Abstract
Zinc-finger nucleases (ZFNs) work as dimers to induce double-stranded DNA breaks (DSBs) at predefined chromosomal positions. In doing so, they constitute powerful triggers to edit and to interrogate the function of genomic sequences in higher eukaryotes. A preferred route to introduce ZFNs into somatic cells relies on their cotransduction with two integrase-defective lentiviral vectors (IDLVs) each encoding a monomer of a functional heterodimeric pair. The episomal nature of IDLVs diminishes the risk of genotoxicity and ensures the strict transient expression profile necessary to minimize deleterious effects associated with long-term ZFN activity. However, by deploying IDLVs and conventional lentiviral vectors encoding HPRT1- or eGFP-specific ZFNs, we report that DSB formation at target alleles is limited after IDLV-mediated ZFN transfer. This IDLV-specific underperformance stems, to a great extent, from the activity of chromatin-remodeling histone deacetylases (HDACs). Importantly, the prototypic and U.S. Food and Drug Administration–approved inhibitors of metal-dependent HDACs, trichostatin A and vorinostat, respectively, did not hinder illegitimate recombination-mediated repair of targeted chromosomal DSBs. This allowed rescuing IDLV-mediated site-directed mutagenesis to levels approaching those achieved by using their isogenic chromosomally integrating counterparts. Hence, HDAC inhibition constitutes an efficacious expedient to incorporate in genome-editing strategies based on transient IDLV-mediated ZFN expression. Finally, we compared two of the most commonly used readout systems to measure targeted gene knockout activities based on restriction and mismatch-sensitive endonucleases. These experiments indicate that these enzymatic assays display a similar performance.
Introduction
Zinc-finger nucleases (ZFNs) consist of a chain of Cys2-His2 artificial zinc-finger motifs fused to the nuclease domain of the type IIS restriction enzyme FokI. The zinc-finger array defines the DNA recognition sequence of the hybrid protein with each of its modules being designed to bind a triplet on the double helix. In general, ZFNs operate as heterodimers that assemble in a tail-to-tail orientation at the target site on opposite DNA strands. The ensuing local dimerization of the FokI domains induces double-stranded DNA breaks (DSBs) at the spacer sequence separating the two ZFN target half-sites (Klug, 2010; Urnov et al., 2010; Rahman et al., 2011). The engagement of the nonhomologous end-joining (NHEJ) and the homology-directed DNA repair pathways at these lesions can yield targeted gene knockouts and chromosomal insertion of exogenous DNA, respectively. As a result, the generation of DSBs at predefined sequences is becoming a powerful strategy to edit in a precise manner the genomes of an increasing number of higher eukaryotes, including those of human cells (Klug, 2010; Urnov et al., 2010; Rahman et al., 2011).
Transient transfection experiments deploying a surrogate plasmid system reporting on nuclease activities indicate that ZFN yields directly correlate with target gene knockout frequencies (Kim et al., 2011). Related to this, methods to optimize the design and the activity of ZFNs are being approached from many different angles. These include the development of the so-called context-dependent zinc-finger array assembly methods as well as FokI variant pairs whose protein–protein interfaces favor the assembly of target-specific heterodimers (Miller et al., 2007; Szczepek et al., 2007; Rahman et al., 2011). Moreover, a simple expedient to increase ZFN-induced gene knockout levels seems to be subjecting target cells to a 3-day hypothermic shock at 30°C (Doyon et al., 2010). Normally, the isolation of ZFNs displaying high specificity and low cytotoxicity requires the screening of various candidates in in vitro assays and in cell lines. Typically, ZFNs can be introduced into these experimental systems as proteins or as ZFN-encoding nucleic acids by using transfection agents. The delivery of ZFNs into a broad range of somatic cell types is of utmost importance for their in-depth evaluation and deployment.
By exploiting the cell infection mechanisms evolved by their parental viruses, viral vectors are among the most efficient gene delivery vehicles available. Moreover, gene product yields are easier to adjust through viral particle-mediated nuclear delivery than via the mass action process resulting from bulk DNA transfections (Zabner et al., 1995; Varga et al., 2005). Lentiviral vectors (LVs) based on the human immunodeficiency virus type-1 (HIV-1) display a particularly attractive number of qualities, including (1) similar transduction of dividing and nondividing cells, (2) rapid production timelines, and (3) broad tropism for mammalian cells through the pseudotyping of their envelops with the vesicular stomatitis virus glycoprotein-G moiety. Another of their features concerns a semirandom chromosomal DNA integration profile, which results in the permanent genetic modification of target cells (Schröder et al., 2002; Wu et al., 2003). Yet in most settings, including that of ex vivo gene therapy research, this capability can be disabled to ensure short-term nuclease expression in proliferating cells and, concomitantly, minimize insertional mutagenesis (Biasco et al., 2012). The disablement of the proviral insertion process is achieved by assembling LVs harboring catalytically inert class I integrase moieties (Philpott and Thrasher, 2007; Wanisch and Yáñez-Muñoz, 2009). The utility of these integrase-defective LVs (IDLVs) is broad since, with the exception of transcription activator-like effector nucleases (Holkers et al., 2013), they can serve as episomal templates for the synthesis of functional genome-modifying enzymes. These enzymes include not only ZFNs (Lombardo et al., 2007, 2011; Gabriel et al., 2011; Provasi et al., 2012) but also site-specific recombinases, transposases, and homing endonucleases from the LAGLIDAGD family (Cornu and Cathomen, 2007; Moldt et al., 2008; Morioka et al., 2009; Staunstrup et al., 2009; Staunstrup and Mikkelsen, 2011).
Although IDLVs exhibit gene delivery activities similar to those of their integration-proficient counterparts, it is often the case that cells transduced with this vector type build up relatively low amounts of the transgene products (see, e.g., Philippe et al., 2006; Cornu and Cathomen, 2007; Wanisch and Yáñez-Muñoz, 2009; Pelascini et al., 2013). Related to this, we have recently reported that histone deacetylases (HDACs) constitute major cellular determinants underlying the weak transcriptional activity of IDLVs in dividing, growth-arrested, and postmitotic cells (Pelascini et al., 2013). In the current work, by deploying IDLV and LV particles encoding monomers of ZFN pairs targeting endogenous or recombinant model alleles, we demonstrate that, compared with matched doses of isogenic LV particles, ZFN yields resulting from IDLV transductions are limiting. Importantly, pharmacological inhibition of classes I, II, and IV HDACs does not compromise NHEJ-mediated repair of chromosomal DSBs created by ZFNs. This finding allowed us demonstrating that HDAC inhibitors can either partially or fully rescue the levels of target gene knockout after IDLV-mediated ZFN delivery.
Materials and Methods
Cells
293T cells, used for vector productions, and HeLa cells, used for vector titrations, were grown in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% and 5% fetal bovine serum (both from Invitrogen), respectively, at 37°C in a humidified-air 10% CO2 atmosphere. The origin of and the culture conditions for the human skeletal muscle progenitor cells (hereinafter referred to as myoblasts), the HeLa-derived H27 indicator cells, and the bone marrow-derived human fetal mesenchymal stem cells (hMSCs) have been previously described (Cudré-Mauroux et al., 2003; Gonçalves et al., 2004, 2011).
Recombinant DNA
The LV transfer plasmids pLV.ZFN-1HPRT and pLV.ZFN-2HPRT, depicted in Fig. 1A, have been described elsewhere (Pelascini and Gonçalves, 2013). The LV transfer constructs pLV.ZFN-1eGFP and pLV.ZFN-2eGFP were made by substituting the ZFN open reading frame (ORF) in pLV.ZFN-1HPRT by those present in the previously described pRK5-derived plasmid pair (Höher et al., 2012). These two pRK5-based plasmids encode obligate heterodimeric ZFNs targeting the eGFP sequence: 5′-ATCCGCCACN6GAGGACGGC-3′.
FIG. 1.
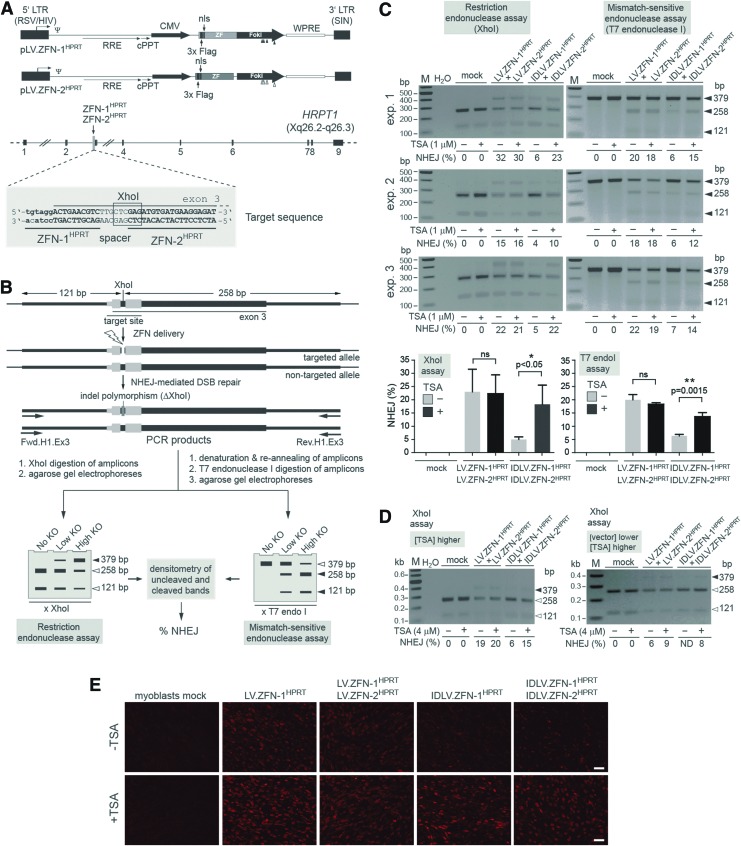
Effect of HDAC inhibition on target gene disruption in human myoblasts after IDLV- versus LV-mediated ZFN delivery. (A) Upper panel: genetic composition of the LV transfer constructs coding for the HPRT1-specific ZFNs. The plasmids pLV.ZFN-1HPRT and pLV.ZFN-2HPRT encode the custom-made nucleases ZFN-1HPRT and ZFN-2HPRT. Their DNA-binding zinc-finger arrays are directed at the upstream end of exon 3 of HPRT1. Close to their N-termini, the ZFNs harbor Flag epitopes and the SV40 large T antigen's nls. Boxes with broken arrow, hybrid 5′LTR consisting of Rous sarcoma virus- and HIV-1-derived sequences; boxes without broken arrow, SIN 3′LTR; ZF, nucleotide sequences coding for the zinc-finger arrays; FokI, sequences encoding mutant FokI nuclease motifs, with the positions of the nucleotide changes engineered to code for residues EELI and QKIK being indicated by solid and open vertical arrowheads, respectively. These FokI variants form obligate target site-specific heterodimers resulting in reduced ZFN-associated cytotoxicity. Ψ, HIV-1 packaging signal; RRE, Rev-responsive element; cPPT, central polypurine tract; WPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. The ZFN ORFs are under the transcriptional control of the CMV immediate-early gene promoter and the polyadenylation signal located within the SIN 3′LTR. The plasmid backbone containing the prokaryotic origin of replication and the β-lactamase gene are not drawn. Lower panel: HPRT1 target locus. The nine exons of this X-linked gene are indicated as solid boxes, and the location of the ZFN-1HPRT and ZFN-2HPRT target sites is pinpointed by the vertical arrow. The HPRT1 segment corresponding to exon 3 is shown in upper case, while that of its contiguous upstream intron sequence is depicted in lower case. The 6 bp sequence flanked by the two ZFN target sites (spacer) partially overlaps with an XhoI recognition site (boxed nucleotides). (B) Schematic representation of the XhoI- and T7 endonuclease I-based genotyping assays employed to measure targeted DSB formation. DNA breaks at the HPRT1 target site can be repaired via the error-prone NHEJ pathway resulting in the introduction of polymorphisms in the form of indels that, in effect, can KO the XhoI recognition sequence. PCR on cellular DNA isolated from cells exposed to ZFN-1HPRT and ZFN-2HPRT should yield amplicons corresponding to targeted and nontargeted alleles. After electrophoresis of XhoI-treated amplicons, the frequencies of DSBs in different target cell populations (i.e., % NHEJ) can be determined through densitometry of XhoI-resistant and XhoI-digested DNA bands (left-hand diagram, solid and open arrowheads, respectively). In parallel or in alternative, amplicons corresponding to targeted and nontargeted alleles can be subjected to a cycle of denaturation and re-annealing of their DNA strands resulting in wild type: mutated heteroduplexes with mismatched base pairs. The T7 endonuclease I by cleaving at DNA sequences with such mismatched based pairs yields low-molecular-weight DNA fragments. After electrophoresis of T7 endonuclease I-treated amplicons, the rate of DSB formation in different target cell populations (i.e., % NHEJ) can be determined through densitometry of full-length and cleaved PCR products (right-hand diagram, open and solid arrowheads, respectively). (C) Analyses of XhoI- and T7 endonuclease I-treated PCR products amplified from genomic DNA of myoblasts transduced with the vectors encoding HPRT1-specific ZFNs. Myoblasts were incubated with 1:1 mixtures of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT. The total vector dose applied was 3.6×107 vgc/ml. During the 16 hr vector transduction period, the myoblast cultures were not exposed (−) or were exposed (+) to 1 μM of TSA. Mock-transduced cells served as negative controls. The genomic DNA was isolated at 48 hr posttransduction, and target gene disruption assays were deployed as outlined in Fig. 1B. The plots in the lower part of the panel correspond to the mean±standard deviation from three independent experiments examined by using XhoI- and T7 endonuclease I-based readouts. ns, not significant. (D) Analyses of XhoI-treated PCR products amplified from genomic DNA of myoblasts transduced with the vectors encoding HPRT1-specific ZFNs. Myoblasts were incubated with 1:1 mixtures of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT. The total vector doses applied were 3.6×107 vgc/ml (left panel) and 2.4×107 vgc/ml (right panel). During the 16 hr vector incubation period, the myoblast cultures were not exposed (−) or were exposed (+) to 4 μM of TSA. The genomic DNA was extracted at 48 hr posttransduction to determine XhoI site disruptions. ND, not detected. (E) Immunofluorescence microscopy analysis of ZFN expression in untreated and TSA-treated myoblasts. Target cells were mock-transduced or were transduced with LV.ZFN-1HPRT and IDLV.ZFN-1HPRT at 1.8×107 vgc/ml each or were cotransduced with 1:1 mixtures consisting of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT at a total vector dose of 3.6×107 vgc/ml. ZFN synthesis was accessed by Flag tag-specific immunofluorescence microscopy at 48 hr posttransduction. Size bars correspond to 200 μm. CMV, cytomegalovirus promoter; DSB, double-stranded DNA break; HDAC, histone deacetylase; IDLV, integrase-defective lentiviral vector; KO, knockout; NHEJ, nonhomologous end-joining; nls, nuclear localization signal; ORFs, open reading frames; PCR, polymerase chain reaction; SIN, self-inactivating; TSA, trichostatin A; ZFN, zinc-finger nuclease. Color images available online at www.liebertpub.com/hgtb
Vector production and titration
The procedures and reagents to generate and purify the HIV-1-based and vesicular stomatitis virus glycoprotein-G-pseudotyped LVs and IDLVs used in the current study have been detailed elsewhere (Pelascini and Gonçalves, 2013; Pelascini et al., 2013). The titers were determined by quantitative polymerase chain reaction (qPCR) and are expressed in terms of vector genome copies per milliliter (vgc/ml) as previously described (Pelascini and Gonçalves, 2013). The titers of vector stocks LV.ZFN-1eGFP, LV.ZFN-2eGFP, IDLV.ZFN-1eGFP, and IDLV.ZFN-2eGFP were 1.36×108, 5.82×107, 1.22×108, and 3.48×107 vgc/ml, respectively, whereas those of LV.ZFN-1HPRT, LV.ZFN-2HPRT, IDLV.ZFN-1HPRT, and IDLV.ZFN-2HPRT were 2.04×108, 1.82×108, 1.89×108, and 1.29×108 vgc/ml, respectively.
Dose–response of HDAC inhibitors on IDLV-transduced cells
Dose–response experiments with HDAC inhibitors on IDLV-transduced cells were performed as follows. Human myoblasts, hMSCs, and HeLa cells were seeded at a density of 1.5×105, 1.5×105, and 2.0×105 cells per well of 24-well plates (Greiner Bio-One), respectively. Twenty-four hours later, myoblasts were mock-transduced or were transduced with 1.2×106 vgc/ml of IDLV.CMV.eGFP (Pelascini et al., 2013). Transductions were performed in the absence or in the presence of trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA) (both from Sigma-Aldrich) at the final concentrations of 0.125, 0.25, 0.5, 0.75, 1.0, 2.0, 4.0, and 8.0 μM. The hMSCs and HeLa cells were mock-transduced or were transduced with 1.35×106 vgc/ml of IDLV.hPGK.eGFP (Pelascini et al., 2013). Transductions were carried out in the absence or in the presence of increasing concentrations of SAHA, that is, 0.125, 0.25, 0.5, 1.0, 2.0, 4.0, 8.0, and 16.0 μM. After a 16 hr incubation period, the cells were washed with phosphate-buffered saline (PBS) and a regular culture medium was added to the cells. At 2 days posttransduction, eGFP expression was quantified by flow cytometry.
Transduction experiments and HDAC inhibitor treatments
Human myoblasts and H27 cells were seeded at a density of 1.5×105 and 2.5×105 cells per well of 24-well plates (Greiner Bio-One), respectively. Twenty-four hours later, myoblasts were mock-transduced or were transduced with LV.ZFN-1HPRT and LV.ZFN-2HPRT or with IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT, whereas H27 cells were mock-transduced or were transduced with LV.ZFN-1eGFP and LV.ZFN-2eGFP or with IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP. All cotransductions were performed at a 1:1 ratio. The myoblast cultures were exposed to a total of 2.4×107 and 3.6×107 vgc/ml, while those of H27 cells were incubated with 6.0×106 vgc/ml. The human myoblasts and the H27 reporter cells were incubated for 16 and 24 hr, respectively, in the absence or in the presence of HDAC inhibitors. The human myoblast cultures were exposed to TSA at the final concentrations of 1 and 4 μM, whereas those of H27 cells were exposed to 3 μM of TSA or to 4 μM of SAHA. At the end of the incubation period, the cells were washed with PBS and a fresh regular culture medium was added. Forty-eight hours posttransduction, myoblasts were harvested to perform an XhoI-based genotyping assay as described in the section Restriction enzyme-based genotyping assays to quantify target gene disruptions, while H27 cells were harvested and expanded in duplicates in the wells of six-well plates (Greiner Bio-One). H27 cells were kept in culture during 8 days with a minimum of three passages. At 10 days posttransduction, a fraction of H27 cultures was used to quantify eGFP knockout levels by flow cytometry. The remaining cells were used to perform eGFP- and Hoechst 33342-directed fluorescence microscopies and a TaqI-based genotyping assay as described in the section Restriction enzyme-based genotyping assays to quantify target gene disruptions.
Measurement of caspase activities in TSA-treated myoblast cultures
To probe the apoptosis-inducing effect of exposing myoblasts for 16 hr to different doses of TSA, the activities of caspase-3/7 were measured by using the Caspase-Glo 3/7 Assay System (Promega). Briefly, human myoblasts were seeded at a density of 2×104 cells per well of 96-well plates (Greiner Bio-One). Twenty-four hours later, the myoblasts were mock-treated or were exposed to TSA at the concentrations specified in the section Dose–response of HDAC inhibitors on IDLV-transduced cells. After a 16 hr incubation period, the cells were washed with PBS, after which the regular culture medium was added onto the washed cells. At 2 days posttreatment, before initiating the caspase activity assay, mock-treated myoblasts were exposed for 3 hr to 1 μM of staurosporine (Sigma-Aldrich). Staurosporine-treated cells were used as positive controls, as this drug is known to induce apoptosis by activating caspase 3 (Kushner et al., 2011). After incubation for 90 min with the proluminescent caspase-3/7 DEVD-aminoluciferin substrate and a thermostable luciferase, the different cell cultures were washed with PBS and lysed as previously detailed (Gonçalves et al., 2010). One-fifth of each 50 μl lysates were processed to measure the caspase-specific luminescent signal according to the manufacturer's protocol. To ensure that the various luminometric measurements were carried out with similar amounts of cellular material, 10 μl lysate samples were used to determine total protein concentrations by using the Bio-Rad Protein Assay (Bio-Rad) following the manufacturer's recommendations. Thus, the final results are expressed in terms of relative light units normalized to protein content on a per-sample basis.
Direct fluorescence microscopy
Target gene knockout in H27 cultures was monitored by direct fluorescence microscopy. Briefly, nuclei were stained with 10 μg/ml Hoechst 33342 (Invitrogen) in the regular medium for 10 min. Subsequently, the cells were washed once with PBS, after which the regular culture medium was added. eGFP- and Hoechst 33342-directed fluorescence microscopies were carried out by using an Olympus IX51 inverse fluorescence microscope (Olympus). Images were captured by an XC30 Peltier-cooled charge-coupled device camera (Olympus) and were archived using Cell^F software (Olympus).
Indirect fluorescence microscopy
The transgene expression profiles in human myoblasts and H27 cells exposed to ZFN-encoding vector particles were accessed as follows. Cultures of myoblasts and H27 indicator cells were not incubated or were incubated with 1 μM of TSA and 3 μM of TSA for 16 hr, respectively. During this period, human myoblasts were exposed to LV.ZFN-1HPRT and IDLV.ZFN-1HPRT at a concentration of 1.8×107 vgc/ml each or were cotransduced with 1:1 mixtures of LV.ZFN-1HPRT and LV.ZFN-2HPRT or of IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT at a total vector dose of 3.6×107 vgc/ml, whereas H27 cells were exposed to LV.ZFN-1eGFP and IDLV.ZFN-1eGFP at a concentration of 3×106 vgc/ml each or were cotransduced with 1:1 mixtures of LV.ZFN-1eGFP and LV.ZFN-2eGFP or of IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP at a total vector dose of 6×106 vgc/ml. Parallel cultures of mock-transduced H27 cells served as negative controls. At 48 hr posttransduction, the target cells were subjected to a previously described immunofluorescence microscopy protocol (Gonçalves et al., 2008). Primary antibodies recognizing a Flag epitope (Sigma-Aldrich; monoclonal anti-Flag M2, clone M2) and an influenza hemagglutinin (HA) epitope (Covance; HA.11, MMS-101R) were used in human myoblasts and H27 cells, respectively. The former antibody was diluted 1:80, whereas the latter was applied at a dilution of 1:3,000. Alexa Fluor 568 goat anti-mouse IgG (H+L; Invitrogen) was used as a secondary antibody diluted 1:500 for both immunostaining protocols. Finally, micrographs were acquired by using the optical instruments and software described under the previous section.
Restriction enzyme-based genotyping assays to quantify target gene disruptions
The gene knockout levels in vector-exposed myoblast and H27 cell cultures were determined as follows. Total cellular DNA was extracted by using the DNeasy Blood & Tissue kit following the manufacturer's instructions (Qiagen). DNA concentrations were measured with the aid of a NanoDrop, and DNA samples were diluted to a final concentration of 20 ng/μl. Sixty nanograms of DNA from myoblasts samples was subjected to PCR using the primers Fwd.H1.Ex3 (5′-TCACTGTATTGCCCAGGTTGGTG-3′) and Rev.H1.Ex3-Rev (5′-GAAAGCAAGTATGGTTTGCAGAGAT-3′) designed to amplify a 379 bp fragment encompassing the ZFN pair cutting site located within exon 3 of HPRT1. The PCR amplifications were performed in 25 μl mixtures consisting of 2 U of GoTaq Flexi DNA polymerase, 1×Colorless GoTaq Flexi Buffer, 3 mM MgCl2 (all from Promega), 400 nM dNTPs (Invitrogen), 200 nM of each primer (Eurofins MWG Operon), and DNase/RNase-free distilled H2O (Invitrogen). PCRs were carried out in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) using the following touchdown cycling conditions. After a 3 min denaturation step at 95°C, the samples were subjected to 10 cycles consisting of 25 sec at 95°C, 25 sec at 63°C, with the annealing temperature decreasing by 0.5°C per cycle, and 25 sec at 72°C. Further amplifications were performed through 25 additional cycles at the annealing temperature of 58°C. The reactions were terminated by a final elongation period of 2 min at 72°C. One hundred nanograms of DNA from H27 cell samples was subjected to PCR using the primers eGFP-Fwd (5′-GCTGAAGGGCATCGACTTC-3′) and eGFP-Rev (5′-ACTTGCTGTAGGTCTGCTTG-3′) designed to amplify a 390 bp DNA fragment encompassing the ZFN pair cutting site located within the recombinant eGFP locus. The PCR amplifications were performed in 50 μl mixtures containing 2.5 U of GoTaq Flexi DNA polymerase, 1× Colorless GoTaq Flexi Buffer, 2 mM MgCl2 (all from Promega), 400 nM dNTPs (Invitrogen), 200 nM of each primer (Eurofins MWG Operon), and DNase/RNase-free distilled H2O (Invitrogen). PCRs were carried out in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) using the following touchdown cycling conditions. After a 3 min denaturation step at 95°C, the samples were subjected to 20 cycles consisting of 25 sec at 95°C, 25 sec at 68°C, with the annealing temperature reaching 58°C after decreasing 0.5°C per cycle, and 25 sec at 72°C. Further amplifications were performed via 20 additional cycles at the annealing temperature of 58°C. The reactions were terminated by a final elongation period of 2 min at 72°C. Eight microliters of PCR mixtures corresponding to the amplification of myoblast and H27 cell DNA was subjected to XhoI and TaqI (both from Fermentas) digestions, respectively. Next, 16 μl samples of restriction enzyme-treated DNA were subjected to electrophoresis through 2% agarose gels stained with ethidium bromide in 1× Tris-Acetate-EDTA (TAE) buffer. Target gene disruption frequencies were determined by measuring DNA fragment densitometries using ImageJ (National Institutes of Health).
T7 endonuclease I-based genotyping assays to quantify target gene disruptions
The DNA regions encompassing the HPRT1 and eGFP target sequences that were subjected to T7 endonuclease I (New England BioLabs) treatments were PCR-amplified as specified under the section Restriction enzyme-based genotyping assays to quantify target gene disruptions. The T7 endonuclease I-based assay was performed essentially as described elsewhere (Mussolino et al., 2011). Briefly, 8 μl PCR samples were first subjected to a heat-denaturing and re-annealing step. Next, the samples were treated with the mismatch-sensitive T7 endonuclease I for 15 min at 37°C and directly subjected to electrophoresis through 2% agarose gels stained with ethidium bromide in 1×TAE buffer. Target gene disruption rates (as measured in terms of the percentage of alleles estimated to have been subjected to NHEJ events) were determined by measuring DNA band densitometries by ImageJ and applying the relationship % NHEJ=100×(1 − [1 − fraction cleaved]1/2). This relationship allows correcting for the fact that at high gene knockout frequencies a nonnegligible proportion of re-annealed DNA molecules will consist of mutated heteroduplex strands (Miller et al., 2007).
Statistics
Data sets were analyzed by using the GraphPad Prism 5 software package and evaluated for significance by applying unpaired two-way Student's t-tests (p<0.05 considered significant).
Sequence analyses of ZFN-targeted alleles
To characterize the small insertions and/or deletions (indels) formed at repaired HPRT1 chromosomal junctions, the XhoI-resistant PCR products corresponding to myoblasts cotransduced with LV.ZFN-1HPRT and LV.ZFN-2HPRT and to 1 μM TSA-treated myoblasts cotransduced with IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT were purified using the GeneJET Gel Extraction Kit (Fermentas). The resulting 379 bp amplicons were ligated into the pGEM-T Easy TA cloning vector (Promega) overnight at 4°C, followed by the transformation of chemically competent cells of E. coli strain DH5α. Positive transformants were identified by PCR using the primers M13-Fwd (5′-GTAAAACGACGGCCAG-3′) and M13-Rev (5′-CAGGAAACAGCTATGAC-3′) (both from Invitrogen). The PCR amplifications were performed in 20 μl mixtures containing 2 U of GoTaq Flexi DNA polymerase, 1× Colorless GoTaq Flexi Buffer, 2 mM MgCl2 (all from Promega), 400 nM dNTPs (Invitrogen), 200 nM of each primer (Eurofins MWG Operon), and DNase/RNase-free distilled H2O (Invitrogen). PCRs were carried out in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) using the following cycling conditions. After a 10 min denaturation step at 95°C to lyse the bacteria, the samples were subjected to 25 cycles consisting of 30 sec at 95°C, 30 sec at 55°C, and 30 sec at 72°C. The reactions were terminated by a final elongation period of 4 min at 72°C. The resulting PCR products were subjected to electrophoresis through a 2% agarose gel stained with ethidium bromide in 1×TAE buffer. Insert-containing plasmids were extracted from 21 independent colonies, and their DNA sequences were determined by Sanger sequencing using the above-mentioned M13 forward primer.
Results
We started by carrying out transduction experiments in human myoblasts to determine the feasibility and the utility of using HDAC inhibitors in targeted mutagenesis experiments based on IDLV-mediated ZFN transfer. To serve as references, we took along in all transduction experiments isogenic chromosomally integrating vector controls whose genome copy numbers matched those of the corresponding IDLVs. Thus, on the basis of the LV transfer plasmids pLV.ZFN-1HPRT and pLV.ZFN-2HPRT (Fig. 1A, upper panel), the vector panel composed of IDLV.ZFN-1HPRT, IDLV.ZFN-2HPRT, LV.ZFN-1HPRT, and LV.ZFN-2HPRT was assembled. These vectors encode ZFNs whose DNA-binding motifs recognize sequences at the upstream end of exon 3 of the X-linked human hypoxanthine phosphoribosyltransferase 1 (HPRT1) locus (Fig. 1A, lower panel). To determine the impact of HDACs on vector-mediated HPRT1 disruption, myoblasts were cotransduced with LV.ZFN-1HPRT and LV.ZFN-2HPRT or with IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT in the presence and in the absence of the prototypic HDAC inhibitor TSA. To detect knocking out of target sequences, cellular DNA was isolated and subjected to restriction enzyme- and T7 endonuclease I-based genotyping assays (Fig. 1B). In these experiments (n=3), controls were provided by mock-transduced myoblasts. As expected, cells that were mock-transduced yielded neither XhoI-resistant nor T7 endonuclease I-sensitive DNA fragments (Fig. 1C). Both readout systems showed that the frequencies of LV-induced HPRT1 disruption were similar in untreated and TSA-treated cells, showing that, under these experimental conditions, target gene knockout levels are not significantly affected by HDAC activity (Fig. 1C). Related to this, these data indicate that transient HDAC inhibition does not compromise illegitimate recombination-mediated repair of site-specific DSBs (Fig. 1C). In contrast, robust IDLV-induced HPRT1 disruption was dependent on HDAC inhibition since target gene knockout frequencies were consistently higher in the presence of the chromatin-remodeling agent (Fig. 1C). Comparable results were obtained after carrying out transduction experiments in myoblast cultures exposed to 4 μM instead of 1 μM of TSA as well as in those incubated with 4 μM of TSA and subjected to a lower dose of vector particles (Fig. 1D). Of note, the latter experimental setup did not result in detectable HPRT1 disruption in cells cotransduced with IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT in the absence of TSA. Immunofluorescence microscopy analysis of ZFN expression in untreated and TSA-treated myoblasts established a direct relationship between the low frequencies of HPRT1 mutagenesis and the limited ZFN yields detected in cultures transduced with the IDLV pair in the absence of HDAC inhibition (Fig. 1E). Moreover, the apoptosis-inducing effect caused by applying to myoblast cultures 1 and 4 μM of TSA for the timeframe of the transduction period proved to be negligible (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/hgtb).
Sequencing of addressed HPRT1 DNA retrieved from cells subjected and not subjected to HDAC inhibition revealed the presence of similar indels at repaired chromosomal junctions (Fig. 2). These indel footprints are consistent with the engagement of the NHEJ DNA repair pathway at ZFN-induced DSBs in cells that were incubated as well as in those that were not incubated with the HDAC-inhibiting agent.
FIG. 2.
Examination by Sanger sequencing of error-prone NHEJ at HPRT1 alleles of human myoblasts transduced with ZFN-encoding vectors. LV clone series: nucleotide sequences of independent XhoI-treated PCR products obtained by using primers Fwd.H1.Ex3 and Rev.H1.Ex3 (Fig. 1B) on DNA from myoblasts cotransduced with LV.ZFN-1HPRT and LV.ZFN-2HPRT. IDLV clone series (ID+): nucleotide sequences of individual XhoI-treated PCR products obtained by deploying the aforementioned primers on DNA from TSA-exposed human myoblasts cotransduced with IDLV.ZFN-1HPRT and IDLV.ZFN-2HPRT. The ZFN target site sequences are boxed, and the spacer nucleotides are in lower case.
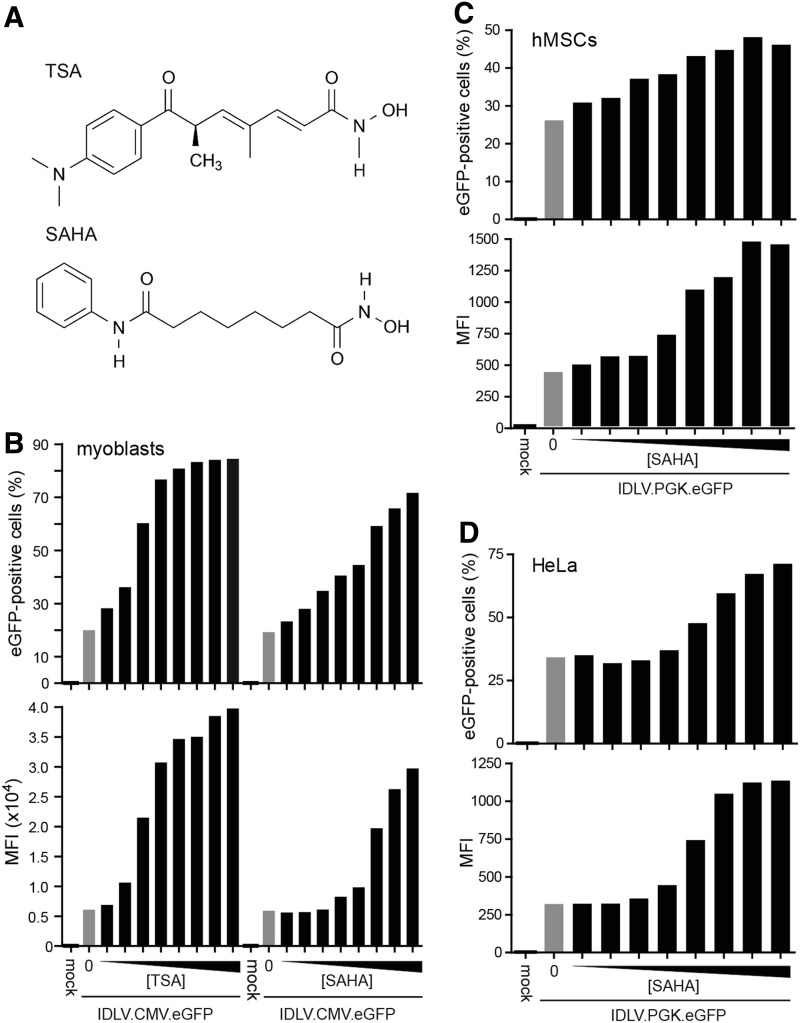
Next, we sought to complement these targeted mutagenesis data by carrying out transduction experiments in HeLa-derived H27 cells. These indicator cells contain a single-copy eGFP allele under the transcriptional control of the human elongation factor 1α gene (EF1A1) promoter (Gonçalves et al., 2004). Therefore, together with eGFP-specific nucleases, they allow measuring reporter gene knockout levels not only through DNA-level assays but also via independent readout systems based on fluorescence microscopy and flow cytometry. In these experiments we tested, in addition to TSA, the HDAC inhibitor SAHA, also known as vorinostat. SAHA, whose chemical structure is depicted next to that of TSA in Fig. 3A, is being utilized to treat refractory cutaneous T-cell lymphoma (Copeland et al., 2010) and to disrupt HIV-1 proviral latency in resting CD4+ T cell reservoirs (Archin et al., 2012). To determine the impact of different concentrations of TSA and SAHA on IDLV-mediated transgene expression, we performed dose–response experiments in cultures of different cell types exposed to the reporter vectors IDLV.CMV.eGFP and IDLV.hPGK.eGFP. Flow cytometric analyses revealed that, at equal small-molecule drug concentrations, TSA yielded higher reporter amounts in treated cultures than those measured in parallel SAHA-exposed cultures. There was, nonetheless, a direct correlation between increasing amounts of SAHA and IDLV.CMV.eGFP-mediated transgene expression (Fig. 3B). This SAHA-dependent transgene upregulation could be recapitulated in hMSCs and in HeLa cells transduced with the IDLV.hPGK.eGFP construct (Fig. 3C and D, respectively). These results suggest that deploying the U.S. Food and Drug Administration (FDA)–approved HDAC inhibitor SAHA may constitute a general pharmacological expedient to improve IDLV-mediated transgene expression.
FIG. 3.
Dose–response of HDAC inhibitors on the transduction levels and transgene product yields resulting from IDLV-mediated gene transfer into human cells. (A) Chemical structures of TSA and SAHA made with the aid of ChemBioDraw version: 12.0d560. (B) Flow cytometric analyses at 48 hr posttransduction of myoblasts (1.5×105 cells/well) exposed for 16 hr to IDLV.CMV.eGFP at 1.2×106 vgc/ml in the absence (gray bars) or in the presence of various doses of TSA or SAHA (solid bars). Mock-transduced myoblasts served to set the threshold between background and eGFP-specific fluorescence. The concentrations of TSA and SAHA applied were 0.125, 0.25, 0.5, 0.75, 1.0, 2.0, 4.0, and 8.0 μM. The flow cytometry data are expressed in terms of the percentage of reporter-positive cells and corresponding mean fluorescence intensity (MFI) values (upper and lower graphs, respectively). (C) Flow cytometric analyses at 48 hr posttransduction of hMSCs (1.5×105 cells/well) exposed to IDLV.hPGK.eGFP at 1.35×106 vgc/ml in the absence (gray bars) or in the presence of different doses of SAHA. Eight different concentrations of SAHA were used with each dose varying by a factor of 2 from a minimum of 0.125 μM to a maximum of 16 μM. Mock-transduced hMSCs served to set background fluorescence levels. (D) Flow cytometric analyses at 48 hr posttransduction of HeLa cells transduced with IDLV.hPGK.eGFP at 1.35×106 vgc/ml in the absence (gray bars) or in the presence of different doses of SAHA. Eight different concentrations of SAHA were used with each dose varying by a factor of 2 from a minimum of 0.125 μM to a maximum of 16 μM. Mock-transduced HeLa cells served to set the threshold between background and eGFP-specific fluorescence. hMSCs, human fetal mesenchymal stem cells; SAHA, suberoylanilide hydroxamic acid; TSA, trichostatin.
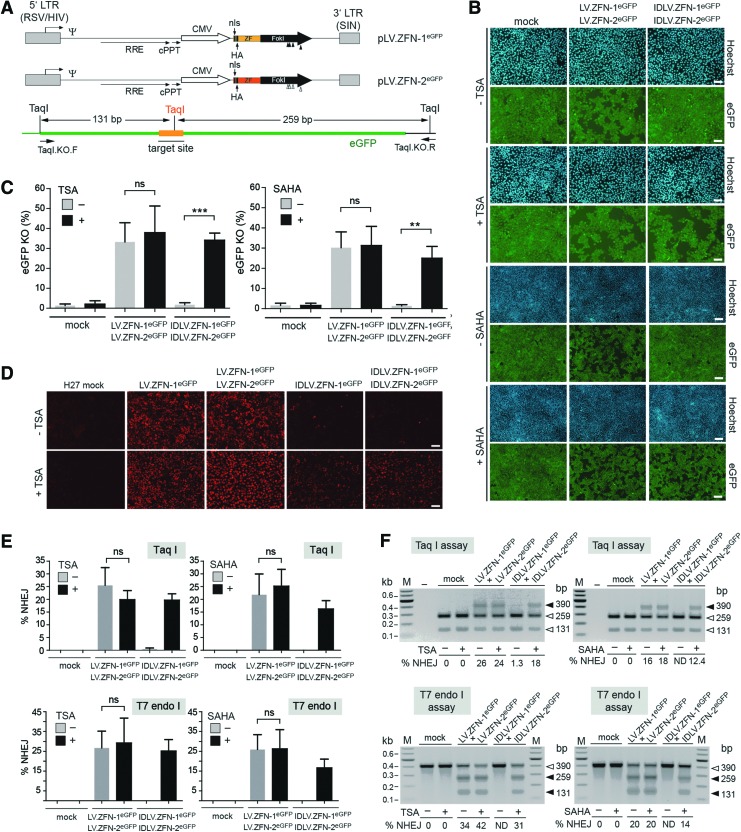
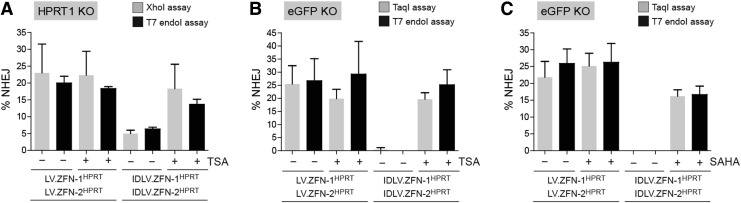
Transduction experiments with the vectors LV.ZFN-1eGFP, LV.ZFN-2eGFP, IDLV.ZFN-1eGFP, and IDLV.ZFN-2eGFP were carried out in H27 indicator cells not exposed and exposed to TSA or to SAHA. These vectors were generated on the basis of the transfer plasmids pLV.ZFN-1eGFP and pLV.ZFN-2eGFP (Fig. 4A, upper panel). Each of these constructs encodes a member of a ZFN pair whose target sequences frame a TaqI recognition site within the eGFP ORF (Fig. 4A, lower panel). The untreated and the HDAC inhibitor-treated H27 cells were mock-transduced or were cotransduced with LV.ZFN-1eGFP and LV.ZFN-2eGFP or with IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP. At 10 days posttransduction, after at least three rounds of subculturing to eliminate the reporter protein from cells bearing disrupted eGFP ORFs, knockout levels were monitored and quantified through live-cell fluorescence microscopy (Fig. 4B) and flow cytometry (Fig. 4C), respectively. The resulting data show that targeted eGFP disruption after LV-mediated ZFN transfer was increased neither by TSA nor by SAHA. On the contrary, eGFP knockout levels were greatly enhanced after IDLV-mediated ZFN transfer in the presence of TSA or of SAHA (Fig. 4B and C). In fact, these target gene disruption levels were similar to those measured in cultures exposed to the LV controls. Of note, in HDAC inhibitor-free cultures transduced with the IDLV pair the frequencies of eGFP-negative cells did not raise above background levels (Fig. 4C). These results correlated with the higher impact of HDAC inhibition on the frequencies of ZFN-expressing cells in cultures exposed to IDLVs when compared with that observed in cultures subjected to LV transductions (Fig. 4D). Furthermore, the eGFP knockout data could be confirmed by genotyping readouts dependent on TaqI restriction fragment length polymorphisms and on the generic T7 endonuclease I assay (Fig. 4E and F). We exploited the fact of having deployed side-by-side assays based on restriction and mismatch-sensitive endonucleases to gauge their relative sensitivities. The cumulative target site genotyping data resulting from the various transduction experiments are depicted in Fig. 5. These results indicate that T7 endonuclease I- and restriction enzyme-based assays display a similar performance.
FIG. 4.
TSA- and SAHA-dependent rescue of target gene knockout levels in reporter H27 cells after IDLV-mediated ZFN delivery. (A) Upper panel: genetic organization of the LV transfer constructs coding for the eGFP-specific ZFNs. The plasmids pLV.ZFN-1eGFP and pLV.ZFN-2eGFP code for the sequence-specific nucleases ZFN-1eGFP and ZFN-2eGFP. The ZFNs contain nls and influenza HA epitopes located close to their N-termini. The FokI positions engineered to encode the heterodimer-forming residues EEAI and QKIV are indicated by solid and open vertical arrowheads, respectively. For a description of regulatory sequences and viral cis-acting elements, see the legend of Fig. 1A. Lower panel: schematic representation of the recombinant chromosomal region targeted by the eGFP-specific ZFNs. TaqI recognition sequences flanking and at the hybrid nucleases' target site (orange line) are indicated together with the expected TaqI DNA fragment sizes resulting from unmodified alleles. The primer set used to amplify this region is also shown. (B) Fluorescence microscopy on monolayers of eGFP-expressing H27 cells. The reporter cells treated or not treated with TSA or SAHA were mock-transduced or were cotransduced with 1:1 mixtures of LV.ZFN-1eGFP and LV.ZFN-2eGFP or of IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP at a total vector dose of 6×106 vgc/ml. H27 cell nuclei in each microscopic field were identified through Hoechst 33342 staining, whereas target gene expression was monitored via eGFP-directed fluorescence microscopy at 10 days posttransduction. Note the dissimilar amounts of eGFP-negative cells. Size bars in micrographs corresponding to TSA- and SAHA-treated H27 cultures are equivalent to 200 and 500 μm, respectively. (C) Flow cytometry-based quantification of eGFP knockout levels in H27 cultures subjected to the aforementioned experimental conditions. The flow cytometric analyses were carried out at 10 days posttransduction. Plots correspond to mean±standard deviation from three independent experiments (D) Immunofluorescence microscopy analysis of ZFN expression in untreated and TSA-treated H27 cells. Target cells were mock-transduced or were transduced with LV.ZFN-1eGFP and IDLV.ZFN-1eGFP at 3×106 vgc/ml each or were cotransduced with 1:1 mixtures consisting of LV.ZFN-1eGFP and LV.ZFN-2eGFP or of IDLV.ZFN-1eGFP and IDLV.ZFN-2eGFP at a total vector dose of 6×106 vgc/ml. ZFN synthesis was accessed by HA-specific immunofluorescence microscopy at 48 hr posttransduction. Size bars correspond to 200 μm. (E) Cumulative results derived from three independent experiments performed on H27 indicator cells deploying the aforementioned experimental conditions. Data shown correspond to mean±standard deviation (F) Representative agarose gel electrophoresis of TaqI- and T7 endonuclease I-treated PCR products amplified from genomic DNA of H27 cells subjected to the aforementioned experimental conditions. HA, hemagglutinin; ND, not detected; vgc, vector genome copies. Color images available online at www.liebertpub.com/hgtb
FIG. 5.
Side-by-side comparison of restriction and mismatch-sensitive endonuclease assays to measure targeted mutagenesis in cells exposed to ZFNs. The cumulative data were gathered from the transduction experiments presented in Fig. 1C (A), in the left-hand panels of Fig. 4E (B), and in the right-hand panels of Fig. 4E (C).
In conclusion, the transient inhibition of cellular metal-dependent HDACs does not interfere with NHEJ-mediated repair of site-specific chromosomal DSBs, constituting, as a result, a valuable approach to induce targeted mutagenesis after the introduction of ZFNs into human cells through IDLV-mediated gene transfer.
Discussion
HDACs catalyze the removal of negatively charged acetyl groups from certain lysine residues of core histones. This change in epigenetic marking contributes to chromatin condensation and ensuing downregulation of cellular gene expression (Berger, 2002; Selvi and Kundu, 2009). Since episomal DNA molecules can likewise acquire chromatin-like structures in cell nuclei, they are also subjected to epigenetic regulatory mechanisms involving posttranslational histone modifications (see, for instance, Riu et al., 2007; Kantor et al., 2009; Ross et al., 2011). We have, in fact, previously demonstrated that, when compared with their chromosomally integrating counterparts, IDLVs are particularly susceptible to HDAC-dependent silencing processes regardless of target cell division rate or vector genetic makeup (Pelascini et al., 2013). Herein, by using matched doses of isogenic ZFN-encoding LV and IDLV sets, we have shown that sequence-directed mutagenesis in human cells after IDLV-mediated gene delivery is suboptimal. Moreover, target gene knockout frequencies achieved by the LV pairs in the presence of HDAC inhibitors were not inferior to those measured in their absence. This suggests that HDAC-interfering approaches neither prevent nor reduce the engagement of error-prone DNA repair mechanisms at genomic DSBs generated by ZFNs. Following from this finding, we demonstrated that small-molecule HDAC inhibitors can serve as adjuvants to either partially or fully rescue the underperformance of IDLVs during ZFN-induced targeted mutagenesis protocols. Thus, taken together, our results support the feasibility and utility of exploiting HDAC-inhibiting methods in gene knockout experimental settings based on IDLV-mediated ZFN transfer. These data are relevant since achieving higher frequencies of gene knockout per IDVL particle should provide for more robust targeted mutagenesis strategies based on these commonly used episomal viral vectors. This is particularly so in light of the persisting difficulties in producing high-titer HIV-1-based vectors for preclinical and clinical testing. More generally, it is also sensible to posit that in many other experimental settings, merely increasing the multiplicities of infection of ZFN-encoding IDLVs will not only be unpractical but will also be counterproductive because of, for instance, vector particle-associated cytotoxicity in target cells (Kang et al., 2002; Kobinger et al., 2004; Ricks et al., 2008).
To test the concept of targeted mutagenesis by combining IDLV-mediated ex vivo delivery of ZFNs and pharmacological interference with HDAC activity, we selected the prototypic and FDA-approved pan-HDAC inhibitors TSA and SAHA, respectively. A luminometric caspase-3/7 activity assay failed to detect significant apoptosis in target cells subjected to broad-spectrum HDAC inhibition under the experimental conditions tested. However, it is known that these small-molecule agents display pleiotropic effects in cells. Although HDAC inhibitors entering phases I and II clinical trials are targeting an increasing number of conditions, the majority of them are directed at life-threatening diseases (Arrowsmith et al., 2012). Among these, givinostat (a.k.a. ITF2357), targeting classes I and II HDACs, showed to preferentially induce apoptosis in hepatocellular carcinoma cells grown in vitro, whereas overt toxic effects were not observed in healthy primary hepatocytes (Armeanu et al., 2005). This HDAC inhibitor was also tested in a phase I clinical study including children suffering from systemic onset juvenile arthritis. Significant therapeutic effects and an excellent safety profile were observed (Vojinovic and Damjanov, 2011).
Despite the above-mentioned promising data, it will be also of considerable interest to probe HDAC-inhibiting molecules whose structures modulate the activity of specific HDAC isoforms (Di Micco et al., 2013). Alternatively, reverse genetic approaches based on RNA interference can be envisioned. Ultimately, these isoform-selective compounds and reagents might help dissecting the epigenetic mechanisms underlying IDLV silencing while displaying lower pleiotropic effects in target cells than those resulting from the use of pan-HDAC inhibitors.
In the current work, we have used pairs of ZFNs specific for sequences embedded within an endogenous housekeeping gene or a constitutively active recombinant expression unit. Hence, the target sites of these ZFN pairs are most probably located in regions with a relatively open chromatin structure. Indeed, the frequencies of LV-induced gene disruption were similar in cells exposed and not exposed to HDAC inhibition, suggesting that chromatin remodeling did not play a significant role in the accessibility of these target loci to the ZFNs. However, it is plausible that loci with a closed chromatin conformation are less prone to targeting such as those whose expression pattern is tissue-specific or subjected to a strict developmental control. Indeed, it is noteworthy mentioning that probing the degree of chromatin packing has been classically approached by using nuclease sensitivity assays. More importantly, there is experimental evidence for the negative role of condensed chromatin on the outcome of nuclease-induced genome engineering efforts (Daboussi et al., 2012; van Rensburg et al., 2013). Therefore, it is conceivable that, owing to higher nuclease concentrations and/or increased accessibility of target sequences, transient exposure of cells to HDAC-inhibiting agents will assist in modifying certain genes in heterochromatic regions.
Mismatch-sensitive endonucleases and restriction enzymes are frequently used as probes to genotype the target sites of programmable nucleases in cellula. Previous experiments have deployed both assays to independently confirm target DNA cleavage upon ZFN expression (Schierling et al., 2012). In most instances, however, only one of the two is used. In the present study, we utilized side-by-side both genotyping assays to determine in a quantitative manner their relative performance. Our data indicate that the T7 endonuclease I-based procedure, compatible with virtually any target site of interest, discloses rates of targeted mutagenesis, which are in the range of those revealed by using less flexible assays dependent on the presence of a restriction enzyme recognition sequence within the spacer of nuclease target sites. These data should be informative to those interested in selecting readout methods for accessing site-specific DSB formation in cells.
In summary, we have established that the inhibition of cellular HDACs constitutes a straightforward methodology to maximize targeted mutagenesis after IDLV-mediated delivery of ZFNs into human cells. Future research should address whether this combinatorial approach based on IDLVs and chromatin modifiers can also be applied in the realm of other genome engineering procedures involving ZFNs or, conceivably, other genome-modifying enzymes (see, e.g., Cornu and Cathomen, 2007; Moldt et al., 2008; Morioka et al., 2009; Staunstrup et al., 2009; Staunstrup and Mikkelsen, 2011).
Supplementary Material
Acknowledgments
The authors thank Josephine M. Janssen for her excellent technical assistance, Prof. Rob Hoeben for his critical reading of the manuscript, Dr. Arnaud Zaldumbide for providing the reagents used in the caspase activities assays, and Dr. David Baker for providing the monoclonal anti-Flag antibody (all from the Department of Molecular Cell Biology of the Leiden University Medical Center, Leiden, The Netherlands). This work was supported by the Prinses Beatrix Spierfonds (W.OR11-18 to M.A.F.V.G.) and by the European Community's 7th Framework Programme for Research and Technological Development (PERSIST—Persisting Transgenesis, Grant agreement number 222878 to T.C. and M.A.F.V.G.).
Author Disclosure Statement
The authors declare no conflicts of interest.
References
- Archin N.M., Liberty A.L., Kashuba A.D., et al. (2012). Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485. Erratum in: Nature 2012, 489, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armeanu S., Pathil A., Venturelli S., et al. (2005). Apoptosis on hepatoma cells but not on primary hepatocytes by histone deacetylase inhibitors valproate and ITF2357. J. Hepatol. 42, 210–217 [DOI] [PubMed] [Google Scholar]
- Arrowsmith C.H., Bountra C., Fish P.V., et al. (2012). Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 [DOI] [PubMed] [Google Scholar]
- Berger S.L. (2002). Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148 [DOI] [PubMed] [Google Scholar]
- Biasco L., Baricordi C., and Aiuti A. (2012). Retroviral integrations in gene therapy trials. Mol. Ther. 20, 709–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland A., Buglio D., and Younes A. (2010). Histone deacetylase inhibitors in lymphoma. Curr. Opin. Oncol. 22, 431–436 [DOI] [PubMed] [Google Scholar]
- Cornu T.I., and Cathomen T. (2007). Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 15, 2107–2113 [DOI] [PubMed] [Google Scholar]
- Cudré-Mauroux C., Occhiodoro T., König S., et al. (2003). Lentivector-mediated transfer of Bmi-1 and telomerase in muscle satellite cells yields a duchenne myoblast cell line with long-term genotypic and phenotypic stability. Hum. Gene Ther. 14, 1525–1533 [DOI] [PubMed] [Google Scholar]
- Daboussi F., Zaslavskiy M., Poirot L., et al. (2012). Chromosomal context and epigenetic mechanisms control the efficacy of genome editing by rare-cutting designer endonucleases. Nucleic Acids Res. 40, 6367–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco S., Chini M.G., Terracciano S., et al. (2013). Structural basis for the design and synthesis of selective HDAC inhibitors. Bioorg. Med. Chem. 21, 3795–3807 [DOI] [PubMed] [Google Scholar]
- Doyon Y., Choi V.M., Xia D.F., et al. (2010). Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods 7, 459–460 [DOI] [PubMed] [Google Scholar]
- Gabriel R., Lombardo A., Arens A., et al. (2011). An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 29, 816–823 [DOI] [PubMed] [Google Scholar]
- Gonçalves M.A.F.V., van der Velde I., Knaän-Shanzer S., et al. (2004). Stable transduction of large DNA by high-capacity adeno-associated virus/adenovirus hybrid vectors. Virology 321, 287–296 [DOI] [PubMed] [Google Scholar]
- Gonçalves M.A.F.V., Swildens J., Holkers M., et al. (2008). Genetic complementation of human muscle cells via directed stem cell fusion. Mol. Ther. 16, 741–748 [DOI] [PubMed] [Google Scholar]
- Gonçalves M.A., Janssen J.M., Holkers M., and de Vries A.A. (2010). Rapid and sensitive lentivirus vector-based conditional gene expression assay to monitor and quantify cell fusion activity. PLoS One 5, e10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves M.A.F.V., Janssen J.M., Nguyen Q.G., et al. (2011). Transcription factor rational design improves directed differentiation of human mesenchymal stem cells into skeletal myocytes. Mol. Ther. 19, 1331–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höher T., Wallace L., Khan K., et al. (2012). Highly efficient zinc-finger nuclease-mediated disruption of an eGFP transgene in keratinocyte stem cells without impairment of stem cell properties. Stem Cell Rev. 8, 426–434 [DOI] [PubMed] [Google Scholar]
- Holkers M., Maggio I., Liu J., et al. (2013). Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 41, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Stein C.S., Heth J.A., et al. (2002). In vivo gene transfer using a nonprimate lentiviral vector pseudotyped with Ross River Virus glycoproteins. J. Virol. 76, 9378–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor B., Ma H., Webster-Cyriaque J., et al. (2009). Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc. Natl. Acad. Sci. USA 106, 18786–18791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Um E., Cho S.R., et al. (2011). Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods 8, 941–943 [DOI] [PubMed] [Google Scholar]
- Klug A. (2010). The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 79, 213–231 [DOI] [PubMed] [Google Scholar]
- Kobinger G.P., Deng S., Louboutin J.P., et al. (2004). Transduction of human islets with pseudotyped lentiviral vectors. Hum. Gene Ther. 15, 211–219 [DOI] [PubMed] [Google Scholar]
- Kushner E.J., MacEneaney O.J., Weil B.R., et al. (2011) Aging is associated with a proapoptotic endothelial progenitor cell phenotype. J. Vasc. Res. 48, 408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Genovese P., Beausejour C.M., et al. (2007). Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25, 1298–1306 [DOI] [PubMed] [Google Scholar]
- Lombardo A., Cesana D., Genovese P., et al. (2011). Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods 8, 861–869 [DOI] [PubMed] [Google Scholar]
- Miller J.C., Holmes M.C., Wang J., et al. (2007). An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785 [DOI] [PubMed] [Google Scholar]
- Moldt B., Staunstrup N.H., Jakobsen M., et al. (2008). Genomic insertion of lentiviral DNA circles directed by the yeast Flp recombinase. BMC Biotechnol. 8, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka Y., Isotani A., Oshima R.G., et al. (2009). Placenta-specific gene activation and inactivation using integrase-defective lentiviral vectors with the Cre/LoxP system. Genesis 47, 793–798 [DOI] [PubMed] [Google Scholar]
- Mussolino C., Morbitzer R., Lütge F., et al. (2011). A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelascini L.P., and Gonçalves M.A.F.V. (2013). Lentiviral vectors encoding zinc-finger nucleases specific for the model target locus HPRT1. Methods Mol. Biol. 1114, 455. [DOI] [PubMed] [Google Scholar]
- Pelascini L.P., Janssen J.M., and Gonçalves M.A.F.V. (2013). Histone deacetylase inhibition activates transgene expression from integration-defective lentiviral vectors in dividing and non-dividing cells. Hum. Gene Ther. 24, 78–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe S., Sarkis C., Barkats M., et al. (2006). Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc. Natl. Acad. Sci. USA 103, 17684–17689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott N.J., and Thrasher A.J. (2007). Use of nonintegrating lentiviral vectors for gene therapy. Hum. Gene Ther. 18, 483–489 [DOI] [PubMed] [Google Scholar]
- Provasi E., Genovese P., Lombardo A., et al. (2012). Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 18, 807–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S.H., Maeder M.L., Joung J.K., et al. (2011). Zinc-finger nucleases for somatic gene therapy: the next frontier. Hum. Gene Ther. 22, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricks D.M., Kutner R., Zhang X.Y., et al. (2008). Optimized lentiviral transduction of mouse bone marrow-derived mesenchymal stem cells. Stem Cells Dev. 17, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riu E., Chen Z.Y., Xu H., et al. (2007). Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol. Ther. 15, 1348–1355 [DOI] [PubMed] [Google Scholar]
- Ross P.J., Kennedy M.A., Christou C., et al. (2011). Assembly of helper-dependent adenovirus DNA into chromatin promotes efficient gene expression. J. Virol. 85, 3950–3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierling B., Dannemann N., Gabsalilow L., et al. (2012). A novel zinc-finger nuclease platform with a sequence-specific cleavage module. Nucleic Acids Res. 40, 2623–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder A.R., Shinn P., Chen H., et al. (2002). HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 [DOI] [PubMed] [Google Scholar]
- Selvi R.B., and Kundu T.K. (2009). Reversible acetylation of chromatin: implication in regulation of gene expression, disease and therapeutics. Biotechnol. J. 4, 375–390 [DOI] [PubMed] [Google Scholar]
- Staunstrup N.H., and Mikkelsen J.G. (2011). Integrase-defective lentiviral vectors—a stage for nonviral integration machineries. Curr. Gene Ther. 11, 350–362 [DOI] [PubMed] [Google Scholar]
- Staunstrup N.H., Moldt B., Mátés L., et al. (2009). Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol. Ther. 17, 1205–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M., Brondani V., Büchel J., et al. (2007). Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 25, 786–793 [DOI] [PubMed] [Google Scholar]
- Urnov F.D., Rebar E.J., Holmes M.C., et al. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 [DOI] [PubMed] [Google Scholar]
- van Rensburg R., Beyer I., Yao X.Y., et al. (2013). Chromatin structure of two genomic sites for targeted transgene integration in induced pluripotent stem cells and hematopoietic stem cells. Gene Ther. 20, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga C.M., Tedford N.C., Thomas M., et al. (2005). Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 12, 1023–1032 [DOI] [PubMed] [Google Scholar]
- Vojinovic J., and Damjanov N. (2011). HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol. Med. 17, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanisch K., and Yáñez-Muñoz R.J. (2009). Integration-deficient lentiviral vectors: a slow coming of age. Mol. Ther. 17, 1316–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li Y., Crise B., et al. (2003). Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 [DOI] [PubMed] [Google Scholar]
- Zabner J., Fasbender A.J., Moninger T., et al. (1995). Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 270, 18997–19007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.