Abstract
Aims: Carbon monoxide (CO)-releasing molecules (CO-RMs) are being developed with the ultimate goal of safely utilizing the therapeutic potential of CO clinically. One such application is antimicrobial activity; therefore, we aimed to characterize and compare the effects of the CO-RM, CORM-3, and its inactivated counterpart, where all labile CO has been removed, at the transcriptomic and cellular level. Results: We found that both compounds are able to penetrate the cell, but the inactive form is not inhibitory to bacterial growth under conditions where CORM-3 is. Transcriptomic analyses revealed that the bacterial response to inactivated CORM-3 (iCORM-3) is much lower than to the active compound and that a wide range of processes appear to be affected by CORM-3 and to a lesser extent iCORM-3, including energy metabolism, membrane transport, motility, and the metabolism of many sulfur-containing species, including cysteine and methionine. Innovation: This work has demonstrated that both CORM-3 and its inactivated counterpart react with cellular functions to yield a complex response at the transcriptomic level. A full understanding of the actions of both compounds is vital to understand the toxic effects of CO-RMs. Conclusion: This work has furthered our understanding of how CORM-3 behaves at the cellular level and identifies the responses that occur when the host is exposed to the Ru compound as well as those that result from the released CO. This is a vital step in laying the groundwork for future development of optimized CO-RMs for eventual use in antimicrobial therapy. Antioxid. Redox Signal. 19, 1999–2012.
Introduction
Carbon monoxide (CO) acts as a respiratory inhibitor by binding to heme groups and preventing electron transfer to O2 (23). CO is also generated within the human body via the breakdown of heme by the enzyme heme oxygenase. The CO generated endogenously fulfills a variety of roles, with properties as diverse as signaling and bactericidal activity (6,9).
However, utilizing CO gas as a therapeutic tool has proven difficult due to its toxic properties and the systemic delivery methods used for administration (5). CO-releasing molecules (CO-RMs) have been developed for over 10 years in an attempt to harness the beneficial properties of CO while minimizing its toxic effects. These CO-RMs have been synthesized with a variety of functions in mind, including targeting and specificity. There is now a wide variety of carbonyl compounds with centers that include ruthenium, iron, manganese, and boron (26,27). Their CO-release rates and kinetics are also diverse with half-lives of seconds to hours. CO-RMs liberate CO in a variety of conditions, including by dissociation in biological buffers (13), by photodissociation (38) and via enzyme activity (33).
Innovation.
Carbon monoxide-releasing molecules (CO-RMs) deliver carbon monoxide (CO) to cells and tissues and show promising results as antimicrobial agents. However, rational exploitation of CO-RMs requires fundamental understanding of their activity, and appreciation of the biological impacts of the CO-RM residue after CO has been liberated. We report that CORM-3 and its CO-depleted form have comprehensive time-dependent effects on the transcript profiles in Escherichia coli particularly in the genes implicated in energy and sulfur metabolism, although the effects of CORM-3 are more profound than those of the inactive residue. Our work highlights the need for integration of chemistry, physiology, and molecular biology before CO-RMs are used clinically.
Ru(CO)3Cl(glycinate), more commonly known as CORM-3, is a water-soluble Ru-containing compound, capable of releasing one CO per molecule with a half-life of ∼2 min in vitro (8,28). CORM-3 is bactericidal against a variety of microbes, including Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus (10,12,13,30,40).
To assess which of the observed effects upon exposure to CORM-3 are a result of CO and which are caused by the Ru-compound devoid of labile CO, it is important to utilize the inactivated compound, iCORM-3, as a control compound. However a number of studies do not utilize an inactivated compound as a control (12,13,30) or use control compounds that are not the inactivated counterpart, but a similar compound where CO has been substituted for another molecule, for example, RuCl2(DMSO)4 (10). In this study, we made iCORM-3 by incubating fresh CORM-3 dissolved in phosphate-buffered saline (PBS) at room temperature for up to 48 h with periodic N2 gas bubbling, after which time very little CO release can be detected (<5% than of the same concentration of CORM-3). This process is thought to allow CORM-3 to release the labile CO, which escapes into the gas phase upon bubbling, leaving a CO-depleted iCORM-3 compound in solution (8). However, mounting literature suggests that CORM-3 is unable to release large quantities of CO under these conditions; rather, the carbonyl compound is altered forming a stable molecule that releases CO with a much slower rate (13,28,37). This inactivation process could produce a very slow CO-releasing tricarbonyl complex (22) or a dicarbonyl compound, which is formed via the release of CO2 from CORM-3 (37). Whichever form is made, we can be sure that this inactivated compound is made directly from the active CORM-3 molecule and is therefore likely to mimic the compound present in vivo after CO has been released.
Despite the ambiguity of its chemical structure, the iCORM-3 compound has been used in studies as a non-CO releasing control for CORM-3 (8,17,34,37) to test the reactivity of the Ru-compound when all labile CO has been removed. In all cases, iCORM-3 was shown to be much less reactive than the CO-RM.
However, no work has been carried out to directly characterize and compare in detail the observed effects of iCORM-3 and CORM-3 on bacteria. This comparison is essential to further our understanding of how these molecules behave at the cellular level and is a vital step in laying the groundwork for future development of optimized CO-RMs for eventual use in antimicrobial therapy.
Results
Growth of E. coli after treatment with CORM-3 or iCORM-3 and intracellular uptake of Ru
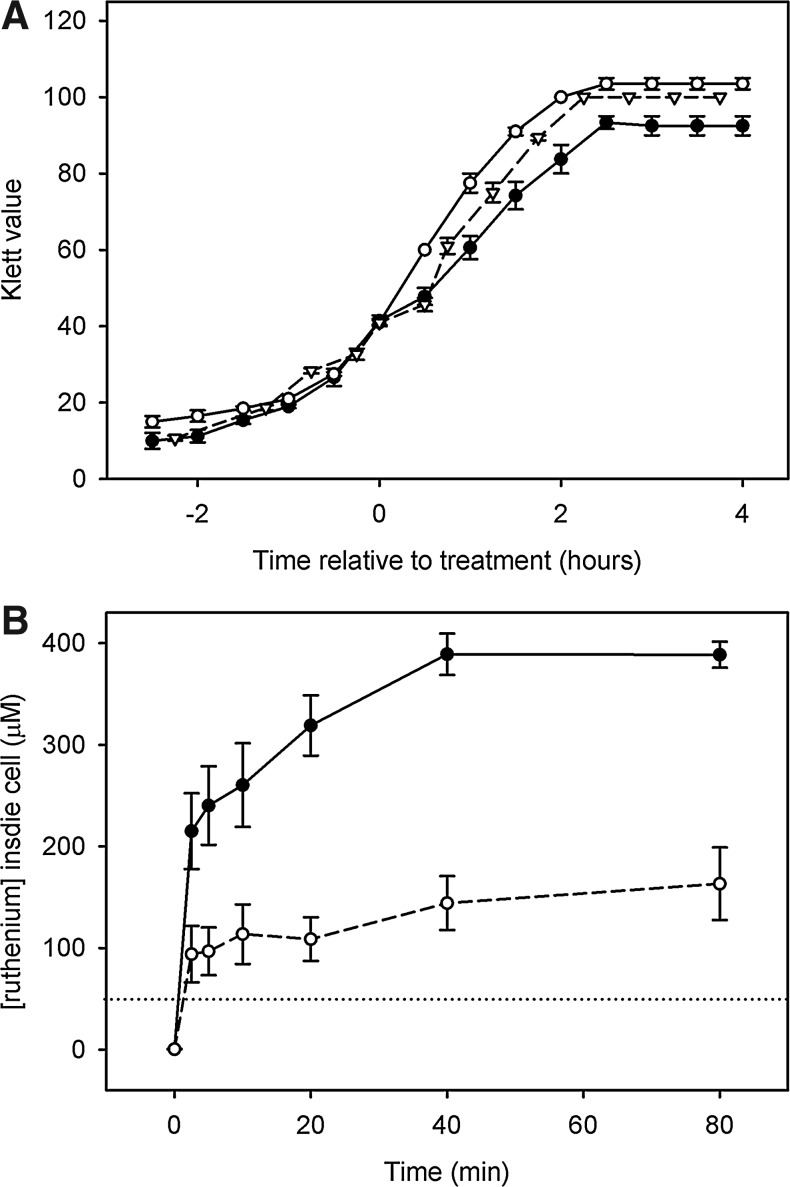
To establish a concentration of CORM-3 that was mildly inhibitory, but not bactericidal, in the Evans growth medium, cells were grown in batch culture to an OD600 ∼0.4, when CORM-3 or iCORM-3 was added to a final concentration of 40 μM. After addition of 40 μM CORM-3, a slight transient reduction in the growth rate was seen (Fig. 1A); this was sufficient to challenge bacterial cells without a significant loss in viability. Addition of 40 μM iCORM-3 resulted in no perturbation of growth.
FIG. 1.
Growth of Escherichia coli in Evans medium treated with Ru(CO)3Cl(glycinate) (CORM-3), inactivated CORM-3 (iCORM-3), or CO-saturated solution and intracellular uptake of the Ru compound. Cells were grown in batch culture to a turbidity of 40 Klett units and treated with 40 μM CORM-3 (closed circles), iCORM-3 (open circles), or CO-saturated solution (open triangles). (A) Growth was monitored every 30 min. N≥2±standard error of the mean (SEM). (B) Samples were taken at 0, 2.5, 5, 10, 20, 40, and 80 min post-treatment, and cell pellets were analyzed by inductively coupled plasma mass spectrometry for Ru content. N≥3±SEM. Dotted horizontal line represents an extracellular concentration of iCORM-3 (40 μM).
Previous work showed that CORM-3 is able to enter growing E. coli cells (10), but this assay was performed at only one time point. To expand upon this, we used inductively coupled plasma mass spectrometry to determine the level and rate of uptake of CORM-3 or iCORM-3 via measurement of the intracellular Ru content of cells over time. This method was chosen for detection of the CORM-3 and iCORM-3, as Ru is absent from untreated cells. CORM-3 was rapidly taken up by the cell as intracellular Ru accumulated at a rate in excess of 85 μM·min−1 over the first 2.5 min after treatment. Cellular Ru levels reached a plateau at ∼40 min (Fig. 1B), indicating that import of the compound had stopped or that an equilibrium was reached between import and export. Analysis of samples exposed to iCORM-3 showed that the concentration of this compound accumulated in cells with an initial rate of ∼40 μM·min−1 and to approximately half the final concentration of CORM-3 (Fig. 1B).
Whole-genome analysis shows distinct responses of E. coli treated with CORM-3 or iCORM-3
To assess how bacterial cells perceive and react to CORM-3 and iCORM-3 over time, transcriptomic analyses of continuously growing cultures were performed after exposure to a final concentration of 40 μM CORM-3 or iCORM-3 at 0 and 100% perceived aerobiosis (2). This is a measure of the extent of O2 deprivation, judged not by residual O2 levels in the chemostat, but by assaying the formation of acetate, a fermentation product that is produced when cells can no longer respire aerobically. Samples were taken immediately before addition of the CO-RM and 2.5, 5, 10, 20, 40, and 80 min after, to gain an understanding of the initial responses and subsequent effects.
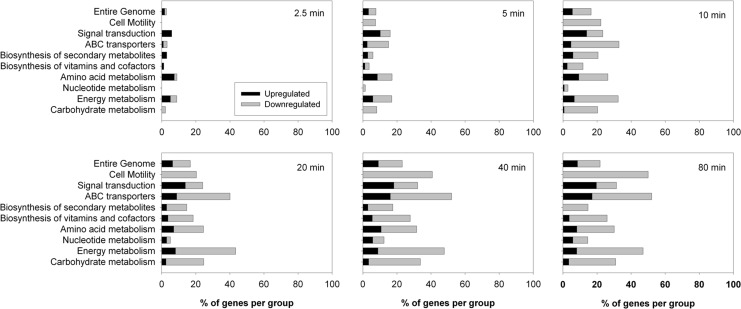
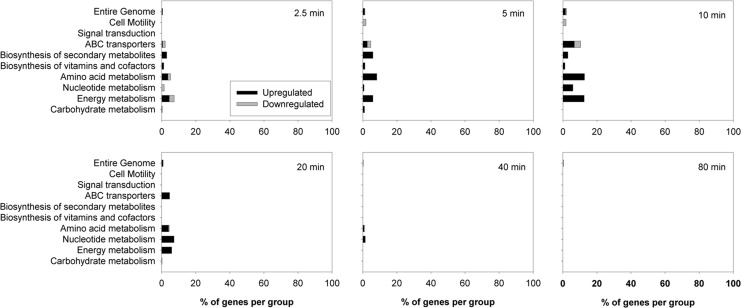
Under aerobic conditions, addition of CORM-3 caused changes in gene expression across a wide variety of functional groups with up to 23% of the entire genome being significantly altered (<0.5-fold or >2-fold change in expression) 40 min after exposure (Fig. 2). Changes in gene expression rose rapidly in the first 10 min, followed by a more gradual increase up to 80 min. In contrast, upon exposure to iCORM-3, there was a much smaller response with a peak of only ∼2% of the entire genome being altered at 10 min, followed by a decline in activity to virtually zero by 40 min (Fig. 3).
FIG. 2.
Expression changes of genes belonging to functional groups in response to CORM-3 exposure under aerobic conditions. Wild-type E. coli cells were grown aerobically in a continuous culture and a defined minimal medium before addition of 40 μM CORM-3. Samples were taken at 0, 2.5, 5, 10, 20, 40, and 80 min after addition of the compound and analyzed transcriptomically. The bars show the percentage of genes belonging to each group that were altered at each time point. Black bars indicate upregulation, and gray bars indicate downregulation.
FIG. 3.
Expression changes of genes belonging to functional groups in response to iCORM-3 exposure under aerobic conditions. Wild-type E. coli cells were grown aerobically in a continuous culture and a defined minimal medium before addition of 40 μM iCORM-3. Samples were taken at 0, 2.5, 5, 10, 20, 40, and 80 min after addition of the compound and analyzed transcriptomically. The bars show the percentage of genes belonging to each group that were altered at each time point. Black bars indicate upregulation, and gray bars indicate downregulation.
In response to both compounds, many genes involved in energy metabolism, amino acid metabolism, and ABC transport were altered. However CORM-3 had much more broad-ranging effects, including alteration of genes involved in cell motility, signal transduction, and carbohydrate metabolism (Fig. 2).
Under anaerobic conditions, there were also changes in genes from a wide variety of functional groups after exposure to CORM-3 (∼24% of the entire genome by 80 min; Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). There were many more genes upregulated under these conditions in contrast to aerobically grown cultures, where the general trend was downregulation. When challenged with iCORM-3 under anaerobiosis, there was also a broad response, but only ∼6% of the genome was significantly altered at its peak (20 min; Supplementary Fig. S2). Genes belonging to broadly the same functional groups were altered by iCORM-3, both aerobically and anaerobically.
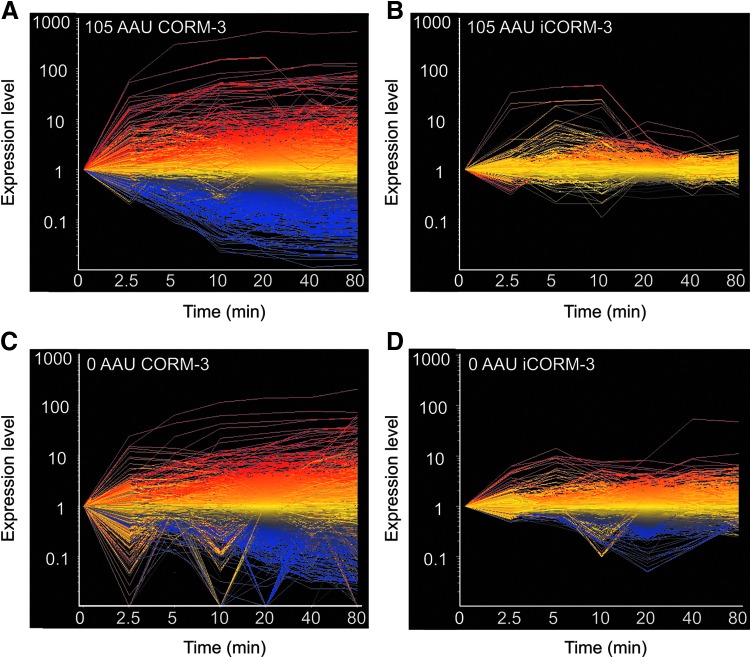
Although the genes altered were from a wide range of functional groups under all conditions, the magnitude of these changes was markedly different between the conditions. CORM-3 under both the aerobic and anaerobic conditions caused much greater alterations in expression of the genes than iCORM-3 (Fig. 4). Full details of all transcriptomic data can be found in the Gene Expression Omnibus database (accession: GSE40811).
FIG. 4.
The patterns of E. coli gene expression after exposure to CORM-3 or iCORM-3 are distinct. The whole-genome expression profiles of E. coli cultures treated with 40 μM CORM-3 (A, C) or iCORM-3 (B, D) are shown over a time course of 2.5, 5, 10, 20, 40, and 80 min. Each line represents a single gene, and those upregulated after 20 min are shown in red; downregulated genes at this time are shown in blue; and genes that do not significantly alter in expression level at 20 min are shown in yellow.
Differential effects of CORM-3 and iCORM-3 under aerobic and anaerobic growth conditions
Gene expression timeseries, generated from microarray analysis, were used to infer transcription factor (TF) activities using the TFInfer statistical software suite (3). Given microarray data from E. coli grown under aerobic or anaerobic conditions and either in the presence or in the absence of CORM-3 or iCORM-3, TFInfer returns the probability distributions for the activity of each TF, as well as the strengths of the TF–target interactions (see the Materials and Methods section).
Here, we are interested in probing the way in which TFs respond to different stimuli. We tested two complementary aspects: which TFs have a similar dynamical response to different stimuli, and which TFs have a significantly different response to different stimuli. To answer the first question, absolute Pearson correlation coefficients were used to compare TF activities in different conditions. Using the probabilistic nature of TFInfer software, we are able to compute a distribution for the (absolute) correlation coefficient between the TF response to CORM-3 and iCORM-3 in aerobic and anaerobic conditions (see Supplementary Data for why the absolute value was used). The TFs were ranked according to their mean absolute correlation coefficient, where a low number indicates that the TF responses had a low correlation between the two conditions, and a number approaching 1 indicates high correlation.
The Pearson correlation comparison results are shown in Table 1 for aerobic growth conditions and Table 2 for anaerobic growth conditions. In Table 1, for example, the activity of ArcA, which represses respiratory metabolism, is uncorrelated upon exposure to CORM-3 and iCORM-3 under aerobic conditions. Further examination reveals that the activity of this TF is increased upon exposure to CORM-3, but to a much lesser extent upon exposure to iCORM-3 (Supplementary Tables 1–4). Anaerobically, however, the response of ArcA to CORM-3 and iCORM-3 shows a much greater correlation (Table 2). In the case of IscR, which is involved in the regulation of the iron–sulfur cluster assembly, there is a similar pattern of behavior when challenged with either CORM-3 or iCORM-3 both aerobically and anaerobically, suggesting that damage to the iron–sulfur clusters could be a consequence of exposure to the CORM-3/iCORM-3 Ru-based compound (Tables 1 and 2).
Table 1.
Absolute Pearson Correlations Comparing Cells Treated with CORM-3 or iCORM-3 Under Aerobic Conditions
| Transcription factor name | Mean absolute Pearson correlation | Standard deviation of correlation |
|---|---|---|
| CRP |
0.079 |
0.059 |
| ArcA |
0.125 |
0.091 |
| IHF |
0.195 |
0.072 |
| FlhDC |
0.197 |
0.134 |
| CusR |
0.212 |
0.139 |
| OmpR |
0.226 |
0.164 |
| HNS |
0.276 |
0.151 |
| EvgA |
0.281 |
0.165 |
| MarA |
0.299 |
0.206 |
| FruR |
0.338 |
0.213 |
| PdhR |
0.341 |
0.227 |
| MhpR |
0.342 |
0.228 |
| YdeO |
0.343 |
0.228 |
| GalR |
0.344 |
0.230 |
| Lrp |
0.345 |
0.110 |
| RstA |
0.352 |
0.230 |
| MalT |
0.352 |
0.207 |
| GlcC |
0.355 |
0.232 |
| IclR |
0.358 |
0.234 |
| NarP |
0.368 |
0.237 |
| Nac |
0.384 |
0.246 |
| GadE |
0.401 |
0.249 |
| GalS |
0.422 |
0.252 |
| DgsA |
0.439 |
0.246 |
| Fur |
0.506 |
0.213 |
| AraC |
0.555 |
0.229 |
| NarL |
0.601 |
0.226 |
| ModE |
0.631 |
0.215 |
| GadWX |
0.723 |
0.156 |
| IscR |
0.734 |
0.126 |
| PhoP |
0.745 |
0.168 |
| GcvA |
0.790 |
0.036 |
| NtrC |
0.814 |
0.093 |
| Fis |
0.823 |
0.097 |
| FNR |
0.867 |
0.071 |
| CpxR | 0.946 | 0.030 |
Transcription factors (TFs) are ranked by their mean absolute Pearson correlation obtained by sampling the TF profiles from the posterior distribution obtained through the inference procedure described in the Materials and Methods section. Listed also is the resulting standard deviation of the absolute Pearson correlation.
CORM-3, Ru(CO)3Cl(glycinate); iCORM-3, inactivated CORM-3.
Table 2.
Absolute Pearson Correlations Comparing Cells Treated with CORM-3 or iCORM-3 Under Anaerobic Conditions
| Transcription factor name | Mean absolute Pearson correlation | Standard deviation of correlation |
|---|---|---|
| NtrC |
0.158 |
0.085 |
| PdhR |
0.179 |
0.128 |
| Nac |
0.186 |
0.139 |
| DgsA |
0.239 |
0.168 |
| CusR |
0.249 |
0.179 |
| Fur |
0.305 |
0.197 |
| NarP |
0.327 |
0.222 |
| AraC |
0.344 |
0.228 |
| YdeO |
0.344 |
0.227 |
| GalR |
0.345 |
0.230 |
| MhpR |
0.345 |
0.228 |
| Lrp |
0.346 |
0.112 |
| IclR |
0.348 |
0.228 |
| ModE |
0.352 |
0.229 |
| FlhDC |
0.376 |
0.160 |
| FruR |
0.380 |
0.138 |
| EvgA |
0.380 |
0.128 |
| GlcC |
0.417 |
0.247 |
| NarL |
0.556 |
0.194 |
| CRP |
0.592 |
0.031 |
| MalT |
0.628 |
0.127 |
| GalS |
0.659 |
0.202 |
| MarA |
0.675 |
0.212 |
| Fis |
0.697 |
0.077 |
| ArcA |
0.735 |
0.056 |
| OmpR |
0.758 |
0.146 |
| CpxR |
0.791 |
0.026 |
| GadE |
0.820 |
0.126 |
| IHF |
0.822 |
0.048 |
| GcvA |
0.865 |
0.062 |
| HNS |
0.867 |
0.043 |
| GadWX |
0.874 |
0.048 |
| RstA |
0.886 |
0.059 |
| IscR |
0.897 |
0.042 |
| FNR |
0.902 |
0.059 |
| PhoP | 0.953 | 0.024 |
TFs are ranked by their mean absolute Pearson correlation obtained by sampling TF profiles from the posterior distribution obtained through the inference procedure described in the Materials and Methods section. Listed also is the resulting standard deviation of the absolute Pearson correlation.
To assess significantly different TF responses across conditions, a modification of the TFInfer algorithm was needed. Briefly, we combined the data from two conditions (e.g., CORM-3 exposure vs. iCORM-3 exposure) to simultaneously infer TF activity in both conditions under the (null) hypothesis that the activity of a given single TF is identical in both conditions. By checking how the model fits the gene expression profiles of the targets of the given TF, we obtain a p-value for any given TF being differentially active under the two different conditions. In this way, we can determine whether the activity of a given TF is significantly different under each condition tested (in this case, CORM-3 vs. iCORM-3) (see the Materials and Methods section and Supplementary Data for more details).
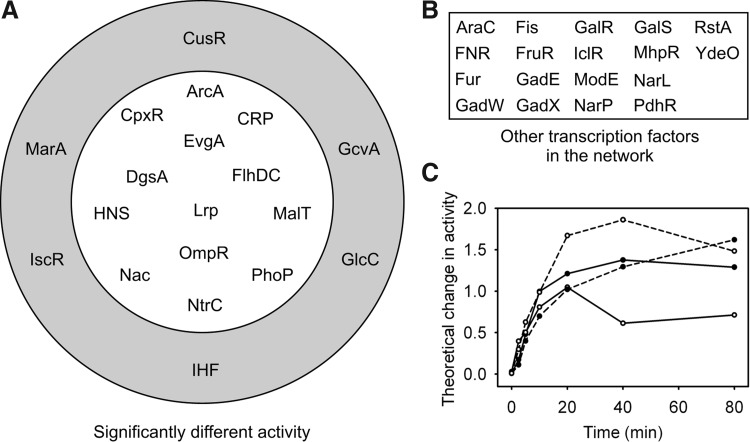
Figure 5 shows the TFs identified as being differentially affected (p<0.05) by exposure to CORM-3 versus iCORM-3 under both the aerobic and anaerobic growth conditions. It can be seen that there are 6 TFs identified as being differentially affected under aerobic conditions only and 13 identified as being differentially affected under both conditions, and that the TFs differentially affected anaerobically are a subset of those affected aerobically.
FIG. 5.
Differential transcription factor (TF) activity of cells exposed to CORM-3 versus iCORM-3 under aerobic or anaerobic conditions. (A) TFs identified as having a significantly different activity (p<0.05) after exposure to CORM-3 or iCORM-3. The outer rings of TFs are those that are identified as differentially active under aerobic growth conditions only. The inner circle contains those TFs identified as differentially active under both aerobic and anaerobic conditions. (B) The remaining TFs that were not identified as differentially active under either aerobic or anaerobic growth conditions. (C) Theoretical change in activity of the TF IscR (as generated by TFInfer) in response to CORM-3 (closed circles) and iCORM-3 (open circles) under aerobic (solid lines) or anaerobic (dashed lines) conditions.
Interestingly, IscR was differentially affected by CORM-3 and iCORM-3 under aerobic conditions in this test. As the activity of this TF was found to correlate under these conditions (Table 1), this suggests that while the response of IscR correlates after exposure to either molecule, that is, its pattern of activity is similar (Tables 1 and 2), the magnitude of that response may differ making the activities significantly different here (Fig. 5C). Another similar example is that of ArcA, mentioned above, which has much higher correlation in anaerobic conditions, but is still identified as having significantly different activity under exposure to CORM-3 or iCORM-3.
Carbohydrate and intermediary energy metabolism is altered in response to CORM-3
In the presence of CORM-3, there was a general trend of downregulation in carbohydrate metabolism and transport genes from ∼10 min post-treatment (Fig. 2). Accompanying this trend was a downregulation of the genes involved in energy metabolism. Many genes involved in oxidative phosphorylation, the reductive carboxylate cycle, and ubiquinone biosynthesis were downregulated (Supplementary Figs. S1, S3, and S4). The formate dehydrogenase-O genes, which rapidly increase in the expression levels in response to a switch from aerobic to anaerobic conditions, were also downregulated under these conditions (1). This indicates that the presence of CORM-3 does not simply cause the cell to shift to an anaerobic form of energy metabolism to bypass the quinol oxidases, known to be inhibited by CO (23). Upon exposure to iCORM-3, carbohydrate transport and metabolism and energy metabolism were largely unaffected (Fig. 3 and Supplementary Figs. S2–S4). Real time PCR (RT-PCR) confirmed that the expression of cyoD, the gene encoding the cytochrome bo terminal oxidase subunit IV, was also downregulated in response to treatment of growing cultures with CO gas at a rate of 50 ml·min−1 (Supplementary Fig. S5), confirming the role of CO-RM-derived CO and CO gas per se in targeting energy metabolism.
ABC transporter expression is altered in response to CORM-3
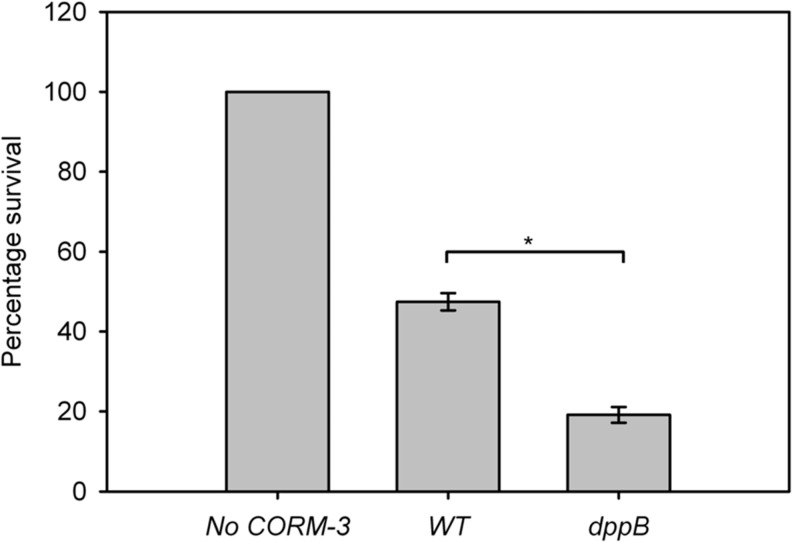
Transport also appears to be a crucial response to CORM-3. Many ABC transporters were downregulated, including the dipeptide permease dppABCDF, responsible for transport of dipeptides into the cell (see GEO: GSE40811). In response to iCORM-3, the downregulation was much less pronounced (with a maximum approximately threefold downregulation). Further, the expression of dppB, in samples exposed to CO gas (50 ml·min−1), was not significantly altered (Supplementary Fig. S5), suggesting that the response of this transporter is probably due to the CORM-3 compound. To determine whether the dipeptide permease was a mechanism by which CORM-3 enters the cell and thus the reason for its apparent downregulation by the bacterium in response to CO-RM, a dppB knockout strain was tested. When challenged with CORM-3, we found that the dppB mutant was more sensitive to CORM-3 treatment than the wild-type strain (Fig. 6), suggesting that not only was the CORM-3 molecule still able to enter the cell but also a loss in function of the dipeptide permease is disadvantageous after CORM-3 treatment. Indeed, CORM-3 entry into the dppB knockout strain was confirmed by Ru analysis after addition of the compound to a final concentration of 40 μM. After 20-min exposure, 227±19 μM Ru was found inside the cells, confirming that CORM-3 passage into the cell is not significantly affected by loss of the dipeptide permease transport system.
FIG. 6.
Mutation of the dipeptide permease transporter causes greater sensitivity to CORM-3. Cultures of WT and dppB were grown in the Evans medium to early-exponential phase before washing and dilution in PBS to ∼106 colony forming units (CFU)·ml−1. Cells were exposed to 0.5 μM CORM-3 and incubated for a further hour before plating serial dilutions to determine the viability as a percentage survival compared to no-CORM-3 treatment. N=3±SEM, * denotes p-value<0.01 as determined by the Student's t-test. CFU, colony forming units.
Several other transporters were upregulated in response to CORM-3, including the sulfate–thiosulfate transport system encoded by cysPUWA, the oligopeptide permease encoded by oppABCDF, the methionine transporter (metQIN), and the glutathione transporter (gsiABCD) (see GEO: GSE40811). There was no significant alteration in expression of these systems upon exposure to iCORM-3, and RT-PCR analysis of samples exposed to bubbling with CO gas showed no significant change in expression of oppB (Supplementary Fig. S5), perhaps suggesting that greater local concentrations of CO must be targeted to cells, as is the case with CORM-3, to elicit a transcriptomic response in these cases.
Cell motility is impaired upon exposure to CO
Expression of the genes involved in cell motility was diminished in response to CORM-3 exposure under aerobic conditions (Fig. 2). In response to iCORM-3, however, the expression of these genes was largely unaffected. To determine whether the decrease in expression after treatment with CORM-3 resulted in reduced motility, swarming assays were performed under normal conditions and under an atmosphere of 50% CO, as previously described (31,39). After 48-h incubation, the mean colony diameters were measured as 9.4±0.5 mm for colonies grown under normal conditions, whereas cells grown in a 50% CO atmosphere had a reduced colony diameter of 6.3±0.6 mm. The Student's t-test, which compares the difference between two means in relation to the variance in the data, revealed that the difference between these data sets is highly significant (p-value<0.001), which suggests that aerobic exposure to CO results in a lowered expression of genes responsible for flagella function and chemotaxis with a corresponding reduction in cell motility.
Sulfur metabolism is a target of CORM-3 and iCORM-3 activity
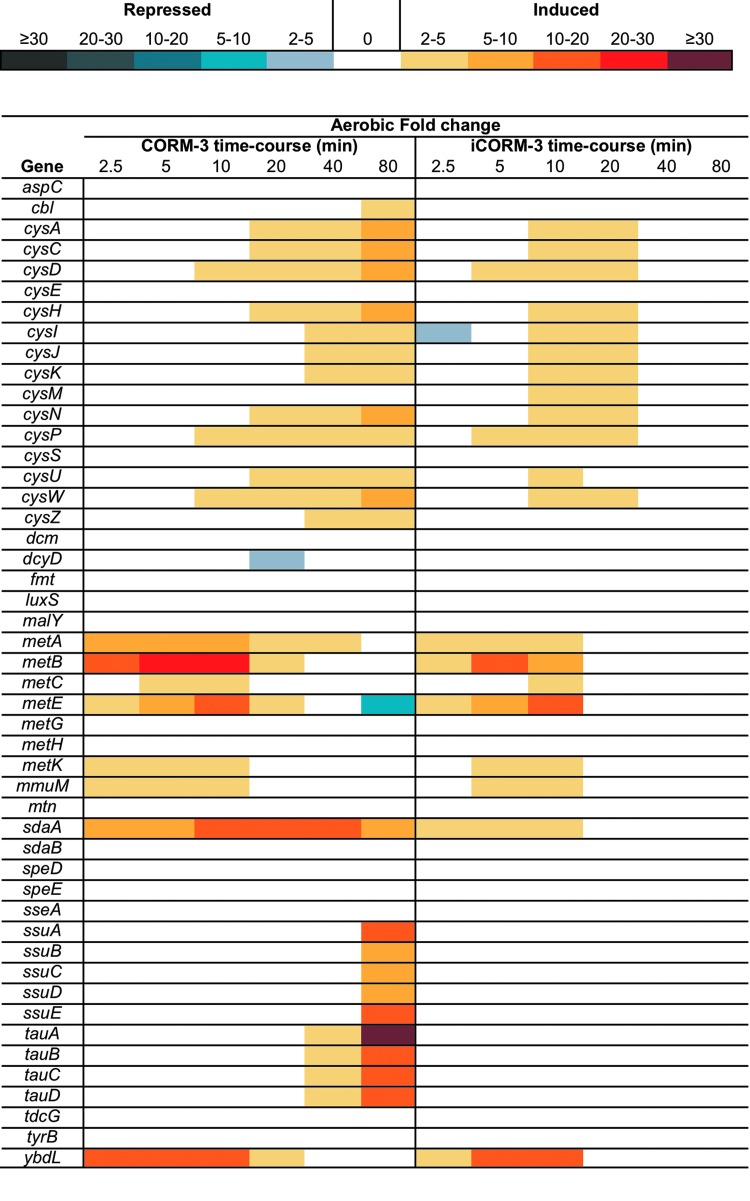
Our data show that sulfate transport and utilization, including the sulfate–thiosulfate transport system and methionine metabolism and transport (cysPUWA and metQIN), were altered upon exposure to CORM-3, and to a lesser extent, to iCORM-3 (Fig. 7 and Supplementary Fig. S6). Increases in the sulfate–thiosulfate, cysteine, and methionine transport systems as well as increases in expression of the cysteine and methionine biosynthesis genes point to a requirement for sulfur and its metabolism in the presence of the Ru compound regardless of the presence or absence of the labile CO. However, an increase in expression of genes that respond to sulfur starvation (ssuABCDE and tauABCD) was observed only in conditions when CORM-3 was administered (Fig. 7 and Supplementary Fig. S6). The tauABCD regulon, responsible for utilization of taurine as a sulfur source, whose expression is regulated by a response to sulfur starvation, is also greatly increased at later time points (2-fold to 65-fold from 10 min). The significance of this is not understood at this time.
FIG. 7.
Differential expression of genes involved in sulfate transport and metabolism. The color-scale bar shows the mean fold changes in individual genes of E. coli grown aerobically in the Evans medium after the addition of 40 μM CORM-3 or iCORM-3.
CORM-3 causes a reduction in free thiol levels
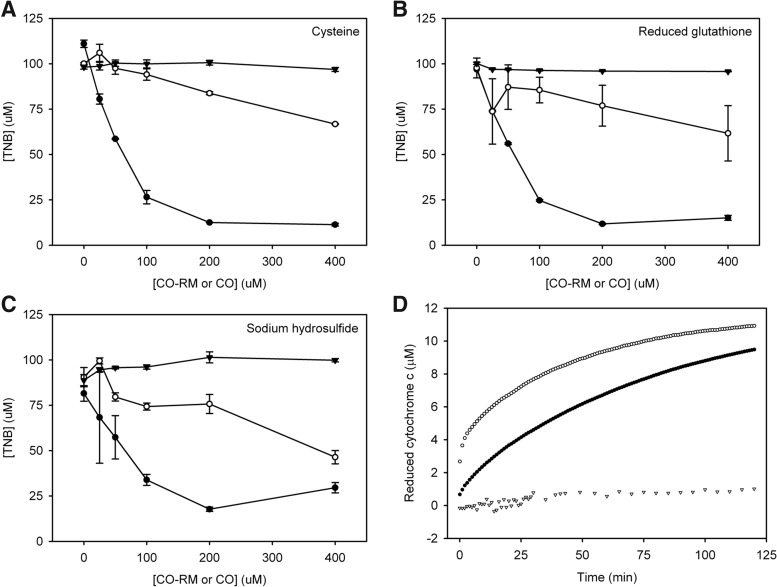
To determine whether CORM-3 was altering free thiol levels in vivo and thus causing the alterations seen in the transcriptomic data of genes that effect thiol metabolism within the cell, assays were carried out whereby reduced glutathione, cysteine, and sodium hydrosulfide were exposed to increasing concentrations of CORM-3, iCORM-3, or CO-saturated solution, and the presence of free thiols was measured by the addition of 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (14). CORM-3 appeared to reduce the number of thiols in solution in all cases (Fig. 8). When iCORM-3 was added in place of CORM-3, the decrease in the level of free thiols was less pronounced, but still significant, suggesting that it is not only CO release that causes the decrease in free thiols in the CORM-3 assays. Indeed, there was no significant effect on the concentration of free thiol groups upon exposure to CO alone (Fig. 8).
FIG. 8.
CORM-3 has a biphasic effect on the presence of free thiols from glutathione, sulfide, and cysteine. CORM-3 (0–400 μM) (closed circles), iCORM-3 (open circles), or CO-saturated solution (closed triangles) was mixed with 100 μM (A) cysteine, (B) reduced glutathione, or (C) sodium hydrosulfide and incubated for 15 min at room temperature; 500 μM 5,5′-dithiobis-(2-nitrobenzoic acid) was added, and the solution incubated for a further 15 min before the absorbance was measured at 412 nm. N=3±SEM. (D) Cytochrome c (20 μM) was added to CORM-3 (1 mM, open symbols) or iCORM-3 (1 mM, closed symbols) in the presence (triangles) or absence (circles) of superoxide dismutase (250 U) in KPi (pH 7.4). The OD550 was measured over time as a measure of the reduction of cytochrome c. N=3, standard errors were≤1 μM.
Previous work has shown that another Ru-based CO-RM, [Ru(CO)3Cl2]2 [CORM-2, (29)], is able to generate reactive O2 species (41). To determine whether similar species are generated by CORM-3 and cause the decrease in free thiol levels in the DTNB assays, we investigated whether CORM-3 and iCORM-3 were able to produce superoxide. We looked for the generation of this species from CORM-3 via the addition of cytochrome c to a solution of CORM-3 in buffer. This assay uses the reduction of cytochrome c by superoxide, which can be measured spectrophotometrically (42). Figure 8D shows a reduction of cytochrome c in the presence of CORM-3, a reaction that is almost entirely abolished by the addition of superoxide dismutase. The addition of iCORM-3 to cytochrome c appears to enhance formation of the reduced species, suggesting that more superoxide is generated by iCORM-3 than CORM-3. However, the superoxide generated by these compounds is very small, only ∼1% of the CORM-3 or iCORM-3 concentration; therefore, the impact this would have on the DTNB assay is negligible.
While CO seems to play no direct role in reduction of free thiol levels, the Ru compound does seem to cause a loss of free thiols available to the DTNB (Fig. 8). It is likely that the thiols bind to Ru, possibly displacing a chloride ion on the CORM-3 and iCORM-3 compounds.
Thiol-containing compounds prevent CORM-3-dependent inhibition of respiration in E. coli membrane particles
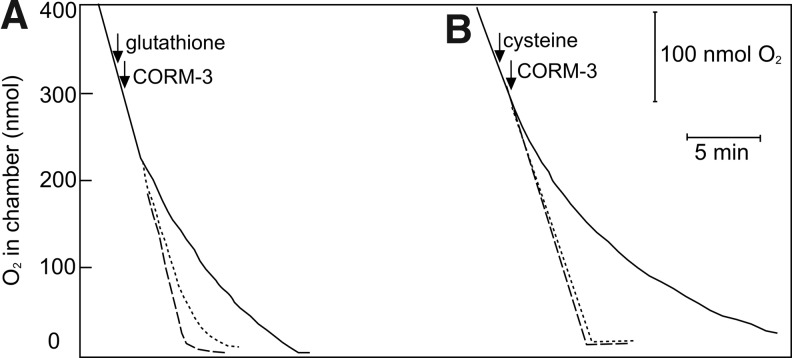
Another possible reason for the alteration in expression of thiol-containing compounds is that they may afford protection to the cell via interaction with CORM-3. One striking target of CORM-3 is respiration via CO binding to the terminal quinol oxidases (10). To assess whether thiol-containing compounds were able to reduce the inhibition of respiration caused by CORM-3, the respiration of the E. coli membrane particles was monitored by measuring O2 consumption in a closed electrode system (18). Inhibition of respiration was observed after the addition of 400 μM CORM-3 to the membrane suspensions; this effect was abolished upon addition of reduced glutathione (200 μM) or cysteine (400 μM) (Fig. 9), which suggests that increasing thiol concentrations may be an effective protective mechanism for cellular respiration.
FIG. 9.
Thiol-containing compounds prevent CORM-3-dependent inhibition of respiration in E. coli membrane particles. E. coli membrane particles were added to the sonication buffer to a final concentration of ∼60 μg·ml−1 inside the O2 electrode chamber. A lid was placed on the chamber, and respiration was initiated by the addition of 6.25 mM nicotinamide adenine dinucleotide. Arrows indicate the addition of (A) 200 μM reduced glutathione or (B) 400 μM cysteine and the addition of 400 μM CORM-3 as described. The dashed lines indicate uninhibited respiration; the solid lines show O2 consumption in the presence of CORM-3, and the dotted lines show O2 consumption in the presence of CORM-3 and the thiol-containing compound. Figure is representative of three biological repeats for each condition.
Discussion
Although previous work has shown that iCORM-3 has much less effect than CORM-3 on biological material (8,17,34,37), very little has been done to elucidate bacterial viability with, and specific responses to, iCORM-3. The data presented herein show that iCORM-3 (made via incubation of CORM-3 in PBS) has virtually no effect on the growth of E. coli, whereas at the same concentration, CORM-3 readily inhibits growth (Fig. 1A). We have shown that iCORM-3 is able to rapidly enter bacterial cells, but only accumulates to half the concentration as CORM-3 (Fig. 1B), indicating that the lack of bactericidal activity afforded by iCORM-3 can only be partially attributed to a decrease in cellular penetration. Further, our transcriptomic and modeling data reveal for the first time that there is a much greater response to CORM-3 than to iCORM-3 over the whole genome, with up to 24% of the genome being altered in response to CORM-3 and as little as 2% altered by the Ru compound devoid of labile CO (Figs. 2 and 3; Supplementary Figs. S1 and S2). This suggests that the majority of changes seen in transcriptomic experiments in the presence of CORM-3 are due to the release of free CO. One notable exception is the gene spy, whose expression is upregulated as much as ∼565-fold upon exposure to CORM-3, but only ∼4.6-fold upon exposure to the same concentration of iCORM-3, and is not significantly altered after continued exposure to CO gas at a rate of 50 ml·min−1. This suggests that exposure to the intact CORM-3 compound is somehow responsible for the changes seen.
Further analysis was undertaken via determination of TF activities under all conditions using probabilistic modeling tools. In Figure 5, one explanation for why the outer ring of TFs is differentially affected by CORM-3 versus iCORM-3 in aerobic growth conditions, but not anaerobic growth conditions, is that the TFs are responding to simulated anaerobic conditions induced by CO from CORM-3. If the cells are already growing under anaerobic conditions, this effect will not be manifested, so these TFs are not differentially affected under anaerobic conditions. The other group of TFs, differentially affected under both aerobic and anaerobic conditions, must be affected by some mechanism other than the simulated anaerobic conditions induced by CO. For example, the other TFs may be direct targets of CO.
The presence of CORM-3 caused a drop in expression of many genes, notably those encoding the dipeptide permease transporter, DppABC. In Salmonella enterica and E. coli, S-nitrosoglutathione is internalized by the DppABC transporter system (11,21). It has been proposed that a periplasmic transpeptidase removes the γ-glutamyl moiety of S-nitrosoglutathione, and that the residual dipeptide, S-nitroso-l-cysteinylglycine, is then transported inward using the Dpp-encoded dipeptide permease (7). Given our transcriptomic data, it was plausible that this transporter could be the route of CORM-3 uptake also. However, when the viability of the dipeptide transport knockout strain was tested in the presence of CORM-3, we found that this strain was more sensitive than the wild type, and that Ru is still able to accumulate inside the cells, suggesting that this is not the mechanism of CORM-3 import. The increased sensitivity could be due to defense against CORM-3 that requires the transport of dipeptides or that this transport system is required to repair the damage caused by the compound. The dipeptide permease is also capable of acting as a heme permease and is involved in endogenous periplasmic heme recycling (25). It could be that this role is vital in protecting the cell from CORM-3 via the shuttling of free heme to form new proteins that will replace the nonfunctional ones containing CO-bound heme.
It has been demonstrated several times in the literature that thiol-containing species abolish bactericidal activity and inhibition of respiration by transition metal-containing CO-RMs in P. aeruginosa and E. coli (12,13,41). In agreement with this, we have shown for the first time that cysteine and reduced glutathione are able to abolish the inhibition of respiration caused by exposure to CORM-3 in E. coli membranes (Fig. 9). It has also been demonstrated that the thiol-containing N-acetylcysteine does not interfere with CO release from CORM-3 in the presence of dithionite (12). However, it is possible that in the presence of membranes alone (where dithionite is not present), the species competing with CO to bind the Ru compound and thus release CO (28) are less competitive, and so interactions between thiol compounds and CORM-3 cause a conformational change that results in slower release of CO and thus an abolition of respiratory inhibition. Indeed, it has been proposed that, after entering blood, CORM-3 and other fac-[Ru(CO)3]2+ compounds react rapidly with plasma proteins, lose one equivalent of CO as CO2, and form protein–Ru(CO)2 adducts that slowly decay with a stepwise loss of CO in contrast to the rapid release seen in a buffer containing dithionite (36,37).
Together, these data highlight the importance of fully characterizing both the compound of interest and any potential inactivated counterpart or product that may be formed, in this case the Ru compound with labile CO removed.
Materials and Methods
Mb-CO assays
Mb-CO assays were performed using a dual-beam Rapid Scanning Monochromator (RMS1000) spectrophotometer (On-Line Instrument Systems, Inc.) as previously described (28).
E. coli strains and growth conditions
E. coli K12 derivative MG1655 was the strain used in all studies. The dppB mutant was obtained from the Keio collection (4,43) and transduced into MG1655 via phage P1 (24,44). Cells were grown in a glucose-limited medium based on that of Evans et al., unless otherwise stated (15) with the following exceptions: 2 mM nitrilotriacetic acid was used as the metal ion chelator replacing citrate; 30 μg·L−1 sodium selenite was added to the medium; and 2–20 mM glucose was used as the carbon source. For batch culture, cells were grown at 37°C and with shaking at 200 rpm in 250- or 500-ml flasks. For continuous culture, cells were grown in an Infors Multifors bioreactor, adapted to fit a Labfors-3 fermentor base unit. Mass flow controllers allowed the correct ratio of air and pure N2 gas (aerobic growth) or 10% O2/90% N2 and pure N2 gases (anaerobic growth) to be mixed to produce the desired O2 input into the chemostat.
Measurement of O2 availability in continuous cultures
Measurement of perceived aerobiosis levels, as perceived by cell cultures within the chemostat, was achieved as previously described (2). Acetate levels were measured using an Enzymatic Bioanalysis kit from R-Biopharm and following the manufacturer's instructions.
CORM-3, iCORM-3, and CO preparation and treatment
CORM-3 was prepared as previously described (8); a 100 mM stock solution was made fresh each day by solubilizing in distilled water, sparged with N2, and kept in the dark. iCORM-3 was prepared by dissolving CORM-3 in PBS at a final concentration of 10 mM and incubating at room temperature for ∼48 h, during which time the solution was sparged periodically with N2 to remove any free CO present. iCORM-3 was assayed for the presence of CO still capable of release by the Mb-CO assay and was used if the resulting Mb-CO formed was <5% of that produced from incubation with CORM-3.
CO-saturated solutions were made by bubbling PBS (pH 7.4) with CO gas for >30 min and used immediately. This gave a stock concentration of CO ∼1 mM.
CORM-3, iCORM-3, or CO-saturated solution were added directly to continuous cultures at a steady-state OD600 ∼0.4.
Ru analysis
Cultures were grown to log-growth phase (OD600 ∼0.4), where 20 ml samples were taken both before and at regular intervals after the addition of 40 μM CORM-3 or iCORM-3. Cells were centrifuged at 5.5 krpm for 20 min in polypropylene tubes, and the pellets washed three times in 0.5% nitric acid. Cell pellets, combined washes, and culture supernatants were assayed in the Chemistry Department, The University of Sheffield, for metal analysis by inductively coupled plasma mass spectrometry (10,19).
Transcriptomic analysis
Samples were removed from chemostat cultures (two biological replicates per condition) into an ice-cold solution of phenol/ethanol in a ratio of 1:19 to rapidly stabilize the RNA. Total RNA was purified using an RNeasy RNA purification kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized using 16 μg of RNA primed with 5 μg Random Primers (Invitrogen). Reaction mixes (30 μl) containing 0.5 mM deoxyadenosine triphosphate, deoxythymidine triphosphate, and deoxyguanosine triphosphate, 0.2 mM deoxycytidine triphosphate (dCTP), and 67 μM Cy-3 or Cy-5-dCTP were incubated at 25°C for 5 min, and then at 50°C overnight with 300 U of Superscript III Reverse Transcriptase (Invitrogen). After synthesis, cDNA was purified using a PCR purification kit (Qiagen).
Procedures for RT-PCR were as described previously (16). For hybridization to the microarray slides, 400 ng cDNA was added to 25 μl 2×Hi-RMP Hybridization Buffer and 5 μl 10×Gene Expression Blocking Agent (Agilent Technologies) to give a 50-μl total volume. Custom Gene Expression Microarray slides each containing 8×15K arrays were purchased from Agilent Technologies. For hybridization, one 8×Gasket Slide was placed inside a Microarray Hybridization Chamber, and the samples were loaded for eight different conditions, followed by placement of the microarray slide over the gasket. After sealing the chamber, the arrays were incubated at 65°C, rotating at 10 rpm for ∼17 h.
After incubation, the slides were washed in Gene Expression Wash Buffer 1 for 1 min, followed by washing in Gene Expression Wash Buffer 2 for 1 min. The slides were dried and scanned in a microarray scanner (Agilent) with subsequent feature extraction and data analysis using GeneSpring GX v7.3.
Modeling TF activities using TFInfer
Using the model in Sanguinetti et al. (35), we express the log-fold change of gene expression under perturbation as the linear combination of TF activities. The formulae and explanation of the terms are included in the Supplementary Data.
The inference procedure used then gives probability distributions for the TF profiles, and TF–gene interaction strengths based on the model and the observed gene expression data, with the means of the distributions providing point estimates for these terms. Although the model is a simplified representation of transcriptional regulation, it is this simplicity that makes it possible to perform large-scale statistical inference, so that one may obtain data-driven estimates of TF activities (20).
A network of 37 TFs and 714 target genes was used for this statistical modeling. This network consists only of the TF–gene interactions that have been confirmed directly by experiments such as binding of purified proteins [e.g., the interaction of CpxR with the spy promoter (32)]. This network is used to exclude potentially spurious TF–gene interactions and uses only the interactions supported by very strong evidence. Two included TFs, GadW and GadX, have the same targets in this restricted representation of the transcriptional network, so have been considered together in the analyses, where we regard the combined contribution of GadW and GadX under the heading GadWX.
Measuring similarity in TF activities between two different conditions
The model above was applied independently to two data sets from two conditions (i.e., exposed to CORMs or exposed to iCORMs). Since the inference procedure gives a probability distribution for TF activity, this implies a probability distribution for the correlation between the activities for a given TF in two different conditions. We calculate the mean and standard deviation of the distribution of the absolute Pearson correlation by sampling the TF activities from the inferred distribution and calculating the absolute Pearson correlation of the samples (see Supplementary Data for details as to why the absolute value of the Pearson correlation was used).
Significance testing for differences in TF activity
The TFInfer model above was modified to perform inference of two sets of TF activities simultaneously, while enforcing the condition that the activity of a given TF must be the same in both conditions. Subtracting the linear combination of TF activities from the gene expression data then leaves (according to the model) independent, normally distributed residuals. Squaring the residuals and summing them gives a chi-squared null distribution against which we can check the residuals from the data to check for deviation from the expected results under the null hypothesis. More details may be found in the Supplementary Data.
In this way, we test for significant differences in TF activity between two different conditions. This is in contrast to the study of the Pearson correlation coefficients, which give some measure of similarity of TF activity.
Motility assays
The swarming behavior was assessed as previously described (31); briefly, 5 μl of liquid culture in the stationary phase was spotted onto 0.3% (w/v) LB/agar. Plates were incubated in sealed anaerobic jars containing a normal or 50% CO gas atmosphere. Colony diameters were measured after incubation for 48 h at 30°C, and the values shown represent the calculated mean±standard deviation from 16 repeats per condition.
Bactericidal assays
Cultures were grown in 20 ml LB medium to an OD600 ∼0.5 at 37°C and with shaking at 200 rpm. Samples were removed, washed twice with PBS (pH 7.4), and diluted at ∼1×106 colony forming units (CFU)·ml−1. The aliquots (5 ml) were treated with the stated concentration of CORM-3 or iCORM-3 and incubated for 1 h at 37°C with gentle shaking. For each sample, a range of serial dilutions were plated onto LB agar and incubated overnight for the determination of cell viability.
Thiol and superoxide assays
For the assay of free thiols, cysteine, reduced glutathione, or sodium hydrosulfide was incubated at room temperature for 15 min with various concentrations (25–400 μM) of CORM-3, iCORM-3, or CO-saturated solution. DTNB (0.5 mM) was subsequently added, and the solution incubated for a further 15 min. The concentration of free thiols was determined by measuring the OD412 using a Jenway spectrophotometer.
To assay for the presence of superoxide, 20 μM cytochrome c was added to 1 mM CORM-3 in KPi (pH 7.8). The OD550 was monitored over time to follow the appearance of reduced cytochrome c. Superoxide dismutase (250 U) was included where indicated.
Respiration measurements
Cells were grown in LB under high aeration at 37°C and with shaking at 250 rpm until late-exponential phase. Cultures were harvested and the membranes prepared as previously described (32).
E. coli membrane particles were resuspended in 2 ml assay buffer (50 mM Tris-HCl, 2 mM magnesium chloride, and 1 mM ethylene glycol tetraacetic acid) to a final concentration of ∼175 μg·ml−1, in a stirred Perspex chamber fitted with a Clark-type polarographic O2 electrode (OXY041A; Rank Bros Ltd., Bottisham, CB25 9DA) held at 37°C (18). The chamber was closed, and respiration initiated by the addition of 6.25 mM nicotinamide adenine dinucleotide. The thiol compound (cysteine, at 400 μM or reduced glutathione at 200 μM) was added to the chamber when the O2 concentration reached ∼165 μM. CORM-3 (400 μM) was added 1 min later. Data were recorded on a chart reader (REC112; Amersham Pharmacia Biotech).
Supplementary Material
Abbreviations Used
- CO-RM
carbon monoxide-releasing molecule
- CORM-3
Ru(CO)3Cl(glycinate)
- dCTP
deoxycytidine triphosphate
- DTNB
5,5′-dithiobis-(2-nitrobenzoic acid) or Ellman's reagent
- iCORM-3
inactivated CORM-3
- KPi
potassium phosphate buffer
- Mb
myoglobin
- PBS
phosphate-buffered saline
- RT-PCR
real time PCR
- SEM
standard error of the mean
- TF
transcription factor
Acknowledgment
This work was supported by the Biotechnology and Biological Sciences Research Council (United Kingdom).
Author Disclosure Statement
B.E. Mann declares a financial interest in Alfama. All other authors declare that no competing financial interests exist.
References
- 1.Abaibou H, Pommier J, Benoit S, Giordano G, and Mandrandberthelot MA.Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J Bacteriol 177: 7141–7149, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeeva S, Hellingwerf KJ, and de Mattos MJT.Quantitative assessment of oxygen availability: perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J Bacteriol 184: 1402–1406, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asif HMS, Rolfe MD, Green J, Lawrence ND, Rattray M, and Sanguinetti G.TFInfer: a tool for probabilistic inference of transcription factor activities. Bioinformatics 26: 2635–2636, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, and Mori H.Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bathoorn E, Siebos DJ, Postma DS, Koeter GH, van Oosterhout AJM, van der Toorn M, Boezen HM, and Kerstjens HAM.Anti-inflammatory effects of inhaled carbon monoxide in patients with COPD: a pilot study. Eur Respir J 30: 1131–1137, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Boczkowski J, Poderoso JJ, and Motterlini R.CO-metal interaction: vital signaling from a lethal gas. Trends Biochem Sci 31: 614–621, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Bowman LAH, McLean S, Poole RK, and Fukuto JM. The diversity of microbial responses to nitric oxide and agents of nitrosative stress: close cousins but not identical twins. In: Advances in Microbial Physiology, Vol 59, edited by Poole RK. London: Elsevier, 2011, pp. 135–219 [DOI] [PubMed] [Google Scholar]
- 8.Clark JE, Naughton P, Shurey S, Green CJ, Johnson TR, Mann BE, Foresti R, and Motterlini R.Cardioprotective actions by a water-soluble carbon monoxide-releasing molecule. Circ Res 93: E2–E8, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Davidge KS, Motterlini R, Mann BE, Wilson JL, and Poole RK.Carbon monoxide in biology and microbiology: surprising roles for the “Detroit perfume.” Adv Microb Physiol 56: 85–167, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Davidge KS, Sanguinetti G, Yee CH, Cox AG, McLeod CW, Monk CE, Mann BE, Motterlini R, and Poole RK.Carbon monoxide-releasing antibacterial molecules target respiration and global transcriptional regulators. J Biol Chem 284: 4516–4524, 2009 [DOI] [PubMed] [Google Scholar]
- 11.De Groote MA, Granger D, Xu YS, Campbell G, Prince R, and Fang FC.Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc Natl Acad Sci U S A 92: 6399–6403, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desmard M, Davidge KS, Bouvet O, Morin D, Roux D, Foresti R, Ricard JD, Denamur E, Poole RK, Montravers P, Motterlini R, and Boczkowski J.A carbon monoxide-releasing molecule (CORM-3) exerts bactericidal activity against Pseudomonas aeruginosa and improves survival in an animal model of bacteraemia. FASEB J 23: 1023–1031, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Desmard M, Foresti R, Morin D, Dagoussat M, Berdeaux A, Denamur E, Crook SH, Mann BE, Scapens D, Montravers P, Boczkowski J, and Motterlini R.Differential antibacterial activity against Pseudomonas aeruginosa by carbon monoxide-releasing molecules. Antioxid Redox Signal 16: 153–163, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Ellman GL.Tissue sulfhydryl groups. Arch Biochem Biophys 82: 70–77, 1959 [DOI] [PubMed] [Google Scholar]
- 15.Evans CGT, Herbert D, and Tempest DW. The Continuous Cultivation of Microorganisms Part 2 Construction of a Chemostat. New York: Academic Press, 1970, pp. 277–327 [Google Scholar]
- 16.Flatley J, Barrett J, Pullan ST, Hughes MN, Green J, and Poole RK.Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J Biol Chem 280: 10065–10072, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Foresti R, Hammad J, Clark JE, Johnson TR, Mann BE, Friebe A, Green CJ, and Motterlini R.Vasoactive properties of CORM-3, a novel water-soluble carbon monoxide-releasing molecule. Br J Pharmacol 142: 453–460, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilberthorpe NJ, and Poole RK.Nitric oxide homeostasis in Salmonella typhimurium—roles of respiratory nitrate reductase and flavohemoglobin. J Biol Chem 283: 11146–11154, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, and Poole RK.Severe zinc depletion of Escherichia coli roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J Biol Chem 284: 18377–18389, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham AI, Sanguinetti G, Bramall N, McLeod CW, and Poole RK.Dynamics of a starvation-to-surfeit shift: a transcriptomic and modelling analysis of the bacterial response to zinc reveals transient behaviour of the Fur and SoxS regulators. Microbiology 158: 284–292, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Jarboe LR, Hyduke DR, Tran LM, Chou KJY, and Liao JC.Determination of the Escherichia coli S-nitrosoglutathione response network using integrated biochemical and systems analysis. J Biol Chem 283: 5148–5157, 2008 [DOI] [PubMed] [Google Scholar]
- 22.Johnson TR, Mann BE, Teasdale IP, Adams H, Foresti R, Green CJ, and Motterlini R.Metal carbonyls as pharmaceuticals? Ru(CO)3Cl(glycinate), a CO-releasing molecule with an extensive aqueous solution chemistry. Dalton Trans 1500–1508, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Keilin D. The History of Cell Respiration and Cytochrome. Cambridge, UK: Cambridge University Press, 1966, pp. 252–268 [Google Scholar]
- 24.Lennox ES.Transduction of linked genetic characters of the host by bacteriophage-PL. Virology 1: 190–206, 1955 [DOI] [PubMed] [Google Scholar]
- 25.Letoffe S, Delepelaire P, and Wandersman C.The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc Natl Acad Sci U S A 103: 12891–12896, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann BE. Carbon monoxide: an essential signalling molecule. In: Medicinal Organometallic Chemistry, edited by Jaouen E. and Metzler-Nolte N. Berlin: Springer-Verlog, 2010, pp. 247–285 [Google Scholar]
- 27.Mann BE. Comp Inorg Chem, II. Oxford: Elsevier (In press) [Google Scholar]
- 28.McLean S, Mann BE, and Poole RK.Sulfite species enhance carbon monoxide release from CO-releasing molecules: implications for the deoxymyoglobin assay of activity. Anal Biochem 427: 36–40, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Motterlini R, Clark JE, Foresti R, Sarathchandra P, Mann BE, and Green CJ.Carbon monoxide-releasing molecules: characterization of biochemical and vascular activities. Circ Res 90: E17–E24, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Nobre LS, Seixas JD, Romao CC, and Saraiva LM.Antimicrobial action of carbon monoxide-releasing compounds. Antimicrob Agents Chemother 51: 4303–4307, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman MS, Corker H, Wu GH, Binet MB, Moir AJG, and Poole RK.Cysteine is exported from the Escherichia coli cytoplasm by CydDC, an ATP-binding cassette-type transporter required for cytochrome assembly. J Biol Chem 277: 49841–49849, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Raffa RG, and Raivio TL.A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol 45: 1599–1611, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Romanski S, Kraus B, Schatzschneider U, Neudoerfl J-M, Amslinger S, and Schmalz H-G.Acyloxybutadiene iron tricarbonyl complexes as enzyme-triggered CO-releasing molecules (ET-CORMs). Angew Chem Int Ed Engl 50: 2392–2396, 2011 [DOI] [PubMed] [Google Scholar]
- 34.Sandouka A, Fuller BJ, Mann BE, Green CJ, Foresti R, and Motterlini R.Treatment with CO-RMs during cold storage improves renal function at reperfusion. Kidney Int 69: 239–247, 2006 [DOI] [PubMed] [Google Scholar]
- 35.Sanguinetti G, Lawrence ND, and Rattray M.Probabilistic inference of transcription factor concentrations and gene-specific regulatory activities. Bioinformatics 22: 2775–2781, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Santos MFA, Seixas JD, Coelho AC, Mukhopadhyay A, Reis PM, Romao CC, and Santos-Silva T.New insights into the chemistry of fac-[Ru(CO)3]2+ fragments in biologically relevant conditions: the CO releasing activity of [Ru(CO)3Cl2(1,3-thiazole)], and the X-ray crystal structure of its adduct with Lysozyme. J Inorg Biochem 117: 285, 2012 [DOI] [PubMed] [Google Scholar]
- 37.Santos-Silva T, Mukhopadhyay A, Seixas JD, Bernardes GJL, Romao CC, and Romao MJ.CORM-3 reactivity toward proteins: the crystal structure of a Ru(II) dicarbonyl-lysozyme complex. J Am Chem Soc 133: 1192–1195, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Schatzschneider U.PhotoCORMs: light-triggered release of carbon monoxide from the coordination sphere of transition metal complexes for biological applications. Inorg Chim Acta 374: 19–23, 2011 [Google Scholar]
- 39.Shepherd M, Sanguinetti G, Cook GM, and Poole RK.Compensations for diminished terminal oxidase activity in Escherichia coli cytochrome bd-II mediated respiration and glutamate metabolism. J Biol Chem 285: 18464–18472, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavares AFN, Nobre LS, and Saraiva LM.A role for reactive oxygen species in the antibacterial properties of carbon monoxide-releasing molecules. FEMS Microbiol Lett 336: 1–10, 2012 [DOI] [PubMed] [Google Scholar]
- 41.Tavares AFN, Teixeira M, Romao CC, Seixas JD, Nobre LS, and Saraiva LM.Reactive oxygen species mediate bactericidal killing elicited by carbon monoxide-releasing molecules. J Biol Chem 286: 26708–26717, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winterbourn CC.Superoxide dismutase-inhibitable reduction of cytochrome-c by the alloxan radical: implications for alloxan cytotoxicity. Biochem J 207: 609–612, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, Datsenko KA, Nakayashiki T, Tomita M, Wanner BL, and Mori H.Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol 5: 335, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanofsky C, and Lennox ES.Transduction and recombination study of linkage relationships among the genes controlling tryptophan synthesis in Escherichia coli. Virology 8: 425–447, 1959 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.