Abstract
Aims: As Candida albicans is the major fungal pathogen of humans, there is an urgent need to understand how this pathogen evades toxic reactive oxygen species (ROS) generated by the host immune system. A key regulator of antioxidant gene expression, and thus ROS resistance, in C. albicans is the AP-1-like transcription factor Cap1. Despite this, little is known regarding the intracellular signaling mechanisms that underlie the oxidation and activation of Cap1. Therefore, the aims of this study were; (i) to identify the regulatory proteins that govern Cap1 oxidation, and (ii) to investigate the importance of Cap1 oxidation in C. albicans pathogenesis. Results: In response to hydrogen peroxide (H2O2), but not glutathione-depleting/modifying oxidants, Cap1 oxidation, nuclear accumulation, phosphorylation, and Cap1-dependent gene expression, is mediated by a glutathione peroxidase-like enzyme, which we name Gpx3, and an orthologue of the Saccharomyces cerevisiae Yap1 binding protein, Ybp1. In addition, Ybp1 also functions to stabilise Cap1 and this novel function is conserved in S. cerevisiae. C. albicans cells lacking Cap1, Ybp1, or Gpx3, are unable to filament and thus, escape from murine macrophages after phagocytosis, and also display defective virulence in the Galleria mellonella infection model. Innovation: Ybp1 is required to promote the stability of fungal AP-1-like transcription factors, and Ybp1 and Gpx3 mediated Cap1-dependent oxidative stress responses are essential for the effective killing of macrophages by C. albicans. Conclusion: Activation of Cap1, specifically by H2O2, is a prerequisite for the subsequent filamentation and escape of this fungal pathogen from the macrophage. Antioxid. Redox Signal. 19, 2244–2260.
Introduction
Candida albicans is the major systemic fungal pathogen of humans causing approximately 400,000 deaths per annum (39). Consequently, there is much interest in the mechanisms employed by this opportunistic pathogen to survive host immune system defences, which includes the generation of toxic reactive oxygen species (ROS). Upon activation of neutrophils and macrophages, the NADPH oxidase (Nox) complex generates superoxide within the phagosome (4), which then rapidly undergoes dismutation to produce hydrogen peroxide (H2O2). Patients with congenital defects that affect the Nox complex, or neutropenia, exhibit enhanced susceptibility to systemic candidiasis (48) indicating the importance of ROS-based mechanisms in the defence against this fungus. However, C. albicans mounts a robust transcriptional response to oxidative stress upon exposure to human blood (14), macrophages (27), and neutrophils (13) and can evade killing by macrophages (26, 27). Furthermore, several studies have reported that the inactivation of ROS-protective enzymes attenuates virulence in systemic models of disease (19, 53). Collectively, such observations indicate that oxidative stress defences are important for survival of C. albicans in the host. However, remarkably little is known regarding the intracellular signaling mechanisms underlying the activation of C. albicans ROS-induced transcriptional responses after phagocytosis, or their importance in promoting the viability of this fungal pathogen against ROS-based immune defences.
Innovation.
The importance of reactive oxygen species-induced transcriptional responses in promoting survival of the major fungal pathogen of humans, Candida albicans, against innate immune defences is unknown. Here we provide new insight into the signaling mechanism by which this pathogen detects the presence of hydrogen peroxide (H2O2) and activates gene expression through the AP-1-like Cap1 transcription factor, and, furthermore, reveal a novel function for the Ybp1 regulatory protein in mediating AP-1-like transcription factor activation. Significantly, we also demonstrate that H2O2-induced activation of Cap1 is vital for C. albicans-mediated killing of macrophages.
In C. albicans, the AP-1-like transcription factor Cap1 is the major regulator of oxidative stress-induced gene expression as determined by transcript profiling and chromatin-immunoprecipitation experiments (52, 55). Cap1 is orthologous to the Yap1 and Pap1 transcription factors in Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively, which have well-studied roles in antioxidant gene expression (33). All three of these transcription factors accumulate in the nucleus after exposure to a range of different oxidizing agents (23, 46, 54), suggesting related mechanisms of regulation. Indeed, studies of Yap1 and Pap1 have revealed that H2O2-induced nuclear accumulation occurs due to the formation of interdomain disulphide bonds between two cysteine rich domains (the n-CRD and c-CRD) found in fungal AP-1-like factors (9, 51). This oxidation event triggers a conformational change which masks the accessibility of a nuclear export sequence located within the c-CRD; thus, allowing Yap1 and Pap1 to accumulate within the nucleus and activate gene expression (23, 46). The glutathione-depleting/modifying agents, diamide and diethylmaleate (DEM), have also been shown to trigger the nuclear accumulation of Yap1 and Pap1. However, this is mediated by the formation of disulphide bonds between closely linked cysteines within the c-CRD in the case of diamide, or direct covalent modification of cysteines by DEM, rather than the formation of interdomain disulphide bonds characteristic of H2O2-treatment (8, 22).
Yap1 and Pap1 are not directly oxidized by H2O2, but instead specific peroxidase enzymes sense and transduce the H2O2 signal to these transcription factors. Intriguingly, the mechanisms regulating this vital post-translational modification of Yap1 and Pap1 are different. For example, Yap1 oxidation requires Gpx3, a glutathione peroxidase (Gpx)-like enzyme (9), whereas oxidation of Pap1 requires the thioredoxin peroxidase activity of an unrelated peroxidase, the 2-Cys peroxiredoxin, Tpx1 (6, 50). Gpx3-mediated activation of Yap1 also requires a second protein, Ybp1, which is exclusive to fungi (49). Although Ybp1 binds Yap1, its function in promoting the H2O2-induced oxidation of Yap1 is unclear. However, one commonly used wild-type S. cerevisiae strain, W303, has a naturally occurring mutant allele of YBP1, ybp1-1, which encodes a truncated Ybp1 protein (37, 49). Interestingly, in such cells, Yap1 oxidation is no longer Gpx3-dependent but mediated albeit less efficiently by the 2-Cys peroxiredoxin, Tsa1, analogous to Pap1 regulation in S. pombe (37). Yap1 activation by diamide and DEM is also independent of Gpx3 function and instead is linked to the direct modification of cysteine residues solely within the c-CRD (3).
Much less is known about Cap1 regulation in C. albicans. For example, although C. albicans Cap1 is oxidized after H2O2 exposure (7), and mutation of the c-CRD impacts on Cap1 regulation (1, 54), nothing is known regarding the mechanisms underlying oxidation of Cap1 by different ROS, or the importance of Cap1-mediated transcriptional responses in allowing C. albicans to contend with ROS-based immune-defences. As the two model yeasts S. cerevisiae and S. pombe execute different mechanisms to regulate oxidation of their respective AP-1-like transcription factors, it is unknown which, if either, mechanism exists in other fungi. Here, we dissect the mechanisms underlying the H2O2-induced oxidation and activation of the Cap1 transcription factor, and investigate the importance of this process in promoting C. albicans survival in macrophages and in different models of infection.
Results
Cap1 function is required for C. albicans-mediated macrophage killing
Previous studies established that C. albicans mounts a transcriptional response upon phagocytosis by macrophages and neutrophils (13, 27), which includes the induction of many genes that are Cap1-regulated in vitro after H2O2 exposure (11, 52). C. albicans can efficiently kill macrophages after phagocytosis, yet whether transcriptional responses to ROS are essential for this phenomenon is unknown. Thus, here, we investigated whether Cap1-induced gene expression is needed to allow C. albicans-mediated macrophage killing. We also compared the impact of loss of Cap1 on macrophage killing with that of the Hog1 stress-activated protein kinase (SAPK), that is also implicated in H2O2-induced gene expression (11), as hog1Δ cells have previously been shown to display increased sensitivity to neutrophil-mediated killing (2).
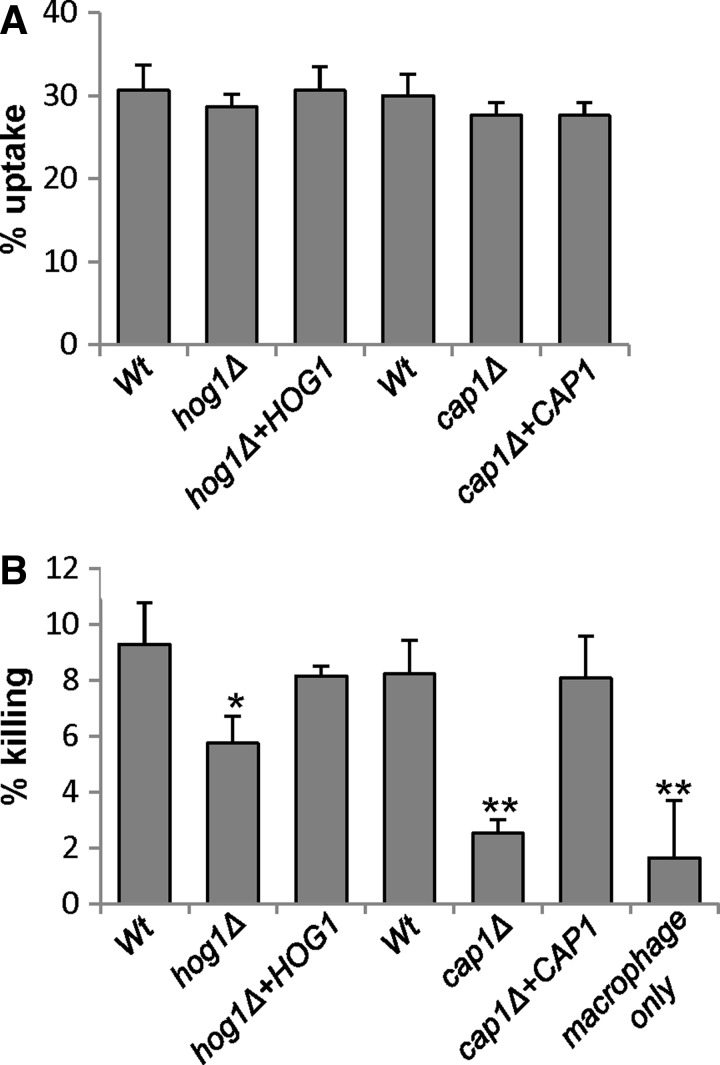
The murine macrophage cell line J774.1 was challenged with cap1Δ and hog1Δ mutant cells and their respective wild-type and reintegrant control strains. Using our standard phagocytosis assay (24, 31), all strains were taken up by macrophages at similar rates (Fig. 1A). Strikingly, cap1Δ cells were extremely defective in macrophage killing compared with wild-type cells (p<0.01), and this defect was rescued upon reintegrating the wild-type CAP1 gene (Fig. 1B). Cells lacking the Hog1 SAPK also displayed impaired macrophage killing compared to wild-type cells (p<0.05), although to a lesser degree than cap1Δ cells (Fig. 1B). This provides the first evidence that Cap1-mediated transcriptional responses to oxidative stress are needed for C. albicans-mediated macrophage killing. Hence, we sought to determine the mechanism underlying oxidant-induced activation of Cap1 to investigate whether this process is vital for C. albicans evasion of macrophages.
FIG. 1.
Cap1 is essential for Candida albicans-mediated killing of macrophages. (A) Percentage uptake of wild-type (Wt, JC21), hog1Δ (JC50), hog1Δ+HOG1 (JC52) and wild type (Wt, JC747), cap1Δ (JC842), cap1Δ+CAP1 (JC807) C. albicans cells by J774.1 macrophages after 3 h coincubation. (B) Percentage of macrophages killed by the hog1Δ and cap1Δ mutants and respective wild-type and reintegrant controls after 3 h coincubation. Data were obtained in triplicate from at least three different experiments by analyzing at least 100 macrophages per well. Analysis of variance (ANOVA) was used to determine statistical significance: *p<0.05; **p<0.01.
Cap1 is oxidized, phosphorylated, and localizes to the nucleus to activate antioxidant gene expression in response to H2O2
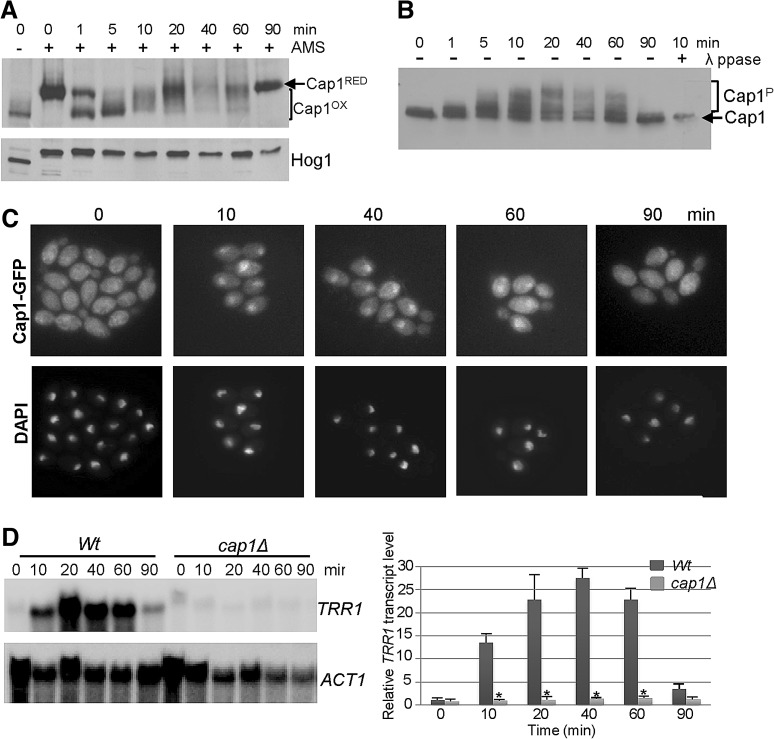
As a first step in dissecting the mechanism underlying Cap1 regulation in C. albicans, we determined the kinetics of Cap1 activation in wild-type cells after treatment with 5 mM H2O2 (Fig. 2). The redox changes in Cap1 after H2O2-exposure were trapped by the thiol alkylating agent AMS (4-acetamido-4′-(idoacetyl)amino)stilbene-2,2′-disulfonic acid) (9), which reacts specifically with SH groups of reduced cysteine residues increasing the molecular weight of thiol-modified proteins by ∼0.5 kDa/cysteine (9). As illustrated in Figure 2A, more mobile oxidized forms of Cap1 occurred immediately post H2O2 treatment and persisted for 1 h. However, the profile of Cap1 oxidation changed over the time course of the experiment. Extensively oxidized forms of Cap1 appeared 1 to 5 min after H2O2 exposure. As such, forms have mobility similar to Cap1 untreated with AMS, which suggests that all six cysteines are oxidized (Fig. 2A). However, 10 to 60 min post H2O2-treatment other lesser oxidized forms of Cap1 are visible, and by 90 min only the reduced form is seen (Fig. 2A). Consistent with oxidation stimulating the nuclear accumulation of Cap1, fluorescent microscopy of cells expressing Cap1-green fluorescent protein (GFP) demonstrated that H2O2 treatment induced nuclear accumulation that was detectable up to 60 min (Fig. 2C). Nuclear accumulation also correlated with the phosphorylation of Cap1, similar to that reported for Yap1 and Pap1 (6, 8), as phosphatase-sensitive slower mobility forms of Cap1-MH were detected within 1 min of H2O2 treatment and these persisted for 60 min (Fig. 2B). Importantly, the timings of oxidation, phosphorylation and nuclear accumulation of Cap1 are consistent with the H2O2-induced expression profile of the Cap1-dependent gene TRR1 (Fig. 2D). Having established the kinetics of various read-outs of Cap1 activation, this then provided the parameters necessary to allow the subsequent identification of regulatory proteins mediating Cap1 activation.
FIG. 2.
Markers of H2O2-induced Cap1 activation. (A) Cap1 is rapidly oxidized after H2O2 exposure. Cap1 oxidation was analyzed by nonreducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting of AMS-modified proteins prepared from cells expressing 2myc-6His-tagged Cap1 (JC948, Cap1-MH), exposed to 5 mM H2O2 for the indicated times. After detection of Cap1, the blot was reprobed with an anti-Hog1 antibody to determine loading. The data shown is representative of four biological replicates. (B) Cap1 is phosphorylated after H2O2 exposure. Lysates from cells expressing Cap1-MH, exposed to 5 mM H2O2 for the indicated times, were enriched on Ni2+-NTA agarose and analyzed by western blotting using an anti-myc antibody. A sample taken at 10 min was also treated with λ–phosphatase to illustrate that the slower migrating forms of Cap1 were due to phosphorylation. The data shown is representative of four biological replicates. (C) Cap1 accumulates in the nucleus after H2O2 exposure. Localization of Cap1 was detected in cells expressing Cap1-GFP (JC1060) before and after exposure to 5 mM H2O2 by fluorescence microscopy. Nuclei were detected by DAPI staining. The data shown is representative of six biological replicates. (D) H2O2-induced TRR1 gene expression is dependent on Cap1. Northern blot analysis of RNA isolated from wild-type (Wt, JC747) and cap1Δ (JC842) cells after exposure to 5 mM H2O2 for the indicated times. Blots were analyzed with a probe specific for the thioredoxin reductase TRR1 gene, and then after stripping the blot with a probe specific for ACT1 (loading control). TRR1 mRNA levels (mean±SEM) relative to the ACT1 loading control, from three independent experiments are shown. ANOVA was used to determine statistical significance: *p<0.025.
Ybp1 mediates H2O2-induced activation of Cap1
To delineate the mechanism underlying H2O2-induced Cap1 oxidation, we examined the C. albicans genome for an orthologue of the S. cerevisiae YBP1 gene, as the presence of a functional Ybp1 protein appears to dictate the mechanism underlying oxidation of AP-1-like transcription factors. For example, in S. cerevisiae cells expressing a wild-type YBP1 allele, oxidation of Yap1 is mediated by the Gpx3-peroxidase in conjunction with Ybp1 (49), whereas cells carrying a naturally occurring mutation of Ybp1 (ybp1-1) use the 2-Cys peroxiredoxin Tsa1, to mediate Yap1 oxidation albeit less efficiently (37, 44, 49). However, in addition to Ybp1, S. cerevisiae also contains a related protein, Ybp2/Ybh1, that does not regulate Yap1 and instead is required for spindle function (36). Sequence analysis of the C. albicans genome revealed only a single homologue, orf19.5034, that exhibits approximately 20% identity to both S. cerevisiae Ybp1 and Ybp2 (49). Hence, to establish whether the function of orf19.5034 in C. albicans is related to Ybp1 and/or Ybp2 in S. cerevisiae, a homozygous null mutant was generated in which both copies of orf19.5034 were deleted. C. albicans orf19.5034Δ cells were much more sensitive to H2O2 than wild-type cells and the reintegrant strain (Fig. 3A), but not to the spindle poisons TBZ or benomyl (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). This result suggested that the function of the gene product encoded by orf19.5034 might be more similar to S. cerevisiae Ybp1 than Ybp2, and thus, we named this gene C. albicans YBP1.
FIG. 3.
Ybp1 and Gpx3 are required for Cap1 activation in response to H2O2. (A) Loss of either Ybp1 or Gpx3 renders cells sensitive to H2O2. 104 and 10-fold dilutions thereof of exponentially growing wild-type (Wt, JC747), cap1Δ (JC842), cap1Δ+CAP1 (JC807), ybp1Δ (JC917), ybp1Δ+YBP1 (JC809), gpx3Δ (JC1317), and gpx3Δ+GPX3 (JC1346) cells were spotted onto agar plates containing the indicated concentrations of H2O2. Plates were incubated at 30°C for 24 h. The data shown is representative of three biological replicates. (B) Cap1 oxidation after H2O2 treatment is dependent on Ybp1 and Gpx3. Cap1 oxidation was measured as described in the Figure 2A legend, in wild-type (Wt, JC948), ybp1Δ (JC954) and gpx3Δ (JC1311) cells expressing Cap1-MH after exposure to 5 mM H2O2 for 10 min. The data shown is representative of four biological replicates. (C) Ybp1 and Gpx3 are required for H2O2-induced nuclear accumulation of Cap1. The localization of Cap1GFP in wild-type (Wt, JC1060), ybp1Δ (JC1054), and gpx3Δ (JC1314) cells was determined by fluorescence microscopy in untreated cells and after exposure to 5 mM H2O2 for 20 min. The data shown is representative of three biological replicates. (D) Cap1 phosphorylation after exposure to H2O2 is dependent on Ybp1 and Gpx3. Cap1 phosphorylation was measured as described in the Figure 2A legend, in wild-type (Wt, JC948), ybp1Δ (JC954) and gpx3Δ (JC1311) cells expressing Cap1-MH after exposure to 5 mM H2O2 for the indicated times. The data shown is representative of three biological replicates. (E) Ybp1 and Gpx3 are required for the H2O2-induced expression of TRR1. Northern blot analysis of RNA isolated from wild-type (Wt, JC747), cap1Δ (JC842), ybp1Δ (JC917), ybp1Δ+YBP1 (JC809), gpx3Δ (JC1317), and gpx3Δ+GPX3 (JC1346) cells after exposure to 5 mM H2O2 for the indicated times. The northern blot was analyzed using specific probes for the Cap1-dependent gene TRR1 and ACT1 (loading control). TRR1 mRNA levels (mean±SEM) relative to the ACT1 loading control, from three independent experiments are shown. ANOVA was used to determine statistical significance: *p<0.025.
When we examined the role of Ybp1 in Cap1 activation, H2O2-induced oxidation of Cap1 was abolished in ybp1Δ cells over a range of H2O2 concentrations (Fig. 3B and Supplementary Fig. S2A). Intriguingly, it also appeared that Cap1 protein levels were lower in cells lacking YBP1 (Fig. 3B and Supplementary Figs. S2A and S3A). Both the H2O2-induced phosphorylation (Fig. 3C), and the nuclear accumulation of Cap1 (Fig. 3D and Supplementary Fig. S2B) were also drastically impaired in ybp1Δ cells, culminating in a severe reduction in H2O2-induced Cap1-dependent gene expression (Fig. 3E and Supplementary Figs. S2C and S3B). Collectively, therefore, these data demonstrate that Ybp1 functions over a range of H2O2 concentrations to promote the oxidation and activation of Cap1 in C. albicans.
Identification of the peroxidase that mediates H2O2-induced Cap1 oxidation
We next turned our attention to the peroxidase that mediates Cap1 oxidation. The 2-Cys peroxiredoxin Tsa1 was found to be dispensable for Cap1 oxidation (Supplementary Fig. S4). Hence, we investigated the potential role of Gpx-like enzymes in Cap1 regulation. Sequence analysis of the C. albicans genome revealed four close homologues of the S. cerevisiae Gpx3 protein, encoded by orf19.85, orf19.86, orf19.87, and orf19.4436 (Supplementary Fig. S5A). Homozygous null mutants of each gene were generated and only cells lacking orf19.4436 were significantly sensitive to H2O2 (Supplementary Fig. S5B). Of the three single gpx mutants characterised in S. cerevisiae only the gpx3Δ mutant, which displays impaired Yap1 oxidation, exhibits significantly increased sensitivity to H2O2 (20). Thus, based on these observations, we named orf19.4436, GPX3, and investigated the potential role of Gpx3 in Cap1 regulation.
Significantly, cells lacking GPX3 phenocopied ybp1Δ cells with regard to H2O2 sensitivity and this could be rescued upon reintegrating GPX3 (Fig. 3A). Furthermore, the H2O2-induced oxidation, phosphorylation, and nuclear accumulation of Cap1 seen in wild-type cells were significantly impaired in cells lacking Gpx3 (Fig. 3B–D). The remaining Gpx-like enzymes were dispensable for Cap1 activation (Supplementary Fig. S5C, D). Consistent with Gpx3 functioning as the peroxidase that activates Cap1, H2O2-induced expression of TRR1 was notably impaired in gpx3Δ cells compared to wild-type and reintegrant cells (Fig. 3E). Thus, taken together, our results indicate that the mechanism underlying H2O2-induced oxidation and thus, activation of Cap1 in C. albicans is similar to that previously reported for Yap1 in S. cerevisiae.
Ybp1 and Gpx3 are dispensable for Cap1 activation in response to glutathione-depleting or glutathione-modifying oxidants
As AP-1-like transcription factors are activated in response to a range of different oxidising agents in addition to H2O2, such as the glutathione-depleting or modifying drugs diamide, DEM and Cd2+, we investigated the specificity of Ybp1 and Gpx3 function in mediating activation of Cap1. While C. albicans cells lacking CAP1 displayed increased sensitivity to diamide, DEM, and Cd2+, gpx3Δ cells did not. Interestingly, cells lacking YBP1 displayed an intermediate stress sensitive phenotype to diamide and Cd2+ (Fig. 4A), which might reflect reduced levels of Cap1 in ybp1Δ mutant cells (see Fig. 3B, Fig.S3A and below). However, consistent with Cap1 activation in response to diamide occurring independently of Gpx1 and Ybp1 in C. albicans, diamide-induced nuclear accumulation of Cap1 was retained in ybp1Δ and gpx3Δ cells (Fig. 4B).
FIG. 4.
Ybp1 and Gpx3 are specifically required for Cap1 activation in response to H2O2. (A) Cap1 mediates stress resistance to more oxidants than Ybp1 and Gpx3. 104 and 10-fold dilutions thereof of exponentially-growing wild-type (Wt, JC747), cap1Δ (JC842), ybp1Δ (JC917), and gpx3Δ (JC1317) cells were spotted onto agar plates containing either 1.5 mM H2O2, 2 mM DEM, 0.5 mM Cd2+, and 2 mM diamide. Plates were incubated at 30°C for 24 h. The data shown is representative of two biological replicates. (B) Ybp1 and Gpx3 are dispensable for diamide-induced Cap1 nuclear accumulation. The localization of Cap1-GFP in wild-type (Wt, JC1060), ybp1Δ (JC1054), and gpx3Δ (JC1314) cells was determined by fluorescence microscopy in untreated cells and after treatment with 5 mM diamide for 20 min. The data shown is representative of two biological replicates.
Ybp1 is required for Cap1 stability and forms a complex with Cap1
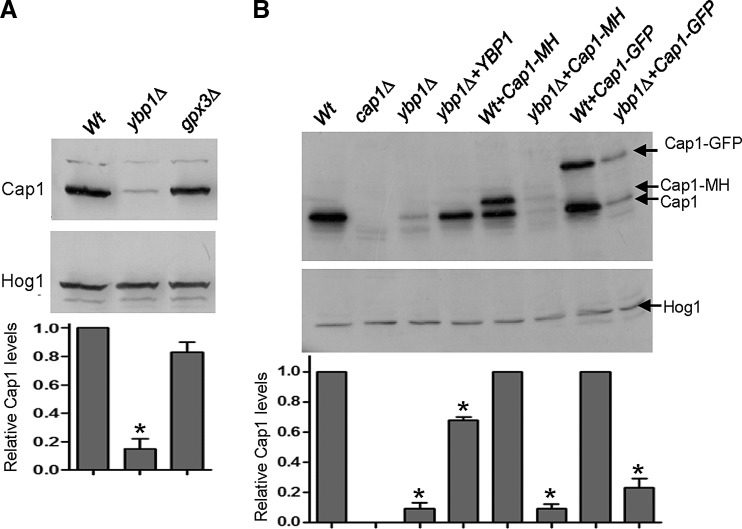
Western blot analyses examining the role of Gpx3 and Ybp1 in Cap1 oxidation indicated that Cap1 levels were significantly lower in ybp1Δ cells than in wild-type and gpx3Δ cells (Supplementary Figs. S2A and S3A and Fig. 3B). To examine this quantitatively, cell extracts were prepared under native conditions and Cap1 levels analysed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting of equivalent amounts of protein using an anti-Cap1 antibody. Consistent with our initial findings, Cap1 protein levels were found to be drastically reduced in ybp1Δ cells compared to wild-type, gpx3Δ, and ybp1Δ+YBP1 cells (Fig. 5A, B). Significantly, the levels of the various tagged versions of Cap1 employed in this study (Cap1-GFP and Cap1-MH) were also depleted in ybp1Δ cells (Fig. 5B).
FIG. 5.
Cap1 protein levels are dependent on Ybp1. (A) Cap1 protein levels are reduced in ybp1Δ cells. 50 μg of whole cell extracts prepared from wild-type (Wt, JC747), ybp1Δ (JC917), and gpx3Δ (JC1317) cells under native conditions were analyzed by SDS-PAGE and western blotting using anti-Cap1 antibodies. The blot was subsequently stripped and reprobed with an anti-Hog1 antibody as a loading control. Relative Cap1 levels (mean±SEM) from three independent biological replicates were quantified using ImageQuant (GE Healthcare). ANOVA was used to determine statistical significance: *p<0.025. (B) Levels of Cap1 in different tagged strains. Whole cell extracts were prepared from wild type (Wt, JC747), cap1Δ (JC842), ybp1Δ (JC917), and ybp1Δ+YBP1 (JC809) cells, wild-type (Wt) and ybp1Δ cells expressing Cap1-MH (JC948/JC954) and wild-type (Wt) and ybp1Δ cells expressing Cap1-GFP (JC1060/JC1054). 30 μg of extract were analyzed by SDS-PAGE and western blotting using anti-Cap1 antibodies. The blot was reprobed with an anti-Hog1 antibody. Relative Cap1 levels (mean±SEM) from three independent biological replicates were quantified using ImageQuant (GE Healthcare). ANOVA was used to determine statistical significance of Cap1 levels between the mutants and respective wild-type strains: *p<0.025.
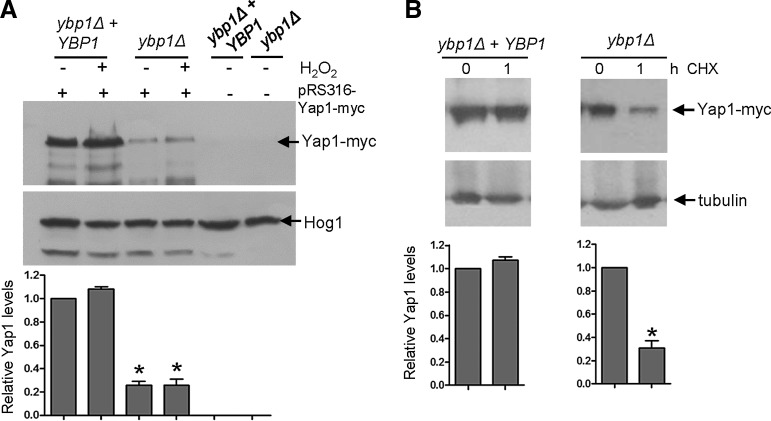
We previously demonstrated that CAP1 gene expression is induced by H2O2 (11). Thus, it was possible that C. albicans Ybp1 is required for the H2O2-induced transcription of CAP1 in a positive feedback mechanism that involves autoregulation by oxidized Cap1. However, RNA analysis revealed only a slight reduction in the H2O2-dependent induction of CAP1 mRNA levels in ybp1Δ cells (Supplementary Fig. S3B). Furthermore, if Ybp1 and Gpx3 mediated oxidation of Cap1 was needed for the Cap1-dependent induction of CAP1 transcription, then it might be expected that gpx3Δ cells would also exhibit reduced Cap1 protein levels, which was not the case (Fig. 5A). However, to directly test whether Ybp1 regulated Cap1 protein levels in a mechanism independent of transcription, CAP1 was placed under the control of the exogenous ACT1 promoter (47) in wild-type, cap1Δ, and ybp1Δ cells. As predicted, ACT1 promoter-driven CAP1 expression restored H2O2 resistance to cap1Δ cells but not to ybp1Δ cells (Fig. 6A). Also, as expected, ACT1-promoter driven CAP1 expression increased Cap1 protein levels in cap1Δ cells (Fig. 6B). In contrast, ectopic expression of CAP1 failed to increase Cap1 protein levels in ybp1Δ cells (Fig. 6B), clearly demonstrating that Ybp1 determines Cap1 levels independently of CAP1-promoter driven transcription.
FIG. 6.
Ybp1 is required for Cap1 stability, but not CAP1 transcription. (A) Ectopic expression of CAP1 does not restore Cap1 activity in ybp1Δ cells. Expression of CAP1 from the ACT1 promoter in ybp1Δ cells does not restore resistance to H2O2. 104 and 10-fold dilutions thereof of exponentially-growing wild-type (Wt), cap1Δ, and ybp1Δ cells containing the vector pACT1 (JC1014, JC1398 and JC1400, respectively), or pACT1-CAP1 (JC1429, JC1388, and JC1390, respectively) were spotted onto agar plates containing increasing concentrations of H2O2. Plates were incubated at 30°C for 24 h. The data shown is representative of three biological replicates. (B) Ectopic expression of CAP1 does not restore Cap1 levels in ybp1Δ cells. Cap1 protein levels are not restored in ybp1Δ cells expressing CAP1 from the ACT1 promoter. 50 μg of whole cell extracts isolated from the strains listed above, were prepared under native conditions and analyzed by SDS-PAGE and western blotting using anti-Cap1 antibodies. The blot was subsequently stripped and reprobed with an anti-Hog1 antibody as a loading control. The data shown is representative of four biological replicates. Relative Cap1 levels were quantified (mean±SEM) from three independent biological replicates using ImageQuant (GE Healthcare). ANOVA was used to determine statistical significance of Cap1 levels between the mutants and respective wild-type strains: *p<0.025. (C) Cap1 levels are reduced in ybp1Δ cells after inhibition of protein translation. Extracts were prepared from wild-type (Wt, JC747) and ybp1Δ (JC842) cells after cycloheximide addition (CHX) for the indicated time. Cap1 levels were determined by western blotting using an anti-Cap1 antibody, after which the blot was reprobed with an anti-Hog1 antibody. Relative Cap1 levels were quantified (mean±SEM) from three independent biological replicates using ImageQuant (GE Healthcare). ANOVA was used to determine statistical significance of Cap1 levels between untreated and cyclohexamide treated cells: *p<0.025.
To determine whether Ybp1 is important for the protein synthesis or stability of Cap1, we next examined the impact of the protein synthesis inhibitor, cycloheximide on Cap1 protein levels, in both wild-type and ybp1Δ cells. Blocking protein translation had no impact on Cap1 protein levels in wild-type cells over the 1 h time course examined, indicating that Cap1 turnover is normally slow (Fig. 6C). In contrast, Cap1 levels were reduced in ybp1Δ cells 1 h after the addition of cycloheximide (Fig. 6C). These data indicate that Ybp1 loss reduces the stability of this transcription factor.
In S. cerevisiae, Ybp1 forms a complex with Yap1 in vivo (49). Based on these results, we reasoned that Ybp1 binding to Cap1 might be important to stabilise Cap1. Hence, to determine whether Ybp1 forms a complex with Cap1 in C. albicans we created strains in which Ybp1 was tagged with 6 His residues and 2-myc epitopes (Ybp1-MH). Ybp1-MH was precipitated from protein extracts from wild-type and gpx3Δ cells before and after treatment with H2O2, and coprecipitation of Cap1 was determined by western blot analysis (Fig. 7A). Ybp1 was found to interact with Cap1 in C. albicans both before and after H2O2 treatment and this interaction occurred independently of Gpx3 (Fig. 7A). However, importantly, as such coprecipitation experiments are performed using whole cell extracts, any compartmentalisation of factors that would prevent such interactions is lost. In S. cerevisiae, Ybp1 is exclusively cytoplasmic (49). Hence, we determined the localization of C. albicans Ybp1 using indirect immunofluorescence. As shown in Figure 7B, Ybp1 is clearly excluded from the nucleus in C. albicans and this cytoplasmic location is maintained after H2O2 treatment. Taken together, these results indicate that Ybp1 interacts with and stabilises the reduced form of Cap1 in the cytoplasm; however, after oxidation Cap1 accumulates in the nucleus and interaction with Ybp1 is presumably lost.
FIG. 7.
Ybp1 is a cytoplasmic protein and interacts with Cap1 in vivo. (A) Ybp1 interacts with Cap1 independently of Gpx3. Extracts were prepared from wild-type (Wt, JC747), cap1Δ (JC842), and wild-type, and gpx3Δ cells expressing 2Myc-6His tagged Ybp1 (YBP1-MH, JC1200; YBP1-MH gpx3Δ, JC1313) before and after exposure to 5 mM H2O2 for 10 min. Ybp1-MH was immunoprecipitated using anti-myc agarose and precipitated proteins and 5% of protein input were then subjected to SDS-PAGE. Coprecipitation of Cap1 was assayed by western blotting using an anti-Cap1 antibody (top panel) and enrichment of Ybp1-MH assayed using anti-Myc antibodies (bottom panel). The data presented are representative of three biological replicates. (B) Ybp1 is located exclusively in the cytoplasm. The localization of 2Myc-6His tagged Ybp1 was determined before and after treatment of JC1200 cells with 5 mM H2O2 for 10 min, by indirect immunofluorescence. The staining pattern of control cells (JC747) in which Ybp1 is untagged is also shown (con). The data presented are representative of two biological replicates.
Ybp1 regulation of AP-1-like transcription factor stability is conserved in S. cerevisiae
Ybp1 has not previously been reported to regulate Yap1 protein levels in S. cerevisiae, although this has not been directly investigated (16, 17, 49). In our previous study, where we demonstrated that Ybp1 was vital for Yap1 oxidation in S. cerevisiae, the published Yap1 oxidation profile indicated that ybp1Δ and ypb1Δ+YBP1 cells might contain similar Yap1 levels (49), although such oxidation profiles were not assessed for equivalent protein loading. Here to directly examine the impact of loss of Ybp1 on Yap1 levels in S. cerevisiae, cell extracts were prepared under native conditions and Yap1 protein levels determined in ybp1Δ cells compared with cells where YBP1 had been reintegrated at the normal locus (ybp1Δ+YBP1). As illustrated in Figure 8A, Yap1 levels are significantly reduced in cells lacking Ybp1. We also examined the impact of cycloheximide treatment on Yap1 protein levels in ybp1Δ and ybp1Δ+YBP1 cells. Similar to that seen in C. albicans (Fig. 6C), blocking protein translation only significantly reduced Yap1 levels in cells lacking YBP1 (Fig. 8B). Collectively, these results indicate a hitherto unidentified role for Ybp1, conserved in both C. albicans and S. cerevisiae Ybp1 proteins, in stabilising their respective AP-1-like transcription factors, Cap1 and Yap1.
FIG. 8.
Ybp1 regulates Yap1 stability in S. cerevisiae. (A) Levels of Yap1 are dependent on Ybp1 in S. cerevisiae. 50 μg of whole cell extracts prepared from ybp1Δ (SR1), or YBP1 reintegrant (SR6) cells containing pRS316-Yap1-Myc (8), expressing Myc-epitope tagged Yap1, untreated or treated with 0.8 mM H2O2 for 30 min were analyzed by SDS-PAGE and western blotting using anti-Myc antibodies. The blot was subsequently stripped and reprobed with an anti-Hog1 antibody as a loading control. The data presented are representative of three biological replicates, and the relative Yap1 levels were quantified (mean±SEM) using ImageQuant (GE Healthcare). ANOVA was used to determine statistical significance of Yap1 levels between the ybp1Δ mutant and the ybp1Δ+YBP1 reconstituted strain: *p<0.025. (B) Yap1 levels are reduced in ybp1Δ cells after inhibition of protein translation. Extracts were prepared from ybp1Δ (SR1), or YBP1 reintegrant (SR6) cells containing pRS316-Yap1-Myc after cycloheximide addition (CHX) for the indicated time. Yap1-Myc levels were determined by western blotting using an anti-myc antibody, after which the blot was reprobed with an antitubulin antibody. The data presented are representative of three biological replicates. Relative Yap1 levels were quantified (mean±SEM) from the three independent experiments using ImageQuant (GE Healthcare). ANOVA was used to determine statistical significance of Yap1 levels between untreated and cyclohexamide treated cells: *p<0.025. Different exposure times were necessary to visualize Yap1 levels in ybp1Δ and YBP1 reintegrant cells.
H2O2-induced activation of Cap1 is important for C. albicans-mediated macrophage killing
Having found that Ybp1 and Gpx3 specifically mediate H2O2-induced activation of Cap1 (Figs. 3 and 4), we next asked whether H2O2-induced activation of Cap1 is important for its role in C. albicans-mediated macrophage killing (Fig. 1B). The macrophage cell line J774.1 was challenged with the cap1Δ, ybp1Δ, and gpx3Δ null mutants. As illustrated in Figure 9A, cells lacking Cap1, or its regulatory proteins Ybp1 and Gpx3, were taken up by macrophages at similar rates as wild-type and reintegrant control strains. Importantly, however, cells lacking either YBP1 or GPX3 displayed a significantly impaired ability to kill macrophages (p<0.01), similar to that seen upon deleting CAP1, and such defects were reversed upon reintegrating the respective wild-type gene (Fig. 9B). These data suggest that H2O2-mediated oxidation of Cap1 is vital for C. albicans macrophage killing. We also investigated whether cells lacking Cap1, Ybp1 or Gpx3 were more sensitive to killing by macrophages. Consistent with the findings above that cap1Δ, ybp1Δ, and gpx3Δ null mutants are less able to kill macrophages, such cells were also more susceptible to macrophage-mediated killing compared to wild-type cells (p<0.002) (Fig. 9C).
FIG. 9.
C. albicans cells lacking Cap1 or its regulators Gpx3 and Ybp1 display impaired killing of macrophages, and are more susceptible to macrophage-mediated killing. (A) Percentage uptake of wild-type (Wt, JC747), cap1Δ (JC842), cap1Δ+CAP1 (JC807), ybp1Δ (JC917), ybp1Δ+YBP1 (JC809), gpx3Δ (JC1317) and gpx3Δ+GPX3 (JC1346) C. albicans cells by J774.1 macrophages after 3 h coincubation. (B) Percentage of macrophages killed by the same cells after 3 h coincubation. Data were obtained in triplicate from at least three different experiments by analyzing at least 100 macrophages per well. ANOVA was used to determine statistical significance: **p<0.01. (C) Percent of C. albicans survival after coincubation with macrophages. Data were obtained in triplicate from three separate biological replicates and ANOVA was used to determine statistical significance: **p<0.01 (D) Micrographs illustrating the defective intracellular hyphal formation after phagocytosis for the cap1Δ, ybp1Δ and gpx3Δ mutant cells in comparison to wild-type and respective reintegrant control cells. Data are representative of hyphal formation in at least 100 macrophages per C. albicans strain analysed. Black arrows indicate the positioning of C. albicans hyphal cells, whereas white arrows indicate nonfilamentous C. albicans cells.
C. albicans-mediated killing of macrophages is thought to be a consequence of hyphae formation, which after elongation, eventually results in the stretching and piercing of the macrophage cell membrane (5). Hence, we next examined whether the inability of cap1Δ, gpx3Δ, and ybp1Δ cells to kill macrophages was due to defects in hyphae formation within the macrophage. Strikingly, micrographs capturing images of these various mutants after phagocytosis clearly demonstrate that the elongated hyphae and subsequent macrophage membrane stretching seen upon phagocytosis of either wild-type or reintegrant strains was absent in macrophages that had taken up cap1Δ, gpx3Δ or ybp1Δ cells (Fig. 9D). Importantly, hyphae formation by these mutants was only impaired within the macrophage, cells which had not been phagocytosed displayed no filamentation defect (for example see cap1Δ panel in Fig. 9D). As Gpx3 and Ybp1 specifically mediate H2O2-induced oxidation and activation of Cap1, these data illustrate that the process of H2O2-stimulated activation of Cap1 is a necessary prerequisite for C. albicans filamentation within the macrophage to facilitate escape from the hostile environment of the phagosome.
H2O2-induced activation of Cap1 is dispensable for C. albicans virulence in a murine model of systemic infection, but is required for C. albicans mediated killing of Galleria mellonella
To investigate whether H2O2-induced activation of Cap1 is important for C. albicans virulence in a systemic model of infection, the virulence of cap1Δ, ybp1Δ, and gpx3Δ null mutants characterised in this study was assessed using the 3 day murine intravenous challenge infection model (29, 30). This model combines weight loss and kidney fungal burden measurements after 3 days of infection to give an “outcome score”. A higher outcome score is indicative of greater weight loss and higher fungal burdens and thus, virulence. Statistical analysis revealed that for all parameters, weight loss, kidney fungal burden and outcome score (Table 1), that there was no significant difference between the wild-type strain, and the cap1Δ, ybp1Δ, and gpx3Δ null mutants and their respective reconstituted strains. Hence, H2O2-induced activation of Cap1 is not required for C. albicans virulence in the murine intravenous challenge model of systemic candidiasis.
Table 1.
Effect of Deleting Cap1, Ybp1, and Gpx3 on Experimental Infection Outcome
| Strain | Kidney burden (log10 CFU/g) | % Weight change | Outcome score |
|---|---|---|---|
| WT (JC747) |
4.7±0.3 |
−1.9±1.7 |
5.6±1.1 |
|
cap1Δ (JC842) |
4.3±0.4 |
−2.2±1.9 |
5.4±1.0 |
|
cap1Δ+CAP1 (JC807) |
4.3±0.4 |
−0.5±2.9 |
4.6±1.5 |
|
ybp1Δ (JC917) |
4.2±0.4 |
−0.2±1.3 |
4.7±0.3 |
|
ybp1Δ+YBP1 (JC809) |
4.2±0.3 |
0.8±3.8 |
3.8±2.1 |
|
gpx3Δ (JC1317) |
4.6±0.3 |
−4.7±1.7 |
7.0±1.1 |
| gpx3Δ+GPX3 (JC1346) | 4.8±0.3 | −4.2±2.2 | 6.9±1.2 |
While unexpected, such findings may be a consequence of directly injecting C. albicans into the bloodstream in such infection models, thereby bypassing any requirement for the fungus to overcome innate immune defences to cross epithelial and endothelial barriers (28). Hence, we also used the Galleria mellonella animal infection model, as this system utilizes phagocytic cells (hemocytes) as part of their host defence (12). By scoring G. mellonella survival after C. albicans infection, we found that cap1Δ cells exhibited a reduction in virulence compared to wild-type cells (p<0.029), whereas there was no difference between wild-type cells and the reintegrant strain (Fig. 10A). In addition, both the ybp1Δ and gpx3Δ mutants also exhibited a significantly slower rate G. mellonella killing compared to wild-type cells (Fig. 10B). These results indicate that H2O2-mediated activation of Cap1 is required for C. albicans pathogenicity in the G. mellonella infection model.
FIG. 10.
C. albicans cells lacking Cap1, Gpx3 or Ybp1 display impaired killing of Galleria mellonella. G. mellonella survival after injection with (A) wild-type (Wt, JC747), cap1Δ (JC842) and cap1Δ+CAP1 (JC807) cells, and (B) wild-type, ybp1Δ (JC917), and gpx3Δ (JC1317) cells. Experiment was performed in duplicate. Survival curves were compared by Kaplan-meier log-rank statistics and p-values are given. ns, not significant; n, number of larvae exposed to a particular strain.
Discussion
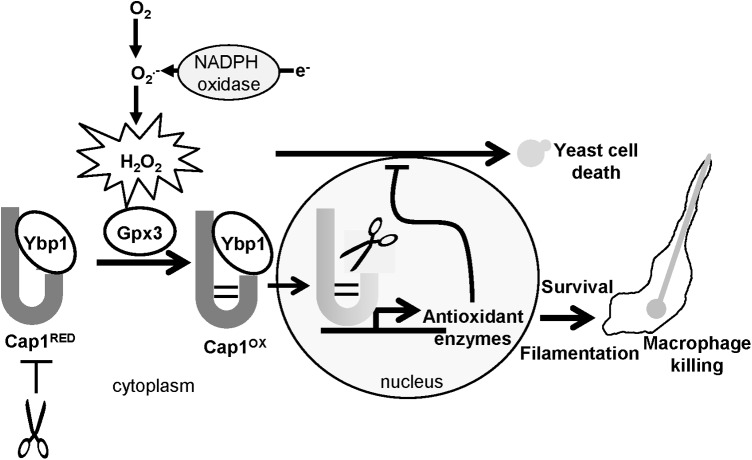
In this study, we show that H2O2-induced activation of Cap1-mediated transcriptional responses is vital for this fungal pathogen to survive phagocytosis by macrophages. Similar to that reported for S. cerevisiae Yap1, H2O2-induced Cap1 oxidation and activation is regulated by a Gpx-like peroxidase, Gpx3, and a cytoplasmic Cap1-interacting protein Ybp1. However, our studies have also uncovered a hitherto undescribed role for Ybp1, as this protein functions to protect cytoplasmic reduced Cap1 and Yap1 from degradation. Importantly, we also show that Gpx3 and Ybp1-mediated Cap1 activation is important to facilitate C. albicans mediated killing of macrophages. A model summarising these major findings is illustrated in Figure 11.
FIG. 11.
Model of Cap1 function in promoting C. albicans-mediated killing of macrophages. Cap1 forms a complex with Ybp1 when in the cytoplasm which functions to prevent degradation of Cap1, and to facilitate H2O2-induced oxidation of Cap1 by the Gpx3 peroxidase. Exposure of C. albicans to H2O2 within the macrophage promotes the Gpx3/Ybp1-mediated oxidation and nuclear accumulation of Cap1. The nuclear accumulation of Cap1 has two consequences: (i) the induction of Cap1-dependent genes with antioxidant functions, which is vital for fungal survival and the subsequent filamentation of C. albicans within the phagosome resulting in macrophage killing, and (ii) the abrogation of the Cap1-Ypb1 interaction, which triggers proteosome-mediated degradation.
In S. cerevisiae, the role of Gpx3 in mediating Yap1 oxidation is well characterised, in that Gpx3 itself is directly oxidized by H2O2, which triggers the formation of a transient disulphide bond between Gpx3 and Yap1 (9). This mixed disulphide is subsequently resolved, leading to the multi-step formation of up to three inter-domain disulphides between the nCRD and cCRD of Yap1 (38), culminating in Yap1 nuclear accumulation and gene activation (9). Cap1 has six cysteine residues of which five are conserved in Yap1, and it is clear that Cap1 also generates a number of different oxidized forms after H2O2 treatment (Fig. 2A). Of the four Gpx-like peroxidases in C. albicans, only that encoded by orf19.4436, which we name Gpx3, mediates H2O2-induced Cap1 oxidation. The basis behind this specificity is not known; however, S. cerevisiae and C. albicans Gpx3 enzymes share regions of similarity which are absent in other Gpx-like peroxidases (Supplementary Fig. S5A), which may underlie the specific function of these enzymes. Gpx3-mediated Yap1 oxidation in S. cerevisiae, also requires the function of Ybp1 (49), and likewise we find that H2O2-mediated Cap1 oxidation is dependent on the orthologue of Ybp1 in C. albicans encoded by orf19.5034.
In contrast to Gpx3, the precise function of Ybp1 in facilitating oxidation of AP-1-like transcription factors is less clear. A recent study in S. cerevisiae, illustrated that the levels of Ybp1 determine the fraction of Yap1 responsive to H2O2-induced oxidation (16). Thus, Ybp1 is required to maintain Yap1 in a structural state that facilitates the formation of the interdomain disulphides within Yap1, which only occur after H2O2 treatment. Our findings reported in this paper provide additional insight into the function of Ybp1 in both C. albicans and S. cerevisiae, as the absence of Ybp1 renders the Cap1 and Yap1 transcription factors unstable. Hence, we propose a model in which Ybp1 binding to Yap1/Cap1 when in the cytoplasm promotes a particular structure of this transcription factor that is both receptive to interdomain disulphide bond formation and is resistant to degradation (Fig. 11). The physiological relevance of Ybp1-mediated stabilisation of Yap1 and Cap1 is likely related to recent reports, which have shown ubiquitin-mediated degradation of nuclear oxidized AP-1-like factors as an important regulatory mechanism (18, 21). We propose that Ybp1 binding to cytoplasmic pools of Yap1 or Cap1 is vital to prevent proteosome-mediated degradation; however, after the nuclear accumulation of Yap1/Cap1 and dissociation from Ybp1 after oxidative stress, such proteins are now sensitive to degradation (Fig. 11). In regard to the oxidative stress-induced nuclear accumulation of Yap1/Cap1, it is important to note that under nonstressed conditions Yap1/Cap1 shuttle in and out of the nucleus. Consistent with this, Cap1 is distributed between the cytoplasm and nucleus in the absence of oxidative stress (Figs. 2C and Fig. 3C). However, as Ybp1 appears to be constitutively cytoplasmic (Fig. 7B), it is possible that Cap1 dissociates from Ybp1 upon nuclear entry both in the presence or absence of oxidative stress. Nonetheless, we propose that it is only upon prolonged nuclear accumulation of Yap1/Cap1 and thus, sustained dissociation from Ybp1 after oxidative stress that triggers the degradation of these AP-1-like factors. Consistent with the hypothesis that Ybp1 binding to AP-1-like transcription factors prevents degradation, we find that treatment of S. cerevisiae ybp1Δ cells with the proteosome inhibitor MG132 results in significantly increased levels of Yap1 (Supplementary Fig. S6). However, we could not reproduce such findings in C. albicans, possibly because the growth conditions optimised to promote MG132 entry into S. cerevisiae (25), do not facilitate entry into this drug-resistant fungus.
In S. cerevisiae, the 2-Cys peroxiredoxin, Tsa1, can stimulate Yap1 oxidation, but only in cells lacking a functional Ybp1 (37, 40). However, Tsa1 is much less effective at mediating Yap1 oxidation compared to Gpx3 and Ybp1 (37, 44). Consistent with residual Tsa1-mediated regulation of Cap1, we see a very low level of Cap1-dependent antioxidant gene expression in ybp1Δ cells (Supplementary Fig. S2). This is more evident at lower levels of H2O2 when Tsa1 is active and not in the inactive sulphinic form (7). This, we predict, underlies our findings that cap1Δ cells are slightly more sensitive to H2O2 than ybp1Δ and gpx3Δ cells. However, attempts to directly determine whether residual Cap1 activation is Tsa1 dependent have been prevented as, despite extensive efforts, we have failed to construct a double ybp1Δtsa1Δ strain, which may be a consequence of other functions of Tsa1 in H2O2 signaling (7). However, it is possible that the residual Gpx3-independent activation of Cap1 is due to H2O2 triggering the formation of hydroxynonenal or other electrolytes, previously shown to be Gpx3-independent potent activators of Yap1 (3).
Significantly, we illustrate in this study that Cap1 plays a vital role in the ability of C. albicans to kill macrophages. As Ybp1 and Gpx3 are also important for C. albicans-mediated macrophage killing, this supports a model in which exposure of C. albicans to H2O2 within the phagosome induces Ybp1/Gpx3-mediated activation of Cap1, which subsequently triggers a transcriptional response that is vital for fungal survival (Fig. 10). Cells lacking CAP1 do display slightly greater impaired macrophage killing compared to ybp1Δ and gpx3Δ cells. This is consistent with the H2O2-sensitive profiles of these mutants and the residual, possibly Tsa1-mediated, Cap1-dependent antioxidant gene expression seen in ybp1Δ and gpx3Δ cells. However, as in vitro, Ybp1 and Gpx3 appear to be major regulators of Cap1 activation within the macrophage. We also provide data that indicates that H2O2-induced activation of Cap1 is necessary to allow the subsequent filamentation and escape of C. albicans from the macrophage. Morphogenetic switching from yeast to hyphal forms has long been recognized to play a major role in C. albicans' ability to kill macrophages (27, 31). However, to the best of our knowledge this is the first report that oxidative stress defences of C. albicans are needed to facilitate filamentation after phagocytosis, and we are actively examining the molecular basis underlying this phenomenon.
In contrast to the vital role of H2O2-induced activation of Cap1 in macrophage survival, we found that this process is dispensable for C. albicans virulence in a murine model of systemic infection. This was unexpected as previous reports have shown that Cap1-dependent genes, such as CTA1 encoding catalase (53) or the thioredoxin encoding TRX1 gene (7), are important for C. albicans survival in systemic models of infection. However, it is possible that Cap1-independent basal levels of such genes may be important for virulence in such models. Moreover, the seemingly differing requirements for Cap1 in promoting C. albicans virulence are consistent with the induction profiles of antioxidant encoding genes in different experimental infection models. For example, while Cap1-dependent antioxidant gene expression is seen after phagocytosis by innate immune cells (10, 13, 27), such genes are not significantly induced during infection of organs after systemic infections (10, 45). This suggests that adaptation to oxidative stress might be critical in the early stages of systemic C. albicans infections, but less important once these infections are established. As C. albicans cells are injected directly into the bloodstream during systemic infection models, this bypasses the innate immune defences, which prevent crossing of epithelial and endothelial barriers (28), and therefore, Cap1-dependent antioxidant gene expression may not be important for promoting fungal survival in such a model. Consistent with this suggestion, we found Cap1 and its regulators Ybp1 and Gpx3 were required for C. albicans virulence in G. mellonella (15), where phagocytosis by haemocytes is an important component of this infection model (12).
Taken together, this study illustrates that Gpx3 and Ybp1 mediated H2O2-induced activation of Cap1 is vital for C. albicans-mediated macrophage killing, survival in macrophages and in an animal model host reliant on ROS-based defence mechanisms. As AP-1-like transcription factors are conserved throughout the fungal kingdom (34), we envisage that such findings will be applicable to other fungal pathogens. Indeed, this is evidenced by the finding that the maize pathogen Ustilago maydis requires Yap1 to survive ROS-mediated early plant defences (32).
Materials and Methods
Strains and growth conditions
The strains used in this study are listed in Table 2, and details of their construction are given in the Supplementary Methods. All strains were growth at 30°C in YPD medium (1% yeast extract, 2% Bacto-peptone, 2% glucose) (41). To stress cells with H2O2, a concentration of 5 mM was employed, as we have previously demonstrated a similar level of C. albicans survival after a 1 h incubation with this dose of peroxide stress (7) to that seen after incubation of wild-type C. albicans with neutrophils for 1 h (13). All tagged versions of Cap1 and Ybp1 generated in this study were functional (Supplementary Fig. S7).
Table 2.
Strains Used in This Study
| Relevant genotype | Source | |
|---|---|---|
|
Candida albicans strains | ||
| JC21 |
ura3:: λ imm434/ura3::λimm434, his1::hisG/his1::hisG CIp20 (URA3, HIS1) |
(42) |
| JC50 |
ura3:: λ imm434/ura3::λimm434, his1::hisG/his1::hisG, hog1::loxP-ura3-loxP, hog1::loxP-HIS-loxP CIp20 |
(42) |
| JC52 |
ura3:: λ imm434/ura3::λimm434, his1::hisG/his1::hisG, hog1::loxP-ura3-loxP, hog1::loxP-HIS-loxP CIp20-HOG1 |
(42) |
| SN148 |
arg4Δ/arg4Δ,leu2Δ/leu2Δ,his1Δ/his1Δ,ura3Δ::imm434/ura3Δ::imm434 iro1Δ::imm434/iro1Δ::imm434 |
(35) |
| JC747 |
SN148+CIp30 (URA3, HIS1, ARG4) |
(7) |
| JC1014 |
SN148+pACT1(URA3) |
This work |
| JC1429 |
SN148+pACT1-CAP1(URA3) |
This work |
| JC948 |
SN148 CAP1-MH-URA3 |
(7) |
| JC1060 |
SN148 CAP1-GFP-URA3 |
(7) |
| JC1200 |
SN148 YBP1-MH-URA3 |
This work |
| JC710 |
cap1::loxP-HIS1-loxP/cap1::loxP-ARG4-loxP |
(7) |
| JC842 |
cap1::loxP-HIS1-loxP/cap1::loxP-ARG4-loxP, CIp20 (HIS1, URA3) |
(7) |
| JC807 |
cap1::loxP-HIS1-loxP/cap1::loxP-ARG4-loxP,CIp20-CAP1 (HIS1, URA3) |
This work |
| JC1398 |
cap1::loxP-HIS1-loxP/cap1::loxP-ARG4-loxP, pACT1(URA3) |
This work |
| JC1388 |
cap1::loxP-HIS1-loxP/cap1::loxP-ARG4-loxP, pACT1-CAP1 (URA3) |
This work |
| JC725 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP |
This work |
| JC917 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP, CIp20 (HIS1, URA3) |
This work |
| JC809 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP,CIp20-YBP1 (HIS1,URA3) |
This work |
| JC954 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP, CA P1-MH-URA3 |
This work |
| JC1054 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP, CAP1-GFP-URA3 |
This work |
| JC1400 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP, pACT1(URA3) |
This work |
| JC1390 |
ybp1::loxP-HIS1-loxP/ybp1::loxP-ARG4-loxP, pACT1-CAP1(URA3) |
This work |
| JC1309 |
gpx3::loxP-HIS1-loxP/gpx3::loxP-ARG4-loxP |
This work |
| JC1317 |
gpx3::loxP-HIS1-loxP/gpx3::loxP-ARG4-loxP CIp20 (HIS1, URA3) |
This work |
| JC1346 |
gpx3::loxP-HIS1-loxP/gpx3::loxP-ARG4-loxP CIp20-GPX3 (HIS1, URA3) |
This work |
| JC1311 |
gpx3::loxP-HIS1-loxP/gpx3::loxP-ARG4-loxP CAP1-MH-URA3 |
This work |
| JC1314 |
gpx3::loxP-HIS1-loxP/gpx3::loxP-ARG4-loxP CAP1-GFP-URA3 |
This work |
| JC1313 |
gpx3::loxP-HIS1-loxP/gpx3::loxP-ARG4-loxP YBP1-MH-URA3 |
This work |
| JC1299 |
orf19.85::loxP-HIS1-loxP/orf19.85::loxP-ARG4-loxP |
This work |
| JC1081 |
orf19.86::loxP-HIS1-loxP/orf19.86::loxP-ARG4-loxP |
This work |
| JC1308 |
orf19.87::loxP-HIS1-loxP/orf19.87::loxP-ARG4-loxP |
This work |
| JC1027 |
tsa1::loxP-HIS1-loxP/tsa1::loxP-ARG4-loxP/tsa1::hisG/tsa1::hisG CIp10 (URA3) |
(7) |
| JC1028 |
tsa1::loxP-HIS1-loxP/tsa1::loxP-ARG4-loxP/tsa1::hisG/tsa1::hisG CIp10-TSA1 (URA3) |
(7) |
| JC998 |
tsa1::loxP-HIS1-loxP/tsa1::loxP-ARG4-loxP/tsa1::hisG/tsa1::hisG CAP1-MH-URA3 |
This work |
| JC1056 |
tsa1::loxP-HIS1-loxP/tsa1::loxP-ARG4-loxP/tsa1::hisG/tsa1::hisG CAP1-GFP-URA3 |
This work |
|
Saccharomyces cerevisiae strains | ||
| SR1 |
MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 ybp1::HIS3 |
(49) |
| SR6 | MATα ade2-1 trp1-1 can1-100 leu2-3,112 his3-11 ura3 ybp1::YBP1 | (49) |
Stress sensitivity tests
Midexponential phase growing cells were diluted in YPD, and approximately 104 cells, and 10-fold dilutions thereof, were spotted in 5 μl onto YPD agar containing the indicated compound. Plates were incubated at 30°C for 24 h.
Cap1 oxidation and phosphorylation assays
Cap1 oxidation was determined as described previously using the thiol alkylating agent AMS (7), with the additional step of alkaline phosphatase (New England Biolabs) treatment of protein lysates (5 units, 37°C for 1 h) before SDS-PAGE. To analyse Cap1 phosphorylation, protein extracts were prepared (42) from C. albicans cells expressing 2-myc 6His tagged Cap1 (Cap1-MH). Cap1-MH was precipitated using Ni2+-NTA agarose (Qiagen), and subjected to SDS-PAGE on 8% gels and western blotting using anti-myc antibodies (9E10; Sigma). In control samples, the Cap1-MH coupled Ni2+-NTA agarose resin was incubated with 200 units of λ protein phosphatase (New England Biolabs) for 30 min at 37°C.
Cap1 and Yap1 protein stability assays
Cap1: C. albicans wild-type (JC747) and ybp1Δ (JC917) cells, were collected by centrifugation and resuspended in fresh YPD media containing 1000 μg/ml cycloheximide (Sigma). Cells were harvested by centrifugation at 0 and 1 h post cycloheximide treatment and snap frozen in liquid nitrogen. Protein extracts were prepared (42), and 50 μg of protein lysate analysed by SDS-PAGE and western blotting using an antibody raised against recombinant Cap1 (49). Yap1: S. cerevisiae ybp1Δ (SR1) and ybp1Δ+YBP1 (SR6) cells transformed with pRS316-Yap1-Myc (8) were treated with 100 μg/ml cycloheximide for 0 and 1 h, and 50 μg of protein lysate analysed by SDS-PAGE and western blotting using anti-myc antibodies. Relative Cap1 and Yap1 levels, in wild-type and ybp1Δ cells, were quantified from three independent experiments using ImageQuant (GE Healthcare).
Cap1-Ybp1 coimmunoprecipitation
Exponentially growing wild-type (JC1200) and gpx3Δ (JC1313) cells expressing Ybp1-MH were harvested before and after treatment with 5 mM H2O2 for 10 min. Protein extracts were prepared (42) and Ybp1-MH immunoprecipitated with anti-myc antibody-coupled agarose (Sigma). After washing, the anti-myc agarose was analysed by SDS-PAGE and western blotting. Ybp1-MH was detected using anti-myc antibodies, and Cap1 was detected using an antibody raised against recombinant Cap1 (54).
Ybp1 localization
Exponentially growing wild-type cells expressing Ybp1-MH (JC1200) were harvested before and after treatment with 5 mM H2O2 for 10 min, and Ybp1-MH localization determined by immunofluorescent staining as previously described (43). The primary antibody used was the monoclonal 9E10 anti-myc antibody (Sigma), at a 1:1000 dilution. The secondary antibody used was a 1:50 dilution of goat anti-mouse FITC-conjugated secondary antibody (Sigma).
Cap1-localization
Cells expressing Cap1-GFP were prepared as described previously (11). 4′,6-diamidino-2-phenylindole and GFP fluorescence were captured using a Zeiss Axioscope, with a 63×oil immersion objective, and Axiovision imaging system.
RNA analysis
Northern blot analyses were performed as described previously (11). Gene-specific probes were amplified by PCR from genomic DNA using oligonucleotide primers specific for ADH7, CTA1, TRR1, and ACT1 (11) and YBP1 (YBP1F and YBP1R). RNA levels were quantified by phosphoimaging (Bio-imaging analyser Fuji Film Bas-1500) and Tina 2.0 software (Raytest).
C. albicans phagocytosis assay
J774.1 macrophages were cultured as described previously (24, 31). C. albicans strains from overnight cultures were added to the J774.1 macrophages at a 1:1 C. albicans/macrophage ratio. After 180 min coincubation, wells were washed twice with 1% (w/v) sterile phosphate buffered saline (PBS) to remove excess cells. C. albicans uptake by macrophages was assessed by light microscopy (Nikon Eclipse TE2000-U microscope with a×40 objective) as described previously (31). Results were expressed as percentage uptake (the percentage of macrophages which have taken up at least one fungal cell). Data were obtained in triplicate from at least 3 different experiments by analyzing at least 100 macrophages per well. One-way analysis of variance (ANOVA) was used to determine statistical significance.
Macrophage killing assays
The macrophage killing assay was conducted under the same conditions as described above for the phagocytosis assay. After removal of excess unbound C. albicans by rigorous washing with 1% (v/v) PBS, killing of macrophages was assessed by trypan blue exclusion (31). A 150-μl sample of trypan blue and 150 μl of 1% PBS were added to cells for 2 min and removed by lightly washing twice with 1% PBS; cells were then fixed with 3% paraformaldehyde. Cells were counted under an inverted light microscope (Nikon Eclipse TE2000-U microscope with 40×objective). Data were obtained in triplicate from at least three separate experiments by analyzing at least 200 macrophages per well. One-way ANOVA was used to determine statistical significance.
Candida killing by macrophage assay
J774.1 macrophages were cultured as described previously (24, 31). These were plated at a density of 2×105 cells in six well plates for 24 h. Exponential growing C. albicans strains were added to the J774.1 macrophages at a 500:1 macrophage/C. albicans ratio or to media without any macrophages. Cells were coincubated overnight and C. albicans colonies were counted. Results are expressed as the percentage of colonies formed in the presence of macrophages compared to those formed in the absence of macrophages. Data were obtained in triplicate from three separate experiments, and one-way ANOVA was used to determine statistical significance.
Murine intravenous challenge model of C. albicans infection
BALB/c female mice (mean body weight 17.3±0.7 g) were housed in groups of 6 with food and water provided ad libitum. C. albicans cells were grown overnight on Sabouraud agar at 30°C, harvested in sterile saline, and cell counts adjusted by hemocytometer to provide a cell suspension to deliver a challenge dose of 3×104 CFU/g body weight. Actual challenge dose was determined from viable counts read 24 h later and was between 2.6×104 and 3.2×104 CFU/g. Mice were infected intravenously via a lateral tail vein. Body weights were recorded daily. 72 h after challenge the animals were weighed, humanely terminated and kidneys removed aseptically. Fungal burdens were measured by viable counts for two half kidneys per animal. Virulence was assessed by fungal kidney burdens at 72 h, and by percent weight change over 72 h, from which an outcome score was calculated (29, 30). Differences were tested statistically by the Mann-Whitney U test. All animal experimentation conformed to the requirements of United Kingdom Home Office legislation and of the Ethical Review Committee of the University of Aberdeen.
G. mellonella survival assay
C. albicans strains were grown in YPD overnight at 30°C. Cells were harvested, washed twice in sterile saline and enumerated by haemocytometer count and resuspended to 6×106 CFU/ml. For each strain, 15–20 G. mellonella larvae were injected with 10 μl (6×104 CFU) of cell suspension into the final left proleg. Larvae were incubated at 37°C and were monitored twice daily. Larvae were defined as dead when they turned grey/black and no longer responded to stimulus. Saline controls and unmanipulated controls all survived to the end of the experiment. Survival curves were compared by Kaplan-meier log-rank statistics.
Supplementary Material
Abbreviations Used
- AMS
4-acetamido-4′-(idoacetyl)amino)stilbene-2,2′-disulfonic acid
- ANOVA
analysis of variance
- CRD
cysteine rich domain
- DAPI
4′,6-diamidino-2-phenylindole
- DEM
diethylmaleate
- GFP
green fluorescent protein
- Gpx
glutathione peroxidase
- H2O2
hydrogen peroxide
- NADPH
nicotinamide adenine dinucleotide phosphate
- Nox
NADPH oxidase
- PBS
phosphate buffered saline
- ROS
reactive oxygen species
- SAPK
stress activated protein kinase
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Wt
wild-type
Acknowledgments
J.Q. thanks Professor Scott Moye-Rowley (University of Iowa) for the gift of the anti-Cap1 antibody and Dr. Despoina Kaloriti (University of Aberdeen) for her help with the Candida killing by macrophage assay. This work was funded by the Wellcome Trust (J.Q./B.A.M./L.P.E./D.M.M.), and a BBSRC DTG-studentship (M.J.P.).
References
- 1.Alarco AM. and Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol 181: 700–708, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arana DM, Alonso-Monge R, Du C, Calderone R, and Pla J. Differential susceptibility of mitogen-activated protein kinase pathway mutants to oxidative-mediated killing by phagocytes in the fungal pathogen Candida albicans. Cell Microbiol 9:1647–1657, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Azevedo D, Tacnet F, Delaunay A, Rodrigues-Pousada C, and Toledano MB. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic Biol Med 35: 889–900, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Babior BM. NADPH oxidase. Curr Opin Immunol 16: 42–47, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Bain JM, Lewis LE, Okai B, Quinn J, Gow NA, and Erwig LP. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet Biol 49: 677–678, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bozonet SM, Findlay VJ, Day AM, Cameron J, Veal EA, and Morgan BA. Oxidation of a eukaryotic 2-Cys peroxiredoxin is a molecular switch controlling the transcriptional response to increasing levels of hydrogen peroxide. J Biol Chem 280: 23319–23327, 2005 [DOI] [PubMed] [Google Scholar]
- 7.da Silva Dantas A, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, and Quinn J. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol Cell Biol 30: 4550–4563, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaunay A, Isnard AD, and Toledano MB. H2O2 sensing through oxidation of the Yap1 transcription factor. Embo J 19: 5157–5166, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaunay A, Pflieger D, Barrault MB, Vinh J, and Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Enjalbert B, MacCallum DM, Odds FC, and Brown AJ. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun 75: 2143–2151, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, and Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell 17: 1018–1032, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fallon J, Kelly J, and Kavanagh K. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol Biol 845: 469–485, 2012 [DOI] [PubMed] [Google Scholar]
- 13.Fradin C, De Groot P, MacCallum D, Schaller M, Klis F, Odds FC, and Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol 56: 397–415, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Fradin C, Kretschmar M, Nichterlein T, Gaillardin C, d'Enfert C, and Hube B. Stage-specific gene expression of Candida albicans in human blood. Mol Microbiol 47: 1523–1543, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Fuchs BB, O'Brien E, Khoury JB, and Mylonakis E. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1: 475–482, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Gulshan K, Lee SS, and Moye-Rowley WS. Differential oxidant tolerance determined by the key transcription factor Yap1 is controlled by levels of the Yap1-binding protein, Ybp1. J Biol Chem 286: 34071–34081, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulshan K, Rovinsky SA, and Moye-Rowley WS. YBP1 and its homologue YBP2/YBH1 influence oxidative-stress tolerance by nonidentical mechanisms in Saccharomyces cerevisiae Eukaryot Cell 3: 318–330, 2004 [DOI] [PMC free article] [PubMed]
- 18.Gulshan K, Thommandru B, and Moye-Rowley WS. Proteolytic degradation of the Yap1 transcription factor is regulated by subcellular localization and the E3 ubiquitin ligase Not4. J Biol Chem 287: 36797–36805, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, and Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology 148: 3705–3713, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Inoue Y, Matsuda T, Sugiyama K, Izawa S, and Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae J Biol Chem 274: 27002–27009, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Kitamura K, Taki M, Tanaka N, and Yamashita I. Fission yeast Ubr1 ubiquitin ligase influences the oxidative stress response via degradation of active Pap1 bZIP transcription factor in the nucleus. Mol Microbiol 80: 739–755, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Kuge S, Arita M, Murayama A, Maeta K, Izawa S, Inoue Y, and Nomoto A. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol Cell Biol 21: 6139–6150, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuge S, Toda T, Iizuka N, and Nomoto A. Crm1 (XpoI) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3: 521–532, 1998 [DOI] [PubMed] [Google Scholar]
- 24.Lewis LE, Bain JM, Lowes C, Gillespie C, Rudkin FM, Gow NA, and Erwig LP. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog 8: e1002578, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Apodaca J, Davis LE, and Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42: 158–162, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, and Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell 90: 939–949, 1997 [DOI] [PubMed] [Google Scholar]
- 27.Lorenz MC, Bender JA, and Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3: 1076–1087, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maccallum DM. Hosting infection: experimental models to assay Candida virulence. Int J Microbiol 2012: 363764, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacCallum DM, Castillo L, Nather K, Munro CA, Brown AJ, Gow NA, and Odds FC. Property differences among the four major Candida albicans strain clades. Eukaryot Cell 8: 373–387, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacCallum DM, Coste A, Ischer F, Jacobsen MD, Odds FC, and Sanglard D. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob Agents Chemother 54: 1476–1483, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenzie CG, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, Gow NA, and Erwig LP. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect Immun 78: 1650–1658, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina L. and Kahmann R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 19: 2293–2309, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moye-Rowley WS. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot Cell 2: 381–389, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolaou E, Agrafioti I, Stumpf M, Quinn J, Stansfield I, and Brown AJ. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol Biol 9: 44, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noble SM. and Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4: 298–309, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohkuni K, Abdulle R, Tong AH, Boone C, and Kitagawa K. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS One 3: e1617, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okazaki S, Naganuma A, and Kuge S. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxid Redox Signal 7: 327–334, 2005 [DOI] [PubMed] [Google Scholar]
- 38.Okazaki S, Tachibana T, Naganuma A, Mano N, and Kuge S. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell 27: 675–688, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Pfaller MA. and Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20: 133–163, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross SJ, Findlay VJ, Malakasi P, and Morgan BA. Thioredoxin peroxidase is required for the transcriptional response to oxidative stress in budding yeast. Mol Biol Cell 11: 2631–2642, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman F. Getting started with yeast. Methods Enzymol 194: 3–21, 1991 [DOI] [PubMed] [Google Scholar]
- 42.Smith DA, Nicholls S, Morgan BA, Brown AJ, and Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 15:4179–4190, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol 41: 19–31, 2001 [DOI] [PubMed] [Google Scholar]
- 44.Tachibana T, Okazaki S, Murayama A, Naganuma A, Nomoto A, and Kuge S. A major peroxiredoxin-induced activation of Yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J Biol Chem 284: 4464–4472, 2009 [DOI] [PubMed] [Google Scholar]
- 45.Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, and Hube B. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol 63: 1606–1628, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, and Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev 12: 1453–1463, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, and Brown AJ. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. Embo J 21: 5448–5456, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vazquez-Torres A. and Balish E. Macrophages in resistance to candidiasis. Microbiol Mol Biol Rev 61: 170–192, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veal EA, Ross SJ, Malakasi P, Peacock E, and Morgan BA. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J Biol Chem 278: 30896–30904, 2003 [DOI] [PubMed] [Google Scholar]
- 50.Vivancos AP, Castillo EA, Biteau B, Nicot C, Ayte J, Toledano MB, and Hidalgo E. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc Natl Acad Sci U S A 102: 8875–8880, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vivancos AP, Castillo EA, Jones N, Ayte J, and Hidalgo E. Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol Microbiol 52: 1427–1435, 2004 [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, Ying K, Chen WS, and Jiang YY. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med 40: 1201–1209, 2006 [DOI] [PubMed] [Google Scholar]
- 53.Wysong DR, Christin L, Sugar AM, Robbins PW, and Diamond RD. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect Immun 66: 1953–1961, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, De Micheli M, Coleman ST, Sanglard D, and Moye-Rowley WS. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol Microbiol 36: 618–629, 2000 [DOI] [PubMed] [Google Scholar]
- 55.Znaidi S, Barker KS, Weber S, Alarco AM, Liu TT, Boucher G, Rogers PD, and Raymond M. Identification of the Candida albicans Cap1p regulon. Eukaryot Cell 8: 806–820, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.