Abstract
Natural products, as major resources for drug discovery historically, are gaining more attentions recently due to the advancement in genomic sequencing and other technologies, which makes them attractive and amenable to drug candidate screening. Collecting and mining the bioactivity information of natural products are extremely important for accelerating drug development process by reducing cost. Lately, a number of publicly accessible databases have been established to facilitate the access to the chemical biology data for small molecules including natural products. Thus, it is imperative for scientists in related fields to exploit these resources in order to expedite their researches on natural products as drug leads/candidates for disease treatment. PubChem, as a public database, contains large amounts of natural products associated with bioactivity data. In this review, we introduce the information system provided at PubChem, and systematically describe the applications for a set of PubChem web services for rapid data retrieval, analysis, and downloading of natural products. We hope this work can serve as a starting point for the researchers to perform data mining on natural products using PubChem.
Keywords: natural products, drug discovery, PubChem, public database, data mining
1 Introduction
Natural products (NPs) have been the most productive source of leads for drug discovery and development. Several biologically active NPs, such as vincristine, irinotecan, etoposide, and paclitaxel, have been extensively explored as drug candidates for clinical applications as well as basic research tools to investigate biological processes [1]. With the advent of combinatorial chemistry, there had been a decreased interest in natural products for NPs-based drug discovery in the 1980s and 1990s. Nevertheless, NPs are regaining attentions recently from pharmaceutical companies and academic institutes. Firstly, the rapid growth in the number of potential therapeutic targets demands the access to novel and diverse chemical libraries. The reservoir of natural products contains abundant chemical novelty and diversity, making it difficult for chemical synthesis technology to replace NPs as a source of new drugs [2, 3]. It is reported that about 40% of the chemical scaffolds of the published NPs are unique and have not been synthesized in laboratory, a crucial driving force for pharmaceutical investigators and companies to target on natural products [4]. Secondly, bioactive NPs are generally small and drug-like, making them more likely to be absorbable and metabolizable by the human body. Hence, the development cost of producing oral medicines derived from NPs is probably much lower than that from combinatorial chemistry [5]; It is noteworthy that NPs also represent an important class of molecules with larger structures and physicochemical properties beyond rule-of-five, which may be better suited for addressing the so-called undruggable target space while still maintaining desirable oral bioavailability [6].
Recent years have witnessed a rapid growth in NPs studies by large-scale screening experiments, which doubtlessly lead to a wealth of information on the bioactivities associated with NPs [7]. Having access to data produced by such experiments can enable many kinds of drug discovery analysis and effective decision making. Indeed, it has become extremely important for creating open access biomedical databases by aggregating disperse yet invaluable data into a standard and integral platform, which would allow researchers to analyze the data by using or developing modern computational methods. Currently, there are a number of public chemical biology databases, such as PubChem [8–10] and ChEMBL [11], providing access to numerous bioactivity data for small molecules. Many of these resources are complementary and vital for drug discovery [12, 13].
PubChem is a public repository for chemical structures and biological activity for small molecules, including a large number of natural products. PubChem offers many tools to facilitate search of chemical structures, bioactivity data, and molecular target information. In this work, to get researchers started by taking advantage of this growing resource, we describe the information framework at PubChem by going through a selected set of basic web tools for searching natural products and linking to their bioactivity data. We hope information gained from PubChem may be beneficial for the natural product research community.
2 A brief overview of PubChem
PubChem (http://pubchem.ncbi.nlm.nih.gov/) [8–10] is an open public repository containing chemical structures and biological properties of small molecules and siRNA reagents deposited by several hundred organizations, with the aim to deliver free and easy access to all deposited data and to provide intuitive data analysis tools. PubChem is hosted at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). PubChem is intensively integrated with Entrez, the NCBI’s primary search engine, with its information content cross-linked to many other biomedical databases, such as PubMed, Gene, Nucleotide, Taxonomy and Protein 3D structure. Figure 1 shows two common gateways for accessing PubChem.
Figure 1.
PubChem gateways. PubChem search can be performed by (a) using the NCBI Entrez interface or (b) using the PubChem home page. The PubChem home page also provides access to a list of tools.
PubChem includes two primary databases, with the Substance database (Accession ID: SID), containing deposited information for small molecules, and the BioAssay database (Accession ID: AID), containing biological test results for deposited substances. PubChem’s third database, the Compound database (Accession ID: CID) is derived purely from the Substance database and contains unique chemical structures and calculated physicochemical properties. As of April 2013, the Substance database contains more than 118 million records, which correspond to over 47 million unique structures in the Compound database. The BioAssay database contains about 650 thousand bioassays and over 200 million of biological test results, which are generated by high-throughput screenings, chemical biology researches, medicinal studies, literature mining projects, and RNAi experiments [9, 10]. The PubChem BioAssay database allows researchers to link the screening results of small molecules to chemical structures, and thousands of protein and gene targets, hence enable researchers to search bioactive compounds, identify structure-bioactivity relationships, construct drug-target network, as well as to derive bioactivity profiles of small molecules and study their polypharmacology properties. Table 1 summarizes a list of web-based PubChem analysis and download tools.
Table 1.
A list of web-based PubChem services and bioactivity analysis tools
3 Search natural products in PubChem
3.1 Use the entrez interface
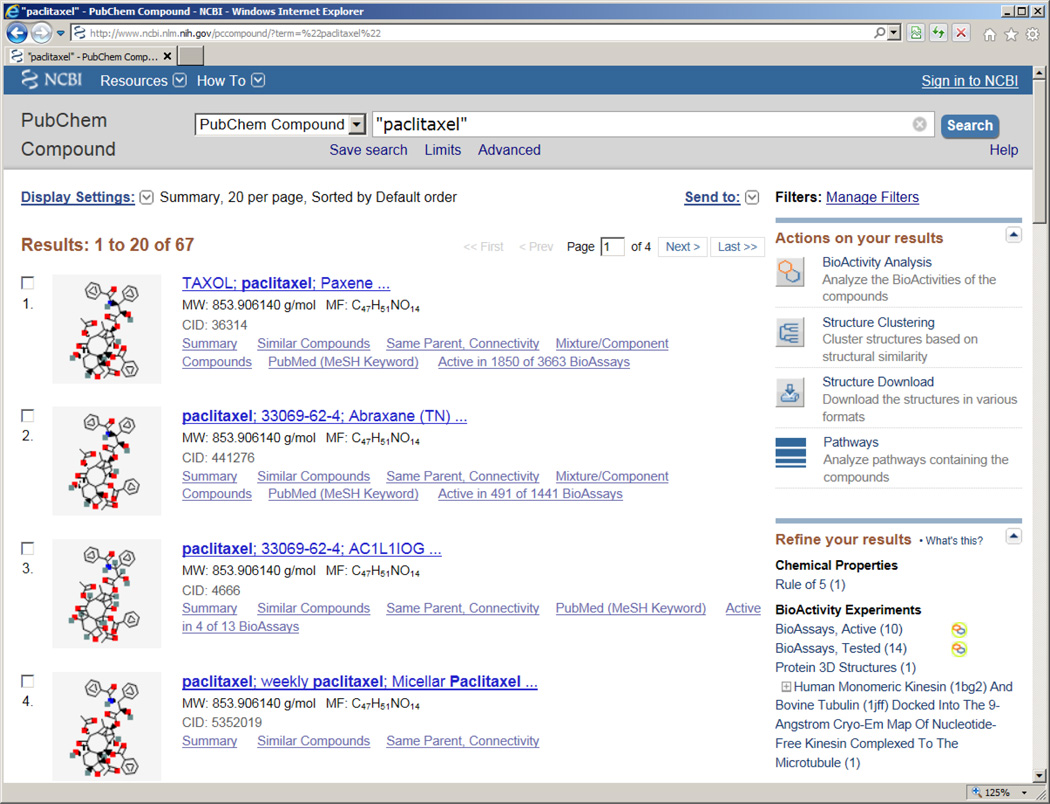
NCBI provides access to all its information resources using the Entrez search engine. The common method to search a natural product of interest is by a keyword (e.g. chemical name). To start with the Entrez system, one needs to first specify a database, such as “PubChem Compound” as shown in Figure 1(a). The PubChem homepage utilizes the Entrez system on the backend, thus provides an exchangeable interface as Entrez itself (Figure 1(b)). Figure 2 documents the summary of the search results against the PubChem Compound database by using the query “paclitaxel”, a mitotic inhibitor isolated from the bark of the Pacific yew tree [14] that is commonly used in cancer chemotherapy.
Figure 2.
Searched results from PubChem Compound database for the query “paclitaxel”.
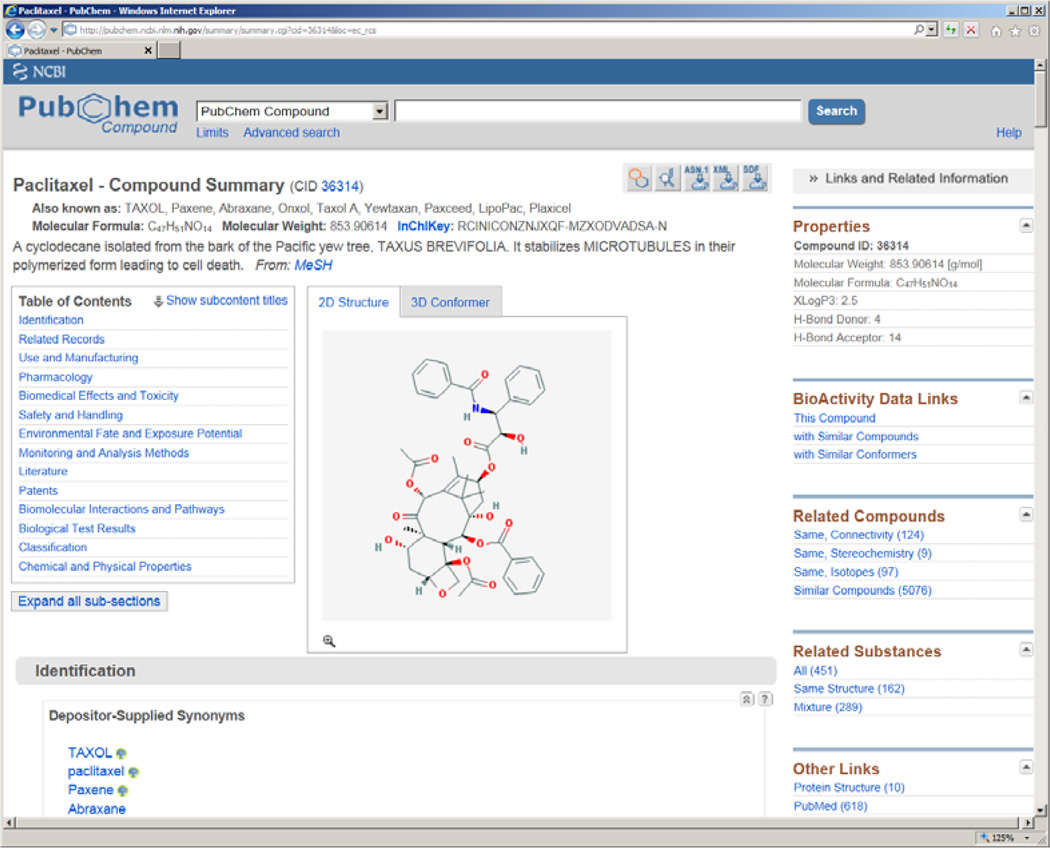
Selective information is shown in this summary view, including chemical structure image, chemical names/synonyms, accession number (CID), molecular weight, molecular formula, and links to a variety of other related information. In addition, there are a number of useful tools under the “Actions on your results” panel. One can analyze bioactivity data (via “BioActivity Analysis”), cluster chemical structures (via “Structure Clustering”), download chemical structures (via “Structure Download”), or link to related pathways (via “Pathways”) for all compounds or selected entries. Likewise, the “Refine your results” panel can be used to retrieve a subset of searched results to one’s interest. For example, the “BioAssay, Active (10)” narrows down the original 67 records to only the ten compounds that are active in biological tests (i.e. associated with active bioactivity data). For a specific record, the user can click on the image of chemical structure and go to the Compound Summary view (Figure 3, http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=36314), which further offers rich contents about the selected compound.
Figure 3.
Compound summary for the natural product paclitaxel (CID: 36314).
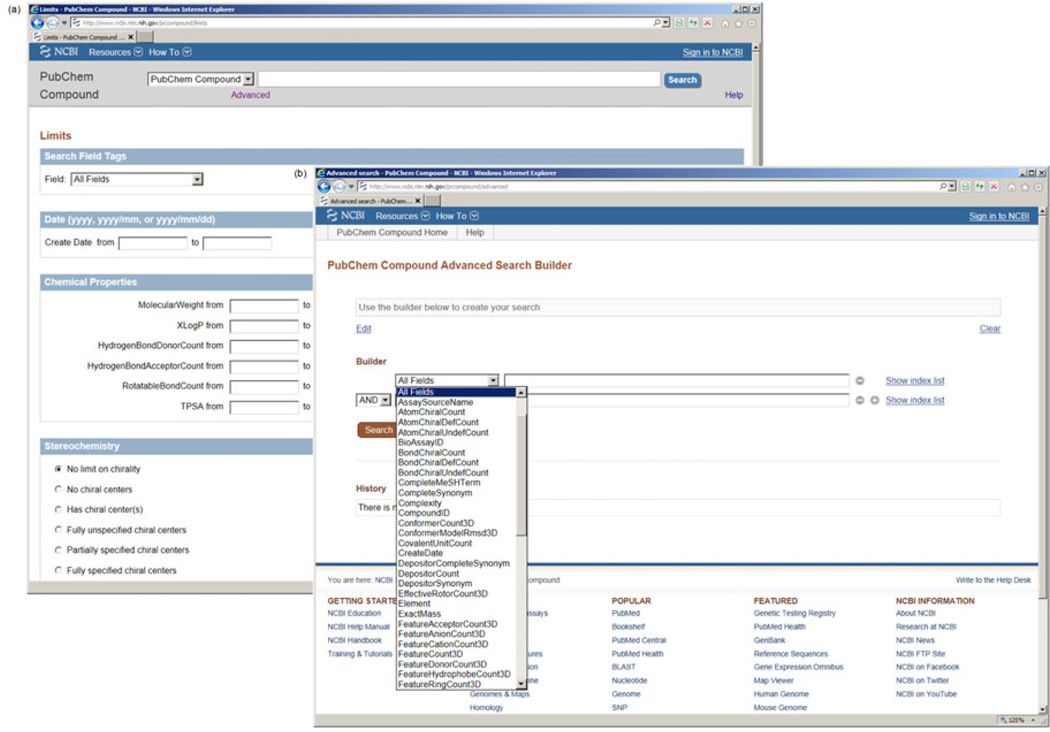
In addition to regular keyword search, Entrez also allows one to construct user-customized query via the “Limits” facility (Figure 4(a), http://www.ncbi.nlm.nih.gov/pccom-pound/limits), an advanced search interface for one to specify and combine search fields based on one’s particular need. For example, one may be interested in retrieving compounds that have been tested in biological experiments. This can be achieved by checking the radio box to the left of “Tested” under the “BioAssays” section in the “Limits” page. The “Advanced” facility (Figure 4(b), http://www.ncbi.nlm.nih.gov/pccompound/advanced), another advanced search interface, shows an easy-to-use query builder and a list of Entrez search history, which in turn can be re-visited or added to the query builder.
Figure 4.
Constructing a user-defined query via the NCBI Entrez’s advanced interfaces. (a) Limits; (b) “Advanced” search facilities.
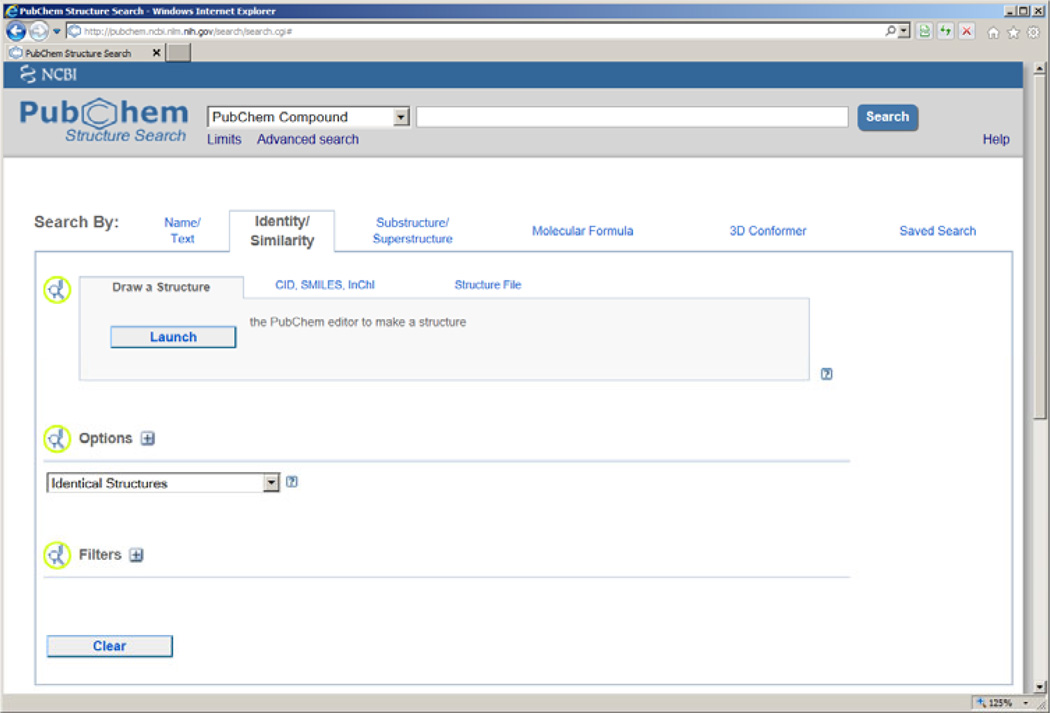
3.2 Use the PubChem structure search interface
The PubChem Structure Search interface (http://pubchem.ncbi.nlm.nih.gov/search/search.cgi) provides a set of powerful functionalities to search chemical records in PubChem that may be relevant to one’s research interest (Figure 5). This interface supports a variety of search options, including synonym search, molecular formula search, and more importantly, chemical identity/similarity search. For instance, if one is interested in searching compounds that are structurally similar to a natural product (e.g. neighbor compounds), one can perform an “Identity/Similarity” search by drawing a chemical structure, entering a SMILES or InChI string, or uploading a file in SD format. Upon the completion of such a structure search, the list of neighbor compounds (if any) will be sent to the Entrez system, from which one can take the full advantage of all the utilities as discussed above including the download function to obtain the chemical structures. Help documentation for performing a structure search at PubChem is available at: http://pubchem.ncbi.nlm.nih.gov/search/help_search.html.
Figure 5.
The PubChem structure search interface.
4 Link to the bioactivities of natural products
Serving as a public repository for small molecule bioactivities via its BioAssay database, PubChem provides an information system storing deposited bioassay records and linking them to tested chemical samples. Hence, searching chemical names, or chemical structures from any of PubChem’s three databases allows one to track down the associated bioactivity information. To this end, PubChem provides web tools for accessing and downloading full bioassay records. It also provides tools to integrate the chemical and biological activity information, hence allows ones to rapidly narrow down to the bioactivity data associated with the query compounds. Taking the paclitaxel search (CID: 36314) as an example, we highlight three common ways for linking compounds to the bioactivity information in this work.
4.1 Link to bioactivity for a single compound
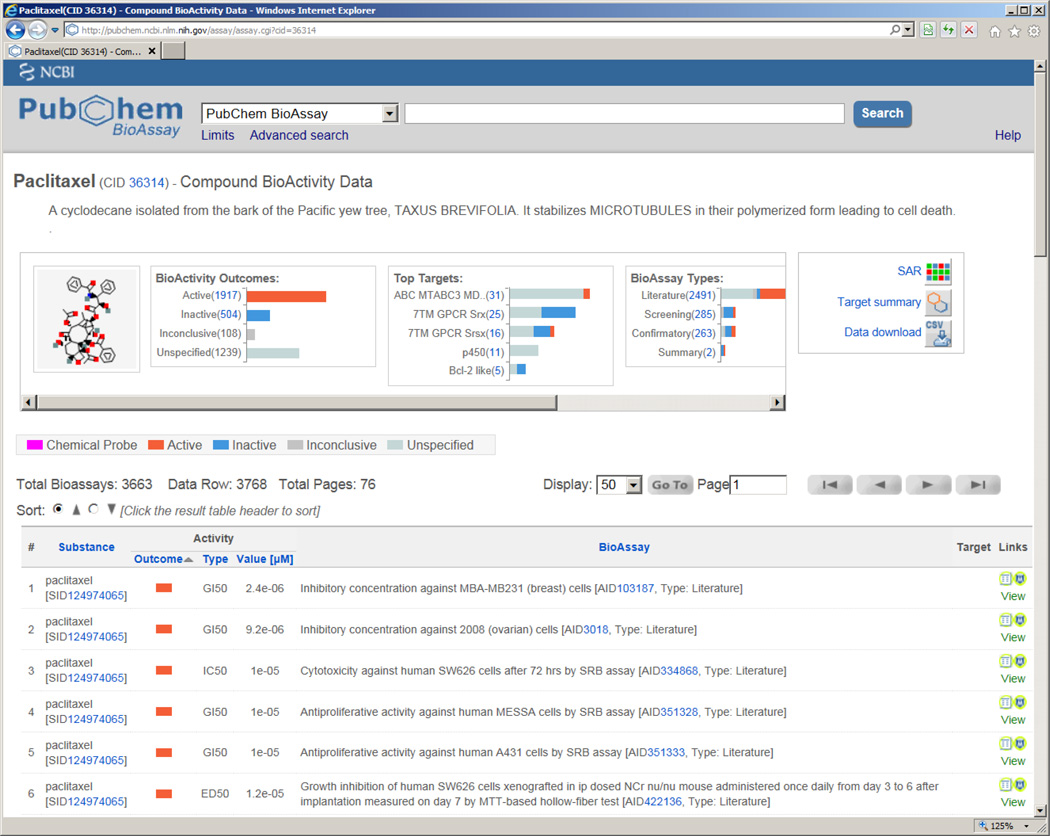
Bioactivity links are provided for each tested PubChem substance (SID) and are aggregated by each substance’s unique chemical structure, i.e. PubChem compound (CID). Such links are directly available when searching compounds in PubChem, which can be accessed via the document summary view. For example, to access to all of the available bioactivity data for paclitaxel represented by CID 36314, one can follow the link “Active in 1850 of 3663 BioAssays” as shown in Figure 2. This operation leads to a summarized data table view (Figure 6, http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?cid=36314), which shows bioactivity outcome (e.g. active or inactive), tested activity value (e.g. potency), molecular target and so on. Full record about the bioassay, which tested the respective chemical sample, can be found in the BioAssay column. Another useful feature of the data table summary view is the graphic filters panel, which shows the number of data records under specific category and thus allows easy access to a subset of interest (e.g. tested compounds against certain protein target). In addition, one always has the freedom of choice to download data by following the “Data download” link.
Figure 6.
Bioassay data summary for paclitaxel (CID: 36314).
4.2 Link to bioactivity for a set of compounds
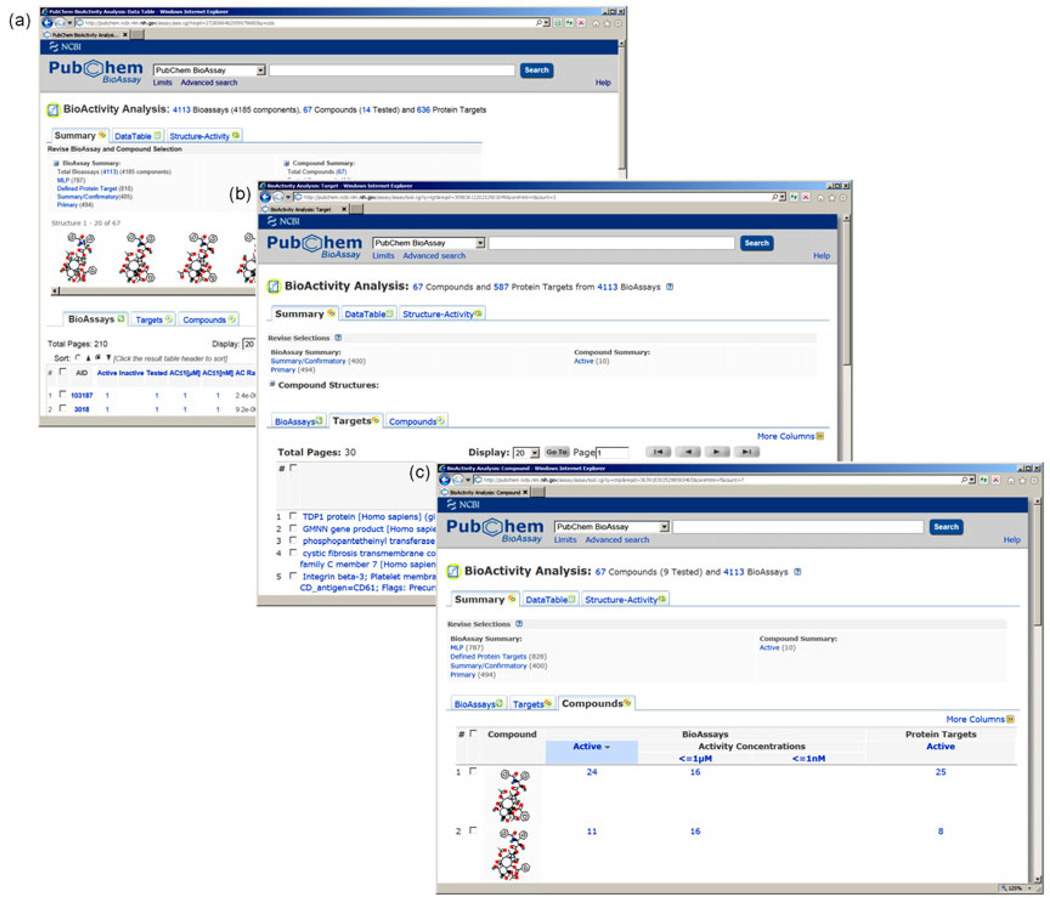
From the document summary view resulted from searching paclitaxel as shown in Figure 2, one may perform the bioactivity analysis for all the 67 compounds or selected records (via check box on the left) identified in this search by following the “BioActivity Analysis” link under the “Actions on your results” section.
The tool is powerful as it provides a comprehensive view of biological activity information available for one or more small molecules (Figure 7(a)). The view linked by the tab “BioAssays” (e.g. BioAssays centric view) groups the tested results for each bioassay record (Figure 7(a)); the “Targets” centric view groups the tested results for each target group of distinct protein sequence (Figure 7(b)); and the “Compounds” centric view groups the test results for each distinct chemical structure (Figure 7(c)). These tools summarize the association between bioassay, compound and target from different points of view meeting a broad range of needs from researchers. For example, the “Target centric view” allows one to focus on specific targets and rapidly collect relevant information such as bioactive compounds identified and information about the screening experiments against a specific target. Furthermore, this service allows users to compare and examine biological outcome across multiple assays, enabling rapid evaluation of target selectivity, locating commonly tested compounds in multiple assays, and selecting and prioritizing a subset data for further analysis, for example, to identify structure-bioactivity relationship (SAR) by clicking the “Structure-Activity” tab.
Figure 7.
Bioactivity analysis tool for a set of compounds. (a) Bioassays centric view; (b) targets centric view; (c) compounds centric view.
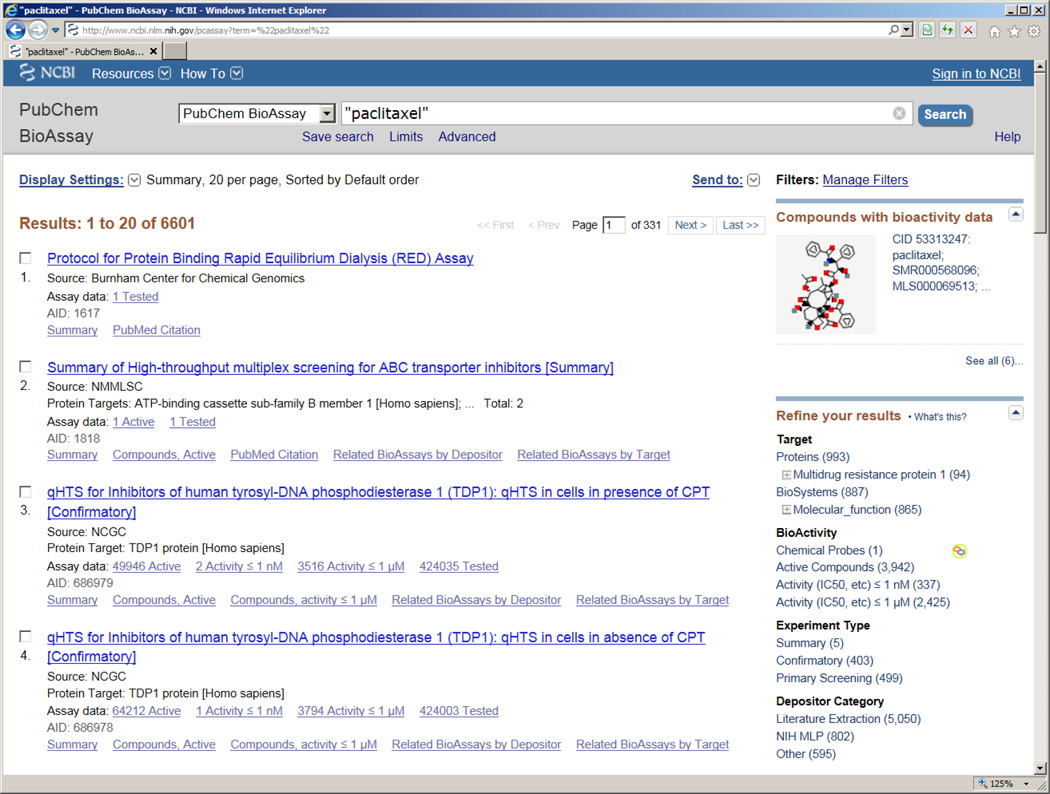
4.3 Link to bioactivity summary from BioAssay search
Chemical names of tested substances in the PubChem Bio-Assay database are indexed for Entrez search. Therefore, searching the BioAssay database using a chemical name, for example paclitaxel, allows one to identify all bioassay records associated with the chemicals (Figure 8). Similar to the search in the PubChem Compound database, this document summary view for the BioAssay search provides a brief summary about the retrieved bioassay records, which in turn link to the full deposited data, including biological results for the searched chemicals. In this particular case of “paclitaxel”, however, Entrez system places a sensor on such query, as a result, one could get all bioactivity data for “paclitaxel” by following the “See all (6)” link shown under the “Compounds with bioactivity data” (Figure 8). The summary view is similar to the one described in section 4.2, hence will not be detailed here.
Figure 8.
Search results for paclitaxel (CID: 36314) from the PubChem Bioassay database.
5 Case study
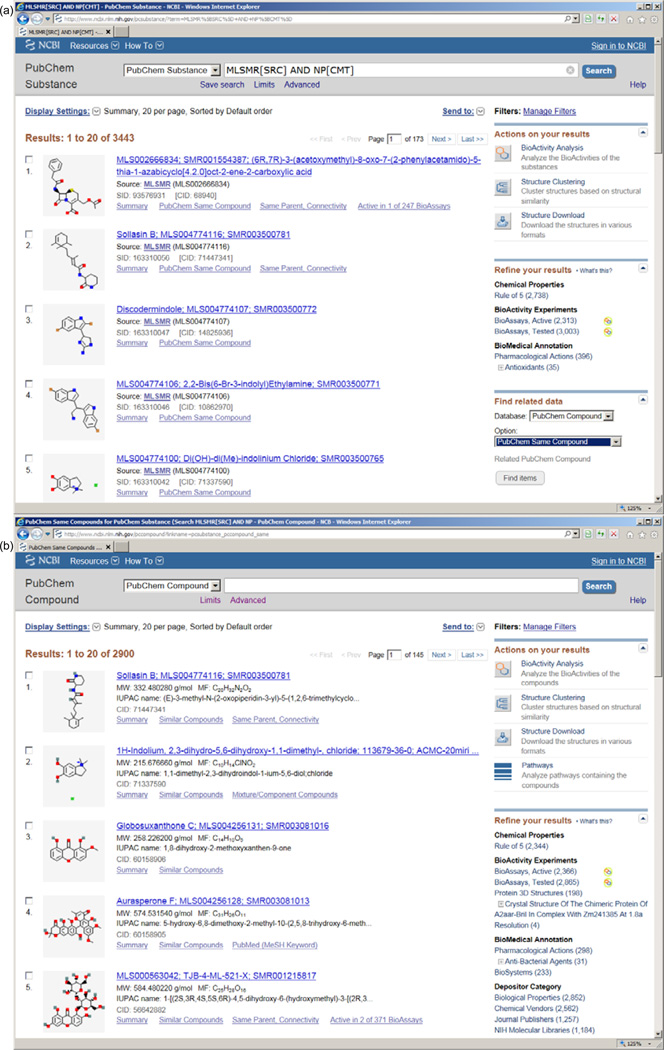
In this review, we highlight a particular set of natural products and the associated bioactivity data in the PubChem database that may have not been paid much attention by the public. They can be retrieved by querying “MLSMR[SRC] AND NP[CMT]” in the PubChem Substance database (http://www.ncbi.nlm.nih.gov/pcsubstance/?term=MLSMR [SRC] AND NP[CMT]) (Figure 9(a)). Currently, it contains 3443 substances in total, which can be linked to 2900 unique chemical structures in the PubChem Compound database (Figure 9(b)). This (e.g. linking to compound accessions) can be accomplished by using the “Find related data” tool (shown at the bottom right of Figure 9(a)) by specifying the “Database” as “PubChem Compound” and the “Option” as “PubChem Same Compound”, respectively. We want to emphasize that, in order to integrate bioactivity data of the same chemical structures from all depositors, compound accessions should be used when retrieving bioassay data. This set of natural products is very informative and insightful because it is a subset of the Molecular Libraries Small Molecular Repository (MLSMR), a central collection of over 400,000 compounds that are screened by all organizations within the screening center network under the NIH Molecular Libraries Program (MLP) [10] to develop tunable small-molecule chemical tools to unravel the functions of genes and protein targets, and other biological systems. In short words, these natural products have been extensively tested among various types of bioassays. We have found that more than 85% (3003 out of 3443) of these natural products have been experimentally tested in over 2000 bioassays against a total of 589 protein targets as well as other molecular targets involved in a wide range of biological pathways. It is notable that 2313 out of the 3003 tested substances are found active in at least one bioassay. Such rich information about these natural products offers a large and valuable pool of bioactivity data for researchers to investigate related fields of their own interests.
Figure 9.
Search results from the PubChem Substance database using the query “MLSMR[SRC] AND NP[CMT]”. (a) 3443 PubChem Substances; (b) 2900 unique PubChem Compounds from 3443 PubChem Substances.
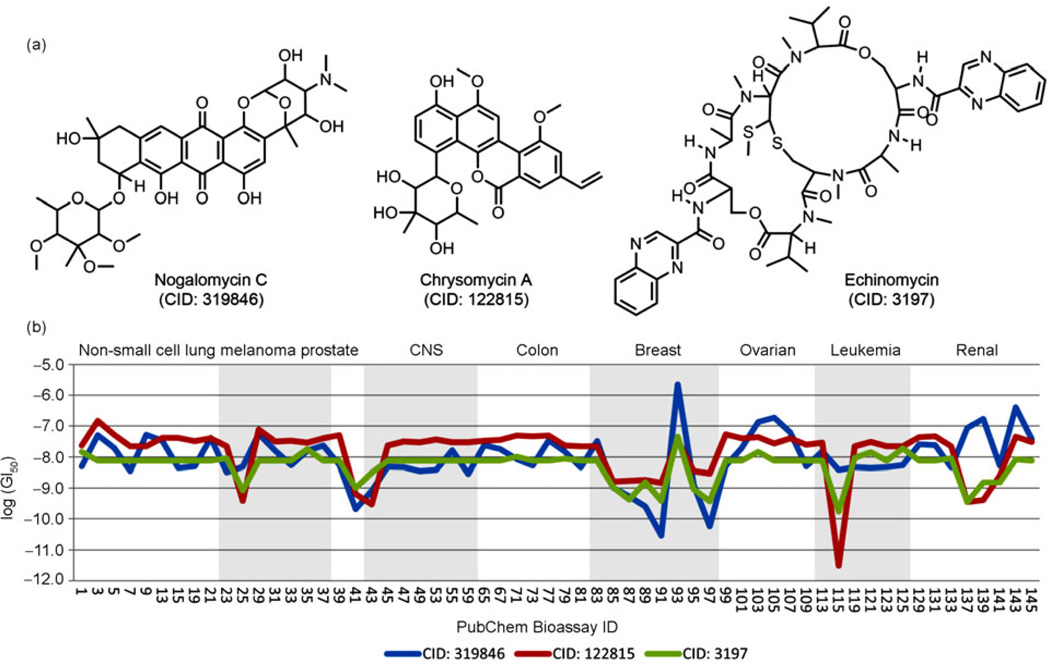
Here, we present a case study to illustrate how one may take advantage of the large-scale bioactivity data deposited in the PubChem database, using three natural products (Nogalomycin C, CID: 319846; Chrysomycin A, CID: 122815 and Echinomycin, CID: 3197). We focused on a subset of bioactivity data deposited by the Developmental Therapeutics Program at the National Cancer Institute of NIH, and generated their bioactivity profiles across the NCI-60 human tumor cell lines employing the approach described in our previous work [15]. The chemical structures of these three natural products and their bioactivity profiles are shown in Figure 10. This analysis shows that these three compounds demonstrate a strong bioactivity for inhibiting Leukemia tumor growth. Furthermore, they share a highly similar bioactivity profiles (Figure 10(b)). Given the significant structural similarity of 0.774 for Nogalomycin C and Chrysomycin A as characterized by Tanimoto coefficient [16] and PubChem fingerprint (ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt), which often indicates strong structure-activity relationships, it may not be surprising to observe such biological profile similarity. On the other hand, Echinomycin is distinct in terms of chemical structure from either Nogaomycin C or Chrysomycin A, with Tanimoto coefficient of merely 0.339 and 0.299, respectively. This is very interesting because it may suggest a novel aspect of the bioactivity profile analysis for scaffold hopping, i.e. searching structurally different compounds with similar biological activities. It is worth noting that, while the NCI-60 human tumor screening bioassays report only growth inhibition data for these compounds but no biological target information is specified, researchers may utilize the PubChem tools described above to identify potential biological targets for these compounds. For example, one may review the bioassay and target data at http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?cid=319846, http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?cid=122815, and http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?cid=3197 respectively. One can also query the three compounds at http://www.ncbi.nlm.nih.gov/pccompound/?term=319846+122815+3197, and then start an analysis by following the “BioActivity Analysis” link. As a step further, one may infer molecular targets for natural products based on the targets known for neighbor compounds, e.g. those that share similar bioactivity profiles. This hypothesis for target prediction has been verified in our previous study [17] and may be particular helpful to natural products based drug discovery and development, where the mechanism of action of an effective natural product is often unclear. It should be emphasized that the generation of bioactivity profile is not limited to the NCI-60 dataset, rather all bioassays in the entire PubChem BioAssay database can be utilized. In fact, there could be many other possibilities for the application of PubChem, such as building predictive models for virtual screening [18], predicting adverse drug reactions [19], constructing BioAssay network [20], developing the topomer Comparative Molecular Field model to guide the design of novel inhibitors [21], visualization of chemical space [22], activity cliffs analysis [23], and many others [24–26]. We expect a wider range of researches to be performed by the community using experimental and computational approaches for further understanding the mechanism of actions of these natural products.
Figure 10.
(a) Chemical structures of three natural products and (b) their bioactivity profiles in the NCI-60 cell lines on nine different organs.
6 Conclusions
Natural products, as potential therapeutic agents, have recently attracted great attention. A large volume of such compounds are deposited in PubChem together with bioactivity data from many screening experiments, which enables researchers not only to retrieve and download chemical structures, but also to use the chemical biology data for target identification and SAR study. In this work, we have shown the typical utilities supported by the NCBI Entrez system and the PubChem information platform for retrieval and data analysis for natural products. In addition, a case study is presented to highlight a subset of natural products in PubChem (as retrieved by http://www.ncbi.nlm.nih.gov/pcsubstance/?term=MLSMR [SRC] AND NP[CMT]) with abundant bioactivity information and illustrate one application of the PubChem resource to investigate the correlation between chemical structures and biological profiles for natural products. The current review may serve as a starting point for the community to further exploit natural products by utilizing the information in PubChem. We anticipate that more researchers will use the PubChem database as a useful resource and tool for drug discovery.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
References
- 1.Liu J, Hu Y, Waller DL, Wang JF, Liu QS. Natural products as kinase inhibitors. Nat Prod Rep. 2012;29:392–403. doi: 10.1039/c2np00097k. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 3.Carlomagno T. NMR in natural products: understanding conformation, configuration and receptor interactions. Nat Prod Rep. 2012;29:536–554. doi: 10.1039/c2np00098a. [DOI] [PubMed] [Google Scholar]
- 4.Shen J, Xu X, Cheng F, Liu H, Luo X, Chen K, Zhao W, Shen X, Jiang H. Virtual screening on natural products for discovering active compounds and target information. Curr Med Chem. 2003;10:2327–2342. doi: 10.2174/0929867033456729. [DOI] [PubMed] [Google Scholar]
- 5.Harvey A. Strategies for discovering drugs from previously unexplored natural products. Drug Discov Today. 2000;5:294–300. doi: 10.1016/s1359-6446(00)01511-7. [DOI] [PubMed] [Google Scholar]
- 6.Koehn FE. Biosynthetic medicinal chemistry of natural product drugs. MedChemComm. 2012;3:854–865. [Google Scholar]
- 7.Calderón AI, Simithy-Williams J, Gupta MP. Antimalarial natural products drug discovery in Panama. Pharm Biol. 2012;50:61–71. doi: 10.3109/13880209.2011.602417. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YL, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang JY, Xiao JW, Zhang J, Bryant SH. An overview of the PubChem bioassay resource. Nucleic Acids Res. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YL, Xiao JW, Suzek TO, Zhang J, Wang JY, Zhou ZG, Han LY, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH. PubChem’s bioassay database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiikkainen P, Franke L. Analysis of commercial and public bioactivity databases. J Chem Inf Model. 2011;52:319–326. doi: 10.1021/ci2003126. [DOI] [PubMed] [Google Scholar]
- 13.Southan C, Várkonyi P, Muresan S. Complementarity between public and commercial databases: New opportunities in medicinal chemistry informatics. Curr Top Med Chem. 2007;7:1502–1508. doi: 10.2174/156802607782194761. [DOI] [PubMed] [Google Scholar]
- 14.Newman DJ, Cragg GM, Snader KM. The influence of natural products upon drug discovery. Nat Prod Rep. 2000;17:215–234. doi: 10.1039/a902202c. [DOI] [PubMed] [Google Scholar]
- 15.Cheng T, Wang Y, Bryant SH. Investigating the correlations among the chemical structures, bioactivity profiles and molecular targets of small molecules. Bioinformatics. 2010;26:2881–2888. doi: 10.1093/bioinformatics/btq550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez JJ. Managing molecular diversity. Chem Soc Rev. 2005;34:143–152. doi: 10.1039/b209064n. [DOI] [PubMed] [Google Scholar]
- 17.Cheng T, Li Q, Wang Y, Bryant SH. Identifying compound-target associations by combining bioactivity profile similarity search and public databases mining. J Chem Inf Model. 2011;51:2440–2448. doi: 10.1021/ci200192v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, Wild DJ. PubChem bioassays as a data source for predictive models. J Mol Graphics Model. 2010;28:420–426. doi: 10.1016/j.jmgm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Pouliot Y, Chiang AP, Butte AJ. Predicting adverse drug reactions using publicly available PubChem bioassay data. Clin Pharmacol Ther. 2011;90:90–99. doi: 10.1038/clpt.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Lushington G, Huan J. The bioassay network and its implications to future therapeutic discovery. BMC Bioinformatics. 2011;12:S1. doi: 10.1186/1471-2105-12-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendt B, Mulbaier M, Wawro S, Schultes C, Alonso J, Janssen B, Lewis J. Toluidinesulfonamide hypoxia-induced factor 1 inhibitors: alleviating drug-drug interactions through use of PubChem data and comparative molecular field analysis guided synthesis. J Med Chem. 2011;54:3982–3986. doi: 10.1021/jm200272h. [DOI] [PubMed] [Google Scholar]
- 22.Awale M, van Deursen R, Reymond J L. MQN-mapplet: Visualization of chemical space with interactive maps of DrugBank, ChEMBL, PubChem, GDB-11, and GDB-13. J Chem Inf Model. 2013;53:509–518. doi: 10.1021/ci300513m. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Maggiora G, Bajorath J. Activity cliffs in PubChem confirmatory bioassays taking inactive compounds into account. J Comput Aided Mol Des. 2013;27:115–124. doi: 10.1007/s10822-012-9632-4. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Wang S, Meng F, Wang J, Zhang Y, Dai E, Yu X, Li X, Jiang W. SM2miR: A database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics. 2013;29:409–411. doi: 10.1093/bioinformatics/bts698. [DOI] [PubMed] [Google Scholar]
- 25.Covell DG. Integrating constitutive gene expression and chemoactivity: Mining the NCI60 anticancer screen. PLoS ONE. 2012;7:e44631. doi: 10.1371/journal.pone.0044631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlich M, Neumann S. MetFusion: Integration of compound identification strategies. J Mass Spectrom. 2013;48:291–298. doi: 10.1002/jms.3123. [DOI] [PubMed] [Google Scholar]