Summary
Background
Current approaches to the detection of colorectal neoplasia associated with inflammatory bowel disease (IBD-CRN) are suboptimal.
Aim
We tested the feasibility of using stool assay of exfoliated DNA markers to detect IBD-CRN.
Methods
This investigation comprised tissue and stool studies. In the tissue study, gene sequencing and methylation assays were performed on candidate genes using tissue DNA from 25 IBD-CRNs and from 25 IBD mucosae without CRN. Mutations on P53, APC, KRAS, BRAF or PIK3CA genes were insufficiently informative, but several aberrantly methylated genes were highly discriminant. In the stool study, we evaluated candidate methylated genes (vimentin, EYA4, BMP3, NDRG4) in a prospective blinded study on buffered stools from 19 cases with known IBD-CRN and 35 age- and sex-matched IBD controls without CRN. From stool-extracted DNA, target genes were assayed by quantitative allele-specific real-time target and signal amplification method.
Results
IBD-CRN cases included 17 with ulcerative colitis (UC) and 2 with Crohn’s disease (CD); 9 had cancer and 10 had dysplasia. Controls included 25 with UC and 10 with CD. Individually, BMP3, vimentin, EYA4, and NDRG4 markers showed high discrimination in stools with respective areas under the ROC curve of 0.91, 0.91, 0.85, and 0.84 for total IBD-CRN and of 0.97, 0.97, 0.95, and 0.85 for cancer. At 89% specificity, the combination of mBMP3 and mNDRG4 detected 9/9 (100%) of CRC and 80% of dysplasia, 4/4 (100%) of high grade and 4/6 (67%) of low grade.
Conclusion
These findings demonstrate feasibility of stool DNA testing for noninvasively detecting IBD-CRN.
Keywords: Stool DNA, inflammatory bowel disease, cancer surveillance, colorectal neoplasms, DNA methylation
Introduction
Patients with inflammatory bowel disease (IBD) are at increased risk of colorectal neoplasia (CRN), including colorectal cancer (CRC).1, 2 Factors known to increase CRC risk in IBD include duration and extent of chronic ulcerative colitis (CUC) or Crohn’s colitis (CD), presence of primary sclerosing cholangitis (PSC), degree of histological activity, and family history of CRC.3–6 To reduce CRC risk, patients with IBD undergo periodic surveillance colonoscopies with multiple random biopsies to detect early visible and occult CRN (dysplasia and cancer).7
Limitations of this colonoscopic surveillance, as currently practiced, include under-sampling with random biopsies, an unknown ideal frequency for performing the surveillance exam, and low grade of evidence for effectiveness.8–10 Some centers use image-enhancing techniques such as chromoendoscopy for surveillance. This has the advantage of identifying more dysplastic lesions by targeted rather than random biopsies,11 but requires special training and sometimes extended endoscopy time. However, regardless of the surveillance technique used, CRN may be missed due to difficulty visualizing neoplastic lesions which are obscured against a background of chronic inflammatory changes.12, 13 Identifying biomarkers that can provide complementary information to colonoscopy could fill an important clinical need in this patient population.
Stool assay of exfoliated molecular markers represents a noninvasive approach that could serve as such an adjunct to colonoscopy.14, 15 While next-generation assay methods have yielded high detection rates for both sporadic CRC and pre-cancer,16–18 stool DNA testing as an approach to neoplasia detection in the IBD population has not been explored.
Tissue-based studies have demonstrated that IBD-CRN is associated with numerous molecular alterations, including acquired mutations in p53,19, 20 APC,21 K-ras,22–24 and BRAF25 as well as aberrant methylation in EYA426, ER, p16, MYOD, p14, E-cadherin, RUNX3, MINT1 and COX-2.27–31. Several other genes, such as BMP3, vimentin (VIM),32 septin 933 and NDRG4,16 are selectively methylated in sporadic CRC but have not been investigated in IBD.
The aims of this investigation were to (1) assess the discriminant value of the mutation markers p53, APC, BRAF, K-ras and PIK3CA and the methylation markers VIM, BMP3, EYA4 and septin 9 for detection of IBD-CRN based on DNA extracted from well-characterized tissue specimens and (2) using the most discriminant tissue markers, prospectively assess the feasibility of stool DNA testing for the detection of premalignant and malignant IBD-CRN.
Methods
Our investigation was approved by the Institutional Review Boards at: Mayo Clinic, Rochester Minnesota, USA; University of Chicago, Chicago Illinois, USA; and Mount Sinai School of Medicine, New York New York, USA.
Tissue Study
Patients
Tissues were identified from a single-center archive of IBD-CRC case and IBD control specimens after confirmation of histologic diagnosis. Cases and controls were matched for age (within a 10-year range), gender, disease duration, anatomic extent (left-sided/extensive) and PSC status (yes/no). DNA was extracted from paraffin-embedded tissues as described.34
Mutation Marker Gene Sequencing
Candidate exons on APC, p53, K-ras, BRAF and PIK3CA were amplified using real-time PCR (see Supplemental Methods).
Real-Time Methylation-Specific PCR (MSP)
After bisulfite treatment, MSP was performed on VIM, BMP3 and septin 9 using Taq polymerase (Invitrogen, Carlsbad, CA) and on EYA4 using SYBR Green master mix (Roche, Mannheim, Germany).
Stool Study
Patients
Case patients with established IBD-CRN were recruited. Those who had undergone endoscopic or surgical treatment of neoplasia or with a history of other neoplasia of the gastrointestinal tract or respiratory system were excluded. Each site recruited IBD control patients undergoing surveillance colonoscopy, matched on age (in 5 year strata) and sex. As anticipated,35 matching on more variables was not possible during prospective enrollment but data was collected on IBD diagnosis (CD/UC/indeterminate), IBD duration, extent of colitis and PSC. IBD activity was assessed by a single expert pathologist, using a previously published protocol.36 After informed consent, participants were given a container and toilet seat mounting bracket kit to collect stools prior to or at least one week after colonoscopy or sigmoidoscopy.37, 38
Sequence-specific gene capture
A 2-gram equivalent of stool supernatant was used for multiplex capture of gene targets (β- actin, VIM, EYA4, BMP3 and NDRG4) by amino conjugated oligonucleotides complementary to target sequences (see Supplemental Methods).39
Assay of Methylated Markers
After capture, target DNA was bisulfite treated and quantitative allele-specific real-time target and signal amplification (QuARTS) reactions were performed on Roche 480 LightCyclers (Indianapolis, IN), as described (see Supplemental Methods).16 EYA4 methylation was assayed by methylation specific PCR, performed on a LightCycler 480 using SYBR Green I Master (Roche) as described.40
Statistical Analysis
Based on a comparison of immunochemical fecal occult blood testing against colonoscopy for the detection of sporadic CRN,41 the feasibility for IBD-CRN detection by stool DNA testing at this initial phase of evaluation was defined a priori as sensitivity for neoplasia >40%. Based on conservative pre-study assumptions, it was estimated that 15 patients in the case group would provide 80% power to distinguish a true sensitivity of 70% from a null value of 40% with a 1-sided one sample proportion test at the 5% level. The distributions of each marker as a continuous variable were compared between cases and controls using the Wilcoxon rank-sum test (JMP v8.0, SAS Institute, Cary NC, USA). Logistic regression was used to calculate receiver operating characteristics (ROC) curves, from which specificity cut-offs were imputed and marker sensitivities (with 95% confidence intervals (CI)) were calculated. To further study the effects of known prognostic factors on marker levels, differences in baseline variables between cases and controls were tested by Chi-square for proportions and Wilcoxon rank-sum for continuous data. When baseline variables were significantly different, case and control marker results were stratified to assess confounding. Univariate and multivariate logistic regression models assessed potential interaction by age, sex and clinical risk factors, including comorbid PSC (yes/no), disease duration (in years) and disease extent (left-sided/extensive). ANOVA was used to assess possible associations between IBD activity (inactive/mild/moderate/severe) and marker levels.
Results
Tissue Study
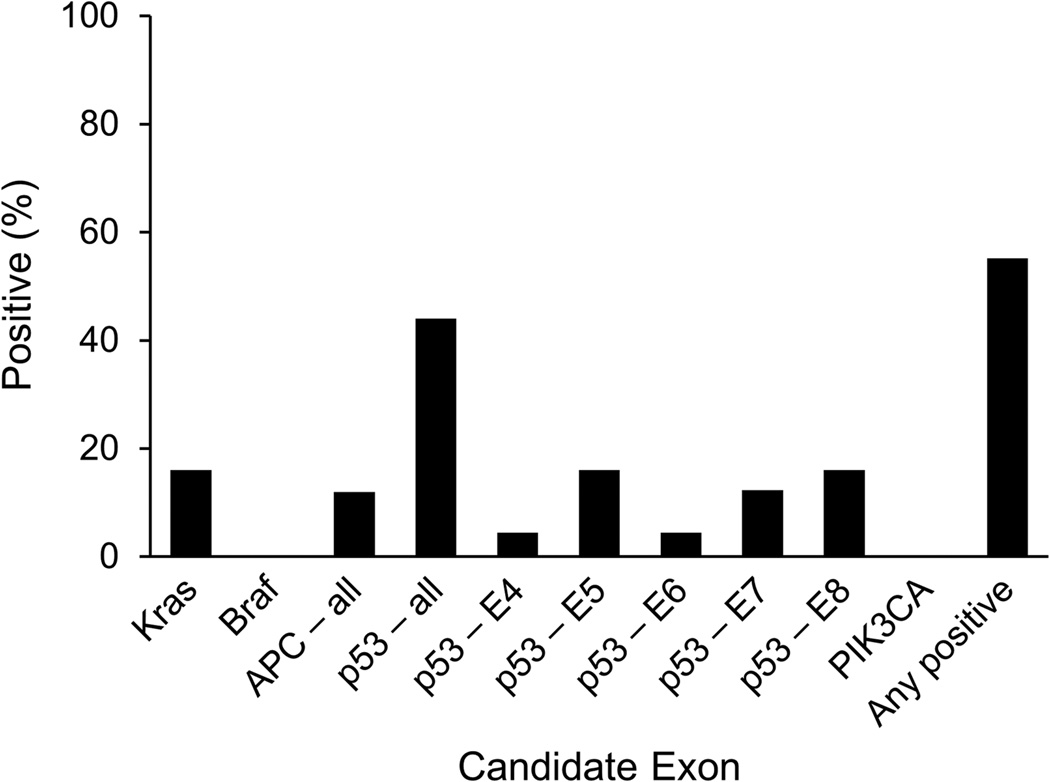
Clinical characteristics were well-matched between cases and controls (Table 1). Figure 1 summarizes the results of DNA sequencing for the case samples. Across 6 APC regions overlapping the mutation cluster region (1, 2, C, N, Y, L2), only 3 mutations were found. Four mutations were found on K-ras. As anticipated, p53 was the most informative marker with 11 mutations detected; however, these were spread out across a wide range of sites on all 5 exons tested. No mutations were identified on BRAF or PIK3CA. While specificity was 100% (no mutations found among control tissues), aggregate sensitivity using all 14 mutation markers combined was only 60%. This is similar to observed rates of DNA mutations assayed from tissues of sporadic CRC and advanced adenomas.42
Table 1.
Patient Characteristics for Tissue Study
| Cases N = 25 |
Controls N = 25 |
|
|---|---|---|
| Male (%) | 16 (64) | 17 (68) |
| Mean age, years (SD) | 52 (14.4) | 50 (11.9) |
| Mean CUC duration, years (SD) | 20.7 (9.2) | 19.9 (8.3) |
| Extensive (%) | 21 (84) | 20 (80) |
| PSC (%) | 4 (16) | 3 (12) |
SD, standard deviation
CUC, chronic ulcerative colitis
PSC, primary sclerosing cholangitis
Cases = Colorectal cancer in CUC, Controls = CUC without neoplasia
Figure 1.
Gene Mutations Detected in Tissue DNA from Inflammatory Bowel Disease Associated Cancers (n=25)
For each of the methylation markers, ROC curves were constructed. Areas under the curve (AUC) were 0.97, 0.87, 0.81 and 0.73 for methylated EYA4 (mEYA4), VIM (mVIM), BMP3 (mBMP3) and Septin 9 respectively. Thus, mEYA4, mVIM and mBMP3 were selected for stool DNA testing. In addition, methylated NDGR4 (mNDRG4) was also selected because of its high discrimination for sporadic CRN in studies performed after the completion of the tissue study.16
Stool Study
Given the high discrimination observed with methylation markers in the tissue study, an analysis of stool from independent sets of cases and controls was performed. Between January 1, 2009 and October 31, 2011, a total of 23 eligible cases and 220 eligible controls were identified and contacted. Nineteen IBD case patients with biopsy-confirmed CRN and 35 IBD control patients without CRN submitted stools (Table 2). Although the proportions of IBD diagnoses and comorbid PSC were not significantly different between the two groups, cases had significantly longer disease duration (p=0.0008) and were significantly more likely to have extensive disease involvement (p=0.01). There was no difference in disease activity between cases and controls (p=0.44).
Table 2.
Patient Characteristics for Stool Study
| Cases N = 19 |
Controls N = 35 |
|
|---|---|---|
| CUC | 17 | 25 |
| Crohn’s disease | 2 | 10 |
| % Male | 63 | 63 |
| Median age, years (range) | 60 (45–72) | 60 (45–77) |
| Median IBD duration, years (range) | 30 (2–50) | 14 (0–45) 1 |
| Extensive (%)2 | 17 (89) | 19 (54)3 |
| IBD Activity4 | ||
| Inactive (%) | 8 (42) | 12 (38) |
| Mild (%) | 9 (45) | 11 (34) |
| Moderate (%) | 1 (5) | 3 (9) |
| Severe (%) | 1 (5) | 6 (19) |
| PSC (%) | 4 (21) | 5 (16) |
p=0.0008
Inflammation proximal to splenic flexure
p=0.01
Disease activity could not be confirmed for 3 control patients
CUC, chronic ulcerative colitis; IBD, inflammatory bowel disease; PSC, primary sclerosing cholangitis.
Cases = IBD with colorectal neoplasia, Controls = IBD without neoplasia
Case neoplasms included 9 cancers with a median size of 2.3 cm (range 0.8 – 5 cm). Six of the 9 (67%) were proximal to the splenic flexure. Median stage43 was I (range I to IIIC). Additional neoplasms included 8 discrete polypoid dysplastic lesions (3 high-grade dysplasia [HGD], 5 low-grade dysplasia [LGD]) with a median size of 2.3 cm (range 1.0 – 6.2) and two flat lesions (1 HGD, 1 LGD) detected on random biopsy (size unknown).
β-actin, a marker of human DNA recovery, amplified in all case and control samples; therefore all patients were included in the analysis. All 4 markers individually showed high discrimination for cancer (Figure 2). AUCs with mBMP3, mVIM, mEYA4 and mNDRG4 were 0.97, 0.97, 0.95 and 0.85, respectively. For IBD-CRN the AUC with mBMP3, mVIM, mEYA4 and mNDRG4 were 0.91, 0.91, 0.85 and 0.84, respectively. For dysplasia, the AUC with mBMP3, mVIM, mEYA4 and mNDRG4 was 0.84, 0.85, 0.75 and 0.77, respectively. Stool assay of mBMP3 alone at 91% specificity was 100% (9/9) sensitive for CRC, 70% (7/10) of dysplasia, and 84% (16/19) sensitive for all CRN (Table 3). At 89% specificity, the combination of mBMP3 and mNDRG4 detected 9/9 (100%) of CRC and 80% of dysplasia, 4/4 (100%) of high grade and 4/6 (67%) of low grade.
Figure 2.
Receiver Operating Characteristics Curve for Detection of Neoplasms by Stool Assay of Methylated Genes. Data plotted for A) BMP3, B) Vimentin, C) EYA4, and D) NDRG4 gene markers. AUC = area under curve; CRC = colorectal cancer.
Table 3.
IBD-Associated Colorectal Neoplasm Detection Rates by Stool Assay of Methylated DNA Markers
| Specificity Cut-off, % | Sensitivity, % (95% CI) |
|||
|---|---|---|---|---|
| mBMP3 | mVIM | mEYA4 | mNDRG4 | |
| CRC1 | ||||
| 94 | 89 (51–99) | 89 (51–99) | 66 (31–91) | 44 (15–77) |
| 91 | 100 (63–100) | 89 (51–99) | 78 (40–96) | 44 (15–77) |
| 89 | 100 (63–100) | 89 (51–99) | 100 (63–100) | 100 (63–100) |
| Neoplasia2 | ||||
| 94 | 68 (43–86) | 68 (43–86) | 53 (29–74) | 37 (17–61) |
| 91 | 84 (60–96) | 68 (43–86) | 63 (39–82) | 37 (17–61) |
| 89 | 84 (60–96) | 68 (43–86) | 74 (48–90) | 74 (48–90) |
| Dysplasia | ||||
| 94 | 50 (20–80) | 50 (20–80) | 40 (14–73) | 30 (8–65) |
| 91 | 70 (35–91) | 50 (20–80) | 50 (20–80) | 30 (8–65) |
| 89 | 70 (35–91) | 50 (20–80) | 50 (20–80) | 50 (20–80) |
CRC = colorectal cancer
Neoplasia = CRC + premalignant dysplasia combined
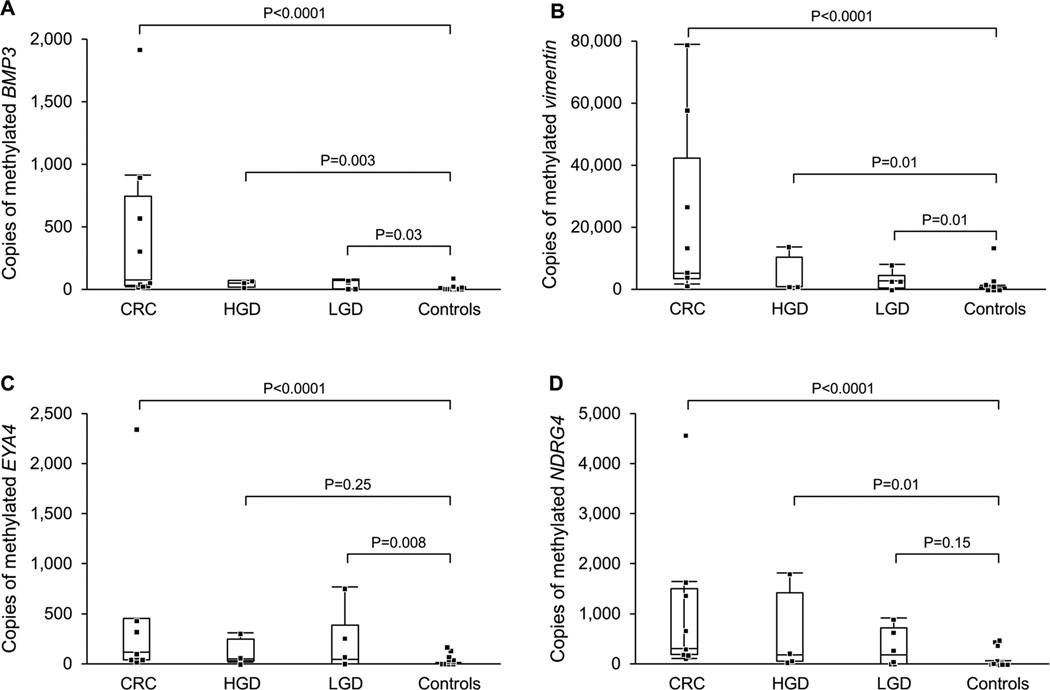
The dynamic range of methylated copy numbers between cases and controls was wide for each stool marker (Figure 3). Among cases, copy numbers of mBMP3, mVIM, mEYA4 or mNDRG4 were not significantly different for proximal versus distal neoplasms (p = 0.58, 0.73, 0.83 and 0.85, respectively). After stratifying for case verus control status, marker levels were not significantly different when comparing patients with CUC and Crohn’s disease.
Figure 3.
Distributions of Methylated Gene Marker Levels in Stools from IBD Cases with Colorectal Neoplasia and from IBD Controls without Neoplasia. Data plotted for A) BMP3, B) Vimentin, C) EYA4, and D) NDRG4 gene markers. CRC = colorectal cancer; HGD = high-grade dysplasia; LGD = low-grade dysplasia.
In multivariate analyses, methylation markers for CRN detection remained significant in models which included age, sex, disease duration, disease extent or the presence of PSC (Supplemental Table 1). ANOVA did not demonstrate any association between markers and disease activity for either cases or controls. Disease duration showed weak correlation with marker levels by univariate linear regression; however, when stratified by case and control status, this association was no longer significant for any of the four methylation markers evaluated. Furthermore, there was no association between anatomic extent of disease (extensive vs. left-sided) and marker levels for either cases or controls.
Discussion
Using DNA methylation markers which were discriminant in tissue, we found that the stool assay achieved high detection rates of both CRC and dysplasia in IBD patients. For example, stool assay of a single informative marker, mBMP3, detected 100% of CRC and 84% of all neoplasms at 91% specificity. Importantly, stool marker levels assayed were unaffected by neoplasm site within the colorectum, as we have observed with sporadic colorectal neoplasia.16 These early results surpassed our predetermined threshold for feasibility.
While corroborative studies are clearly needed, our data suggest the potential usefulness of stool DNA testing to inform the frequency and rigor of colonoscopic surveillance. A non-invasive test which could be performed without bowel cleansing in a patient’s own home might improve compliance with surveillance, which is currently poor, even among high-risk patients.44, 45 Algorithms incorporating stool DNA as a complement to colonoscopy could potentially lengthen the interval between surveillance examinations in marker-negative patients, which could also reduce the high cost of surveillance endoscopy.46 Conversely, a patient with a positive stool DNA test may benefit from colonoscopy at shorter surveillance intervals and/or using enhanced imaging techniques such as chromoendoscopy.
The tissue study based on well-matched cases and controls showed that methylation markers are highly discriminant for IBD-CRN. These tissue findings corroborate our previous observations with mBMP3 and mEYA4.26
Prior studies of methylation markers in IBD-CRN have focused on tumor suppressor genes.27, 29, 30, 47, 48 While BMP3 and NDRG4 are known tumor suppressors in CRC,16, 49, 50 the biology need not be fully understood before a marker is clinically useful. The roles of EYA4 and VIM in IBD-carcinogenesis are unclear, and these genes are aberrantly methylated in other tissues as well.51–53 Studies have also demonstrated that mVIM is a sensitive stool marker for sporadic CRC.54, 55
Our study has several limitations. First, this was a case-control study sized to assess early feasibility of stool DNA testing for detection of IBD-CRN. While sample size was sufficient to meet the central aim, the power to evaluate sub-classes among covariates and marker combinations was limited. Larger studies are needed to achieve greater precision for the sensitivity estimates. Additionally, only 2 endoscopically inapparent (flat) dysplastic lesions were available for analysis, limiting inferences that can be drawn about discrimination for this endpoint. Nevertheless, the high sensitivity achieved by the combination of mBMP3 and mNDRG4 warrants further study, particularly when considering that these two markers have proved complementary for detection of sporadic CRN in studies with large sample sizes.16–18 Methylated BMP3 detected more neoplasms in stool compared to tissue, which could reflect a high prevalence of sporadic-type CRN in IBD patients.56 Second, while cases and controls in the stool study were well-matched on most variables, cases had a longer median duration of IBD and were more likely to have extensive disease. Accordingly, disease duration was further evaluated in stratified comparisons and multivariate models and was not found to significantly influence marker levels. Other parameters of disease severity, including anatomic extent of inflammation, degree of inflammation, and presence of concomitant PSC, also had no effect on stool marker levels. Last, conventional colonoscopy with non-targeted biopsies was used as the criterion standard, and this approach lacks sensitivity for CRN as currently practiced for IBD surveillance.12, 13 Control patients who tested positive for methylation markers might therefore have had falsely negative colonoscopies, which would have affected specificity estimates. Underscoring the imperfect nature of colonoscopy, two of the 9 CRC cases with positive stool results in this study were missed on colonoscopy and diagnosed only after colectomy.
These early results in tissue and stool represent an important first step in the evaluation of stool DNA as a noninvasive tool for detection of CRN in IBD patients. Further studies are needed to corroborate and expand these novel findings. Particularly, prospective cohort studies conducted in the IBD surveillance setting will help determine how this noninvasive tool might improve colonoscopy yield and patient outcomes, and potentially lower healthcare costs.
Supplementary Material
Acknowledgements
Funding was provided by a grant from the Charles Oswald Foundation. Dr. Kisiel was supported by the Maxine and Jack Zarrow Family Foundation of Tulsa Oklahoma. ALL authors approved the final version of the article, including the authorship list.
Footnotes
Authorship Statement:
John B. Kisiel, MD (Guarantor of the article, Study design, data acquisition, analysis, drafting manuscript)
Tracy C. Yab (Data acquisition, analysis of data)
Fareeda Taher Nazer Hussain (Data acquisition, analysis of data)
William R. Taylor (Data acquisition, analysis of data)
Megan M. Garrity-Park (Study design, data acquisition, analysis of data)
William J. Sandborn, MD (Critical revision of manuscript, conceptual input)
Edward V. Loftus, Jr., MD (Critical revision of manuscript, conceptual input)
Bruce G. Wolff, MD (Data acquisition, critical revision of manuscript, conceptual input)
Thomas C. Smyrk, MD (Pathology interpretation, data acquisition, critical revision of manuscript)
Steven H. Itzkowitz, MD (Conceptual input, study supervision, manuscript review)
David T. Rubin, MD (Conceptual input, study supervision, manuscript review)
Hongzhi Zou, MD, PhD (Assay design and data analysis)
Douglas W. Mahoney (Statistical analysis)
David A. Ahlquist, MD (Study conceptual design, data analysis, drafting and editing of manuscript, obtained funding, study supervision)
Disclosures:
Mayo Clinic is a minor equity investor in and has licensed intellectual property to Exact Sciences. Consistent with Mayo Clinic policy, Drs. Kisiel and Ahlquist, Mr. Taylor, and Ms. Yab could share in potential future equity or royalties. Dr. Itzkowitz is on the Scientific Advisory Board of Exact Sciences Corporation.
REFERENCES
- 1.Jess T, Loftus EV, Jr, Velayos FS, et al. Risk of intestinal cancer in inflammatory bowel disease: a population-based study from olmsted county, Minnesota. Gastroenterology. 2006;130(4):1039–1046. doi: 10.1053/j.gastro.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 2.Howe HL, Wu X, Ries LA, et al. Annual report to the nation on the status of cancer, 1975–2003, featuring cancer among U.S. Hispanic/Latino populations. Cancer. 2006;107(8):1711–1742. doi: 10.1002/cncr.22193. [DOI] [PubMed] [Google Scholar]
- 3.Itzkowitz SH, Harpaz N. Diagnosis and management of dysplasia in patients with inflammatory bowel diseases. Gastroenterology. 2004;126(6):1634–1648. doi: 10.1053/j.gastro.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 4.Cairns SR, Scholefield JH, Steele RJ, et al. Guidelines for colorectal cancer screening and surveillance in moderate and high risk groups. Gut. 2010;59(5):666–689. doi: 10.1136/gut.2009.179804. (update from 2002). [DOI] [PubMed] [Google Scholar]
- 5.Colonoscopic Surveillance for Prevention of Colorectal Cancer in People with Ulcerative Colitis, Crohn's Disease or Adenomas. London: National Institute for Health and Clinical Excellence (UK); 2011. [PubMed] [Google Scholar]
- 6.Nuako KW, Ahlquist DA, Mahoney DW, Schaid DJ, Siems DM, Lindor NM. Familial predisposition for colorectal cancer in chronic ulcerative colitis: a case-control study. Gastroenterology. 1998;115(5):1079–1083. doi: 10.1016/s0016-5085(98)70077-0. [DOI] [PubMed] [Google Scholar]
- 7.Farraye FA, Odze RD, Eaden J, Itzkowitz SH. AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology. 2010;138(2):746–774. 774 e1–774 e4. doi: 10.1053/j.gastro.2009.12.035. quiz e12-3. [DOI] [PubMed] [Google Scholar]
- 8.Karlen P, Kornfeld D, Brostrom O, Lofberg R, Persson PG, Ekbom A. Is colonoscopic surveillance reducing colorectal cancer mortality in ulcerative colitis? A population based case control study. Gut. 1998;42(5):711–714. doi: 10.1136/gut.42.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loftus EV., Jr Does monitoring prevent cancer in inflammatory bowel disease? J Clin Gastroenterol. 2003;36(5 Suppl):S79–S83. doi: 10.1097/00004836-200305001-00014. discussion S94-6. [DOI] [PubMed] [Google Scholar]
- 10.Collins PD, Mpofu C, Watson AJ, Rhodes JM. Strategies for detecting colon cancer and/or dysplasia in patients with inflammatory bowel disease. Cochrane database of systematic reviews (Online) 2006;(2) doi: 10.1002/14651858.CD000279.pub3. CD000279. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian V, Mannath J, Ragunath K, Hawkey CJ. Meta-analysis: the diagnostic yield of chromoendoscopy for detecting dysplasia in patients with colonic inflammatory bowel disease. Alimentary Pharmacology & Therapeutics. 2011;33(3):304–312. doi: 10.1111/j.1365-2036.2010.04525.x. [DOI] [PubMed] [Google Scholar]
- 12.Connell WR, Lennard-Jones JE, Williams CB, Talbot IC, Price AB, Wilkinson KH. Factors affecting the outcome of endoscopic surveillance for cancer in ulcerative colitis. Gastroenterology. 1994;107(4):934–944. doi: 10.1016/0016-5085(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 13.Lim CH, Dixon MF, Vail A, Forman D, Lynch DA, Axon AT. Ten year follow up of ulcerative colitis patients with and without low grade dysplasia. Gut. 2003;52(8):1127–1132. doi: 10.1136/gut.52.8.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351(26):2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 15.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128(1):192–206. doi: 10.1053/j.gastro.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 16.Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142(2):248–256. doi: 10.1053/j.gastro.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlquist DA, Taylor WR, Mahoney DW, et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2012;10(3):272–277. doi: 10.1016/j.cgh.2011.10.008. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lidgard GP, Domanico M, Bruinsma JJ, et al. An Optimized Multi-Marker Stool Test for Colorectal Cancer Screening: Initial Clinical Appraisal. Gastroenterology. 2012;142(5) Suppl 1:S–770. [Google Scholar]
- 19.Taylor HW, Boyle M, Smith SC, Bustin S, Williams NS. Expression of p53 in colorectal cancer and dysplasia complicating ulcerative colitis. The British journal of surgery. 1993;80(4):442–444. doi: 10.1002/bjs.1800800411. [DOI] [PubMed] [Google Scholar]
- 20.Lashner BA, Shapiro BD, Husain A, Goldblum JR. Evaluation of the usefulness of testing for p53 mutations in colorectal cancer surveillance for ulcerative colitis. Am J Gastroenterol. 1999;94(2):456–462. doi: 10.1111/j.1572-0241.1999.877_f.x. [DOI] [PubMed] [Google Scholar]
- 21.Odze RD, Brown CA, Hartmann CJ, Noffsinger AE, Fogt F. Genetic alterations in chronic ulcerative colitis-associated adenoma-like DALMs are similar to non-colitic sporadic adenomas. The American journal of surgical pathology. 2000;24(9):1209–1216. doi: 10.1097/00000478-200009000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Bell SM, Kelly SA, Hoyle JA, et al. c-Ki-ras gene mutations in dysplasia and carcinomas complicating ulcerative colitis. British journal of cancer. 1991;64(1):174–178. doi: 10.1038/bjc.1991.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holzmann K, Klump B, Borchard F, et al. Comparative analysis of histology, DNA content, p53 and Ki-ras mutations in colectomy specimens with long-standing ulcerative colitis. Int J Cancer. 1998;76(1):1–6. doi: 10.1002/(sici)1097-0215(19980330)76:1<1::aid-ijc1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Hirota Y, Tanaka S, Haruma K, et al. pS2 expression as a possible diagnostic marker of colorectal carcinoma in ulcerative colitis. Oncol Rep. 2000;7(2):233–239. [PubMed] [Google Scholar]
- 25.Aust DE, Haase M, Dobryden L, et al. Mutations of the BRAF gene in ulcerative colitis-related colorectal carcinoma. Int J Cancer. 2005;115(5):673–677. doi: 10.1002/ijc.20925. [DOI] [PubMed] [Google Scholar]
- 26.Osborn NK, Zou H, Molina JR, et al. Aberrant methylation of the eyes absent 4 gene in ulcerative colitis-associated dysplasia. Clin Gastroenterol Hepatol. 2006;4(2):212–218. doi: 10.1016/j.cgh.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Issa J-PJ, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated Age-related CpG Island Methylation in Ulcerative Colitis. Cancer research. 2001;61(9):3573–3577. [PubMed] [Google Scholar]
- 28.Sato F, Shibata D, Harpaz N, et al. Aberrant methylation of the HPP1 gene in ulcerative colitis-associated colorectal carcinoma. Cancer research. 2002;62(23):6820–6822. [PubMed] [Google Scholar]
- 29.Wheeler JM, Kim HC, Efstathiou JA, Ilyas M, Mortensen NJ, Bodmer WF. Hypermethylation of the promoter region of the E-cadherin gene (CDH1) in sporadic and ulcerative colitis associated colorectal cancer. Gut. 2001;48(3):367–371. doi: 10.1136/gut.48.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrity-Park MM, Loftus EV, Jr, Sandborn WJ, Bryant SC, Smyrk TC. Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am J Gastroenterol. 2010;105(7):1610–1619. doi: 10.1038/ajg.2010.22. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T, Kobunai T, Ikeuchi H, et al. RUNX3 copy number predicts the development of UC-associated colorectal cancer. International journal of oncology. 2011;38(1):201–207. [PubMed] [Google Scholar]
- 32.Zou H, Harrington JJ, Shire AM, et al. Highly methylated genes in colorectal neoplasia: implications for screening. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2686–2696. doi: 10.1158/1055-9965.EPI-07-0518. [DOI] [PubMed] [Google Scholar]
- 33.Grutzmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS ONE. 2008;3(11):e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrity-Park MM, Loftus EV, Jr, Bryant SC, Sandborn WJ, Smyrk TC. Tumor necrosis factor-alpha polymorphisms in ulcerative colitis-associated colorectal cancer. Am J Gastroenterol. 2008;103(2):407–415. doi: 10.1111/j.1572-0241.2007.01572.x. [DOI] [PubMed] [Google Scholar]
- 35.Fletcher RH, Fletcher SW. Clinical Epidemiology. 4 ed. Lippincott Williams & Wilkins; 2005. [Google Scholar]
- 36.Gupta RB, Harpaz N, Itzkowitz S, et al. Histologic inflammation is a risk factor for progression to colorectal neoplasia in ulcerative colitis: a cohort study. Gastroenterology. 2007;133(4):1099–1105. doi: 10.1053/j.gastro.2007.08.001. quiz 1340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou H, Harrington JJ, Klatt KK, Ahlquist DA. A sensitive method to quantify human long DNA in stool: relevance to colorectal cancer screening. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1115–1119. doi: 10.1158/1055-9965.EPI-05-0992. [DOI] [PubMed] [Google Scholar]
- 38.Olson J, Whitney DH, Durkee K, Shuber AP. DNA stabilization is critical for maximizing performance of fecal DNA-based colorectal cancer tests. Diagn Mol Pathol. 2005;14(3):183–191. doi: 10.1097/01.pas.0000176768.18423.7e. [DOI] [PubMed] [Google Scholar]
- 39.Kisiel JB, Yab TC, Taylor WR, et al. Stool DNA testing for the detection of pancreatic cancer: Assessment of methylation marker candidates. Cancer. 2011 doi: 10.1002/cncr.26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kisiel JB, Yab TC, Taylor WR, et al. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer. 2012;118(10):2623–2631. doi: 10.1002/cncr.26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129(2):422–428. doi: 10.1016/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 42.Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Annals of internal medicine. 2008;149(7):441–450. doi: 10.7326/0003-4819-149-7-200810070-00004. W81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edge SBB DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th ed. New York: Springer; 2010. [Google Scholar]
- 44.Velayos FS, Liu L, Lewis JD, et al. Prevalence of colorectal cancer surveillance for ulcerative colitis in an integrated health care delivery system. Gastroenterology. 2010;139(5):1511–1518. doi: 10.1053/j.gastro.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 45.Vienne A, Simon T, Cosnes J, et al. Low prevalence of colonoscopic surveillance of inflammatory bowel disease patients with longstanding extensive colitis: a clinical practice survey nested in the CESAME cohort. Alimentary Pharmacology & Therapeutics. 2011;34(2):188–195. doi: 10.1111/j.1365-2036.2011.04711.x. [DOI] [PubMed] [Google Scholar]
- 46.Rubenstein JH, Waljee AK, Jeter JM, Velayos FS, Ladabaum U, Higgins PD. Cost effectiveness of ulcerative colitis surveillance in the setting of 5-aminosalicylates. The American journal of gastroenterology. 2009;104(9):2222–2232. doi: 10.1038/ajg.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato F, Harpaz N, Shibata D, et al. Hypermethylation of the p14(ARF) gene in ulcerative colitis-associated colorectal carcinogenesis. Cancer research. 2002;62(4):1148–1151. [PubMed] [Google Scholar]
- 48.Moriyama T, Matsumoto T, Nakamura S, et al. Hypermethylation of p14 (ARF) may be predictive of colitic cancer in patients with ulcerative colitis. Diseases of the colon and rectum. 2007;50(9):1384–1392. doi: 10.1007/10350-007-0302-x. [DOI] [PubMed] [Google Scholar]
- 49.Loh K, Chia JA, Greco S, et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer. 2008;47(6):449–460. doi: 10.1002/gcc.20552. [DOI] [PubMed] [Google Scholar]
- 50.Melotte V, Lentjes MH, van den Bosch SM, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. Journal of the National Cancer Institute. 2009;101(13):916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- 51.Zou H, Osborn NK, Harrington JJ, et al. Frequent methylation of eyes absent 4 gene in Barrett's esophagus and esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2005;14(4):830–834. doi: 10.1158/1055-9965.EPI-04-0506. [DOI] [PubMed] [Google Scholar]
- 52.Yang Wu DJ, Yab TC, Taylor WR, et al. Aberrant Gene Methylation in the Neoplastic Progression of Barrett's Esophagus: Identification of Candidate Diagnostic Markers. Gastroenterology. 2011;140(5) Supplement 1:S-222. [Google Scholar]
- 53.Moinova H, Leidner RS, Ravi L, et al. Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21(4):594–600. doi: 10.1158/1055-9965.EPI-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itzkowitz SH, Jandorf L, Brand R, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5(1):111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 55.Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol. 2008;103(11):2862–2870. doi: 10.1111/j.1572-0241.2008.02088.x. [DOI] [PubMed] [Google Scholar]
- 56.Odze RD, Farraye FA, Hecht JL, Hornick JL. Long-term follow-up after polypectomy treatment for adenoma-like dysplastic lesions in ulcerative colitis. Clin Gastroenterol Hepatol. 2004;2(7):534–541. doi: 10.1016/s1542-3565(04)00237-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.