Abstract
Despite diverging ~365 million years ago, tetrapod limbs and pectoral fins express similar genes that could be regulated by shared regulatory elements. In this study, we set out to analyze the ability of enhancers to maintain tissue specificity in these two divergent structures. We tested 22 human sequences that were previously reported as mouse limb enhancers for their enhancer activity in zebrafish (Danio rerio). Using a zebrafish enhancer assay, we found that 10/22 (45 %) were positive for pectoral fin activity. Analysis of the various criteria that correlated with positive fin activity found that both spatial limb activity and evolutionary conservation are not good predictors of fin enhancer activity. These results suggest that zebrafish enhancer assays may be limited in detecting human limb enhancers, and this limitation does not improve by the use of limb spatial expression or evolutionary conservation.
Keywords: Enhancers, Fin, Limb, Mesenchyme, ZPA, AER
Introduction
The divergence of the modern tetrapod from a shallow water fish is estimated to have occurred approximately 365 million years ago (MYA), while the last common ancestor for the mammalian and teleost (zebrafish) lineages dates to over 450 MYA (Kumar and Hedges 1998). Paired fins are considered homologous structures of vertebrate limbs because of shared developmental components (Yano and Tamura 2013). Data has shown that many genes are shared in fin and limb development but have different regulation and expression. Furthermore, regulatory elements have been shown to control spatiotemporal development of appendages and are suggested to have a causative role in the morphological changes from the fin to the limb (Mercader 2007).
Enhancers involved in early developmental regulation can be evolutionarily conserved (Pennacchio et al. 2006). In addition, there are also reports suggesting that evolutionary conservation is not always a good filter in detecting functional enhancers in different species (McGaughey et al. 2008; Blow et al. 2010). A lingering question is how well orthologous enhancers are conserved in terms of their function among divergent species. Ritter and colleagues (2010) found that at least 30 % of the conserved sequences that they tested were positive for enhancer activity when using human-specific enhancers in mouse and zebrafish. Another recent study found this number to be even lower, with only 17 % of human sequences that were conserved to fish displaying similar enhancer activity in mouse and zebrafish (Ariza-Cosano et al. 2012). This study also suggested that most differences in enhancer activity could be due to changes in gene expression of that particular model. Here, we set out to analyze the ability of mammalian limb enhancers to maintain tissue specificity in the developing zebrafish pectoral fin.
Materials and methods
Limb enhancer selection and cloning
Limb enhancers were primarily selected from the VISTA enhancer browser (Visel et al. 2007) and named in this article using the browser’s nomenclature. In addition, we also selected the zone of polarizing activity (ZPA)-regulating sequence (ZRS) enhancer, which is expressed in the ZPA of the developing limb (Lettice et al. 2003). A total of 22 human candidate sequences were cloned with Gateway Technology (Invitrogen) or the In-Fusion HD Cloning System (Clontech) into the E1b-Tol2-GFP vector (Li et al. 2009) and were sequence verified (Quintara Biosciences) for the presence of the proper insert.
Zebrafish enhancer assays
Wild-type AB zebrafish were mated, and embryos were collected at the one-cell stage for microinjection; 125 ng from each of the E1b-Tol2-GFP vectors carrying the candidate enhancer elements was injected along with 175 ng of Tol2 transposase mRNA (Li et al. 2009), to facilitate genomic integration into these embryos using standard procedures (Fisher et al. 2006). All enhancer candidates were injected in at least two different injection days to make sure that embryo quality, injection mix, or injector did not compromise the enhancer assay. Green fluorescent protein (GFP) activity was monitored at 24, 48, and 72 h post-fertilization (hpf). For each construct, at least 50 live embryos were annotated up to 72 hpf, and enhancer candidates were scored as positive fin enhancers upon pectoral fin GFP activity of ≥20 % (pectoral fin GFP activity/total live embryos) at either time point. All animal work was approved by the UCSF Institutional Animal Care and Use Committee protocol number AN084690.
Results and discussion
Limb enhancer selection
In order to test the fin activity of various limb enhancers, we selected previously characterized limb enhancers. The VISTA enhancer browser (Visel et al. 2007) currently has 139 human sequences (hs) that tested positive for limb activity in embryonic day (E) ll.5 mouse embryos. We classified these enhancers based on their expression pattern in the developing mouse limb. Their limb activity pattern was defined as follows: whole mesenchyme, intermediate mesenchyme, partial mesenchyme, apical ectodermal ridge (AER), and ZPA (Online resource 1). We chose 18 human elements for our subsequent zebrafish enhancer assays by selecting those that were mainly expressed in the limb and that demonstrated strong limb activity (based on the number of embryos showing limb activity versus total LacZ-stained embryos). Since the AER is an important signaling center for proper distal limb and fin outgrowth (Mercader 2007), we also chose an additional three AER-expressing elements (hs483, hs1112, and hs1442) that also had activity in additional tissues (brain and craniofacial). In addition to elements from the VISTA enhancer browser, we also selected the ZRS element, which regulates Sonic hedgehog (Shh) expression in the ZPA during limb development (Lettice et al. 2003). Combined 22 elements were selected for our zebrafish enhancer assays (Online resource 2).
Differences in enhancer activity
The human sequence of these enhancers was cloned into the E1b-Tol2-GFP zebrafish enhancer assay vector (Li et al. 2009) and microinjected into one-cell stage zebrafish embryos using standard procedures (Fisher et al. 2006). Although the pectoral fin only becomes visible after 28 hpf (Sordino et al. 1995; Mercader 2007), we looked for GFP activity at 24, 48, and 72 hpf for all tissues. Out of the 22 tested sequences, ten (45 %) showed positive pectoral fin enhancer activity, defined as ≥20 % of live embryos with consistent GFP activity at any single time point (Table 1, Online resource 3). Ritter and colleagues (2010) achieved a 30 % success rate of finding positive human enhancer activity in zebrafish and a similar 30 % success rate when testing the orthologous zebrafish sequences in zebrafish. By analyzing highly conserved human regulatory elements in mouse and fish, Ariza-Cosano and colleagues (2012) found that less than 17 % of tissue-specific enhancers showed functional conservation in zebrafish. This study also utilized six limb enhancers from the VISTA enhancer browser (hs200, hs259, hs312, hs335, hs609, and hs774) (Visel et al. 2007), finding two (hs312 and hs774) of the six (33 %) to be expressed in the fin, which is less than our current results. We also tested hs259 and hs774 and report that both have positive GFP activity in the fin at 72 hpf. It is worth noting that there were differences between our study and the aforementioned studies. Ritter et al. (2010) and Ariza-Cosano et al. (2012) selected sequences based on conservation between human and fish, while we focused on a specific and divergent tissue, fin/limb, and only half of the tested sequences were conserved between human and fish. In addition, a different minimal promoter (gata2a) was used in the study of Ariza-Cosano et al. (2012), and fish were only annotated from 24 to 48 hpf in both studies (Ritter et al. 2010). In this study, four of the positive enhancers, hs259, hs774, hs1109, and hs1430, were negative for enhancer activity at 48 hpf, but positive at 72 hpf (Table 1, Online resource 2). These differences could provide rationale as to why the frequency rates are slightly higher than previous findings.
Table 1.
Summary of results for the human limb enhancers tested in zebrafish. Elements in the peach box represent 48–72 hpf positive pectoral fin enhancers. Elements in the blue box represent sequences that were negative for pectoral fin expression at 48–72 hpf. Expression patterns are the following: whole mesenchyme (WM), partial mesenchyme (PM), intermediate mesenchyme (IM), apical ectodermal ridge (AER), and zone of polarizing activity (ZPA). “Fin” denotes GFP expression in the pectoral fin at either 48 or 72 hpf. “Other” denotes that expression of ≥20 % was observed in another tissue. If the assayed sequence was conserved between human and fish as determined by either the UCSC Genome Browser or the ECR Browser, it has a green check mark
| Element | Mouse Pattern | Zebrafish Pattern | Fish Conservation | ||
|---|---|---|---|---|---|
| Fin | Other | ||||
| POSITIVE | hs243 | AER/PM | ✓ | ✓ | ✓ |
| hs259 | IM | ✓ | ✓ | x | |
| hs280 | PM | ✓ | ✓ | ✓ | |
| hs741 | WM | ✓ | ✓ | ✓ | |
| hs774 | WM | ✓ | ✓ | ✓ | |
| hs919 | WM | ✓ | ✓ | ✓ | |
| hs1109 | WM | ✓ | ✓ | x | |
| hs1430 | WM | ✓ | ✓ | x | |
| hs1447 | PM | ✓ | ✓ | x | |
| hs1452 | WM | ✓ | ✓ | x | |
| Element | Mouse Pattern | Zebrafish Pattern | Fish Conservation | ||
|---|---|---|---|---|---|
| Fin | Other | ||||
| NEGATIVE | hs72 | WM | x | x | ✓ |
| hs483 | AER | x | ✓ | ✓ | |
| hs516 | WM | x | ✓ | x | |
| hs1112 | AER | x | ✓ | ✓ | |
| hs1422 | AER | x | x | ✓ | |
| hs1432 | WM | x | x | ✓ | |
| hs1442 | AER | x | x | x | |
| hs1455 | IM | x | ✓ | x | |
| hs1463 | PM | x | x | x | |
| hs1473 | WM | x | x | x | |
| hs1487 | WM | x | ✓ | x | |
| ZRS | ZPA | x | ✓ | ✓ | |
Spatial limb activity patterns are not good predictors for conserved enhancer activity
We next looked to see if there was any correlation between the spatial limb activity and fin enhancer activity. All of the sequences with positive fin expression displayed mouse mesenchymal limb activity. Mesenchymal cells make up most of the cell population during limb/fin development and are important constituents of both fin and limb development, which could be one of the reasons for the human limb mesenchyme enhancers being expressed in the fin (Mercader 2007). However, 7 of the 12 sequences that were negative for fin enhancer activity also had some kind of mesenchymal expression, suggesting that this is not a reliable filter in identifying human sequences that could function as fin enhancers.
Only one of the five AER enhancers, hs243, displayed pectoral fin expression. hs243 was the only AER enhancer to also have mesenchyme expression in E11.5 mouse embryos, which could be the cause for it having pectoral fin enhancer activity (Online resource 3). The fin AER can only be detected up to 36 hpf, and thereafter, it changes to the apical fold (AF). The AF and AER, although being homologous structures, have two different fates; the AF becomes a part of the mature fin, while the AER is barely detectable in adult limbs (Mercader 2007). This difference in developmental fates may serve as rationale for why four of the five tested AER enhancers did not show expression in the fin. To verify that we did not miss pectoral fin enhancer expression in the time points when AER can be detected in the fin, we also analyzed all five AER enhancers for their expression at 28, 32, and 36 hpf. hs243 was the only element positive for pectoral fin expression at 32 and 36 hpf (Online resource 2), suggesting that its fin expression is not correlated with the AER to AF shift, since it is also a later pectoral fin enhancer. It is also worth noting that while the ZRS enhancer is conserved to fish and expressed in the ZPA, it did not show pectoral fin expression at any of the assayed time points including 28, 32, and 36 hpf (Online resource 2).
Sequence differences may cause nonspecific enhancer expression
All ten positive fin enhancers had enhancer activity in other tissues (Table 1, Online resource 2). In addition, seven of the elements that tested negative for pectoral fin expression were enhancers in other tissues (hs483, hs516, hs1112, hs1445, hs1487, and ZRS) (Table 1, Fig. 1b, c). Of the three AER enhancers with expression in another tissue (hs483, hs1112, and hs1442), only hs1112 showed reporter expression in a similar tissue as in mouse (midbrain). The ZRS, while being negative for pectoral fin, showed forebrain enhancer activity at 48 hpf (Fig. 1d). Overall, our results imply that limb-specific enhancers are expressed in other tissues, not exclusively in the pectoral fin during zebrafish development. These results are line with a previous report that also observed various activity domains for mammalian enhancers in zebrafish (Ariza-Cosano et al. 2012).
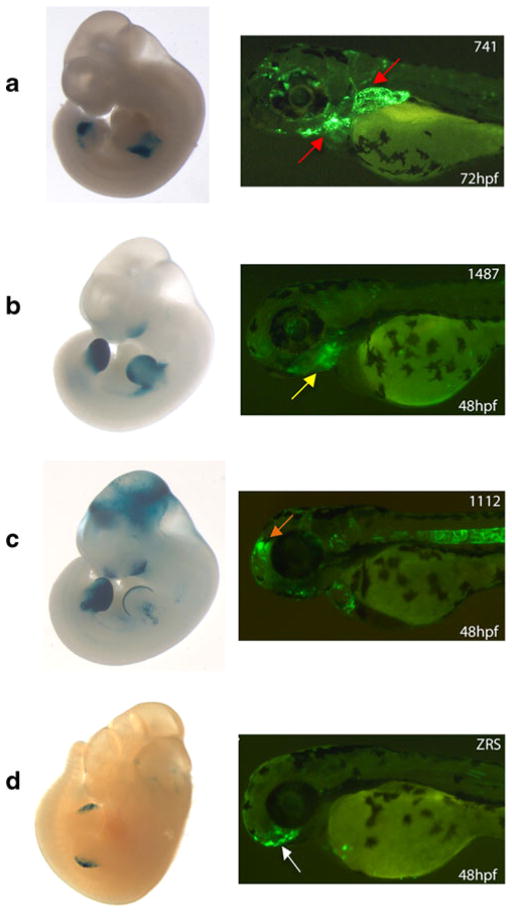
Fig. 1.
Human limb enhancer expression in mice and zebrafish. a hs741 shows mouse enhancer expression in the limb mesenchyme at E11.5 and zebrafish pectoral fin expression (red arrow) and pharyngeal arch expression at 72 hpf. b hs1487 has mouse enhancer expression in the limb mesenchyme and partial expression in the branchial arch (two out of five available embryos in the VISTA Enhancer Browser). This sequence only shows pharyngeal arch enhancer expression (yellow arrow) in zebrafish at 48 and 72 hpf, here depicted at 48 hpf. c hs1112 is expressed in the AER and mouse forebrain, midbrain, and hindbrain at E11.5. In zebrafish, this sequence shows midbrain (orange arrow) and notochord expression at 48 hpf. d The ZRS enhancer shows activity in the mouse ZPA at E11.5. In zebrafish, this sequence has enhancer activity in the forebrain (white arrow) at 48 hpf. Mouse embryo pictures for elements hs741, hs1487, and hs1112 were obtained from the VISTA Enhancer Browser (Visel et al. 2007). The ZRS enhancer was generated in our lab and reported in the study of Laurell et al. (2012)
Conservation of mammalian limb enhancers to fish
We next assessed whether there was a correlation between evolutionary conservation of mammalian elements to fish species and positive pectoral fin enhancer activity. Analysis of evolutionary conservation using the University of California, Santa Cruz (UCSC) Genome Browser (Kent et al. 2002) and the Evolutionary Conserved Region (ECR) Browser (Ovcharenko et al. 2004) found that 11 of the 22 tested sequences had human–fish conservation. Five of the 11 conserved elements (45 %) had pectoral fin enhancer expression (Table 1, Online resource 2). However, 5 of the 10 positive fin elements (50 %) were also not conserved to fish, which may suggest that conservation is not a more efficient filter to identify functionally conserved regulatory elements.
Although different in structure and regulatory machinery, zebrafish can serve as an efficient model to study predicted regulatory elements from other organisms. Our results show that neither sequence conservation nor spatial limb patterns substantially increase success rates in identifying human sequences that drive expression in the zebrafish fin. Similar assays will be needed for other tissues in order to better assess the functional utility of zebrafish in detecting mammalian enhancers.
Supplementary Material
Acknowledgments
We would like to thank the Ahituv Lab for helpful comments on the manuscript. This work was supported in part by grants from the National Institutes of Child Health and Human Development (NICHD) (number R01HD059862) and the National Human Genome Research Institute (NHGRI) (number R01HG005058). NA is also supported in part by NHGRI grant number 1R01HG006768, the National Institute of General Medical Science (NIGMS) award number GM61390, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) award number 1R01DK090382, the National Institute of Neurological Disorders and Stroke (NINDS) award number 1R01NS079231 and by a grant from the Simons Foundation (SFARI no. 256769). BMB was supported in part by a grant from the NIGMS/IRACDA 2K12GM081266.
Footnotes
Communicated by Matthias Hammerschmidt
Electronic supplementary material The online version of this article (doi:10.1007/s00427-013-0453-9) contains supplementary material, which is available to authorized users.
Contributor Information
Betty M. Booker, Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, CA, USA. Institute for Human Genetics, University of California, San Francisco, CA, USA
Karl K. Murphy, Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, CA, USA. Institute for Human Genetics, University of California, San Francisco, CA, USA
Nadav Ahituv, Email: nadav.ahituv@ucsf.edu, Department of Bioengineering and Therapeutic Sciences, University of California, San Francisco, CA, USA. Institute for Human Genetics, University of California, San Francisco, CA, USA.
References
- Ariza-Cosano A, Visel A, Pennacchio LA, Fraser HB, Gomez-Skarmeta JL, Irimia M, Bessa J. Differences in enhancer activity in mouse and zebrafish reporter assays are often associated with changes in gene expression. BMC Genomics. 2012;13:713. doi: 10.1186/1471-2164-13-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42(9):806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1(3):1297–1305. doi: 10.1038/nprot.2006.230. 10.1038/nprot. 2006.230. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392(6679):917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Laurell T, Vandermeer JE, Wenger AM, Grigelioniene G, Nordenskjold A, Arner M, Ekblom AG, Bejerano G, Ahituv N, Nordgren A. A novel 13 base pair insertion in the sonic hedgehog ZRS limb enhancer (ZRS/LMBR1) causes preaxial polydactyly with triphalangeal thumb. Hum Mutat. 2012;33(7):1063–1066. doi: 10.1002/humu.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12(14):1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Li Q, Ritter D, Yang N, Dong Z, Li H, Chuang JH, Guo S. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev Biol. 2009;337(2):484–495. doi: 10.1016/j.ydbio.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey DM, Vinton RM, Huynh J, Al-Saif A, Beer MA, McCallion AS. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18(2):252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev Growth Differ. 2007;49(6):421–437. doi: 10.1111/j.1440-169X.2007.00942.x. 10.1111/j.1440 169X.2007.00942.x. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32(Web server issue):W280–286. doi: 10.1093/nar/gkh35532/suppl_2/W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter DI, Li Q, Kostka D, Pollard KS, Guo S, Chuang JH. The importance of being cis: evolution of orthologous fish and mammalian enhancer activity. Mol Biol Evol. 2010;27(10):2322–2332. doi: 10.1093/molbev/msq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375(6533):678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved noncoding sequences. Nature. 2006;444(7118):499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35(Database issue):D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Tamura K. The making of differences between fins and limbs. J Anat. 2013;222(1):100–113. doi: 10.1111/j.1469-7580.2012.01491.x. 10.1111/j.1469-7580.2012. 01491.×. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.