 loop of single Rpt subunits. Degradation activities under saturating conditions were measured relative to reconstituted proteasome containing wild-type (WT) recombinant base. Errors were estimated to be ±10% (s.d.) of the mean WT value based on repeat measurements (n = 3 technical replicates). The circular diagrams are alternative representations, with the line thickness corresponding to the observed degradation activities for a mutation in the respective subunit.
All Ar-Φ-loop mutants exhibited peptidase-gate opening activities at 65–85% of the wild-type level, with the exception of mutant Rpt2, which had 38% activity (Table 1). Rpt2 occupies the lowest position in the apo staircase of ATPases, and docking the PAN crystal structure into the Rpt2 EM density indicates that its Ar-Φ loop is located close to the core-particle interface, where direct or indirect contacts with the N-termini of α-subunits may affect gating. However, despite this defect in gate-opening activity, the Ar-Φ-loop mutation in Rpt2 did not lower the base affinity for the core particle (KD = 138 nM versus KD = 127 nM for wild-type base).
Proteasomes reconstituted with the Ar-Φ mutant base variants degraded substrate at substantially different rates (Fig. 4a). The biggest defects were observed for mutations in Rpt4 and Rpt2, which are localized clockwise-next to the top subunit in the substrate-free and substrate-engaged spiral staircase, respectively. That the ranking of defects for Ar-Φ-loop mutations appears shifted clockwise by one subunit relative to the defects observed for Walker-B mutants likely results from the fact that loop mutations primarily act in cis on a subunit's ability to translocate, whereas eliminating ATP-hydrolysis may affect conformational changes in the clockwise-next subunits. This would be consistent with the rigid-body model recently proposed for ClpX47,48, in which the small AAA+ subdomain of one subunit and the large AAA+ subdomain of its clockwise-next neighbor move as one unit to propel substrate through the pore. ATP hydrolysis in a particular subunit might thus drive movements of the clockwise-next rigid body, which includes the substrate-interacting Ar-Φ loop of the neighboring subunit. For the proteasome, only the rigid body between Rpt3 and Rpt4 at the top of the spiral staircase appears to be present in the absence of substrate9, while the remaining rigid bodies form during substrate-induced conformational changes20. Those rigid bodies within the substrate-engaged ATPase ring could thus cause the ranking of Rpt contributions to appear shifted by one subunit when comparing Ar-Φ-loop and Walker-B mutants.
Our data qualitatively coincide with results from a recent study using endogenous 26S proteasomes from yeast, demonstrating a comparable ranking of degradation defects for individual Ar-Φ-loop mutations22, with the exception of Rpt2. The fact that no single Ar-Φ-loop mutation completely abrogated degradation may indicate that multiple Rpts simultaneously interact with substrate during engagement and translocation, as suggested for the bacterial unfoldase ClpX40. Furthermore, removal of the Ar-Φ-loop tyrosine may not fully abolish pore-loop function and thus cause much weaker defects than a Walker-B mutation, which traps the entire subunit in a static ATP-bound state.
The pore-2 loop, located below the Ar-Φ loop in the central channel and directly adjacent to the Walker-B motif, has been implicated in substrate binding and unfolding as well as peptidase interaction and the control of ATPase activity in ClpX41,47,49. A recent study on the archaeal homohexameric unfoldases Cdc48 and PAN suggested that the pore-2 loop also interacts with the proteasomal core particle50. As the importance of the pore-2 loop for 26S-proteasome function is still unknown, we replaced the highly conserved aspartate with asparagine (DN) in the pore-2 loops of individual Rpts, with Rpt2 requiring a glutamate to asparagine mutation in this position. Base variants with a single pore-2-loop mutation hydrolyzed ATP up to three-fold faster than wild type, with the exception of Rpt1, which showed a 25% decrease (Table 1). The increased ATPase rates are reminiscent of previous observations for ClpX49 and may result from induced structural changes in the pore-2 loop that affect the adjacent Walker-B portion of the ATPase active site. Proteasomes reconstituted with pore-2-loop mutants exhibited a wide range of degradation rates (Fig. 4b), with the most severe defect observed for Rpt3, located at the top of the substrate-free staircase. Changes in substrate degradation did not directly correlate with changes in ATP hydrolysis, indicating that, similar to our observations for the Ar-Φ loop, pore-2-loop mutations may interfere with substrate interactions and thus result in futile ATP-hydrolysis events. The largest discrepancies between ATPase and degradation activities were observed for pore-2-loop mutations in Rpt3 and Rpt4, corroborating that Rpt subunits at the top of the pre-engaged staircase are particularly important for substrate processing, presumably by enabling efficient engagement. The degradation defects do not appear to be a consequence of compromised gate opening, as all pore-2-loop mutants show robust peptidase-gate opening activity, ranging from 84% to 192% compared to wild-type base (Table 1). The substantial variations in gate opening indicate that pore-2 loops are involved in unfoldase-peptidase communication and the Rpt C-terminal tails are not the sole determinants of the interaction between these subcomplexes. Our results thus further emphasize the functional differences between individual Rpts.
Three-fold symmetry at the base-core interface
In contrast to the half-ring asymmetry of individual Rpt contributions to substrate degradation, previous studies suggested a three-fold symmetry for the base-core interaction, in which the C-terminal tails of Rpt2, Rpt3, and Rpt5 dock into core-particle pockets9,10. Only these subunits contain the conserved C-terminal HbYX (hydrophobic/tyrosine/unspecified) motif that is critical for inducing core-particle gate opening7,17,18. The three remaining tails have been postulated to stabilize the complex7, but recent crosslinking, structural, and biochemical studies provided further conflicting results on which C-terminal tails mediate core-particle binding9,20,33,51. Moreover, it has been suggested that the nucleotide state of a Rpt subunit affects the conformation of its C-terminal tail, thereby modulating peptidase interaction52. Individual Rpt subunits are thus expected to contribute differently to base-core interactions, but a detailed model for the specialized roles of C-terminal tails in peptidase binding, gate opening, and substrate transfer is lacking.
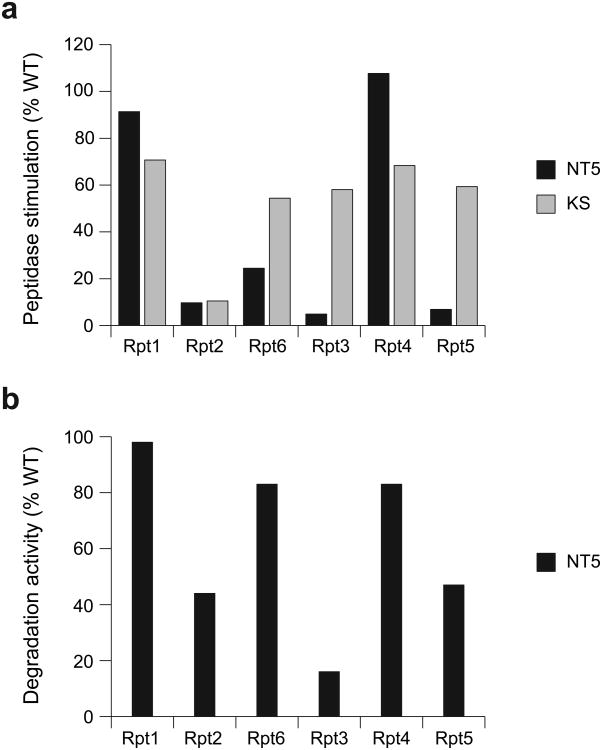
Our recombinant expression and in-vitro reconstitution system allows truncation of the Rpt C-terminal tails to quantitatively characterize their contributions to stability and function of the base-core complex. Consistent with previous studies, we found that the HbYX-containing tails of Rpt2, Rpt3, and Rpt5 were crucial for gate opening of the core particle. These three tails were independently and nearly equally important, as their individual truncation reduced gate-opening activity of the base by 90–95% (Fig. 5a, Table 1). Surprisingly, removing the tail from Rpt6, which lacks an HbYX motif, also decreased gate opening by 75%, whereas truncating Rpt1 and Rpt4 had little effect.
Figure 5. Rpt C-terminal tails contribute to gate opening differentially and in a nucleotide-independent manner.
(a) Base variants with either a C-terminal tail truncation (NT, black) or an “empty state” Walker-A KS mutation (KS, gray) in individual Rpt subunits were assessed for their ability to stimulate gate opening of the core particle relative to wild-type (WT) base. (b) Substrate degradation activities for proteasomes reconstituted with base subcomplexes containing C-terminal tail truncations of single Rpt subunits. Degradation of a polyubiquitinated GFP fusion substrate was monitored by the loss of fluorescence. Degradation rates under saturating conditions were measured relative to reconstituted proteasome containing wild-type (WT) recombinant base. Errors were estimated to be ±10% (s.d.) of the mean WT value based on repeat measurements (n = 3 technical replicates).
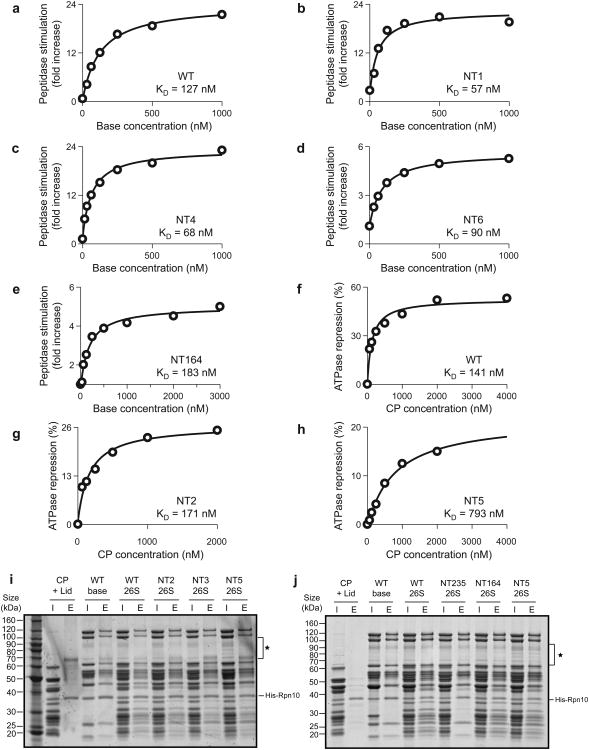
Titration experiments revealed that the C-terminal tails of Rpt1, Rpt4, and Rpt6 do not contribute to the stability of the base-core complex. The three individual tailless mutants and the triple mutant bound to core particle with KD values similar to or even lower than the wild-type value (Fig. 6), which may indicate that those tails cause steric clashes or compete with neighboring HbYX-containing tails for designated binding pockets. In contrast, truncating HbYX tails strongly affected peptidase binding. Tail truncations in Rpt2 and Rpt5 lowered the affinity for peptidase approximately 1.3-fold and 5.6-fold, respectively. Base lacking all three HbYX tails did not bind the peptidase (Fig. 6j). Affinity measurements for the base with tailless Rpt3 were impeded by the lack of a quantitative readout, as this mutant neither triggered core-gate opening nor exhibited core-mediated repression of ATPase activity.
Figure 6. Affinities of core-peptidase binding for base variants lacking individual C-terminal tails.
Core-particle (CP) binding to base mutants with truncated C-terminal tails was assessed by titration measurements in peptidase-stimulation and ATPase-repression assays. (a-e) For base variants competent to stimulate gate opening (NT1, NT4, NT6, NT164), binding affinity was measured by titrating increasing amounts of base into a gate-opening assay containing 25 nM CP. (f-h) CP binding to base variants that did not exhibit significant peptidase stimulation (NT2, NT5) was assessed by titrating increasing amounts of CP into an NADH-coupled ATPase assay containing 100 nM base. CP-mediated repression of base ATPase activity yielded bindings curves and KD values. (I and j) Nickel affinity pull-down assays examining CP binding to base variants lacking individual or multiple HbYX-containing tails. Untagged lid and CP subcomplexes were pulled down using a His6 tag on Rpt3 (base). Lanes are labeled I (input) and E (elution), and nonspecific contaminant bands are labeled (|). Removal of individual HbYX-containing tails (Rpt2, Rpt3, Rpt5) did not abolish base binding to CP. The NT164 base variant exhibited CP binding comparable to WT base, whereas removal of the three HbYX-containing tails (NT235) abrogated CP binding. The NT235 base mutant also exhibited reduced lid binding, consistent with the fact that purified base and lid subcomplexes did not form stable regulatory particle when reconstituted in the absence of CP (unpublished data, R.B.). This observation was made even for WT base and lid subcomplexes independent of whether they were isolated from endogenous or recombinant sources.
Proteasomes reconstituted with saturating amounts of base lacking a single tail from Rpt2, Rpt3, or Rpt5 showed decreased substrate degradation at 44%, 16%, or 47% of the wild-type rate (Fig. 5b), which is likely a consequence of gate-opening defects. Base with tailless Rpt6 supported degradation at greater than 80% of the wild-type rate. Robust degradation despite compromised gate-opening activity, as observed for Rpt6, may occur because translocation by the base pushes substrate through a partially opened gate, whereas the gate-opening assay reports solely on peptide diffusion into the peptidase.
Interaction of base and core particle to open the gate is known to be ATP-dependent and differentially stimulated by various ATP-analogs4,34,53. It has been postulated for some homohexameric unfoldases, including PAN and HslU, that ATP hydrolysis may drive conformational changes in the C-terminal tails of individual subunits and thereby regulate nucleotide-dependent binding and gating interactions with the peptidase7,34,54. Based on this model, an ATP-bound subunit would have an exposed C-terminal tail that interacts with the core particle, whereas an empty- or ADP-state subunit would have its tail buried in an occluded conformation. It was further proposed that at any given time the 26S proteasome has only two Rpt subunits, arranged in para position across the ring, in an ATP-bound state with C-terminal tails exposed for peptidase interaction52.
We tested whether peptidase interaction of an Rpt tail in fact depends on the nucleotide state of the particular subunit. According to this model, removing a given tail should resemble trapping that subunit in an empty-state conformation. We therefore prevented nucleotide binding to individual Rpts by mutating the conserved lysine to serine (KS) in their Walker-A motifs (Supplementary Note 1). We produced base variants with single-subunit KS mutations and found that only mutation of Rpt2 caused severe base-assembly defects. Characterization of the other KS mutants demonstrated that trapping a particular Rpt subunit in an empty state has much weaker effects on gate-opening activity than truncating its C-terminal tail (Fig. 5a). Base containing a KS mutation in Rpt3 or Rpt5 retained 58% or 59% of wild-type gate-opening activity, whereas the corresponding tailless mutants showed only 5% or 7% activity. Trapping Rpt6 in an empty state preserved 54% of wild-type activity, whereas removing its tail reduced gate opening to 25%. Moreover, all base variants with single KS mutations exhibited some degree of misassembly, such that the measured gate-opening activities represent only lower bounds. Thus, contrary to previous models, the C-terminal tails are accessible for peptidase binding independent of the nucleotide state of individual Rpt subunits, even though the base-core interaction depends on a global ATP-bound conformation of the Rpt ring.
The Walker-A mutants were not suited to qualitatively analyze the contribution of individual Rpt subunits to substrate processing, as they caused universally severe degradation defects (Table 1). In contrast to fixing subunits in an ATP-bound state with a Walker-B mutation, empty-state subunits potentially influence the hexamer conformation or compromise subunit communication required for processive ATP-hydrolysis in the ring.
Discussion
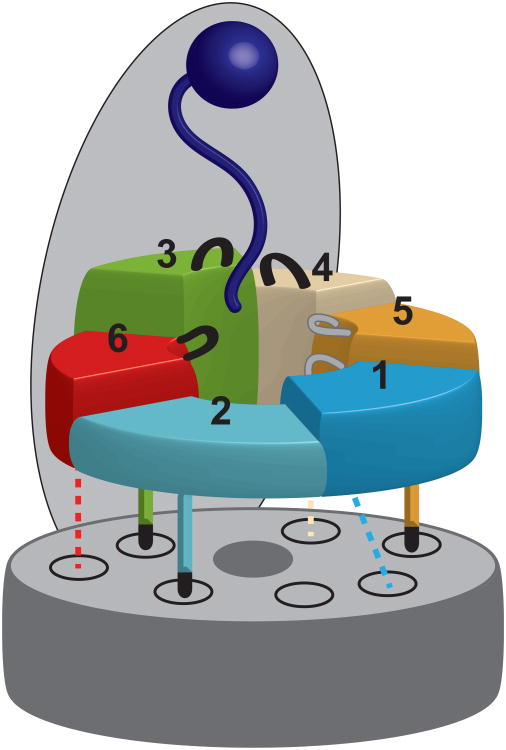
Our mutational studies using heterologously expressed base subcomplex and in-vitro reconstituted 26S holoenzymes revealed that substrate degradation by the proteasome relies on distinct functional asymmetries with strongly non-equivalent contributions of individual Rpts. While a static three-fold symmetry determines the interactions between the base ATPase ring and the core particle, substrate degradation seems to depend on asymmetric spiral-staircase arrangements of the ATPases in which subunits close to the pore entrance play crucial roles in substrate engagement (Fig. 7).
Figure 7. Model for the contribution of individual ATPase subunits to substrate processing and core particle interaction.
The substrate (dark blue) is bound to the proteasome lid and positioned for ATP-dependent processing by the heterohexameric ATPase ring of the base. Rpt6 (red), Rpt3 (green), and Rpt4 (beige), which are located at the top of the pre-engaged spiral staircase of ATPase subunits, play a critical role in substrate degradation. In contrast, Rpt5 (gold), Rpt1 (light blue), and Rpt2 (cyan) are less crucial. The C-terminal tails of the ATPases are docked into their cognate binding pockets in the core particle α-ring (gray). Tails that are crucial for binding and gate opening (Rpt2, Rpt3, Rpt5) are shown as solid lines with black termini, representing the HbYX motif. The remaining three tails that do not contribute to the stability of the base-core interaction (Rpt1, Rpt4, Rpt6) are shown as dashed lines.
Our data demonstrate a clear three-fold symmetry at the base-core interface, in which the HbYX - containing C-terminal tails of the alternating subunits Rpt2, Rpt3, and Rpt5 are critical for both binding the core particle and triggering gate opening (Fig. 7). The tails of the interjacent Rpt1, Rpt4, and Rpt6 are dispensable for the base-peptidase interaction, and Rpt1 and Rpt4 are largely irrelevant for gate opening. The apparent function of the Rpt6 tail in core-particle gating may result from Rpt6's role in ensuring the correct register of the heterohexameric base atop the α-ring of the core particle. Recent structural and functional data suggest that Rpt6 plays a role in base-core assembly and its C-terminus is the only non-HbYX tail that docks into just one specific α pocket33,51. It is thus possible that Rpt6's tail prevents the neighboring HbYX-containing tails from mispairing with this pocket.
Interactions of the three HbYX-containing tails with the peptidase seem static and independent of the nucleotide state of the particular Rpt subunits. This finding contradicts previous models proposing that the Rpt C-terminal tails change conformation during the ATP-hydrolysis cycle and thus lead to a “wobbling” of the base atop the peptidase7,34,52. Earlier structural studies demonstrated that in the absence of ATP several AAA+ hexamers adopt a “lockwasher” conformation55-57 that likely prevents binding to the planar surface of a peptidase. The ATP-dependence of the base-core interaction thus seems to originate not from conformational changes of individual tails, but rather from the global geometry of the ATPase ring, which must be at least partially nucleotide-bound to facilitate peptidase docking.
Eliminating ATP hydrolysis or introducing Ar-Φ-loop mutations in single Rpts revealed that the lid-facing half of the ATPase ring is particularly crucial for substrate degradation. These key Rpts occupy top positions in the spiral-staircase arrangement adopted by the large AAA+ subdomains in the absence of substrate (Fig. 7), and they must undergo ATP-hydrolysis-induced conformational changes to engage an incoming substrate and initiate translocation. The Rpt hexamer then transitions to an alternative, more planar staircase arrangement, in which Rpt1 occupies the top position and rigid-body interfaces are formed between most subunits20. Our data suggest that ATP hydrolysis in Rpt1 is more important than in neighboring subunits to drive substrate translocation, possibly because Rpt1 triggers a burst of coordinated hydrolysis events around the ring. Nevertheless, individual Rpts may contribute more equally to translocation once substrate is engaged.
Spiral-staircase arrangements have been previously observed in several homohexameric motors of the AAA+ and RecA families, and it has been suggested that individual ATPase subunits successively transition through the different vertical registers of the spiral to drive substrate translocation47,57-60. However, the apparently rigid spiral-staircase configurations observed for the proteasomal ATPase ring in the absence and presence of substrate, together with the biochemical data presented here, contradict stepwise successive progression of individual Rpts through the different registers. Instead, the proteasome base relies on differential subunit contributions prescribed by the regulatory particle's asymmetric architecture. Conformational changes in the top subunits of a pre-engaged staircase thread an incoming substrate, before the ATPase ring switches to an engaged staircase arrangement, in which subunits contribute more equally to protein translocation.
Although all mutant base complexes in this study were characterized by in-vitro assays that may or may not wholly recapitulate the complexity of in-vivo conditions, our results agree with and substantially extend earlier findings, suggesting non-equivalent contributions of individual Rpt subunits to substrate processing by the 26S proteasome21-23. Protein degradation by the proteasome deviates from models previously proposed for other AAA+ unfoldases, pointing toward major differences between the operating principles of homo- and heterohexameric AAA+ motors. Future experiments will have to address to what extent these motor designs rely on different strategies for substrate engagement, generation of mechanical force for unfolding, and translocation of the polypeptide chain into a peptidase for degradation.
Online Methods
Recombinant base expression and purification
Thirteen subunits were cloned into three Novagen vectors including pCOLA-1 (FLAG-Rpt1, Rpt2, His6-Rpt3, Rpt4, Rpt5, Rpt6), pETDuet-1 (Rpn1, Rpn2, Rpn13), and pACYCDuet-1 (Nas2, Nas6, Hsm3, Rpn14). Each subunit was preceded by a T7 promoter and all plasmids contained one T7 terminator at the end of the multiple cloning sites. Genes for rare tRNAs were also included in the pACYCDuet-1 plasmid to account for differences in codon usage between yeast and E. coli. Point mutations in individual ATPase subunits were generated by PCR using pETDuet-1 plasmids containing individual Rpt subunits, which were then used for amplification and substitution into the wild type hexamer in pCOLADuet-1. Base expression strains were generated by co-transforming the pETDuet-1, pCOLA-1 and pACYCDuet-1 plasmids into E. coli BL21-star (DE3) cells. The base subcomplex was produced by growing the expression strain to OD600=0.6–0.8 and inducing with 1mM isopropyl-b-D-thiogalactopyranoside overnight at 18°C. Cells were harvested by centrifugation at 5000 rpm for 15 min, resuspended in nickel buffer (25 mM HEPES pH 7.6, 100 mM NaCl, 100 mM KCl, 10 % glycerol, 10 mM MgCl2, 0.5 mM EDTA, 20 mM imidazole) supplemented with 2 mg ml−1 lysozyme, protease inhibitors and benzonase. Cells were lysed by freeze-thaw and sonication on ice for 1 min 30 sec in 15 sec bursts. Lysate was clarified by centrifugation at 15,000 rpm at 4°C for 30 min. A two-step affinity purification of the base subcomplex was performed using Ni-NTA agarose (Qiagen) to select for His6-Rpt3 and anti-FLAG M2 resin (Sigma-Aldrich) selecting for FLAG-Rpt1. 0.5 mM ATP was present in all purification buffers. The Ni-NTA and anti-FLAG M2 columns were eluted with nickel buffer containing 250 mM imidazole or 0.15 mg ml−1 3×FLAG peptide, respectively. The Flag column eluate was concentrated using a 30,000 MWCO concentrator (Amicon) and run on a Superose6 gel filtration column (GE Healthcare) equilibrated with gel filtration buffer (60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 % glycerol, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 0.5 mM ATP).
Purification of yeast holoenzyme and subcomplexes
Yeast holoenzyme, core particle, base and lid subcomplexes were purified from S. cerevisiae essentially as previously described61. Frozen yeast cells were lysed using a Spex SamplePrep 6870 Freezer/Mill. Holoenzyme was purified from a yeast strain containing FLAG-Rpn11. Lysed cells were resuspended in lysis buffer containing 60 mM HEPES pH 7.6, 100 mM NaCl, 100 mM KCl, 10 % glycerol, 5 mM MgCl2, 0.5 mM EDTA, 0.2 % NP-40 and ATP regeneration mix (5 mM ATP, 0.03 mg ml−1 creatine kinase, 16 mM creatine phosphate). Holoenzyme was bound to anti-FLAG M2 resin and washed with wash buffer (60 mM HEPES pH 7.6, 100 mM NaCl, 100 mM KCl, 10 % glycerol, 5 mM MgCl2, 0.5 mM EDTA, 0.1 % NP-40, 0.5 mM ATP). Holoenzyme was eluted with 0.15 mg ml−1 3×FLAG peptide and further purified by gel filtration using a Superose-6 column with gel filtration buffer (see above). Lid and base subcomplexes were isolated from FLAG-Rpn11 or FLAG-Rpn2 yeast strains, respectively, and purified by exposure to a 1 M NaCl wash while bound to anti-FLAG M2 resin. Base purification buffers included 0.5 mM ATP. Core particle was purified from a 3×FLAG-Pre1 yeast strain using a 500 mM salt wash. All subcomplexes underwent size exclusion chromatography using a Superose-6 column as described above.
Yeast strains
Yeast lid and holoenzyme were purified from strain YYS40 (genotype MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1 Rpn11::Rpn11-3×FLAG(HIS3), source Y. Saeki). Core particle was prepared either from strain RJD1144 (genotype MATa his3Δ200 leu2-3,112 lys2-801 trpΔ63 ura3-52 PRE1-FLAG-6×HIS::Ylpac211(URA3) source R. Deschaies) or strain yAM14 (genotype MATa ade2-1 his3-11,15 leu2-3,112 trip1-1 ura3-1 can1-100 bar1 PRE1::PRE1-3×FLAG(KanMX), this study).
Native gel electrophoresis
Analysis of proteasome holoenzyme and subcomplexes by native gel was performed as described previously61. Assembly reactions were incubated for 15 min at 23°C with 5 mM ATP, followed by electrophoresis on a 3.5% native polyacrylamide gel. Electrophoresis was conducted at 4°C with stirring and running buffer containing 0.5 mM ATP. The gel was overlaid with developer solution (running buffer with 100 nM Suc-LLVY-AMC peptide and 0.02% SDS) and incubated at 30°C for 10 minutes prior to imaging. Fluorescence imaging was performed using a Typhoon scanner (GE Healthcare) and followed by Coomassie staining.
ATPase and peptidase stimulation assays
ATPase activity was quantified using an NADH-coupled ATPase assay. 500 nM base was incubated with 1× ATPase mix (3 U ml−1 pyruvate kinase, 3 U ml−1 lactate dehydrogenase, 1 mM NADH, 7.5 mM phosphoenol pyruvate) at 30°C. Absorbance at 340 nm was monitored for 900 sec at 10 sec intervals using a UV-Vis Spectrophotometer (Agilent). Peptidase stimulation was monitored by following the increase in fluorescence resulting from cleavage of a fluorogenic peptide substrate62, Suc-LLVY-AMC (Boston Biochem), using a QuantaMaster spectrofluorimeter (PTI). 50 nM core particle was incubated with saturating base subcomplex in the presence of an ATP regeneration system (5 mM ATP, 16 mM creatine phosphate, 6 mg ml−1 creatine phosphokinase) and 50 μM Suc-LLVY-AMC. Titration experiments were conducted using either 25 nM core particle and increasing amounts of base (peptidase stimulation) or 100 nM base and increasing amounts of core particle (ATPase activity). KD values were extracted by fits to a simple binding curve using Grafit (Erithacus Software).
Multiple and single turnover GFP substrate degradation
Proteasome holoenzyme was reconstituted from core particle, lid, base and Rpn10. A GFP-titinV15P-cyclin-PY fusion protein was modified in vitro with a polyubiquitin chain using Uba1, Ubc1, Rsp5 and wild-type ubiquitin. Degradation reactions were performed at 30°C in gel filtration buffer (60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 10 % glycerol, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 0.5 mM ATP) supplemented with an ATP regeneration system. Details for the gel-based degradation assay can be found in the legend of Supplementary Fig. 2. Single and multiple turnover degradation activities were monitored by the loss of GFP fluorescence (excitation 467 nm; emission 511 nm) using a QuantaMaster spectrofluorimeter (PTI). Multiple turnover degradation experiments were performed with 50 nM reconstituted holoenzyme under Vmax conditions (saturating base, lid and Rpn10) with 2 μM substrate. Excess base, lid, and Rpn10 did not affect the observed degradation rate (see Supplementary Fig. 3 for lid and Rpn10). For single turnover degradation, 2 μM holoenzyme was reconstituted from equimolar concentrations of subcomplexes and mixed with 100 nM substrate. Single turnover degradation traces fit with double exponential decay curves using Grafit (Erithacus Software)
Affinity pulldowns
Base subcomplexes were mixed with core particle, lid and Rpn10 in nickel buffer with 1× ATP regeneration system at 23°C for 15 min. All components were present at a concentration of 900 nM in the reaction, which was then incubated with 5 μl of magnetic Dynabeads (Invitrogen) at 23°C for 15 min. The beads were washed three times with nickel buffer supplemented with 0.05% NP-40 and 0.5 mM ATP. Bound proteins were eluted with nickel buffer containing 500 mM imidazole. Pulldown samples were run on a 10% SDS-PAGE gel, stained with Sypro Ruby and imaged using a Typhoon scanner (GE Healthcare).
Supplementary Material
Acknowledgments
We thank the members of the Martin lab for helpful discussions. E.J.W. acknowledges support from the NSF Graduate Research Fellowship. This research was funded in part by the Searle Scholars Program (A.M.), the US National Institutes of Health grant R01-GM094497-01A1 (A.M.), and start-up funds from the Molecular and Cell Biology Department at University of California, Berkeley (A.M.).
Footnotes
Author Contributions: R.B., E.E., E.J.W., and A.M. designed experiments, expressed and purified proteasome constructs, performed biochemical experiments, and analyzed data. R.B. and A.M. prepared the manuscript.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–39. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–7. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 4.Liu CW, et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–23. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 7.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–44. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomko RJ, Jr, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem Biophys. 2011;60:13–20. doi: 10.1007/s12013-011-9178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–91. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A. 2012;109:14870–5. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamazaki J, et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–36. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–7. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 13.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 14.Sakata E, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2012;109:1479–84. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–20. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 16.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–8. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YC, DeMartino GN. C termini of proteasomal ATPases play nonequivalent roles in cellular assembly of mammalian 26 S proteasome. J Biol Chem. 2011;286:26652–66. doi: 10.1074/jbc.M111.246793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–22. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sledz P, et al. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc Natl Acad Sci U S A. 2013;110:7264–9. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol. 2013;20:781–8. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. Embo J. 1998;17:4909–19. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erales J, Hoyt MA, Troll F, Coffino P. Functional asymmetries of proteasome translocase pore. J Biol Chem. 2012;287:18535–43. doi: 10.1074/jbc.M112.357327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YC, Li X, Thompson D, Demartino GN. ATP-binding by proteasomal ATPases regulates cellular assembly and substrate-induced functions of the 26S proteasome. J Biol Chem. 2012 doi: 10.1074/jbc.M112.424788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SH, Moon JH, Yoon SK, Yoon JB. Stable incorporation of ATPase subunits into 19 S regulatory particle of human proteasome requires nucleotide binding and C-terminal tails. J Biol Chem. 2012;287:9269–79. doi: 10.1074/jbc.M111.316208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler A, et al. The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol Cell. 2001;7:1143–52. doi: 10.1016/s1097-2765(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 26.Park S, et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–70. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar B, Kim YC, DeMartino GN. The C terminus of Rpt3, an ATPase subunit of PA700 (19 S) regulatory complex, is essential for 26 S proteasome assembly but not for activation. J Biol Chem. 2010;285:39523–35. doi: 10.1074/jbc.M110.153627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J Biol Chem. 2009;284:24891–903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–99. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–5. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–13. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko T, et al. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–25. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Park S, et al. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature. 2013;497:512–6. doi: 10.1038/nature12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DM, et al. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–98. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 35.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–27. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 37.Weibezahn J, Bukau B, Mogk A. Unscrambling an egg: protein disaggregation by AAA+ proteins. Microb Cell Fact. 2004;3:1. doi: 10.1186/1475-2859-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomez EB, Catlett MG, Forsburg SL. Different phenotypes in vivo are associated with ATPase motif mutations in Schizosaccharomyces pombe minichromosome maintenance proteins. Genetics. 2002;160:1305–18. doi: 10.1093/genetics/160.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–84. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 40.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–51. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008;29:441–50. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada-Inagawa T, Okuno T, Karata K, Yamanaka K, Ogura T. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J Biol Chem. 2003;278:50182–7. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- 43.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–67. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–69. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park E, et al. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem. 2005;280:22892–8. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- 46.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–41. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–56. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glynn SE, Nager AR, Baker TA, Sauer RT. Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine. Nat Struct Mol Biol. 2012;19:616–22. doi: 10.1038/nsmb.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barthelme D, Sauer RT. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc Natl Acad Sci U S A. 2013;110:3327–32. doi: 10.1073/pnas.1300408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian G, et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011;18:1259–67. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–38. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Demartino GN. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J. 2009;421:397–404. doi: 10.1042/BJ20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seong IS, et al. The C-terminal tails of HslU ATPase act as a molecular switch for activation of HslV peptidase. J Biol Chem. 2002;277:25976–82. doi: 10.1074/jbc.M202793200. [DOI] [PubMed] [Google Scholar]
- 55.Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–70. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- 56.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–52. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 57.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 58.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–34. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–5. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 60.Costa A, et al. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–7. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leggett DS, Glickman MH, Finley D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol Biol. 2005;301:57–70. doi: 10.1385/1-59259-895-1:057. [DOI] [PubMed] [Google Scholar]
- 62.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–62. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.