Abstract
Human cytomegalovirus (CMV) is responsible for approximately 40,000 congenital infections in the United States each year. Congenital CMV disease frequently produces serious neurodevelopmental disability, as well as vision impairment and sensorineural hearing loss. Development of a CMV vaccine is therefore considered to be a major public health priority. The mechanisms by which CMV injures the fetus are complex and likely include a combination of direct fetal injury induced by pathologic virally-encoded gene products, an inability of the maternal immune response to control infection, and the direct impact of infection on placental function. CMV encodes gene products that function, both at the RNA and the protein level, to interfere with many cellular processes. These include gene products that modify the cell cycle; interfere with apoptosis; induce an inflammatory response; mediate vascular injury; induce site-specific breakage of chromosomes; promote oncogenesis; dysregulate cellular proliferation; and facilitate evasion of host immune responses. This minireview summarizes current concepts regarding these aspects of the molecular virology of CMV and the potential pathogenic impact of viral gene expression on the developing fetus. Areas for potential development of novel therapeutic intervention are suggested for improving the outcome of this disabling congenital infection.
Keywords: Cytomegalovirus, congenital CMV infection, immune evasion, pathogenesis, vaccine
INTRODUCTION
Cytomegalovirus (CMV) infections are ubiquitous in humans and usually asymptomatic in the immunocompetent host [1, 2]. However, some populations are at risk for serious CMV-associated disease. The medical importance of CMV is most significant in immunocompromised hematopoietic stem cell and solid organ transplant patients; individuals with human immunodeficiency virus (HIV) infection; and pregnant women [3-5]. Of particular concern is the problem of congenital CMV infection. CMV is the most common congenital viral infection in the developed world, occurring in to 0.4-2.3% of all newborns, depending upon the population studied [6]. The overall birth prevalence of congenital CMV infection is estimated to be 0.64% but varies considerably among different study populations. Factors associated with an increased risk of congenital CMV infection include non-white race, low socioeconomic status, premature birth, and the maternal CMV seroprevalence rate in the particular population under study, with higher seroprevalence rates being associated with increased incidence of congenital infection [7]. The public health impact of congenital CMV infection is striking; more children suffer from long-term sequelae as a result of congenital CMV infection than either trisomy 21 or fetal alcohol syndrome [8]. Sequelae of congenital CMV infection are particularly severe for the developing central nervous system (CNS) and include mental retardation, cerebral palsy, neurodevelopmental disorders, and sensorineural hearing loss (SNHL) [9]. Symptomatic and disabling congenital CMV can follow both primary and recurrent maternal infection [10-12] but it is generally believed that primary CMV infections are transmitted more frequently to the fetus, and are more likely to cause fetal injury, than are recurrent infections [13, 14]. This observation has helped to drive the effort to develop a CMV vaccine that might, if effective in conferring robust immunity to women of childbearing age, be able to interrupt maternal-fetal transmission and the attendant sequelae of resulting infection [15].
The pathogenesis of fetal injury in the setting of congenital CMV infection is poorly understood. Pathology associated with acute CMV infection has been attributed to lytic virus replication, with ultimate end-organ damage occurring either secondary to virus-mediated cell death, or from host inflammatory responses targeting virus-infected cells [16]. Factors that contribute to fetal injury include the timing of infection relative to the gestational age of the fetus [17]; the maternal immune status [13]; the extent of CMV-induced placental injury [18]; the magnitude of the viral load in the amniotic fluid [19]; the induction of host genes (fetal and/or placental) that occurs in response to infection and the pathogenic effects these gene responses may mediate [20]; and, possibly, the genotype of the particular strain(s) of CMV infecting the fetus [21]. In recent years, considerable effort has been expended to explore potential mechanisms by which specific CMV gene products may mediate pathogenesis. This improved understanding of the molecular and cellular biology of CMV infection may, in turn, yield novel approaches for future therapies. In this minireview, the molecular virology of CMV is reviewed, with an emphasis on the current level of understanding of potential virally-mediated molecular mechanisms of injury at the cellular level. CMV-encoded genes that perturb the cell cycle, modify apoptosis, modulate the immune response, and interact with cellular function are discussed. Future prospects for targeted interventions, particularly those that might protect the fetus, are suggested.
CMV: BASIC VIROLOGY
CMV is a member of the Herpesviridae family of viruses. It is one of the 8 human herpesviruses (HHV) [22]. All of these viruses are double-stranded, DNA viruses; all are capable of inducing chronic or latent disease in the infected host; and all share similarities in virion morphology and genome organization. CMV, designated as HHV-5, is a member of the betaherpesvirus subfamily of the Herpesviridae; this subfamily also includes the roseolaviruses, HHV-6 and HHV-7, which are responsible for the clinical syndrome of exanthem subitum (roseola infantum) in young children [22].
Virion Morphology and Composition
CMV, like other Herpesviridae, is an enveloped virus with a double-stranded DNA genome. The three distinct regions of the CMV virus particle include: an icosahedral capsid; the tegument layer; and an outer lipid envelope. The morphology of CMV is demonstrated in the electron microscopy (EM) studies shown in Fig. (1). The capsid, which comprises 162 capsomere subunits arranged in an icosahedral symmetry, houses the viral genome, and is classically highly electron-dense when imaged by EM [23]. In the virus particle, the capsid is surrounded by the tegument, a protein-rich layer containing several of the dominant targets of the T-lymphocyte response to infection, including a 65-kilodalton (kDa) phosphoprotein (pp) referred to as pp65 [24, 25]. Surrounding the tegument is the envelope layer. The envelope contains several virally-encoded glycoproteins (g), including protein complexes designated as the gB complex, the gM/gN complex, and the gH/gL/gO complex. CMV-seropositive individuals mount an immune response characterized by neutralizing antibodies that target these glycoproteins; hence, recombinant forms of these proteins have emerged as potential candidates for subunit vaccine development [15, 26-28].
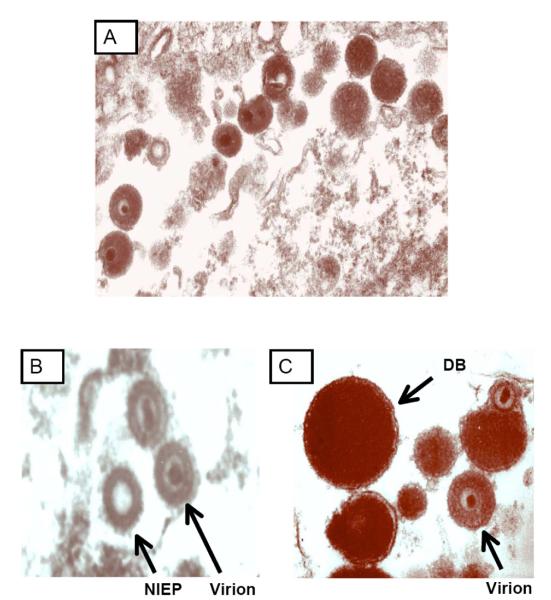
Fig. (1).
Electron microscopic visualization of morphology of cytomegalovirus particles. Human foreskin fibroblasts were inoculated with the Toledo strain of cytomegalovirus and, following 7 days of incubation, cells were fixed with paraformaldehyde, stained with phosphotungstic acid, and visualized by EM (University of Minnesota). A) Image demonstrating cell-associated virus particles [virions and dense bodies (DB)]. B) Comparison of mature virion and non-infectious enveloped particle (NIEP, arrow), which contains capsid proteins but no DNA and is hence replication-defective. C) Comparison of virions and DB (arrow). In contrast to virions, DB are larger, contain no viral DNA and no capside proteins, and are non-infectious.
In addition to serving as targets of the humoral immune response, these glycoproteins also play a central role in the binding and entry of CMV into cells. CMV employs at least two distinct, cell-type specific mechanisms of cell entry. Entry of CMV into endothelial and epithelial cells is mediated by endocytosis in a pH-dependent fashion; in contrast, entry into fibroblasts is non-endocytic, and pH-independent [29]. CMV fibroblast entry is believed to be initiated by binding of virion-associated gB to an as-yet incompletely characterized cell surface receptor, followed by fusion with the cell membrane in a process that requires a complex of three other glycoproteins: gH, gL, and gO [29]. In contrast to the model of CMV entry for fibroblasts, endocytic entry of CMV into endothelial and epithelial cells requires a complex of gH, gL, and three other proteins encoded by a region of the CMV genome referred to as the UL128-131 gene locus [30], a region of the genome discussed in further detail in the next section of this review. Emerging insights into the nature of these protein/cell surface interactions has resulted in development of new therapeutic strategies for prevention of CMV infection, based on vaccines and immunotherapies that could, in principle, block cell entry, particularly at epithelial surfaces where primary infections are typically acquired.
During the process of viral replication, a variety of types of CMV particles are generated. In cell culture, CMV infection leads to the assembly and release of, in addition to infectious virions, noninfectious defective particles termed “dense bodies” (DB), so designated because of their characteristic highly electron-dense appearance when imaged by EM. Another type of body, designated as a “noninfectious enveloped particle” (NIEP), is also generated during viral replication Fig. (1) [31, 32]. The structure and protein composition of NIEPs are comparable to those of virions, but they lack DNA and are therefore not infectious [31]. DBs are enveloped spherical structures that lack capsid proteins and DNA [32]. They consist mainly of viral tegument proteins and glycoproteins. In cell culture, the biology of DBs mimics that of infectious virus, since DBs enter cells efficiently and deliver their protein components intracellularly. Purified DBs are highly immunogenic when used to immunize animals, and sera from seropositive individuals are immunoreactive with these particles. For these reasons, DBs have been proposed as a novel CMV vaccine candidate [33]. In principle, DBs could induce a broad range of humoral and cellular immune responses to CMV tegument and glycoproteins, but, in contrast to live virus vaccines, would theoretically demonstrate a more favorable safety profile, given their lack of DNA and inability to replicate.
Recent work has demonstrated that CMV particles exhibit additional levels of complexity extending beyond the structural elements of the nucleocapsid, tegument, and envelope. Using CMV gene array technology, a novel class of viral RNA transcripts, termed virion RNAs, has recently been identified in infectious virions [34, 35]. These RNAs, which are packaged during virion assembly, are delivered to the host cell immediately on infection, potentially allowing viral gene products to be expressed in an infected cell before any viral transcription or host immune response occurs. The role of virion RNAs in pathogenesis is unknown at this time, but these transcripts may represent novel targets for therapeutics, such as anti-sense RNAs.
The CMV virion was initially characterized by analyses that were focused on identification of protein products that were, in turn, predicted by the nucleotide sequence of the virus. However, the advent of proteomic technologies has facilitated a more comprehensive analysis of the proteins present in CMV virions and DBs, and has yielded some surprising findings. A proteomic analysis of CMV virions using mass spectrometry was recently undertaken [36]. This study facilitated identification of viral and cellular proteins present in purified viral particles and dense bodies and allowed determination of the stoichiometry of the various protein species. A total of 71 CMV-encoded proteins and, intriguingly, over 70 host proteins were identified; interestingly, host proteins included cellular structural proteins, enzymes, and signal transduction molecules. DBs were found to contain only 29 viral proteins and reduced quantities of cellular proteins were present, compared to virions. These results underscore the complex interplay that exists between CMV and the cell it infects. More evidence for this complexity has been noted in recent spatial-temporal proteomic analyses that identified multiple cellular trafficking tracts involved in virion maturation, requiring host cell binding partners in the endosomal sorting complex and clathrin pathways [37, 38]. Considerable future work remains to characterize the virion proteome, to elucidate the process of virion assembly, and to define the relationship of the viral proteome to pathogenesis. Recently, an elegant analysis of protein-protein interactions contributing to CMV virion assembly was performed using yeast two-hybrid analysis. Over 1000 pairwise combinations for binary interaction were assessed in the two-hybrid assay, and multiple interactions involving tegument and capsid proteins were identified [39]. Continued examination of these protein-protein interactions should provide insights into the mechanisms of virion assembly and maturation, which in turn may yield novel intervention strategies.
CMV: VIRAL GENOME AND MOLECULAR GENETICS
Genome Structure, Organization, and Replication
CMV has the largest genome of any human viral pathogen. The CMV genome consists of approximately 235 kilobase pairs (kbp) of DNA, existing in a complex conformation [40]. The genome is organized as two regions of unique sequences, the unique long (UL) and unique short (US) areas, flanked by two sets of terminal and internal inverted repeat sequences (TRL/IRL) and (IRS/TRS) Fig. (2). The presence of these two unique regions bracketed by repeats allows generation of four potential isomeric conformations for a single unit length genome, depending upon the relative positioning of the unique segments during genome packaging into virions. This feature of CMV genome structure, in turn, likely contributes to the generation of genetic diversity among progeny virions following DNA replication.
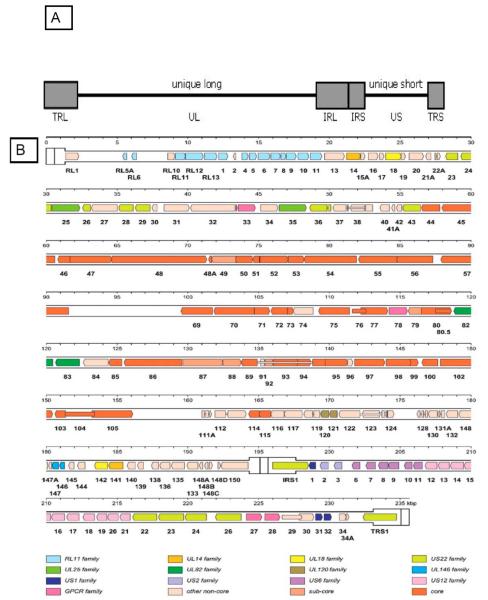
Fig. (2).
Schematic of CMV Genome. A) Structure of the CMV genome. The CMV genome is organized as two regions of unique sequences, unique long (UL) and unique short (US), flanked by two sets of inverted repeats (TRL/IRL) and (IRS/TRS) (light shaded boxes). UL - Unique long; US - Unique short; TRL - Terminal repeat long; IRL - Internal repeat long; inverted repeat of TRL; TRS -Terminal repeat short; IRS - Internal repeat short; inverted repeat of TRS. This genome structure allows one of four potential isomeric configurations to be represented in any given infectious virus particle. Figure originally published in [41] and used with permission. B) Genetic map of wild-type CMV. Related groupings of protein-encoding regions are indicated by colored arrows, with gene family nomenclature indicated below. Many CMV genes are spliced and these introns are indicated as narrow white bars. Colors also delineate whether genes are conserved in all Herpesviridae (core genes) or between the Betaherpesviridae (CMV, HHV6, HHV7) and Gammaherpesviridae (Epstein-Barr virus, HHV-8; sub-core genes). A group of 22 ORFs (ORFs ~133-150 in the UL region of the genome) are prone to rapid deletion/mutation/rearrangement when CMV is subjected to laboratory passage in fibroblast cell culture: this represents the so-called ULb’ region of the genome (see text for details). Figure reproduced from [42] and used with permission, Copyright 2000, National Academy of Sciences, U.S.A.
By convention, CMV genes are designated numerically, left-to-right, based upon the sequence of open reading frames (ORFs) identified in the UL and US genome segments. Viral gene expression is complex and highly regulated through interactions between virally-encoded proteins and host cellular factors [40]. The replication cycle of CMV is divided temporally into three regulated phases: the immediate early (IE) phase, the early phase, and the late phase. IE messenger RNA (mRNA) is transcribed within the first few hours after infection of the cell and these transcripts encode IE proteins that modulate both host and viral gene expression, including transactivators of transcription of other viral gene promoters. Early gene products include DNA replication proteins and other structural proteins. Late gene products are typically transcribed at ~72 hours post-infection and include structural proteins and other proteins involved in virion assembly, structure, and egress.
Originally, the designation of CMV ORFs was based on the DNA sequence analysis of a highly laboratory-adapted strain of CMV, the AD169 strain [42]. One of the most important advances in the study of CMV genetics came with the discovery that laboratory-adapted strains of CMV, such as AD169 and another lab-adapted strain, the Towne strain, undergo significant mutations, deletions, and rearrangements during the process of tissue culture adaptation [41]. In recent years, detailed DNA sequence comparisons of the Towne, AD169, and a less-attenuated strain of CMV, the Toledo strain, have been performed. The Toledo genome was discovered to contain a DNA segment of approximately 13 kbp that was not found in the Towne genome and a segment of approximately 15 kbp that was not found in the AD169 genome. These novel sequences were located at the unique long (UL)/internal repeat (IRL) boundary, within the L component of the viral genome, and contained multiple potential ORFs that were not present in AD169. This region of the genome was designated the ULb’ region. A total of 22 previously-unrecognized ORFs (denoted UL133 to UL154) were identified in this “wild-type” region of the CMV genome Fig. (2b). These observations suggested that the true number of genes carried by wild-type CMV was much higher than those originally identified by the sequencing of laboratory-adapted strains and that genes had been overlooked because of the large-scale deletion and rearrangement of the ULb’ region that occurs during laboratory passage of virus isolates. The potential selective advantage driving the deletion of these genes during passage of virus in fibroblast cells is unknown. Subsequent studies have compared the patterns of genomic deletions and rearrangements present in the AD169 and Towne strains of CMV [44] and carefully analyzed the genomes of unpassaged clinical isolates. These studies have suggested that the true number of genes encoded by CMV is 165-173 [45-48]. The continued study of the molecular biology of low-passage “clinical isolates” of CMV is a high-priority area for future work. Increasingly, it is becoming clear that there are not only extensive sequence variation amongst CMV genomes, but also great functional variation in genome structure. These variations include genome size, inversion, orientation, and coding potential, and occur even within the most highly conserved genes. Some of the factors contributing to this divergence include nucleotide polymorphism, DNA strand composition asymmetry, variation in codon usage, and variability in the rate of evolution in conserved genes from strain to strain [49, 50]. The application of techniques that allow DNA sequencing to be performed directly from clinical specimens containing CMV [51], such as urine or saliva from a congenitally-infected child, should enable the collection of more authentic data regarding the CMV genome. This, in turn, is likely to yield new insights into the pathogenicity of heretofore uncharacterized virus genes.
A schematic representation of the CMV genome is illustrated in Fig. (2b) [46]. The UL133-151 genes in this figure correspond to the ULb’ region, the region of the genome that is subject to deletion and rearrangement during passage of CMV in cell culture. Interestingly, the laboratory-passaged strain, Towne, was used as a vaccine against CMV in a number of clinical trials prior to the complete characterization of its genome structure [15]. Its relative lack of therapeutic efficacy as a vaccine may relate, at least in part, to some of the mutations and genomic rearrangements that occur during tissue culture attenuation, possibly due to deletion of genes that encode proteins that are important in protective immunity. The putative role that many of the specific CMV genes and gene families play in pathogenesis is summarized in the following sections of this review. However, since the function of many CMV ORFs remains unknown, this understanding is, at best, a very incomplete one.
Molecular Mechanisms of Pathogenesis: Effects of CMV on Cell Biology
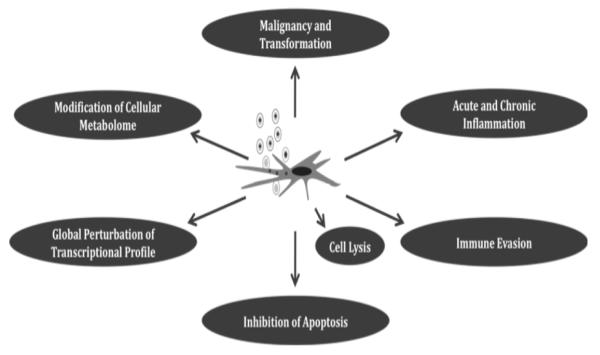
Injury to the fetus is mediated by a complex balance of the pathogenic effects of viral infection and the maternal immune response to infection. What is known about the potential role of virally-encoded gene products in mediating this injury? In the absence of an animal model to study congenital human CMV infection (discussed below), most studies have, by necessity, been performed in in vitro systems, by definition in the absence of a compensatory immune response. The following section provides a brief overview of work from many laboratories that has advanced our knowledge of viral genes and pathogenesis, summarizing some of the in vitro models employed. In some circumstances the putative pathologic effect of a CMV-encoded gene is believed to be mediated directly by that gene product; in other situations, CMV infection leads to downstream effects on cellular gene expression that then may potentially lead to pathologic outcomes, as illustrated Fig. (3). In many circumstances, CMV gene products mediate more than one pathogenic effect and there is considerable overlap across these categories.
Fig. (3).
Impact of CMV infection on cellular physiology, gene expression, and function. Following CMV infection, a variety of fates potentially await the infected cell, depending on the target organ and embryological state of the cell. These include transformation/dysregulation of cellular replication; immune activation and/or immune evasion; modification of apoptosis mechanisms and perturbation of cell cycle regulation; changes in cellular metabolism; and global perturbations in transcriptional and proteomic profiles. Cell death and lysis may also occur following infection. The synergy of these effects on the infected cell likely mediates much of the pathology and pathogenesis associated with congenital infection.
Modification of Cellular Apoptosis Mechanisms by CMV
Programmed cell death, or apoptosis, is a mechanism whereby damaged or infected cells are eliminated from tissue by an “auto-destructive pathway.” The apoptotic process is essential for the elimination of damaged or poorly-developed cells during organogenesis and is also considered a critical defense mechanism to eliminate virus-infected cells from the host. Apoptosis plays a central role in embryogenesis. Spatial and temporal regulation of cellular apoptotic machinery is essential for normal development and homeostasis of the developing fetus in all multicellular organisms [52]. Thus, any impact of CMV infection on cellular apoptotic pathways could play a potential role in teratogenic events occurring during in utero infection.
Two predominant pathways mediate apoptosis in the mammalian cell [52, 54]: the intrinsic pathway, which triggers cellular sensor proteins such as p53 and initiates a cascade of biochemical signals leading to the mitochondrial release of cytochrome C, and the extrinsic pathway, which is activated by external signals primarily involving the immune system. Activation of the extrinsic pathway leads to phosphorylation of receptor death domains, as found in the tumor necrosis factor (TNF) receptor family and Fas, by their respective ligands. Apoptotic signals, both intrinsic and extrinsic, converge to induce the activation of the caspase proteases, leading to proteolysis of the cell architecture, metabolic derangement, genomic fragmentation, and, eventually, cell death.
CMV has evolved several mechanisms to delay the intrinsic apoptotic signaling pathway. The viral IE proteins, IE1 (IE72) and IE2 (IE86), are known for their ability to inhibit apoptosis [55]. CMV-mediated inhibition of late apoptotic events is associated with sequestering of the tumor suppressor protein, p53, by IE2 [56]. CMV encodes two other genes that suppress apoptosis in the late replication phase. UL36 encodes an inhibitor of caspase 8 activation (vICA), and UL37 (exon 1) encodes a viral mitochondria-localized inhibitor of apoptosis (vMIA) [57-59]. The vICA protein inhibits apoptosis by binding to procaspase 8 and preventing its activation to an active form, while vMIA inhibits apoptosis by interacting with Bax (a pro-apoptotic molecule) and sequestering it within the mitochondrial membrane in an inactive form, a process that requires association of vMIA with internal lipid rafts in the endoplasmic reticulum and mitochondria-associated membranes [60]. CMV replication requires expression of vMIA and deletion of the UL37 gene in a recombinant virus can only be overcome by inhibiting cellular apoptotic responses using caspase inhibitors or by supplying other anti-apoptotic proteins to the cell [61]. Interestingly, an anti-apoptotic function has recently been attributed to another CMV gene product, but it is an RNA molecule, and not a protein, that mediates this effect. This RNA, the 2.7β (TRL4/IRL4) RNA, binds mitochondrial respiratory complex I, helping to maintain mitochondrial ATP production late in viral infection and preventing formation of death-induced reactive oxygen species [62].
Why does CMV encode such elaborate mechanisms to inhibit apoptosis? The presumed benefit of the anti-apoptosis mechanisms encoded by CMV may relate in part to the slow replication cycle of the virus. Inhibition of apoptotic mechanisms presumably helps sustain this slow replication cycle and may promote the propensity of CMV to establish chronic or latent infection. The impact of inhibition of apoptosis for the fetus could be quite profound and this could help explain elements of CMV-induced teratogenesis, since apoptosis is, as noted, a critical element of normal fetal development and organogenesis. A better understanding of these mechanisms could conceivably lead to targeted cellular therapies to correct dysregulated fetal development. Establishment of improved animal model system to study CMV pathogenesis will also shed light on pathogenesis. CMVs are highly species-specific and human CMV will not infect laboratory animals [63]. Remarkably, it has recently been shown that viral modulation of apoptosis contributes substantially to the species-specificity of CMVs. If apoptosis is blocked, murine and rat cytomegaloviruses are able to replicate in human cells [64]. Mutations in another murine CMV coding region, the M112-113 locus, have also been shown to help override the interspecies barrier, apparently through mechanisms other than modulation of apoptosis [65]. Potentially, the continued elucidation of the mechanisms that dictate the species-specificity of CMVs could be of great value in developing future models in which human CMV could be used to infect animals - a breakthrough that, in turn, could allow preclinical efficacy evaluation of human CMV-specific vaccines and antivirals.
CMV Infection and the Cell Cycle
Like apoptosis, cell-cycle regulation plays a critical role in fetal development. This is particularly important for the developing CNS. Recent findings in rodents and primates indicate that regulation of the cell cycle, specifically of the G1 phase, has a crucial role in controlling area-specific rates of neuron production and the generation of normal neuron architecture [66]. Intriguingly, CMV infection has been shown in a number of studies to have a striking impact on cell cycle progression. In addition to their role in inhibiting apoptosis, CMV IE gene products have been shown to modify cell cycle progression. IE proteins, IE72 and IE86, as well as proteins encoded by the UL82 and UL69 genes, alter cell cycle progression in human fibroblasts by interacting with cell cycle regulatory proteins. In quiescent fibroblasts, CMV infection activates cell cycle regulatory proteins at early time points (6-12 hours) post-infection and accelerates cell cycle progression from G0/G1 to early S phase. At later stages of infection, when viral DNA replication is initiated, progression of cellular DNA synthesis is inhibited and the cell cycle is arrested at the G2/M stage. After CMV-mediated arrest, chromosome segregation and cytokinesis are then blocked [67-71]. Recently, a role for the major tegument protein, pp65 (UL83 gene product) has also been suggested in the arrest of cell proliferation in G1/S, since the accumulation of this protein in the nucleolus is very prominent in G1 and G1/S, but very minimal in S or G2/M [72].
The virally-mediated inhibition of cellular DNA synthesis appears to be critically important for ensuring viral replication, since introduction of a mutation in the IE86 protein results in a recombinant CMV that is unable to block cell cycle progression. As a result, this recombinant virus replicates very poorly in cell culture [73]. What is the presumed benefit to CMV conferred by the presence of viral genes that modify the cell cycle? Modulation of cell cycle progression in infected cells may enable CMV to maximize the availability of the cellular DNA replication machinery, without competition from the cellular genome for the same resources [74]. This interplay between CMV and the cell cycle may impact the cell types infected or injured in the developing fetus. In the developing brain, CMV productively infects astrocytes and neural precursor cells, cell types known for their ability to undergo cell division in vivo. However, neurons, a terminally differentiated cell, are not permissive to productive viral infection [75].
Although CMV mediates cell-cycle arrest, the virus must also allow a mechanism that allows newly replicated viral genomes to egress from the nuclear lamina during viral DNA replication, in order to complete the viral life cycle. To ensure that this occurs, CMV encodes a protein, the UL97 gene product, which mimics the cellular protein, Cdc2/cyclin-dependent kinase 1, a kinase that normally triggers the entry of cells into mitosis. This cellular kinase is normally responsible for breaking down the nuclear lamina during mitosis [76, 77]. Thus, the UL97 gene product provides a mechanism allowing the completion of viral replication, in spite of the overall effect of cell cycle arrest mediated by infection. The identification of the ability of the UL97 protein to break down the nuclear lamina identifies UL97 as a potential novel antiviral drug discovery target, since inhibition of this process would greatly impair the ability of CMV to complete its replication cycle.
Impact of CMV on Chromosomes
One of the more intriguing observations in recent years regarding the impact of CMV infection on the biology of the cell has been the discovery that CMV infection may leads to chromosomal breakage. These studies provide a provocative hypothesis relating to the putative role of CMV as a teratogen [42, 78, 79]. In a study using human fibroblasts, infection with CMV during the S-phase of the cell cycle resulted in two specific chromosome 1 breaks, at positions 1q42 and 1q21. Purified virions, and not infected cell supernatants alone, were responsible for the effect, which could be blocked by co-incubation of virus with neutralizing antibody. UV-inactivated virus was as efficient as untreated virus in inducing specific damage to chromosome 1, suggesting a requirement for viral adsorption/penetration, but not viral gene expression [80]. The implications of chromosomal breakage or injury to the developing fetus are self-evident. Provocatively, in initial studies identifying injury to chromosome 1, two loci were identified near the break point that were of particular interest with respect to potential molecular mechanisms of CMV-induced birth defects: DFNA7 and USH2A. The DFNA7 gene has been linked to the inheritance of an autosomal dominant, nonsyndromic, progressive form of hearing loss [81]. Perturbation of the DFNA7 gene caused by CMV-induced breakage could conceivably be linked to the development of progressive SNHL. The USH2A gene, physically closest to the most prevalent CMV-induced break described in cell culture, encodes a protein important in the pathogenesis of Usher’s Syndrome Type II, an autosomal recessive disorder responsible for both SNHL and blindness [82, 83]. Subsequent fine-mapping techniques pinpointed CMV-induced genetic damage to 1q23.3, within a maximal region of 37 kb. A breakpoint was noted between DFAN7 and another hearing impairment locus, DFNA49. This breakpoint was noted to be in close proximity to the MPZ gene previously shown to be involved in autosomal dominant Charcot-Marie-Tooth syndrome with auditory neuropathy [84]. These observations suggest a possible relationship between these chromosomal breaks and the CMV-induced sequelae of SNHL and visual impairment. To date, the precise CMV gene(s) responsible for these chromosomal breaks have not been identified. However, a recent study in stably-transfected cells suggested that some CMV-induced chromosomal aberrations could be attributed to a specific CMV gene, UL76, although the chromosome(s) targeted in these cells were not specified [85].
Impact of CMV on Cellular Metabolism
In addition to study of the viral proteome, mass spectrometry has been employed to examine the impact of CMV infection on the cellular proteome. In a study of the impact of CMV infection on cellular metabolism [86], mass spectrometry was employed to measure the levels of intracellular metabolites at multiple time points after infection of fibroblasts. These studies demonstrated that, as the infection progressed, the levels of metabolites involved in the glycolysis pathway, the citric acid cycle, and pyrimidine nucleotide biosynthesis markedly increased. Peak levels of multiple metabolites during infection far exceeded those observed during normal cellular growth and homeostasis, indicating that CMV induces a distinct metabolic program. In a followup study [87], a flux measurement approach based on mass spectrometry was used more precisely to quantify changes in metabolic activity following viral infection. Significant increases occurred in flux through the tricarboxylic acid cycle and fatty acid biosynthesis pathway. Intriguingly, pharmacological inhibition of fatty acid biosynthesis in turn suppressed the replication of CMV. The concept of targeting the CMV-induced perturbations on cellular metabolism as an antiviral strategy was further validated in additional recent studies of the impact of infection on the glycolytic pathway. CMV infection was found to target calmodulin-dependent kinase kinase 1 (CaMKK1) expression, increasing the levels of CaMKK1 mRNA and protein. Inhibition of CaMKK severely attenuated production of viral progeny and blocked viral DNA replication [88]. Pharmacologic inhibition of CMV-induced fatty acid synthesis and/or activation of glycolysis may represent novel antiviral intervention strategies for control of CMV infection, and future metabolic profiling may identify additional metabolic targets that could be exploited for development of antiviral therapies.
CMV Genes Impact the Host Immune Response: Evasion and Inflammation
Throughout the course of its co-evolution with the human host, CMV has developed a number of mechanisms that modify the interaction between the virus and the immune system. Many of these immune modulation genes are homologs of normal cellular immune effectors, and are believed to have been ‘hijacked’ from the host genome. Some of these gene products interfere with host innate and/or adaptive immune responses, toward a presumed goal of promoting immune evasion; others appear to play a pro-inflammatory role, seemingly to promote inflammation that, in turn, could facilitate widespread dissemination CMV in the infected host. These functions are summarized in Table 1, and are briefly reviewed below.
Table 1.
Cytomegalovirus Immune Modulation Genes
| Cytomegalovirus Immune Modulation Genes | |
|---|---|
| CMV Gene(s) | Putative Immunomodulatory Function |
|
UL16
UL18 UL142 |
Homologs of MHC Class I molecules; promote natural killer (NK) cell evasion through binding of NK-inhibitory ligands, down-regulation of NK-activating ligands |
|
UL40
UL83 UL141 |
NK cell evasion genes; promote expression of non-classical HLA receptors; binding and inhibition of NK activating proteins; down-regulation of CD155 (activating receptor) |
|
TRS1
IRS1 |
dsRNA binding proteins; prevent activation of PKR following CMV infection; evasion of interferon response |
| UL111A | IL-10 cytokine homolog; possible down-regulation of host inflammation to evade immune clearance |
|
UL146
UL147 |
Homologs of C-X-C (alpha) chemokines; possibly function to promote widespread dissemination of infection in host |
|
UL128
UL130 |
Homologs of C-C (beta) chemokines; immune function unclear; ancillary proteins involved in viral entry into epithelial cells |
|
UL33
UL78 US27 US28 |
Homologs of G-protein coupled receptors; ligands may include multiple chemokines and cytokines; signaling molecules; may function as “chemokine sinks” to sequester immune activation proteins, compatible with an immune evasion role |
|
US2
US3 US6 US11 |
Genes interfering with MHC class I antigen processing; prevent development of cytotoxic T-cell response to CMV through retention of MHC in ER; facilitation of degradation of MHC; downregulation of MHC expression; immune evasion function |
| UL144 | Tumor necrosis factor receptor homolog; possible role modifying cytokine response to TH2 polarization; immune evasion |
| UL112 | MicroRNA; functions at RNA not protein level; NK cell evasion |
Modulation of Cytokine and Chemokine Responses
The CMV genome encodes several proteins that have the functional of cytokines, as well as chemokines (CKs; low-molecular weight “chemotactic cytokines” with characteristic highly conserved cysteine motifs [89]). The best-characterized cytokine homolog is a CMV homolog of inter-leukin (IL)-10, encoded by the UL111A gene. Although the CMV IL-10 has only limited homology (27%) to human IL-10 [90] it can, like human IL-10, induce a potent anti-inflammatory response [91-93]. CMV IL-10 may, by down-regulating host inflammation, diminish the immune response to infection. The host immune response, in turn, may target CMV IL-10 as a countermeasure; it was recently demonstrated that 28% of sera from CMV-seropositive blood donors contain antibody to viral IL-10, and that these antibodies not only potently inhibited the binding of CMV IL-10 to cellular receptors, but also specifically inhibited CMV IL-10-induced signaling through the JAK-STAT kinase pathway [94]. CMV IL-10 may contribute to fetal pathogenesis by virtue of an effect on placental development, where it impairs trophoblast invasiveness; impaired placentation, in turn, may compromise normal fetal growth and development [95, 96].
In addition to the IL-10 homolog, CMV encodes two ORFs with homology to the “CXC” family of CK [89]: these are UL146 and UL147 [97]. Notably, these genes are encoded within the ULb’ region of the CMV genome, a region (as already noted) that is associated with enhanced pathogenesis of the virus but is also subject to deletion and rearrangements during cell culture passage Fig. (2b) [41]. The functionality of these CKs and their role in the in vivo contribution to pathogenesis contributed by these ORFs is unclear, although the UL146 protein has been shown to be able to induce chemotaxis and intracellular calcium release in neutrophils [97]. It has been suggested that these proteins may function to promote neutrophil-mediated dissemination of CMV in the infected host. Other CMV genes have been noted to have homology to the CCCK family, in particular the UL128 and UL130 genes; however, the true function of these proteins appears to be related to their ability to form complexes with CMV glycoproteins that mediate CMV entry into endothelial and epithelial cells, and not through any intrinsic CK activity [30, 98].
Finally, as well as encoding cytokine and CK homologs, the CMV genome also encodes homologs of their cellular receptors. The best-characterized putative cytokine receptor is a member of the TNF receptor superfamily encoded by the UL144 gene [98-100], another gene found in the ULb’ region of the genome. The natural ligand for this receptor remains undefined, although the protein does up-regulate NF-kappaB expression and enhances expression of the CK receptor, CCL22 [101]. Since CCL22 attracts T-helper (Th)2 cells, UL144 could function as an anti-inflammatory effector, compatible with a role as an immunoevasive protein. CK receptors are members of the G-protein coupled receptor (GPCR) family [89] and there are at least 4 potential GPCRs encoded by the CMV genome: UL33, UL78, US27, and US28 [102, 103]. Of these, the US28 gene is the best studied [104]. US28 can bind ligands from multiple different CK classes and this, in turn, can trigger calcium mobilization, compatible with a putative functional role of US28 as a signaling molecule. However, the role of US28 in the virus life cycle or in pathogenesis remains unclear. US28 may exist to sequester host CKs (functioning as a “CK sink”) in order to promote evasion of the immune response; alternatively, US28 may function to activate signaling pathways that contribute to viral replication or spread, compatible with a pro-inflammatory function.
Modulation of Innate Immunity
One of the most important effectors of innate immune responses to CMV infection is the natural killer (NK) cell. NK cells play a critical role in cytotoxic killing of virus-infected cells at very early time points post-infection. NK responses depend upon the relative contributions of inhibitory receptors and activating receptors expressed on the cell surface. CMV has evolved several genes that interact with NK-mediated pathways, exploiting both inhibitory and activating pathways. One of the first such identified proteins in HCMV, gpUL18, is encoded by the gene UL18 and appears to exert its NK evasion effect by binding the NK cell inhibitory receptor, LIR-1, with a higher affinity than the host major histocompatability complex (MHC) class I molecule [105-107]. Recently, it has been shown that cells expressing another HCMV MHC class I homolog, the product of the UL142 gene, are protected from NK cell lysis [108]. The NK evasion effect is related to the ability of gpUL142 to down-regulate an NK activating ligand, MICA, that is a ligand for the activating NK cell receptor, NKG2D [109]. A number of other CMV genes, include UL40, UL83, and UL141, also appear to function as NK cell evasins [110].
CMV also encodes a number of functions that interfere with type 1 interferons (IFNα and IFNβ), a critical aspect of the host innate immune defense against viral infection [111]. The IE1 (IE72) and IE2 (IE86) proteins interfere, through distinct mechanisms, with induction of transcription of IFN-responsive genes, and with activation of the IFNβ promoter, respectively [112-114]. CMV also expresses proteins that interact with IFN-induced gene products. Among the most effective of these immune evasion functions are associated with the proteins encoded by TRS1 and IRS1. These proteins interfere with the protein kinase R (PKR) IFN-induced antiviral defense pathway. PKR is activated by double-stranded RNA (dsRNA), which is produced during CMV genome replication. In the presence of dsRNA, PKR undergoes a conformational change, dimerizes, autophosphorylates, and then phosphorylates the α subunit of eukaryotic initiation factor 2α, leading to inhibition of translation initiation and of viral replication [115, 116]. The CMV-encoded proteins TRS1 and IRS1 are capable of binding dsRNA, hence promoting evasion of the PKR pathway of host innate defense [117, 118].
Evasion of Adaptive Immunity
Although innate immune responses represent the “first line of defense” against CMV infection, long-term disease control depends upon T cell-mediated immune responses, particularly cluster designation (CD)8+ T cell responses. To counteract these responses, CMV encodes genes that prevent T cell control of infection through interference of both the MHC class I and class II antigen presentation pathways [119]. Notably, most of these genes map to the US region of the CMV genome and have characteristics of glycoproteins. They are resident to the endoplasmic reticulum (ER) of the CMV-infected cell. The US3 gene encodes a protein that binds MHC class I molecules and retains them in the ER, in the process preventing antigen presentation to CD8+ T cells [120, 121]. The US2 and US11 gene products bind to MHC I molecules, leading to their translocation from the ER to the cytoplasm, where they undergo proteasome-mediated degradation [122, 123]. The US6 gene encodes a protein that downregulates MHC class I expression [124, 125]. Down-regulation of MHC class I expression could make infected cells susceptible to NK cell mediated-lysis, which (as noted above) is a likely explanation for why CMV encode class I homologs as putative NK evasins [105-107].
Intriguingly, it has been suggested that MHC class I downregulation may play a role in fetal pathogenesis at the level of the fetal cytotrophoblast, the cell layer separating the maternal and fetal circulation in the placenta [126]. Fetal cytotrophoblasts do not express the “classical” HLA antigens, HLA-A and HLA-B, but express the “non-classical” class I HLA-G and HLA-C antigens, which inhibit NK-mediated cell lysis, in the process protecting the placental-fetal unit from being rejected as a foreign allograft. Notably, CMV US3 and US6 can down-regulate the cell-surface expression of both HLA-G and HLA-C in trophoblast cells; if such down-regulation resulted in increased NK lysis of placental trophoblast, this observation could provide a mechanistic explanation underlying CMV-related fetal injury [126]. However, when recombinant CMVs with deletions in these genes were tested in an ex vivo model of trophoblast differentiation and invasion, HLA-G expression continued to be down-regulated even in the absence of US3 and US6, implying that another viral function is responsible [127].
CMV-Encoded microRNAs
Like many viruses, CMV encodes a number of microRNAs (miRNAs). The miRNAs are approximately 22 nucleotide non-coding transcripts that mediate gene expression at the post-transcriptional level [128-130]. To date, approximately 15 miRNAs have been identified in CMV-infected cells. Many cellular processes, including apoptosis, cell cycle events, and immune response, are regulated by miRNAs. Recently, a novel mechanism of NK cell evasion was attributed to a CMV miRNA. As previously noted, one way in which CMV inhibits immune cell activation is by inhibiting NK cell receptor-activating ligands. One such example is the protein product of the UL142 gene, which downregulates the activating ligand, MICA [109]. It was recently demonstrated that another NK cell activating ligand, MICB, is downregulated by CMV, and this effect is mediated by a miRNA encoded by the UL112 locus [131-133]. Other CMV miRNAs appear to regulate viral gene expression and replication in cell culture [134, 135]. In rodent models of CMV infection, miRNAs play a role in tissue persistence and latency in the salivary gland, possibly mediated by an immunoevasive effect [136, 137]. Another study recently identified multiple cellular targets for a specific CMV miRNA, miR-US25-1. This miRNA was observed to bind target sites in the untranslated leader regions of many cellular mRNAs, in the process mediating significant reduction in gene expression. Interestingly, many of the genes targeted by this miRNA are associated with cell cycle control, suggesting a previously unrecognized mechanism by which CMV modulates the cell cycle [138]. Adding complexity to this rapidly emerging field of study is the observation that CMV not only encodes its own miRNAs, but also alters expression of cellular miRNAs, in the process facilitating viral replication in the CMV-infected cell [139]. Continued study of viral and host miRNAs is likely to lead to development of new oligonucleotide-based therapeutic approaches designed to modify the impact these sequences play on CMV replication and cellular physiology [140].
CMV-Encoded Molecular Mechanisms of Vasculopathy and Vascular Injury
Another mechanism by which CMV infection may mediate injury to the host is through infection of the vasculature. A number of vascular diseases have been hypothesized to be caused by CMV infection [141, 142]. Vascular diseases that have been associated with CMV include atherosclerosis, restenosis (typically occurring following coronary artery angioplasty), and transplant vascular sclerosis. The pathogenesis of these disease associations is uncertain. It has been hypothesized that these diseases are the result of mechanical and/or immune-mediated injury, followed by inflammation. Accordingly, CMV-encoded immune modulation genes have been implicated as potential mediators for endovascular injury and inflammation. This hypothesis holds that CMV infection plays a role in the migration of smooth muscle cells (SMC) from the vessel media to the vascular intima, which is followed, in turn, by cellular proliferation and inflammation. This sequence of events is theorized ultimately to lead to narrowing of the blood vessel, with the resultant compromise in tissue perfusion promoting pathologic consequences.
One CMV gene theorized to play a role in CMV-induced endovascular infection and injury is the product of the US28 gene. As already noted [104], US28 is a GPCR that interacts with a number of CK ligands. In functional studies, US28 mediates vascular SMC migration, compatible with its proposed role in CMV-associated vascular injury [143]. CMV-induced cellular proliferation may be driven in part by interactions between IE proteins and the p53 protein. Apoptosis of SMC can be induced by p53 but the CMV IE2 protein can protect these cells from p53-mediated apoptosis [144]. Moreover, the IE2 protein can also induce SMC proliferation and migration, suggesting another mechanism by which CMV infection and gene expression could promote a proproliferative effect [145]. CMV infection also stimulates the synthesis of IL-6, which upregulates the anti-apoptotic protein, survivin, in endothelial cells; this sequence may in turn play a role in neo-angiogenesis and in the acceleration of vascular diseases such as atherosclerosis and transplant vasculopathy [146].
Although these studies suggest mechanisms by which CMV infection could contribute to vascular injury and disease, what is the relevance to congenital CMV pathogenesis? Although most vascular diseases, particularly atherosclerosis, evolve over decades, vascular compromise could be of great potential importance for the health of the placenta and fetus. Vascular insufficiency due to CMV-mediated injury of endothelial cells could, in turn, compromise perfusion to the developing fetus or to the placenta, resulting in hypoxia and growth restriction. In an animal model of CMV pathogenesis, end-organ pathology in the setting of experimental infection was associated with significant disseminated vascular pathology, which evolved rapidly over a period of only a few weeks following experimental infection [147]. Vascular injury compromising perfusion of the fetal brain at critical windows of development is associated with the fetal brain disruption sequence, a syndrome characterized by multiple CNS anomalies. Conceivably, CMV-induced vascular injury could play a major role in this sequence; indeed, congenital CMV infection has been implicated as a causative factor in previous reports of this syndrome in the literature [148].
CMV-Encoded Molecular Mechanisms of Latency, Persistence and Oncogenesis
In addition to exploiting the immune response in such a way as to facilitate establishment of a persistent infection, cleared slowly (if at all) by the host immune response, CMV also appears to encode specific gene products that result in the establishment of a latent infection, resulting in a quiescent virus reservoir ready to be reactivated following appropriate provocative stimuli. The bone marrow appears to be the site where CMV latency is established [149], although the cell type where the latent viral genome is maintained remains incompletely characterized. Latent CMV genome has been described residing in both CD14+ monocytes and CD33+ myeloid precursor cells [150, 151]. Allogeneic stimulation has been successful in inducing CMV reactivation in CD14+ monocytes in the presence of T lymphocyte cytokines, providing strong experimental evidence for this cell as a site for latency [152, 153]. On the other hand, there is evidence for latency-specific transcription from CD33+ progenitor cells, both in the context of in vivo and in vitro infection. Interestingly, these RNAs are transcribed in an “antisense” orientation relative to the IE transcription unit [154, 155].
In addition to these cell types, CMV also infects CD34+ hematopoietic progenitor cells and CMV DNA can be detected in CD34+ cells from healthy seropositive individuals [156]. Recently, an in vitro model to study CMV latency in these primitive hematopoietic progenitor cells was described [157]. In the context of this model, CMV gene expression in CD34+ cells during long-term bone marrow culture has been noted to be markedly different from that observed following infection in fibroblasts. Using this model, it was shown that low-passage strains of CMV, but not laboratory-adapted strains, could establish latent infection, suggesting that the ULb’ region of the CMV genome encoded a latency-associated gene. Further work demonstrated that this gene was the UL138 gene. UL138 encodes a ~21 kDa protein that localizes to the Golgi apparatus as an integral type I membrane protein, where it may contribute to latency by mediating some as-yet undefined aspect of protein trafficking [158, 159]. The implications of understanding the role this gene product plays in latency extend to the issue of congenital CMV, which is known to occur in women with longstanding immunity to the virus, due either to re-infection with a new strain, or reactivation of a latent strain [10-12]. Novel antivirals targeting this gene could be very valuable in preventing such reactivation events.
Many of the viral gene products that impact latency, cell cycle regulation, and cellular proliferation have been invoked in hypotheses linking CMV infection to oncogenesis. CMV has been linked to several malignancies, including colon cancer, malignant glioma, Hodgkin’s lymphoma, and cervical and prostate cancer [160]. As previously noted, CMV IE2 protein interacts with the tumor suppressor protein, p53 [144]. Given the central role of p53 in the pathogenesis of many human cancers [161], these interactions suggest molecular mechanisms by which CMV could trigger malignant transformation of cells. Notably, an interaction of the CMV UL97 gene product with another cellular tumor suppressor protein, the retinoblastoma gene product (pRB), has recently been identified [162]. This interaction is of potential interest with respect to oncogenesis, given the wellknown interaction of the human papillomavirus type 16 (HPV-16) E7 gene with pRB, and the role that this interaction plays in the pathogenesis of cervical cancer [163]. Interestingly, HPV-16 E7 can complement a laboratory-generated mutant of CMV from which the UL97 gene has been deleted [164], suggesting that these disparate viruses have evolved similar genetic mechanisms to exploit and modify this regulator of cellular proliferation.
Other CMV genes have been implicated in the molecular pathogenesis of malignancy. These include the UL82 gene product, which binds to pRB and degrades it, stimulating cellular DNA synthesis [165, 166]. One of the first events following CMV infection in cell culture is the induction of proto-oncogenes, such as c-myc, and it has been postulated that this event could play a role in the pathogenesis of CMV-induced cancers [167]. Transforming activity has been mapped to other regions of the CMV genome. Within one such region, referred to as the mtrII locus, a 79 amino acid ORF was identified that was essential for maintenance of the cellular transformation phenotype [168, 169]. Subsequent studies demonstrated that this ORF was, in fact, the first exon of the CMV IL10 homolog [68]. Although the issue of whether CMV is an important co-factor in the pathogenesis of malignancies remains unsettled, these putative oncogenic/transforming gene products could be relevant to the problem of congenital CMV infection. It is possible that some of the putative mechanisms of oncogenesis could play a role in teratogenesis of the CMV-infected fetus, although to date there is no direct evidence that this is the case.
Virus Strain Variation
Given the plethora of putative pathogenesis determinants encoded by CMV, it is remarkable in many ways that the majority of congenitally-infected infants are not damaged by infection. As noted, the factors that impact the likelihood that an infected infant will be affected by CMV include: the timing of infection in utero [17]; the maternal immune status [13]; the status of the placenta [18]; and, possibly, the viral load in the maternal and fetal compartments [19]. Another variable that may impact the prognosis of congenital infection may be viral strain variation. It has been hypothesized that some clinical strains of CMV are intrinsically more harmful to the fetus, based on variability observed in genes implicated in viral pathogenesis. Experimental evidence to support this hypothesis is conflicting. Some subtypes of CMV classified on the basis of their UL144 (TNF receptor homolog) sequence were described as being more likely to be associated with symptomatic disease in newborns [21], irrespective of the viral load. On the other hand, three subsequent studies were unable to confirm any association with UL144 genotype and the outcome of congenital infection [170-172]. No differences in clinical outcome could be attributed to variants of the CK homologs, UL146 and UL147, when genotypes based on variation in these genes were compared in congenitally-infected infants [172, 173]. When genotypes based on the sequence heterogeneity observed in the envelope glycoprotein gene, gN (UL73), were compared, congenital infection with one genotype, gN-1, was statistically associated with an improved prognosis with respect to long-term neurodevelopmental sequelae [174]. In another study of infants with congenital infection, the distribution of genotypes for the gB glycoprotein gene (UL55) showed significant differences, depending upon the disease classification observed, but no information was reported on long-term neurodevelopmental sequelae [175]. A report of the use of artificial neural network technology for predicting the outcome of congenital CMV infection on the basis of sequence data from four genes identified specific nucleotide sequences that were highly predictive of outcome [176]. If it can be confirmed that virus strain variation has a substantive impact on clinical outcomes in congenitally-infected infants, important implications for vaccine design could emerge. The application of high-throughput techniques for sequencing CMV genomes directly from clinical specimens (and hence obviating the need for passage of viral strains in tissue culture prior to sequence analysis) should aid in resolving this unanswered question [51].
Animal Models of Congenital Infection and Fetal Pathogenesis
Although in vitro systems have documented many putative mechanisms by which CMV infection interferes with cellular gene expression and cell biology, the authentication of these effects as critical events in the in vivo pathogenesis of congenital CMV has been difficult to establish. The primary challenge, as noted, is the species-specificity of CMVs, which makes it unfeasible to evaluate the pathogenesis of human CMV in an animal model [63]. This limitation has been overcome to some extent in studies of transgenic mice expressing the human CMV major IE promoter sequences as a transgene driving expression of a reporter gene, lac Z. In these studies, the tissue distribution of IE-mediated gene expression was defined in the developing CNS; target sites of gene expression included the cerebral cortex, cerebellum, and eighth cranial nerve [177]. Human CMV replicates in fetal thymic explants maintained in SCID/hu mice and this has been exploited as an in vivo model for testing antiviral therapies [178].
Although these experimental approaches have been useful, they do not authentically model the pathogenesis of CMV infection. Studies of bona fide pathogenesis, therefore, have typically required the study of species-specific CMVs in their respective animal hosts. The pathology of fetal infection with rhesus macaque CMV mimics that observed in infants [179], although the cost of primate studies limits the practical usefulness of this model. Among rodent models, murine cytomegalovirus (MCMV) has provided a useful model system in which to study fetal neuropathogenesis [180, 181] and teratogenesis [182, 183]. A limitation of MCMV, however, is its inability to cross the mouse placenta and establish infection of the fetus. In contrast to MCMV, the guinea pig CMV is capable of establishing fetal infection, and this model has been valuable in the study of CMV vaccines and antiviral therapies, as well as the pathogenesis of fetal infection [184]. A key advance in the study of pathogenesis of CMV in all such models has been the ability to clone and maintain CMV genomes as infectious bacterial artificial chromosomes in Escherichia coli [185-187], allowing the powerful mutagenesis techniques available in prokaryotic systems to be applied to the generation of recombinant viruses with key mutations in pathogenesis genes. This approach has allowed the validation of the in vivo importance of many putative pathogenesis genes in animal models [188, 189] and this information in turn should help direct future, novel interventional studies designed to prevent CMV-associated injury in the newborn infant.
SUMMARY AND PERSPECTIVE
In summary, CMV has evolved highly complex sets of functions that have a profound effect on the biology of the cell and, presumably, on the human host. These effects, remarkably, are mediated not only by transcription of genes that modulate metabolism, senescence, cell cycle control, persistence, oncogenesis, and immune functioning, but also are mediated by regulatory and structural RNAs. From these experimental observations, hypotheses have been generated regarding the molecular pathogenesis of CMV-related injury and teratogenesis in the developing fetus. These include disruption of normal developmental pathways due to modifications of apoptotic and cell cycle pathways, altered perfusion of developing organ systems secondary to CMV-mediated endovascular injury, teratogenic CMV-mediated chromosomal injury, placental insufficiency with its attendant fetal compromise, and others. In spite of these remarkable advances in our understanding of how CMV interacts with the host at the cellular and molecular level, the precise mechanism(s) of CMV-related pathogenesis for the fetus remain largely unknown. Even less clear is what interventions can be anticipated to be of benefit to improve the outcome for the fetus, as evidenced by the fact that no randomized controlled trials exist, to date, supporting the use of any therapeutic interventions aimed at preventing CMV transmission or improving CMV-related outcomes for the fetus [190]. Even as much-needed vaccines and antivirals move forward in clinical trials, there is an important need to develop better model systems to study CMV-mediated injury of the developing fetus. Insights from such studies will in turn lead to novel interventions to help address this major public health problem.
Acknowledgments
Supported by NIH grants R01HD044864 and R01HD038416.
Footnotes
DISCLOSURES The author reports no conflicts of interest relevant to this publication.
REFERENCES
- [1].Weller TH. The cytomegaloviruses: ubiquitous agents with protean clinical manifestations. I. N. Engl. J. Med. 1971;285(4):203–214. doi: 10.1056/NEJM197107222850406. [DOI] [PubMed] [Google Scholar]
- [2].Weller TH. The cytomegaloviruses: ubiquitous agents with protean clinical manifestations. II. N. Engl. J. Med. 1971;285(5):267–274. doi: 10.1056/NEJM197107292850507. [DOI] [PubMed] [Google Scholar]
- [3].Boeckh M, Nichols WG, Papanicolaou G, Rubin R, Wingard JR, Zaia J. Cytomegalovirus in hematopoietic stem cell transplant recipients: current status, known challenges, and future strategies. Biol. Blood Marrow Transplant. 2003;9(9):543–558. doi: 10.1016/s1083-8791(03)00287-8. [DOI] [PubMed] [Google Scholar]
- [4].Jacobsen MA, Mills J. Serious cytomegalovirus disease in the acquired immunodeficiency syndrome (AIDS): clinical findings, diagnosis, and treatment. Ann. Intern. Med. 1988;108(4):585–594. doi: 10.7326/0003-4819-108-4-585. [DOI] [PubMed] [Google Scholar]
- [5].Nigro G. Maternal-fetal cytomegalovirus infection: from diagnosis to therapy. J. Matern. Fetal Neonatal. Med. 2009;22(2):169–174. doi: 10.1080/14767050802609767. [DOI] [PubMed] [Google Scholar]
- [6].Nyholm J, Schleiss MR. Prevention of maternal cytomegalovirus infection: current status and future prospects. Int. J. Wom. Health. 2010;2:23–35. doi: 10.2147/ijwh.s5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 2007;17(4):253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- [8].Cannon MJ, Finn Davis K. Washing our hands of the congenital cytomegalovirus disease epidemic. BMC Public Health. 2005;5:70–77. doi: 10.1186/1471-2458-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cheeran MC, Lokensgard JR, Schleiss MR. Neuropathogenesis of congenital cytomegalovirus infection: disease mechanisms and prospects for intervention. Clin. Microbiol. Rev. 2009;22(1):99–126. doi: 10.1128/CMR.00023-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 2001;344(18):1366–1371. doi: 10.1056/NEJM200105033441804. [DOI] [PubMed] [Google Scholar]
- [11].Yamamoto AY, Mussi-Pinhata MM, Boppana SB, Novak Z, Wagatsuma VM, Oliveira PD, Duarte G, Britt WJ. Human cytomegalovirus reinfection is associated with intrauterine transmission in a highly cytomegalovirus-immune maternal population. Am. J. Obstet. Gynecol. 2010;202(3):297, e1–e8. doi: 10.1016/j.ajog.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ross SA, Arora N, Novak Z, Fowler KB, Britt WJ, Boppana SB. Cytomegalovirus reinfections in healthy seroimmune women. J. Infect. Dis. 2010;201(3):386–389. doi: 10.1086/649903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fowler KB, Stagno S, Pass RF, Britt WJ, Boll TJ, Alford CA. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N. Engl. J. Med. 1992;326(10):663–667. doi: 10.1056/NEJM199203053261003. [DOI] [PubMed] [Google Scholar]
- [14].Revello MG, Gerna G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 2002;15(4):680–715. doi: 10.1128/CMR.15.4.680-715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bernstein DI. Vaccines for cytomegalovirus vaccines. Infect. Disord. Drug Targets. 2011;11 doi: 10.2174/187152611797636695. ???-??? [DOI] [PubMed] [Google Scholar]
- [16].Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- [17].Pass RF, Fowler KB, Boppana SB, Britt WJ, Stagno S. Congenital cytomegalovirus infection following first trimester maternal infection: symptoms at birth and outcome. J. Clin. Virol. 2006;35(2):216–220. doi: 10.1016/j.jcv.2005.09.015. [DOI] [PubMed] [Google Scholar]
- [18].Fisher S, Genbacev O, Maidji E, Pereira L. Human cytomegalovirus infection of placental cytotrophoblasts in vitro and in utero: implications for transmission and pathogenesis. J. Virol. 2000;74(15):6808–6820. doi: 10.1128/jvi.74.15.6808-6820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lazzarotto T, Varani S, Guerra B, Nicolosi A, Lanari M, Landini MP. Prenatal indicators of congenital cytomegalovirus infection. J. Pediatr. 2000;137(1):90–95. doi: 10.1067/mpd.2000.107110. [DOI] [PubMed] [Google Scholar]
- [20].Challacombe JF, Rechtsteiner A, Gottardo R, Rocha LM, Browne EP, Shenk T, Altherr MR, Brettin TS. Evaluation of the host transcriptional response to human cytomegalovirus infection. Physiol. Genomics. 2004;18(1):51–62. doi: 10.1152/physiolgenomics.00155.2003. [DOI] [PubMed] [Google Scholar]
- [21].Arav-Boger R, Battaglia CA, Lazzarotto T, Gabrielli L, Zong JC, Hayward GS, Diener-West M, Landini MP. Cytomegalovirus (CMV)-encoded UL144 (truncated tumor necrosis factor receptor) and outcome of congenital CMV infection. J. Infect. Dis. 2006;194(4):464–473. doi: 10.1086/505427. [DOI] [PubMed] [Google Scholar]
- [22].Schleiss MR. Persistent and recurring viral infections: the human herpesviruses. Curr. Probl. Pediatr. Adolesc. Health Care. 2009;39(1):7–23. doi: 10.1016/j.cppeds.2008.10.003. [DOI] [PubMed] [Google Scholar]
- [23].Chen DH, Jiang H, Lee M, Liu F, Zhou ZH. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology. 1999;260(1):10–16. doi: 10.1006/viro.1999.9791. [DOI] [PubMed] [Google Scholar]
- [24].Landini MP, La Placa M. Humoral immune response to human cytomegalovirus proteins: a brief review. Comp. Immun. Microbiol. Infect. Dis. 1991;14(2):97–105. doi: 10.1016/0147-9571(91)90124-v. [DOI] [PubMed] [Google Scholar]
- [25].Kern F, Bunde T, Faulhaber N, Kiecker F, Khatamzas E, Rudawski IM, Pruss A, Gratama JW, Volkmer-Engert R, Ewert R, Reinke P, Volk HD, Picker LJ. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 2002;185(12):1709–1716. doi: 10.1086/340637. [DOI] [PubMed] [Google Scholar]
- [26].Navarro D, Lennette E, Tugizov S, Pereira L. Humoral immune response to functional regions of human cytomegalovirus glycoprotein B. J. Med. Virol. 1997;52(4):451–459. [PubMed] [Google Scholar]
- [27].Rasmussen L, Matkin C, Spaete R, Pachl C, Merigan TC. Antibody response to human cytomegalovirus glycoproteins gB and gH after natural infection in humans. J. Infect. Dis. 1991;164(5):835–842. doi: 10.1093/infdis/164.5.835. [DOI] [PubMed] [Google Scholar]
- [28].Mach M, Kropff B, Dal Monte P, Britt W. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73) J. Virol. 2000;74(24):11881–11892. doi: 10.1128/jvi.74.24.11881-11892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 2006;80(2):710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA. 2005;102(50):18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130(1):118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- [32].Pepperl S, Munster J, Mach M, Harris JR, Plachter B. Dense bodies of human cytomegalovirus induce both humoral and cellular immune responses in the absence of viral gene expression. J. Virol. 2000;74(13):6132–6146. doi: 10.1128/jvi.74.13.6132-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mersseman V, Böhm V, Holtappels R, Deegen P, Wolfrum U, Plachter B, Reyda S. Refinement of strategies for the development of a human cytomegalovirus dense body vaccine. Med. Microbiol. Immunol. 2008;197(2):97–107. doi: 10.1007/s00430-008-0085-2. [DOI] [PubMed] [Google Scholar]
- [34].Bresnahan WA, Shenk T. A subset of viral transcripts packaged within human cytomegalovirus particles. Science. 2000;288(5475):2373–2376. doi: 10.1126/science.288.5475.2373. [DOI] [PubMed] [Google Scholar]
- [35].Sarcinella E, Brown M, Tellier R, Petric M, Mazzulli T. Detection of RNA in purified cytomegalovirus virions. Virus Res. 2004;104(2):129–137. doi: 10.1016/j.virusres.2004.03.008. [DOI] [PubMed] [Google Scholar]
- [36].Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, 2nd, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 2004;78(20):10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tandon R, AuCoin DP, Mocarski ES. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol. 2009;83(20):10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. A targeted spatial-temporal proteomic approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell Proteomics. 2010;9(5):851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Phillips SL, Bresnahan WA. Identification of binary interactions between human cytomegalovirus virion proteins. J. Virol. 2011;85(1):440–447. doi: 10.1128/JVI.01551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields’ Virology. 5th ed Vol. 1. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 2701–2772. [Google Scholar]
- [41].Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996;70(1):78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bystrevskaya VB, Lobova TV, Smirnov VN, Makarova NE, Kushch AA. Centrosome injury in cells infected with human cytomegalovirus. J. Struct. Biol. 1997;120(1):52–60. doi: 10.1006/jsbi.1997.3897. [DOI] [PubMed] [Google Scholar]
- [43].Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA, 3rd, Kouzarides T, Martignetti JA, Preddie E, Satchwell SC, Tomlinson P, Weston KM, Barrell BG. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- [44].Bradley AJ, Lurain NS, Ghazal P, Trivedi U, Cunningham C, Baluchova K, Gatherer D, Wilkinson GW, Dargan DJ, Davison AJ. High-throughput sequence analysis of variants of human cytomegalovirus strains Towne and AD169. J. Gen. Virol. 2009;90(pt. 10):2375–2380. doi: 10.1099/vir.0.013250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003;84(pt. 1):17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- [46].Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004;85(pt. 5):1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- [47].Murphy E, Shenk T. Human cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 2008;325:1–19. doi: 10.1007/978-3-540-77349-8_1. [DOI] [PubMed] [Google Scholar]
- [48].Jung GS, Kim YY, Kim JI, Ji GY, Jeon JS, Yoon HW, Lee GC, Ahn JH, Lee KM, Lee CH. Full genome sequencing and analysis of human cytomegalovirus strain JHC isolated from a Korean patient. Virus Res. 2011;156(1-2):113–120. doi: 10.1016/j.virusres.2011.01.005. [DOI] [PubMed] [Google Scholar]
- [49].Brondke H, Schmitz B, Doerfler W. Nucleotide sequence comparisons between several strains and isolates of human cytomegalovirus reveal alternate start codon usage. Arch. Virol. 2007;152(11):2035–2046. doi: 10.1007/s00705-007-1026-x. [DOI] [PubMed] [Google Scholar]
- [50].Wang A, Ren L, Abenes G, Hai R. Genome sequence divergences and functional variations in human cytomegalovirus strains. FEMS Immunol. Med. Microbiol. 2009;55(1):23–33. doi: 10.1111/j.1574-695X.2008.00489.x. [DOI] [PubMed] [Google Scholar]
- [51].Cunningham C, Gatherer D, Hilfrich B, Baluchova K, Dargan DJ, Thomson M, Griffiths PD, Wilkinson GW, Schulz TF, Davison AJ. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 2010;91(pt. 3):605–615. doi: 10.1099/vir.0.015891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Adachi-Yamada T, O’Connor MB. Mechanisms for removal of developmentally abnormal cells: cell competition and morphogenetic apoptosis. J. Biochem. 2004;136(1):13–17. doi: 10.1093/jb/mvh099. [DOI] [PubMed] [Google Scholar]
- [53].Benedict CA. Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factor Rev. 2003;14(3-4):349–357. doi: 10.1016/s1359-6101(03)00030-3. [DOI] [PubMed] [Google Scholar]
- [54].Benedict CA, Norris PS, Ware CF. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 2002;3(11):1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- [55].Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 1995;69(12):7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tsai HL, Kou GH, Chen SC, Wu CW, Lin YS. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J. Biol. Chem. 1996;271(7):3534–3540. [PubMed] [Google Scholar]
- [57].Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED, Kieff ED, Mocarski ES, Chittenden T. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA. 1999;96(22):12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Reboredo M, Greaves RF, Hahn G. Human cytomegalovirus proteins encoded by UL37 exon 1 protect infected fibroblasts against virus-induced apoptosis and are required for efficient virus replication. J. Gen. Virol. 2004;85(pt. 12):3555–3567. doi: 10.1099/vir.0.80379-0. [DOI] [PubMed] [Google Scholar]
- [59].Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA. 2001;98(14):7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Williamson CD, Zhang A, Colberg-Poley AM. The human cytomegalovirus protein UL37 exon 1 associates with internal lipid rafts. J. Virol. 2011;85(5):2100–2111. doi: 10.1128/JVI.01830-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].McCormick AL. Control of apoptosis by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 2008;325:281–295. doi: 10.1007/978-3-540-77349-8_16. [DOI] [PubMed] [Google Scholar]
- [62].Zhao J, Sinclair J, Houghton J, Bolton E, Bradley A, Lever A. Cytomegalovirus beta 2.7 RNA transcript protects endothelial cells against apoptosis during ischemia/reperfusion injury. J. Heart Lung Transplant. 2010;29(3):342–345. doi: 10.1016/j.healun.2009.09.006. [DOI] [PubMed] [Google Scholar]
- [63].Kern ER. Pivotal role of animal models in the development of new therapies for cytomegalovirus infections. Antiviral Res. 2006;71(2-3):164–171. doi: 10.1016/j.antiviral.2006.05.018. [DOI] [PubMed] [Google Scholar]
- [64].Jurak I, Brune W. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 2006;25(11):2634–2642. doi: 10.1038/sj.emboj.7601133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schumacher U, Handke W, Jurak I, Brune W. Mutations in the M112/M113-coding region facilitate murine cytomegalovirus replication in human cells. J. Virol. 2010;84(16):7994–8006. doi: 10.1128/JVI.02624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007;8(6):438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- [67].Bresnahan WA, Boldogh I, Thompson EA, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224(1):150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- [68].Jault FM, Jault JM, Ruchti F, Fortunato EA, Clark C, Corbeil J, Richman DD, Spector DH. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 1995;69(11):6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 1996;70(12):8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Song YJ, Stinski MF. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA. 2002;99(5):2836–2841. doi: 10.1073/pnas.052010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Du G, Dutta N, Lashmit P, Stinski MF. Alternative splicing of the human cytomegalovirus major immediate-early genes affects infectious-virus replication and control of cellular cyclin-dependent kinase. J. Virol. 2011;85(2):804–817. doi: 10.1128/JVI.01173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Arcangeletti MC, Rodighiero I, Mirandola P, De Conto F, Covan S, Germini D, Razin S, Dettori G, Chezzi C. Cell-cycle-dependent localization of human cytomegalovirus UL83 phosphoprotein in the nucleolus and modulation of viral gene expression in human embryo fibroblasts in vitro. J. Cell. Biochem. 2011;112(1):307–317. doi: 10.1002/jcb.22928. [DOI] [PubMed] [Google Scholar]
- [73].Petrik DT, Schmitt KP, Stinski MF. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 2006;80(8):3872–3883. doi: 10.1128/JVI.80.8.3872-3883.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Castillo JP, Kowalik TF. Human cytomegalovirus immediate early proteins and cell growth control. Gene. 2002;290(1-2):19–34. doi: 10.1016/s0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- [75].Lokensgard JR, Cheeran MC, Gekker G, Hu S, Chao CC, Peterson PK. Human cytomegalovirus replication and modulation of apoptosis in astrocytes. J. Hum. Virol. 1999;2(2):91–101. [PubMed] [Google Scholar]
- [76].Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009;5(1):e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Prichard MN. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 2009;19(4):215–229. doi: 10.1002/rmv.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Luleci G, Sakizli M, Gunalp A. Selective chromosomal damage caused by human cytomegalovirus. Acta. Virol. 1980;24(5):341–345. [PubMed] [Google Scholar]
- [79].AbuBakar S, Au WW, Legator MS, Albrecht T. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Environ. Mol. Mutagen. 1988;12(4):409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- [80].Fortunato EA, Dell’Aquila ML, Spector DH. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2000;97(2):853–858. doi: 10.1073/pnas.97.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Fagerheim T, Nilssen O, Raeymaekers P, Brox V, Moum T, Elverland HH, Teig E, Omland HH, Fostad GK, Tranebjaerg L. Identification of a new locus for autosomal dominant non-syndromic hearing impairment (DFNA7) in a large Norwegian family. Hum. Mol. Genet. 1996;5(8):1187–1191. doi: 10.1093/hmg/5.8.1187. [DOI] [PubMed] [Google Scholar]
- [82].Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Moller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J. Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science. 1998;280(5370):1753–1757. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- [83].Sumegi J, Wang JY, Zhen DK, Eudy JD, Talmadge CB, Li BF, Berglund P, Weston MD, Yao SF, Ma-Edmonds M, Overbeck L, Kelley PM, Zabarovsky E, Uzvolgyi E, Stan-bridge EJ, Klein G, Kimberling WJ. The construction of a yeast artificial chromosome (YAC) contig in the vicinity of the Usher syndrome type IIa (USH2A) gene in 1q41. Genomics. 1996;35(1):79–86. doi: 10.1006/geno.1996.0325. [DOI] [PubMed] [Google Scholar]
- [84].Nystad M, Fagerheim T, Brox V, Fortunato EA, Nilssen O. Human cytomegalovirus (HCMV) and hearing impairment: Infection of fibroblast cells with HCMV induces chromosome breaks at 1q23.3, between loci DFNA7 and DFNA49-Both involved in dominantly inherited, sensorineural, hearing impairment. Mutat. Res. 2008;637(1-2):56–65. doi: 10.1016/j.mrfmmm.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Siew VK, Duh CY, Wang SK. Human cytomegalovirus UL76 induces chromosome aberrations. J. Biomed. Sci. 2009;16:107. doi: 10.1186/1423-0127-16-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2(12):e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 2008;26(10):1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].McArdle J, Schafer XL, Munger J. Inhibition of calmodulin-dependent kinase kinase blocks human cytomegalovirus-induced glycolytic activation and severely attenuates production of viral progeny. J. Virol. 2011;85(2):705–714. doi: 10.1128/JVI.01557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Le Y, Gong W, Shen W, Li B, Dunlop NM, Wang JM. A burgeoning family of biological mediators: chemokines and chemokine receptors. Arch. Immunol. Ther. Exp. (Warsz) 2000;48(3):143–150. [PubMed] [Google Scholar]
- [90].Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc. Natl. Acad. Sci. USA. 2000;97(4):1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold ME, Schall TJ. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 2002;76(3):1285–1292. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Raftery MJ, Wieland D, Gronewald S, Kraus AA, Giese T, Schönrich G. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J. Immunol. 2004;173(5):3383–3391. doi: 10.4049/jimmunol.173.5.3383. [DOI] [PubMed] [Google Scholar]
- [93].Chang WL, Baumgarth N, Yu D, Barry PA. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 2004;78(16):8720–8731. doi: 10.1128/JVI.78.16.8720-8731.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].de Lemos Rieper C, Galle P, Pedersen BK, Hansen MB. Characterization of specific antibodies against cytomegalovirus-encoded IL-10 produced by 28% of the CMV-seropositive blood donors. J. Gen. Virol. 2011;92(Pt7):1508–1518. doi: 10.1099/vir.0.028738-0. [DOI] [PubMed] [Google Scholar]
- [95].Roth I, Fisher SJ. IL-10 is an autocrine inhibitor of human placental cytotrophoblast MMP-9 production and invasion. Dev. Biol. 1999;205(1):194–204. doi: 10.1006/dbio.1998.9122. [DOI] [PubMed] [Google Scholar]
- [96].Yamamoto-Tabata T, McDonagh S, Chang HT, Fisher S, Pereira L. Human cytomegalovirus interleukin-10 downregulates metalloproteinase activity and impairs endothelial cell migration and placental cytotrophoblast invasiveness in vitro. J. Virol. 2004;78(6):2831–2840. doi: 10.1128/JVI.78.6.2831-2840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA. 1999;96(17):9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 2004;78(18):10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Benedict CA, Butrovich KD, Lurain NS, Corbeil J, Rooney I, Schneider P, Tschopp J, Ware CF. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J. Immunol. 1999;162(12):6967–6970. [PubMed] [Google Scholar]
- [100].Lurain NS, Kapell KS, Huang DD, Short JA, Paintsil J, Winkfield E, Benedict CA, Ware CF, Bremer JW. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J. Virol. 1999;73(12):10040–10050. doi: 10.1128/jvi.73.12.10040-10050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Poole E, King CA, Sinclair JH, Alcami A. The UL144 gene product of human cytomegalovirus activates NFkappaB via a TRAF6-dependent mechanism. EMBO J. 2006;25(18):4390–4399. doi: 10.1038/sj.emboj.7601287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Vischer HF, Vink C, Smit MJ. A viral conspiracy: hijacking the chemokine system through virally encoded pirated chemokine receptors. Curr. Top. Microbiol. Immunol. 2006;303:121–154. doi: 10.1007/978-3-540-33397-5_6. [DOI] [PubMed] [Google Scholar]
- [103].Beisser PS, Lavreysen H, Bruggeman CA, Vink C. Chemokines and chemokine receptors encoded by cytomegaloviruses. Curr. Top. Microbiol. Immunol. 2008;325:221–242. doi: 10.1007/978-3-540-77349-8_13. [DOI] [PubMed] [Google Scholar]
- [104].Vomaske J, Nelson JA, Streblow DN. Human cytomegalovirus US28: a functionally selective chemokine binding receptor. Infect. Disord. Drug Targets. 2009;9(5):548–556. doi: 10.2174/187152609789105696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11(5):603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- [106].Yang Z, Bjorkman PJ. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc. Natl. Acad. Sci. USA. 2008;105(29):10095–10100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Occhino M, Ghiotto F, Soro S, Mortarino M, Bosi S, Maffei M, Bruno S, Nardini M, Figini M, Tramontano A, Ciccone E. Dissecting the structural determinants of the interaction between the human cytomegalovirus UL18 protein and the CD85j immune receptor. J. Immunol. 2008;180(2):957–968. doi: 10.4049/jimmunol.180.2.957. [DOI] [PubMed] [Google Scholar]
- [108].Wills MR, Ashiru O, Reeves MB, Okecha G, Trowsdale J, Tomasec P, Wilkinson GW, Sinclair J, Sissons JG. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 2005;175(11):7457–7465. doi: 10.4049/jimmunol.175.11.7457. [DOI] [PubMed] [Google Scholar]
- [109].Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 2006;346(1):175–181. doi: 10.1016/j.bbrc.2006.05.092. [DOI] [PubMed] [Google Scholar]
- [110].Powers C, DeFilippis V, Malouli D, Früh K. Cytomegalovirus immune evasion. Curr. Top. Microbiol. Immunol. 2008;325:333–359. doi: 10.1007/978-3-540-77349-8_19. [DOI] [PubMed] [Google Scholar]
- [111].Marshall EE, Geballe AP. Multifaceted evasion of the interferon response by cytomegalovirus. J. Interferon Cytokine Res. 2009;29(9):609–619. doi: 10.1089/jir.2009.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Paulus C, Krauss S, Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. USA. 2006;103(10):3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, Hayward GS, Ahn JH. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 2008;82(21):10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Taylor RT, Bresnahan WA. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 2005;79(6):3873–3877. doi: 10.1128/JVI.79.6.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].DeFilippis VR. Induction and evasion of the type I interferon response by cytomegaloviruses. Adv. Exp. Med. Biol. 2007;598:309–324. doi: 10.1007/978-0-387-71767-8_22. [DOI] [PubMed] [Google Scholar]
- [116].Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: recent insights and therapeutic opportunities. Biochem. Pharmacol. 2008;75(3):589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- [117].Child SJ, Hakki M, De Niro KL, Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 2004;78(1):197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J. Virol. 2009;83(9):4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Jones TR, Hanson LK, Sun L, Slater JS, Stenberg RM, Campbell AE. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 1995;69(8):4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Ahn K, Angulo A, Ghazal P, Peterson PA, Yang Y, Fruh K. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA. 1996;93(20):10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jones TR, Wiertz EJ, Sun L, Fish KN, Nelson JA, Ploegh HL. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA. 1996;93(21):11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Wiertz EJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, Rapoport TA, Ploegh HL. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384(6608):432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- [123].Wiertz EJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84(5):769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- [124].Ahn K, Gruhler A, Galocha B, Jones TR, Wiertz EJ, Ploegh HL, Peterson PA, Yang Y, Fruh K. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 1997;6(5):613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- [125].Hengel H, Koopmann JO, Flohr T, Muranyi W, Goulmy E, Hammerling GJ, Koszinowski UH, Momburg F. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity. 1997;6(5):623–632. doi: 10.1016/s1074-7613(00)80350-7. [DOI] [PubMed] [Google Scholar]
- [126].Jun Y, Kim E, Jin M, Sung HC, Han H, Geraghty DE, Ahn K. Human cytomegalovirus gene products US3 and US6 down-regulate trophoblast class I MHC molecules. J. Immunol. 2000;164(2):805–811. doi: 10.4049/jimmunol.164.2.805. [DOI] [PubMed] [Google Scholar]
- [127].Jones TR, Muzithras VP. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J. Virol. 1992;66(4):2541–2546. doi: 10.1128/jvi.66.4.2541-2546.1992. 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Dölken L, Pfeffer S, Koszinowski UH. Cytomegalovirus microRNAs. Virus Genes. 2009;38(3):355–364. doi: 10.1007/s11262-009-0347-0. [DOI] [PubMed] [Google Scholar]
- [129].Fannin Rider PJ, Dunn W, Yang E, Liu F. Human cytomegalovirus microRNAs. Curr. Top. Microbiol. Immunol. 2008;325:21–39. doi: 10.1007/978-3-540-77349-8_2. [DOI] [PubMed] [Google Scholar]
- [130].Grey F, Nelson J. Identification and function of human cytomegalovirus microRNAs. J. Clin. Virol. 2008;41(3):186–191. doi: 10.1016/j.jcv.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. 2010;11(9):806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- [133].Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod’homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, Wang EC, Griffin CA, Davison AJ. Modulation of natural killer cells by human cytomegalovirus. J. Clin. Virol. 2008;41(3):206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Stern-Ginossar N, Saleh N, Goldberg MD, Prichard M, Wolf DG, Mandelboim O. Analysis of human cytomegalovirus-encoded microRNA activity during infection. J. Virol. 2009;83(20):10684–10693. doi: 10.1128/JVI.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Meyer C, Grey F, Kreklywich CN, Andoh TF, Tirabassi RS, Orloff SL, Streblow DN. Cytomegalovirus microRNA expression is tissue specific and is associated with persistence. J. Virol. 2011;85(1):378–389. doi: 10.1128/JVI.01900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Dölken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, Koszinowski UH, Jonjic S, Pfeffer S. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog. 2010;6(10):e1001150. doi: 10.1371/journal.ppat.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog. 2010;6(6):e1000967. doi: 10.1371/journal.ppat.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 2008;82(18):9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Moens U. Silencing viral microRNA as a novel antiviral therapy? J. Biomed. Biotechnol. 2009;2009:419539. doi: 10.1155/2009/419539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Hansson GK, Robertson AK, Söderberg-Nauclér C. Inflammation and atherosclerosis. Annu. Rev. Pathol. 2006;1:297–329. doi: 10.1146/annurev.pathol.1.110304.100100. [DOI] [PubMed] [Google Scholar]
- [142].Chen R, Xiong S, Yang Y, Fu W, Wang Y, Ge J. The relationship between human cytomegalovirus infection and atherosclerosis development. Mol. Cell Biochem. 2003;249(1-2):91–96. [PubMed] [Google Scholar]
- [143].Streblow DN, Söderberg-Nauclér C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson JA. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99(5):511–520. doi: 10.1016/s0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- [144].Tanaka K, Zou JP, Takeda K, Ferrans VJ, Sandford GR, Johnson TM, Finkel T, Epstein SE. Effects of human cytomegalovirus immediate-early proteins on p53-mediated apoptosis in coronary artery smooth muscle cells. Circulation. 1999;99(13):1656–1659. doi: 10.1161/01.cir.99.13.1656. [DOI] [PubMed] [Google Scholar]
- [145].Zhou YF, Yu ZX, Wanishsawad C, Shou M, Epstein SE. The immediate early gene products of human cytomegalovirus increase vascular smooth muscle cell migration, proliferation, and expression of PDGF beta-receptor. Biochem. Biophys. Res. Commun. 1999;256(3):608–613. doi: 10.1006/bbrc.1999.0387. [DOI] [PubMed] [Google Scholar]
- [146].Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, Caposio P. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood. 2011;117(1):352–361. doi: 10.1182/blood-2010-06-291245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Persoons MC, Stals FS, van dam Mieras MC, Bruggeman CA. Multiple organ involvement during experimental cytomegalovirus infection is associated with disseminated vascular pathology. J. Pathol. 1998;184(1):103–109. doi: 10.1002/(SICI)1096-9896(199801)184:1<103::AID-PATH964>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- [148].Corona-Rivera JR, Corona-Rivera E, Romero-Velarde E, Hernández-Rocha J, Bobadilla-Morales L, Corona-Rivera A. Report and review of the fetal brain disruption sequence. Eur. J. Pediatr. 2001;160(11):664–667. doi: 10.1007/s004310100813. [DOI] [PubMed] [Google Scholar]
- [149].Mendelson M, Monard S, Sissons P, Sinclair J. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 1996;77(pt. 12):3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- [150].Taylor-Wiedeman J, Sissons JG, Borysiewicz LK, Sinclair JH. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 1991;72(pt. 9):2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- [151].Hahn G, Jores R, Mocarski ES. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA. 1998;95(7):3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Söderberg-Nauclér C, Fish KN, Nelson JA. Interferon-gamma and tumor necrosis factor-alpha specifically induce formation of cytomegalovirus-permissive monocyte-derived macrophages that are refractory to the antiviral activity of these cytokines. J. Clin. Invest. 1997;100(12):3154–3163. doi: 10.1172/JCI119871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Söderberg-Nauclér C, Streblow DN, Fish KN, Allan-Yorke J, Smith PP, Nelson JA. Reactivation of latent human cytomegalovirus in CD14 (+) monocytes is differentiation dependent. J. Virol. 2001;75(16):7543–7554. doi: 10.1128/JVI.75.16.7543-7554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Kondo K, Kaneshima H, Mocarski ES. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. USA. 1994;91(25):11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Kondo K, Xu J, Mocarski ES. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA. 1996;93(20):11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].von Laer D, Meyer-Koenig U, Serr A, Finke J, Kanz L, Fauser AA, Neumann-Haefelin D, Brugger W, Hufert FT. Detection of cytomegalovirus DNA in CD34+ cells from blood and bone marrow. Blood. 1995;86(11):4086–4090. [PubMed] [Google Scholar]
- [157].Goodrum FD, Jordan CT, High K, Shenk T. Human cytomegalovirus gene expression during infection of primary hema topoietic progenitor cells: a model for latency. Proc. Natl. Acad. Sci. USA. 2002;99(25):16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Goodrum F, Reeves M, Sinclair J, High K, Shenk T. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood. 2007;110(3):937–945. doi: 10.1182/blood-2007-01-070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Petrucelli A, Rak M, Grainger L, Goodrum F. Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J. Virol. 2009;83(11):5615–5629. doi: 10.1128/JVI.01989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Söderberg-Nauclér C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J. Intern. Med. 2006;259(3):219–246. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- [161].Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 2009;1(5):a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, Kern ER. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 2008;82(10):5054–5067. doi: 10.1128/JVI.02174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Helt AM, Galloway DA. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24(2):159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- [164].Kamil JP, Hume AJ, Jurak I, Münger K, Kalejta RF, Coen DM. Human papillomavirus 16 E7 inactivator of retinoblastoma family proteins complements human cytomegalovirus lacking UL97 protein kinase. Proc. Natl. Acad. Sci. USA. 2009;106(39):16823–16828. doi: 10.1073/pnas.0901521106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kalejta RF, Shenk T. Proteasome-dependent, ubiquitin independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. USA. 2003;100(6):3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell Biol. 2003;23(6):1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Boldogh I, AbuBakar S, Albrecht T. Activation of protooncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247(4942):561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- [168].Muralidhar S, Doniger J, Mendelson E, Araujo JC, Kashanchi F, Azumi N, Brady JN, Rosenthal LJ. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J. Virol. 1996;70(12):8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Doniger J, Muralidhar S, Rosenthal LJ. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin. Microbiol. Rev. 1999;12(3):367–382. doi: 10.1128/cmr.12.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Picone O, Costa JM, Chaix ML, Ville Y, Rouzioux C, Leruez-Ville M. Human cytomegalovirus UL144 gene polymorphisms in congenital infections. J. Clin. Microbiol. 2005;43(1):25–29. doi: 10.1128/JCM.43.1.25-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Mao ZQ, He R, Sun M, Qi Y, Huang YJ, Ruan Q. The relationship between polymorphisms of HCMV UL144 ORF and clinical manifestations in 73 strains with congenital and/or perinatal HCMV infection. Arch. Virol. 2007;152(1):115–124. doi: 10.1007/s00705-006-0826-8. [DOI] [PubMed] [Google Scholar]
- [172].Heo J, Petheram S, Demmler G, Murph JR, Adler SP, Bale J, Sparer TE. Polymorphisms within human cytomegalovirus chemokine (UL146/UL147) and cytokine receptor genes (UL144) are not predictive of sequelae in congenitally infected children. Virology. 2008;378(1):86–96. doi: 10.1016/j.virol.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Virology. 2008;383(1):168–171. doi: 10.1016/j.virol.2008.10.007. Erratum in: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Arav-Boger R, Foster CB, Zong JC, Pass RF. Human cytomegalovirus-encoded alpha-chemokines exhibit high sequence variability in congenitally infected newborns. J. Infect. Dis. 2006;193(6):788–791. doi: 10.1086/500508. [DOI] [PubMed] [Google Scholar]
- [174].Pignatelli S, Dal Monte P, Rossini G, Lazzarotto T, Gatto MR, Landini MP. Intrauterine cytomegalovirus infection and glycoprotein N (gN) genotypes. J. Clin. Virol. 2003;28(1):38–43. doi: 10.1016/s1386-6532(02)00236-6. [DOI] [PubMed] [Google Scholar]
- [175].Jin H, Wang X, Li S. Human cytomegalovirus glycoprotein B genotype correlates with different symptoms of infected infants. Intervirology. 2007;50(3):219–223. doi: 10.1159/000100564. [DOI] [PubMed] [Google Scholar]
- [176].Arav-Boger R, Boger YS, Foster CB, Boger Z. The use of artificial neural networks in prediction of congenital CMV outcome from sequence data. Bioinform. Biol. Insights. 2008;2:281–289. doi: 10.4137/bbi.s764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Koedood M, Fichtel A, Meier P, Mitchell PJ. Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J. Virol. 1995;69(4):2194–2207. doi: 10.1128/jvi.69.4.2194-2207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Mocarski ES, Bonyhadi M, Salimi S, McCune JM, Kaneshima H. Human cytomegalovirus in a SCID-hu mouse: thymic epithelial cells are prominent targets of viral replication. Proc. Natl. Acad. Sci. USA. 1993;90(1):104–108. doi: 10.1073/pnas.90.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Barry PA, Lockridge KM, Salamat S, Tinling SP, Yue Y, Zhou SS, Gospe SM, Britt WJ, Tarantal AF. Nonhuman primate models of intrauterine cytomegalovirus infection. ILAR J. 2006;47(1):49–64. doi: 10.1093/ilar.47.1.49. [DOI] [PubMed] [Google Scholar]
- [180].Tsutsui Y, Kosug I, Kawasaki H. Neuropathogenesis in cytomegalovirus infection: indication of the mechanisms using mouse models. Rev. Med. Virol. 2005;15(5):327–345. doi: 10.1002/rmv.475. [DOI] [PubMed] [Google Scholar]
- [181].Tsutsui Y, Kosugi I, Kawasaki H, Arai Y, Han GP, Li L, Kaneta M. Roles of neural stem progenitor cells in cytomegalovirus infection of the brain in mouse models. Pathol. Int. 2008;58(5):257–267. doi: 10.1111/j.1440-1827.2008.02221.x. [DOI] [PubMed] [Google Scholar]
- [182].Baskar JF, Stanat SC, Sulik KK, Huang ES. Murine cytomegalovirus-induced congenital defects and fetal maldevelopment. J. Infect. Dis. 1983;148(5):836–843. doi: 10.1093/infdis/148.5.836. [DOI] [PubMed] [Google Scholar]
- [183].Baskar JF, Peacock J, Sulik KK, Huang ES. Early-stage developmental abnormalities induced by murine cytomegalovirus. J. Infect. Dis. 1987;155(4):661–666. doi: 10.1093/infdis/155.4.661. [DOI] [PubMed] [Google Scholar]
- [184].Schleiss MR. Nonprimate models of congenital cytomegalovirus (CMV) infection: gaining insight into pathogenesis and prevention of disease in newborns. ILAR J. 2006;47(1):65–72. doi: 10.1093/ilar.47.1.65. [DOI] [PubMed] [Google Scholar]
- [185].Adler H, Messerle M, Koszinowski UH. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 2003;13(12):111–121. doi: 10.1002/rmv.380. [DOI] [PubMed] [Google Scholar]
- [186].Borst EM, Crnkovic-Mertens I, Messerle M. Cloning of beta-herpesvirus genomes as bacterial artificial chromosomes. Methods Mol. Biol. 2004;256:221–239. doi: 10.1385/1-59259-753-X:221. [DOI] [PubMed] [Google Scholar]
- [187].Ruzsics Z, Koszinowski UH. Mutagenesis of the cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 2008;325:41–61. doi: 10.1007/978-3-540-77349-8_3. [DOI] [PubMed] [Google Scholar]
- [188].Cicin-Sain L, Bubi I, Schnee M, Ruzsics Z, Mohr C, Jonji S, Koszinowski UH. Targeted deletion of regions rich in immune-evasive genes from the cytomegalovirus genome as a novel vaccine strategy. J. Virol. 2007;81(24):13825–13834. doi: 10.1128/JVI.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Crumpler MM, Choi KY, McVoy MA, Schleiss MR. A live guinea pig cytomegalovirus vaccine deleted of three putative immune evasion genes is highly attenuated but remains immunogenic in a vaccine/challenge model of congenital cytomegalovirus infection. Vaccine. 2009;27(31):4209–4218. doi: 10.1016/j.vaccine.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].McCarthy FP, Giles ML, Rowlands S, Purcell KJ, Jones CA. Antenatal interventions for preventing the transmission of cytomegalovirus (CMV) from the mother to fetus during pregnancy and adverse outcomes in the congenitally infected infant. Cochrane Database Syst. Rev. 2011;3 doi: 10.1002/14651858.CD008371.pub2. CD008371. [DOI] [PMC free article] [PubMed] [Google Scholar]