Abstract
Here we present approaches for using multi-elemental imaging (specifically synchrotron X-ray fluorescence microscopy, SXRF) in ionomics, with examples using the model plant Arabidopsis thaliana. The complexity of each approach depends on the amount of a priori information available for the gene and/or phenotype being studied. Three approaches are outlined, which apply to experimental situations where a gene of interest has been identified but has an unknown phenotype (Phenotyping), an unidentified gene is associated with a known phenotype (Gene Cloning) and finally, a Screening approach, where both gene and phenotype are unknown. These approaches make use of open-access, online databases with which plant molecular genetics researchers working in the model plant Arabidopsis will be familiar, in particular the Ionomics Hub and online transcriptomic databases such as the Arabidopsis eFP browser. The approaches and examples we describe are based on the assumption that altering the expression of ion transporters can result in changes in elemental distribution. We provide methodological details on using elemental imaging to aid or accelerate gene functional characterization by narrowing down the search for candidate genes to the tissues in which elemental distributions are altered. We use synchrotron X-ray microprobes as a technique of choice, which can now be used to image all parts of an Arabidopsis plant in a hydrated state. We present elemental images of leaves, stem, root, siliques and germinating hypocotyls.
Keywords: Metallomics, ionomics, SXRF, gene characterization, elemental homeostasis
Introduction
1) Ionomics
Ionomics is defined as the study of the quantitative and simultaneous measurement of the elemental composition of living organisms, and changes in this composition in response to physiological stimuli, developmental state, and genetic modification1. Ionomics encompasses metallomics, which is concerned with the analysis of the entirety of metal and metalloids, with identification of their binding ligands within a cell or tissue type2. Approximately one third of all proteins are associated with a metal ion, with the most common elements being the micronutrients zinc (Zn) and iron (Fe) and the macronutrients calcium (Ca) and magnesium (Mg) (Protein Data Bank, http://www.rcsb.org).
The work described here is primarily concerned with characterizing genes that regulate elemental homeostasis in plants, and use the model plant Arabidopsis thaliana (L.) Heynh. (Mouse-ear cress). Arabidopsis is a member of the mustard family (Brassicaceae), which includes agronomically important species such as cabbage, broccoli and rapeseed. Similar approaches to those described here are equally applicable to other model systems, for which a range of genetic and genomic tools is available. Arabidopsis is used as a model plant for the study of cell and molecular biology of flowering plants, but resources are also available for grasses such as rice (Oryza sativa L.)3, and legumes such as Medicago trunculata and soybean (Glycine max)4. A wide range of plant species has been imaged via synchrotron X-ray fluorescence, including hyperaccumulator plants and agronomic crop species. Interested readers are referred to5, 6 for further information.
The distribution of essential and potentially toxic elements to cells and organelles for assimilation depend on transport proteins: membrane-intrinsic proteins that allow specific ions to pass through under tight homeostatic regulation. Increases in the physiological demand for a particular element prompt a cascade of genetic signals, changing gene expression and resulting in an increase in transport proteins in the target membrane. This increases trafficking of the target element across the membrane, causing changes in the cellular or subcellular localization of the target elements. Changes in elemental localization on a cellular and even sub-cellular level can be imaged by elemental mapping (for a review of techniques see5, 7. The ideal techniques for mapping elemental localization in plant tissues are those that preclude the need for sample preservation (which can alter the distribution and/or abundance of elements) and that also allow simultaneous collection of a suite of biologically relevant elements. Synchrotron X-ray fluorescence mapping has emerged as a technique of choice for gene characterization studies5, and is rapidly adapting to the needs of the life sciences community7, 8.
2) Applications of ionomics research
An understanding of ion homeostasis in plants has applications in biofortification, food safety and environmental remediation. Biofortification uses traditional plant breeding or genetic engineering to increase the amount and/or bioavailability of nutrients in the edible portions of crop plants9-15. Essential mineral nutrients such as Fe, Zn and Ca have been targeted for biofortification as a way of making them more accessible to the rural poor, where supplementation is often unsuccessful. Additionally, selenium (Se) is a target of biofortification in Europe and the UK following a decline in the Se status of Europeans16. Wheat is no longer imported from the Se-enriched soils of North America, and European and UK soils lack this essential micronutrient. Traditionally associated with women and children living in poor, rural areas, micronutrient malnutrition affects more than half of the world’s population17. Further, an increasing body of data indicates that the Western diet is particularly nutrient-poor as a result of the predominance of polished cereal grains (where the nutrient-rich bran and germ are removed) and processed foods18.
Issues of food safety center around the uptake of non-essential, potentially toxic elements by crop plants. Approaches similar to those used in biofortification can be used to develop plants with inhibited transport of specific elements in to their edible tissues, such as seeds and leaves. For example, the discovery of accumulation of the metalloid arsenic (As) in rice (Oryza sativa L.) grown both in the As-contaminated soils of Bangladesh and South East Asia19, and the U.S. and Europe20. Rice has an enhanced ability to accumulate As compared other cereal crops, such as wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.)21, as a result of differential regulation of silicon (Si) transporters, through which trivalent inorganic As gains access. The substrate specificity of membrane transporters plays a pivotal role in uptake of non-essential elements: uptake of many contaminant elements arises from the non-specificity of nutrient transporters. Examples include the uptake of cadmium (Cd) via Fe transporters22, and uptake of 137Cs via potassium (K) transporters23.
Environmental remediation using plants to recover metal(loid)s while improving soil quality is known as phytoextraction24. The study of metal tolerance in plants25 revealed that certain plant species could thrive in soils with metal concentrations toxic to the majority of other species, adapting via a number of strategies, one of which included hyperaccumulation26, 27. Previous studies show that ectopic expression of vacuolar metal transporters can increase plant tolerance to toxic elements28-31. This has led to the idea that insertion of genes conferring these extraordinary metal tolerances into the genome of high-biomass plants will allow the development of plants designed to clean up contaminated soils, by removing metal contaminants from the rhizosphere into easily harvestable tissue32, 33. Further, the availability of technologies that convert plant biomass into energy and the recovery of metals from ash suggest this is an area ripe for commercial exploitation.
3) Using SXRF in ionomics
Studying ion homeostasis in plants requires measurement of elemental accumulation under various experimental conditions and, in particular, determination of elemental distribution within and between plant organs. Although ion sensitive probes such as fluorophores and histological stains can be used to image the distribution of a limited suite of elements and can aid in our understanding of how elemental distribution is affected by changes in gene expression34-38, elemental imaging such as SXRF microscopy offers a number of advantages.
SXRF beamlines are designed to apply the technique of X-ray fluorescence analysis in a spatially resolved manner with high detection sensitivity and with minimal sample preparation. SXRF uses characteristic X-rays emitted from atoms excited by an external source (in this case synchrotron X-rays) for elemental analysis. Detection sensitivity is dictated by the beamline design (which varies with scientific focus and facility), the characteristics of the sample and the specific optical configuration used for a particular experiment. For some elements detection can be in the ppb range, and detection limits of 10-100 ppm for heavy and transition elements are not uncommon. Detection sensitivity is a function of atomic number due to differences in fluorescence yield, which decreases with decreasing atomic number, and the fact that less energetic X-ray emissions are more easily absorbed by the sample, the air path and sample/detector filters39. It can be difficult to quantify elements lighter than sodium (Na) due to low X-ray yields. The more significant restriction for plant scientists is absorption of characteristic X-ray both by the sample itself and by the air path between the sample and detector. Given that most instruments are designed to allow the researcher to analyze materials under ambient conditions, typically in air or a low absorption environment such as helium, and given the types of sources and optics most commonly used, the elements broadly accessible to SXRF analysis of biological tissues fall between P and Cd in the Periodic Table using characteristic X-rays from the K electron shell for these elements, and between Y and Pu when using fluorescence from the L electron shell (http://www.cxro.lbl.gov/PDF/X-Ray-Data-Booklet.pdf). Therefore a very large portion of the periodic table can be investigated, depending on concentrations, potential spectral overlaps and beamline configuration.
Because X-ray microprobes rely on the X-ray fluorescence process for identification and quantification of specific elements and because these energy levels are distinct for each atom, analysis via SXRF at the appropriate energy shows the distribution and abundance of an element regardless of the chemical form, location and binding ligand of that element. The escape depth, (the depth from which characteristic X-rays can escape the sample and reach the detector) is well characterized and is a function of X-ray energy and the X-ray attenuation of the sample (with thickness in biological materials generally being the most important component). As long as the incident X-ray beam energy is above an elements’ binding energy and the characteristic X-ray have sufficient energy to escape the sample and reach the detector; SXRF imaging collects data for multiple elements simultaneously. This multi-elemental capability is critical from an ionomic standpoint, because concurrent elemental responses can implicate responsible genes, if they correspond to the targets of a particular membrane transporter. Generally, SXRF requires little to no chemical sample preservation. The biggest consideration is in using trace element-free substrates and keeping the sample motionless during analysis, without causing scattered radiation from mounting materials or obstructing the beam path. Analysis times have shortened by an order of magnitude in recent years, as a result of the use of high brightness sources that provide 1000× more flux, advances in detector technology, software processing and stage design. Concurrently, the move toward more brilliant, higher flux synchrotrons has made beam damage the more prominent challenge for fresh tissue analysis. In addition to faster scanning, sample stages designed to maintain a humid or cold environment for fresh biological tissues are particularly useful.
4) Complementary tools
The Ionomics Hub (iHub)40 is a searchable online database for elemental profiles, with 175,074 samples from Arabidopsis thaliana shoots and seeds already analyzed, which includes 12,942 unique lines (natural accessions and mutants). The database can be searched by specific lines, gene name, or by the element of interest, including Ca, K, Mg, phosphorous (P), sulfur (S), rubidium (Rb), strontium (Sr), boron (B), cobalt (Co), copper (Cu), Fe, lithium (Li), manganese (Mn), molybdenum (Mo), nickel (Ni), Se, Zn, As, Cd and sodium (Na). iHub also has databases for rice (Oryza sativa) soybean (Glycine max) and yeast. For the gene cloning approach described here, the iHub is a valuable starting point for finding genes potentially responsible for a particular elemental phenotype.
Exploring Arabidopsis genotypes and/or phenotypes using SXRF
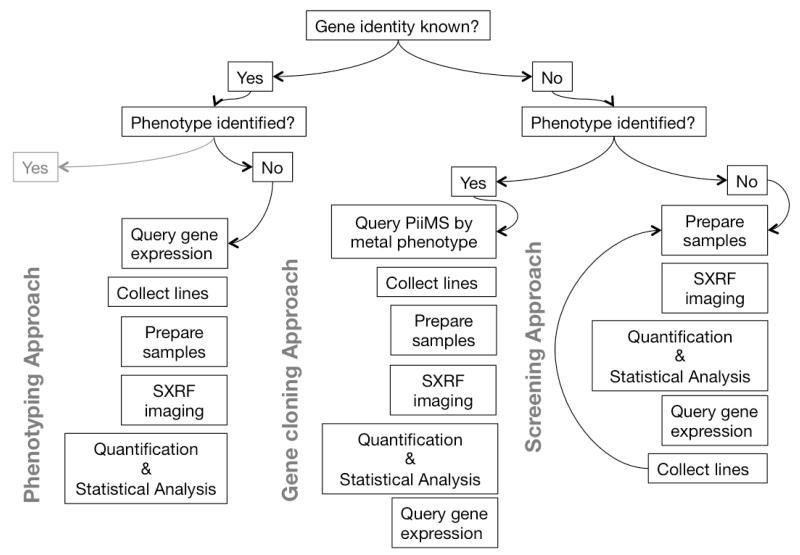
A key assumption of using spatially resolved elemental analysis to characterize genes is that the target element(s) coincide spatially with the activity of the protein of interest. Co-localization was observed for the gene vacuolar iron transporter 1 (VIT1)41, where expression in the endodermal layer of the Arabidopsis embryo42 coincided with the distribution of Fe. This co-localization has a physiological basis, in that gene expression, protein synthesis and the resultant elemental redistribution occurred in close proximity on a cellular level. This allowed us to develop a series of workflows for using SXRF as a tool in ionomics, using the co-localization assumption to generate hypotheses about gene expression, directing gene searches to a particular tissue layer, developmental stage or cell organelle. These workflows are summarized in Figure 1, and are described below along with data as examples of the possible outcomes of combining SXRF with a priori information.
Figure 1.
A summary of workflows for using elemental imaging in gene functional characterization, based on the extent of a priori information.
Although the approaches described here make use of this assumption, there are exceptions. Membrane bound transport proteins such as HMA2/HMA4, that are responsible for xylem loading in to the roots of A. thaliana and A. halleri43, 44 are an example where expression and activity of a gene/protein can result in changes in elemental concentrations in distant sites, because HMA2/HMA4 transport into the xylem leads to Zn translocation to shoots. Further, of the limited examples of SXRF use as a metallomics tool in the literature, work has shown that it need not be restricted to membrane-integral transport proteins45. In this case, SXRF was used to show overexpression of cytoplasmically localized nicotianamine synthase enzymes in the rice endosperm, which resulted in a higher abundance of Fe and Zn in this tissue.
Approach 1: Phenotyping
This example uses VIT1 as an example for the Phenotyping Approach. Here we demonstrate how use of two online transcriptomic databases led directly to the analysis of the embryo, silique and seed as the most likely location for detection of an Fe distributional phenotype in WT and vit1-1 loss of function lines (Figure 2).
Figure 2.
Phenotyping approach: applies when the identity of the gene is known, but there is no information on its physiological function or phenotypic changes caused by altered gene expression (i.e., loss-of-function or over-expression). As an example, we show how publicly available transcriptomic data on AtVIT1 would lead to seed analysis and the finding the AtVIT1 is responsible for Fe localization in the vasculature. AtVIT1 expression based on microarray data available on Genevestigator (A) and eFP browser (B) databases shows high expression in the embryo (A), and at the maturation stage (B). Elemental maps of Fe in the organs where expression is highest (indicated by red boxes on panels A and B) shows distributional phenotypes in the embryo (C) and developing siliques (D). (C) Transversal imaging of whole seeds shows Fe localization in association with the vasculature in wild-type (WT), but mislocalized in vit1-1 loss-of-function mutant (C, upper panels). Fe distribution in endodermal cell layer in wild-type (WT) and vit-1-1 (C mid panel) and in single cells (C, lower panel). Colourbars show the minimum (lower) and maximum (higher) values scaled between WT and vit1-1, and scale bars are from 100 μm, 10 μm and 2 μm from top to bottom. (D) Fe (red), Zn (green) and Mn (blue) localization in full siliques during seed filling. Upper panel shows siliques 7 days DAF (embryogenesis); lower panel shows siliques after 12 DAF (maturation).
1) Query gene expression to select sample organ(s)
Although the indirect effects of the function of a particular gene may manifest in loss-of-function mutants or overexpression lines in the form of growth defects such as slow germination or early flowering, mapping the elemental distribution in the plant organ(s) in which the gene of interest is maximally expressed is intuitive. For the model plant Arabidopsis, there are a number of publicly available transcriptomic databases where gene expression can be queried in terms of organ, tissue, cell type, developmental stage and treatment accumulation. Websites such as Genevestigator46, and the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi, Winter et al 2007) allow the user to explore expression metadata from many microarray experiments. In our example, we analyzed the expression pattern of VIT1 (AT2G01770). In Genevestigator, we used wild-type data only, with linear expression level values, and showed this in a scatterplot with the anatomy data (Figure 2A). In eFP browser, we used an absolute scale and 300 as signal threshold for all pictographs, and showed Seed and Developmental Map data. High expression should determine preferential sites for SXRF analyses. Specific datasets that are not integrated to user-friendly databases might also be interesting. These are often deposited under accession number in repositories like Gene Expression Omnibus (GEO47 http://www.ncbi.nlm.nih.gov/geo/).
2) Generate/order transgenic lines
Once the expression site of the candidate gene is determined, knockout or partial loss-of-function mutants (i.e., little to no expression of the gene) can be obtained or generated for comparative SXRF analyses with wild-type. Extensive collections of T-DNA insertion lines of Arabidopsis with known flanking sequence tags (FST) are found at Arabidopsis Biological Resource Center (ABRC; http://abrc.osu.edu/). These lines are often already homozygous for the knockout mutation, and can be easily ordered. For candidate genes that are not interrupted in any readily available line, knockdown mutants (i.e., silenced by RNAi or artificial microRNA48, 49) can be generated. Overexpression and/or tissue-specific expression of the gene of interest in the mutant A. thaliana line background could be also generated to complement the loss-of-function phenotyping.
3) Sample preparation
a) Embedding or in vivo analysis
The need for and type of sample preparation should always be established in consultation with the beamline scientist for the particular synchrotron beamline to be used in the experiment. Hard X-ray microprobe beamlines using detectors that operate in an ambient environment have the advantage of being suitable for the analysis of hydrated samples or liquids. The use of low absorption environment enclosures around the sample and the detector optimize detector capture of low energy characteristic X-rays such as P, S, Ca, and Si, but place limitations on maintaining the hydrated condition of the sample during analysis.
While SXRF provides a significant benefit by allowing analysis of whole, unsectioned samples, in order to take advantage of focused beam dimensions that are now routinely are under 1 μm, samples should be sectioned rather thinly, especially where three-dimensional analysis (tomography) is not available. The penetrative nature of the X-rays means that a researcher may be interrogating material at hundreds of micrometers depth if the characteristic X-rays are of sufficiently high energy, making the resultant elemental images difficult to interpret. The use of resin embedding is generally not recommended because of the risk of redistribution and alteration of the speciation of elements of interest during dehydration and resin infiltration, and the likelihood of contamination with elements associated with resin embedding preparation. Chemical sample preparation should always be supported with in vivo studies demonstrating a lack of preparation artifacts. Figure 2C shows a comparison between in vivo analysis (Fe in whole seeds), and analysis of resin embedded tissues of an Arabidopsis embryonic root tissue. In the former, samples were prepared as described in Punshon et al,50. In this case, the analysis focused on Fe tightly bound within vacuoles, and was not disrupted by the sample preparation technique.
b) Sectioning and sample thickness
Analysis of sectioned tissue via SXRF imaging has the added advantage of presenting a known and uniform thickness of material for analysis, which in turn allows more accurate calculations of abundance and escape depth for each element of interest; the latter providing an understanding of how many tissue layers are being represented in an elemental map. Whether sectioned or unsectioned, determining sample thickness is typically essential in elemental mapping. Sample thickness measurements not made during sectioning can often be determined by focusing on the sample mounting material and sample surface or can be measured directly using incident X-rays arsorption by the sample and assuming a reasonable X-ray attenuation coefficient.
4) Mounting samples for in vivo analysis
Where live or fresh tissues can be analyzed directly, sample stabilization for the duration of analysis is needed. Under ideal circumstances, plants should be grown in identical conditions to those under which they will be analyzed, to prevent responses to sudden changes in humidity, light and temperature. Growth of plants on agar plates, although a common laboratory practice, is generally not ideal, because of the stress caused by rapid transition of plants from the high humidity of the plates to the ambient humidity in the beamline hutch. Although growth in soil can present contamination issues, it is possible to distinguish the characteristic elemental spectra of the experimental soil used to exclude it from any anomalous distributions, and forced air canisters can be used to remove soil particulates from plants tissues prior to analysis. Plants can conceivably be germinated and soil-grown to the desired stage in smaller containers that can be placed directly on a tomography or XY stage sample holder. Agar-grown plants can be acclimated to ambient humidity levels prior to analysis, by gradually removing growth container lids for several hours preceding their analysis.
Plant organs such as leaves, roots and flattened cereal grains can be imaged in two dimensions by mounting on an XY stage. Kapton™ polyamide adhesive tape or conventional double-sided tape can be used for sample mounting, although the adhesives can contain high amounts of Br, S and Cl. For very moist samples such as root tips, Kapton™ or Ultralene™ film (available in thicknesses down to 4 μm) can be applied over the sample, sealing it onto the adhesive tape. Under these conditions, detached leaves and roots remain stable for periods of analysis exceeding one hour. Where root architecture needs to be preserved, growth of seedlings on agar covered in nylon mesh allows root removal without damage.
Unsectioned plant organs such as stems and larger grains can be imaged using a tomography or XYθ stage. Computed X-ray tomography, analogous to methods employed in conventional medical CAT scans, can provide a spatially resolved image of the distribution of elements in plants even for samples that are thick relative to the focused X-ray beam. Reconstruction software for these beamlines allows researchers to remove overlapping layers of tissue in silico in three dimensions (e.g. Chimera51).
Sample support structures such as phenolic foams that can be impregnated with water are useful in tomography of hydrated tissue sections such as roots and stem. Although modern X-ray microprobes go to great lengths to provide highly collimated beams with minimal scatter, some amount of scattered radiation around the focused beam is inevitable. As such, it is best for sample mounts for tomography to be metal-free even though the X-ray beam does not penetrate them directly.
5) Analysis
a) Analyzing fluorescence spectra
Most SXRF beamlines collect X-ray fluorescence using solid state energy dispersive detectors. The most common detectors used today typically provide energy resolutions between 170-200 eV for the elements of interest to plant biologists. Although spectral overlaps at these resolutions are generally distinct, careful analysis of the collected spectrum obtained at each pixel of the SXRF image or tomogram is important to ensure correct identification of spectral peaks52. Modern beamlines today, driven by improved electronics, faster networks and larger data storage capacity, also typically acquire full energy dispersive data for each pixel in the image, even when images are megapixel in size or larger. Collection of whole fluorescence spectra has the advantage of being accessible for reconstructing all elements within the energy range, which is useful when unexpected elemental associations are revealed by analysis.
b) Spectral overlap
Analysis of the energy of fluorescent X-rays highlights several important overlaps to be aware of during plant tissue analysis. Potential overlaps can be numerous and are dependent on the relative concentrations in the sample being interrogated, the incident beam energy being used (whether a particular element being is being excited at that energy) and the energy resolution of the detector being utilized at the beamline. It is advisable to discuss any potential interferences that may be encountered with the beamline scientists beforehand. That being said, in looking at plant materials a certain set of spectral interferences (given the most commonly used energy dispersive detectors at modern beamlines and the elements most likely to be observed) are particularly noteworthy to keep in mind (Table 2). As an example, the close proximity of the emission lines of the abundant macronutrient K (the potassium K-edge binding energy is 3.61 keV, its Kα1 emission energy is 3.31 keV and Kβ1 is 3.59 keV) to the L emission lines for Cd (L3-edge binding energy 3.54 keV, Lα13.13, Lβ13.32 keV) make analysis of Cd challenging. Particularly problematic is the fact that, due to its toxicity, one is typically attempting to visualize Cd at very low concentrations in a sample that likely contains 1000’s of ppm of K. Ways to get around this issue include using a beamline that can tune the incident beam energy to excite the Cd L3 at 3.54 keV without exciting K fluorescence at 3.61 keV, or to use a beamline that can tune the incident beam energy above 26.7 keV so that Cd can be analyzed at the K-edge. An example of the latter is given by Fukuda et al. 53, who used high-energy X-rays (37 keV) to reveal the subcellular distribution of Cd in the Cd-hyperaccumulator Arabidopsis halleri ssp. gemmifera, and used the Cd Kα1 emission line at 23.17 keV. Likewise, osmium tetroxide (OsO4), a common biological stain used in transmission electron microscopy has a Lβ1 fluorescence line which overlies the Kα1 of arsenic (As). Generally plant tissue analyzed for As cannot be prepared via traditional electron microscopy techniques, or in laboratories regularly using OsO4. The close proximity of spectral peaks of Fe and Mn, Ni and Cu and Cu and Zn mean that peak fitting is often needed when one or other of these elements is enriched, to check that spectral overlap of the Kβ peaks does not cause artifacts in the quantification of the neighboring element. In cases where the Kβ peak of an element impinges upon the Kα peak of another, simple mathematical calculations can be used to subtract the counts.
Table 2.
List of likely spectral overlaps in SXRF analysis of biological tissue, assuming a 170 eV spectral width (typical of silicon drift detectors).
| Element | Electron Binding Energy |
Potential spectral overlaps (assuming 200 eV resolution) |
|---|---|---|
| P | 2145 (K) | Generally no significant natural overlaps on Kα, but with silicon-based detectors Ca Kα escape peak can significantly overlap. |
| S | 2472 (K) | Generally no significant natural overlaps on Kα, but with silicon-based detectors Ca Kβ escape peak can significantly overlap. |
| K | 3608 (K) | Overlap on Kα with Ar Kβ from air excitation |
| Ca | 4039 (K) | Overlap on Kα with K Kβ |
| Ti | 4966 (K) | Generally no significant overlaps on Kα |
| V | 5465 (K) | Overlap on Kα with Ti Kβ |
| Cr | 5989 (K) | Overlap on Kα with V Kβ |
| Mn | 6539 (K) | Overlap on Kα with Cr Kβ if Cr concentrations high, some potential minor overlap with Fe Kα if Fe concentrations significantly higher than Mn |
| Fe | 7112 (K) | Overlap on Kα with Mn Kβ |
| Co | 7709 (K) | Overlap on Kα with Fe Kβ |
| Ni | 8333 (K) | Generally no significant overlaps on Kα |
| Cu | 8979 (K) | Generally no significant overlaps on Kα |
| Zn | 9659 (K) | Generally no significant overlaps on Kα |
| As | 11867(K) | Overlap on Kα with Pb Lα1 |
| Se | 12658(K) | Generally no significant overlaps on Kα |
| Rb | 15200(K) | Generally no significant overlaps on Kα |
| Sr | 16106 (K) | Generally no significant overlaps on Kα, potential Fe Kα pileup overlap at very high Fe concentrations |
| Mo | 20000 (K) / 2520 (L3) | Overlap on Lα with S Kα |
| Ag | 25514 (K) / 3351 (L3) | Overlap on Lα with Ar Kα from air excitation |
| Cd | 26711 (K) / 3538 (L3) | Overlap on Lα with Ar Kβ from air excitation |
c) Experimental design
The sub-micron nature of SXRF mapping means that much smaller elemental masses can be analyzed in comparison with bulk techniques. During bulk analysis, such as inductively coupled plasma mass spectroscopy (ICP-MS) a much higher elemental mass is needed to obtain a volume-averaged metal concentration, which suppresses natural variation in elemental content. For example, ICP-MS can accurately determine metal concentration in samples weighing less than 1 mg, which in the case of Arabidopsis seed equates to approximately 50-100 seeds. In SXRF, small numbers of samples are compared and quantified. The influence of microclimate variations during plant growth and natural variation can have a confounding effect when analysis is at a sub-micron scale. When imaging metal distribution phenotypes, a wild-type grown alongside the mutant of interest should always be analyzed, and confirmation of phenotypes by replicated analysis of plants grown at different times should be standard practice. In general, bulk elemental analysis of plant material should be conducted to determine whether elemental concentrations fall within the detection limits for the particular beamline used for SXRF analysis, but can also be used to check for batch-to-batch variation in growth conditions.
6) Standards-based data quantification
Standard-based quantification of normalized fluorescence count data in to either μg cm−2 or μg g−1 require the analysis of standard reference materials during each beamline experiment. In XRF analysis generally, a standards based approach for quantification works best when standards are matrix-matched to the unknowns and where both standards and unknowns are ideally homogenous and isotropic. Unfortunately, for X-ray microprobes standards that are matrix matched to the samples being analyzed and are homogeneous at the micrometer scale are few. Certified thin-film standards that consist of trace element enriched glasses and monolayers deposited on to polycarbonate filters or silicon nitride windows are often available at synchrotron beamlines and are generally analyzed prior to the beginning of the experiment. This allows the user to measure the fluorescence response during each experiment. Measurement of sample density is required when converting μg cm−2 into μg g−1, and measurement of sample thickness is required for quantification. Standard-based quantification is provided as an additional feature in the beamline-specific software (e.g. Sam’s Microanalysis Toolkit: SMAK54, or as a separate program (e.g. NRLXRF 55-57).
7) Results
Results from Genevestigator and eFP browser point to a role for VIT1 in seed and embryo development (Figure 2A and 2B), suggesting these organs and growth stages are the best starting point for elemental imaging. Previous studies in yeast had provided a great deal of information about the function of VIT1 on a cellular level, as a vacuolar influxer of Fe and Mn41. However seed from loss of function mutant vit1-1 did not show reduction in Fe or Mn. According to Genevestigator, VIT1 transcript accumulation is low, but an expression peak in A. thaliana organs and tissues is found in the embryo (Figure 2A). The expression level is considered to be medium (the highest observed for this particular gene) in the embryo, which accounts for seed and silique medium levels as shown (Figure 2A).
The eFP browser data shows that VIT1 expression changes throughout seed development. According to one dataset (Figure 2B, upper panel) VIT1 transcripts start to accumulate at the linear cotyledon stage, and peaks during embryo maturation and greening. The second dataset (Figure 2B, lower panel) shows higher VIT1 expression at the torpedo and walking stick stages, but lower later on during maturation.
Based on publicly available databases of transcriptomics, seeds would be the organ/tissue of choice for SXRF. Thus, wild-type Col-0 and vit1-1 seeds were analyzed by SXRF tomography (41; Figure 2C). In wild-type, Fe is localized close to the vasculature of the cotyledons and the radicle, more precisely in the endodermal layer. In vit1-1, Fe localization is disrupted, being associated with the abaxial surface of cotyledons and outside the endodermis in the radicle (Figure 2C). High resolution subcellular imaging (Figure 2C) showed localization of Fe in large subcellular bodies in WT, compared to numerous bodies in vit1-1 containing comparatively lower abundances of Fe. Studies in yeast and in plants showed localization of VIT1 to the vacuolar membrane41, therefore it is assumed that these subcellular bodies are the vacuoles of quiescent endodermal cells of the embryo.
The comparative elemental composition of soil-grown WT and vit1-1 siliques during embryonic development was studied by SXRF imaging at 7 and 12 days after flowering (Figure 2D). This showed that transport of both Fe and Zn to developing embryos was delayed in the vit1-1 loss of function mutant.
Approach 2: Gene cloning
This is a theoretical workflow, based on the assumptions made in the first tested approach, and has no example published in the scientific literature. For the purpose of this review, we have generated an example using the workflow stages described. It is based on the knowledge that metal concentrations vary amongst A. thaliana natural accessions. SXRF techniques can help to dissect which cells, groups of cells or subcellular organelles contribute to the variation in metal concentration detected by volume-averaged methods such as ICP-MS in whole organs and, once a site is determined, help to identify the molecular players involved in metal-specific accumulation at that site. Again, this approach works on the assumption that gene product and metal distribution are co-localized.
Workflow stages:
1) Query ionomicsHub/ Ionomics Atlas database
In order to find variability for the selected phenotype, the publicly available ionomicsHub (iHub) website (www.ionomicshub.org,40 has the ionomics profile of leaves from 348 A. thaliana natural accessions, and seed profile for a subset of 96 accessions. Thus, this approach is more suited to phenotypes found in one of these two organs. Contrasting accessions that contain high and low concentrations of specific or multiple elements should be identified. For the purpose of this workflow example, we used two accessions with contrasting concentrations of Ca in their leaves: Ull2-5 (74,119 μg/g) and TOU-E-11 (59,344 μg/g) (Figure 3). In comparison, the upper and lower Ca concentrations in all accessions are 76,247 μg/g and 47,851 μg/g (Cvi-0 and Sei-0 accessions, respectively). Ideally, it would be preferable to find accessions with a range of concentrations in addition to simply the highest and lowest, as the phenotype is likely to be quantitative.
Figure 3.
Gene cloning approach: Using SXRF imaging to find genes responsible for tissue-specific metal accumulation within organs. (A) Calcium elemental maps of Arabidopsis natural accessions Ull2-5 and TOU-E-11, isolated from an iHub ionomic screen for high and low leaf Ca (74,119.49 μg/g and 59,343.65 μg/g, respectively), to identify differences in Ca distribution or abundance in ecotypes with contrasting Ca profiles revealed the trichome as a sink for Ca. Maps were conducted at the Stanford Synchrotron Radiation Lightsource (SSRL) BL2-3 at 11 keV, using a 2 × 2 μm beam, 7 μm steps and 50 ms dwell. Region of Interest (ROI) data extraction showing (B) total intensity of Ca per trichome (mean ± SE) and (C) percentage of total leaf Ca localized to trichomes and other leaf tissue.
2) Collect lines/natural accessions
A. thaliana natural accessions are available and can be ordered at ABRC (http://abrc.osu.edu/), as described in Approach 1.
3) Imaging and analysis
Imaging should be performed as described in workflow 1. Considering our example, 2D mapping of leaves would be the method of choice. One of the primary reasons for conducting spatially resolved elemental analysis is to statistically compare the elemental profile of cellular or subcellular features, often too small for isolation or extraction via other means. Thus, selection of the data from a specific area within an elemental map, and comparison with similar areas in other maps is a basic post-processing function. Beamline software allows the user to extract the data from a particular feature54 and conduct descriptive statistical analysis. Frequently used tests include correlation or regression between elements of interest, and analysis of variance or t-tests between samples. As shown (Figure 3), abundance differences can easily be picked out. In most cases, the user will be able to identify the specific anatomical feature by comparison of elemental images with micrographs collected from the sample. In situations where the user in unaware of elemental features, cluster recognition analysis – supplied with many of the beamline software applications, allow statistically distinct features to be picked out.
4) Query gene expression databases for likely candidates
The ability to find site-specific metal accumulation opens the possibility to clone genes involved in determining the observed distribution. ICP-MS measurements from natural accessions were used with Mendelian genetics to clone genes responsible for Cd58, Co59 and Mo60 accumulation in leaves. Although a similar method has the potential to be combined with SXRF, this technique is not currently sufficiently high-throughput. Depending on the beamline and leaf size, at present an elemental map can generally take between anywhere from 3 to 12 hours to collect, making it hard to analyze more than 10 to 20 genotypes, although mapping speed will increase in future. Still, when the metal accumulation site is determined, one can switch to a higher throughput technique. For example, trichomes from several natural accessions could be isolated61 and measured for metal concentration by a volume-averaged method such as ICP-MS.
A more feasible option is to take advantage of publicly available gene expression databases, as described in Approach 1, to find candidate genes for phenotypes found by SXRF imaging. Cell type/tissue-specific microarray dataset should be analyzed to find transcripts that are enriched in the metal accumulation sites of interest. Both Genevestigator and eFP browser have comprehensive transcriptomic datasets allowing researchers to generate lists of site-specific expressed genes. In our example, transcripts that show high expression in trichomes but lower expression in leaves should be considered61. At this point, knowledge of element homeostasis is necessary, as the candidate gene selection would be based on function description. Although not available, site-specific gene expression from natural variation combined with site-specific metal localization quantification by SXRF would have the potential to give the best candidate genes in the future.
5) Generate loss-of-function/ silenced lines and/or test gene expression in natural variation
The function of good candidate genes could be tested in loss-of-function mutants, which would be comparatively analyzed with WT plants to determine the effects of the absence of the gene product on site-specific metal localization. Mutant lines ordering/generation and SXRF should be performed as described in approach 1. Another possibility is that the variation observed in distinct accessions is explained by expression level. To test this, the simplest choice would be RT-qPCR. Higher or lower gene expression would be correlated with high/low metal accumulation. Considering our example, trichomes could be prepared and tested as described61. Once correlation is found, genomic loci sequencing and fusion with GUS would provide further evidence for different alleles involvement in metal abundance variation.
6) Results
Ca is localized in the trichome cells as already described in the literature62, 63. Mapping leaves shows that Ca is indeed localized in trichomes in both accessions, and that the high Ca accession Ull2-5 has a higher Ca concentration in its trichomes compared to the low Ca accession TOU-E-11 (Figure 3A). By using region of interest (ROI) analyses, it is possible to dissect the contributions of trichomes to the total Ca abundance in each leaf. After ROI selection, individual trichome information was obtained, such as total intensity, mean, standard deviation for all elements detected in that area, as well as number of pixels. The same data can be obtained from leaf surface (which does not consider trichomes) and for the whole leaf. This information allows Ca abundance quantification in each trichome of a leaf, as well as partitioning the total Ca present in whole leaf into trichome and non-trichome fractions. The trichomes of high Ca accession Ull2-5, compared to low Ca accession TOU-E-11, showed higher Ca abundance (Figure 3B). The partitioning also shows that Ull2-5 has a higher percentage of its Ca in trichomes compared to TOU-E-11 (25% to 16%, respectively; Fig 3C).
As mentioned, approach 2 does not have a published example. To find candidate genes for Ca abundance variation observed in Figure 3A, both Genevestigator and eFP browser have comprehensive transcriptomic datasets allowing researchers to generate lists of site-specific expressed genes, such as trichomes61. In our example, transcripts that show high expression in trichomes and code for proteins of families already related to Ca homeostasis such as Ca transporters or Ca-binding proteins would be more likely candidates.
For the purpose of this example, a trichome-specific dataset61, 64 is best suited to find candidate genes. We searched for calcium transporters that are highly expressed in trichomes compared to leaves after trichome removal. Table 1 shows the selected genes that are increased in trichomes (Figure 4). Although not tested, these proteins would be good candidates for Ca localization in trichomes.
Table 1.
Candidate genes related to calcium transport highly expressed in trichomesa.
| AGI Locus | Annotation | Expression Ratiob |
|---|---|---|
| AT1G53210 | Sodium/calcium exchanger family protein / calcium-binding EF hand family protein |
2.2 |
| AT3G22910 | Calcium-transporting ATPase, plasma membrane-type, putative / Ca(2+)− ATPase, putative (ACA13) |
65 |
| AT4G03560 | ATTPC1 (TWO-PORE CHANNEL 1); calcium channel/ voltage-gated calcium channel |
1.97 |
| AT5G57110 | ACA8 (AUTOINHIBITED CA2+ −ATPASE, ISOFORM 8); calcium-transporting ATPase/ calmodulin binding |
4 |
Figure 4.
Screening approach: Rapid surveying for differences in elemental distributions and/or abundance using sample arrays. (A) Elemental image of macro and micronutrient elements in an Arabidopsis thaliana seed array showing wild type ecotype Columbia-0 and vit1-1 (vacuolar iron transporter 1 loss of function mutant) seeds. Col-0 seeds were obtained from the same plant. (B) Within plant seed-to-seed variation in total elemental abundances is assessed. Individual seed abundance for K, Ca, Cu, Zn, Fe and Mn is shown.
Approach 3: Screening
Accepting the assumptions that in many cases gene expression and metal distributional phenotypes spatially coincide, it follows that a screening approach, where neither the identity nor the change in phenotype associated with loss-of-function of a gene are known, would potentially yield information. In this approach, imaging elemental distribution in tissues grown and prepared under identical conditions are screened, often in an array formation (Figure 4), at relatively low spatial resolutions, so that any distributional or abundance differences can be picked out.
Workflow stages:
Analysis via SXRF: In this example the sample matrix consists of an array of seeds, which would ideally be collected from of a range of lines grown under identical conditions, such as high or low metal concentrations, but here are shown for Arabidopsis wild-type Columbia-0. Because they are naturally dehydrated, seeds have been used to test the screening approach, but as sample preservation and stabilization techniques improve, it is anticipated that leaves, roots or even whole seedlings can be arrayed and screened.
Quantification and post-processing analysis: The appearance of distributional or abundance variations are easy to pick out from arrays compared to single samples, and thus the variety or similarity in responses can be identified easily in an array. The involvement of a particular cell type or tissue layer in the response can be derived visually, or quantified using statistical comparison of isolated regions of interest. General reductions in elemental abundances can be easily established using region of interest analysis. In this example, vit1-1 mutant seed, known to change Fe localization, was included to show localization differences.
Query gene expression databases for likely candidates: Online transcriptomic databases can be used to survey gene expression at a specific growth stage and in a specific organ. As shown in Figure 2, some browsers will provide specific tissue layer expression information. Thus, a response that is consistently found in a particular tissue layer can be used to provide a list of candidate genes. Alternately, more direct measurement of mRNA in extracted cells can be carried out, involving laser capture microdissection65.
Generate/order loss-of-function mutant lines
Growth conditions: Exposure of loss-of-function or overexpression lines to growth conditions known to produce a phenotype.
Imaging and analysis: In this case, the imaging would test the hypothesis that the phenotype is no longer evident in loss-of-function lines, or an enhanced phenotype in over-expression lines to provide functional evidence for the gene responsible.
Discussion
Limitations
The approaches described here hinge upon spatial co-localization of gene expression and the resultant modification in metal distribution. Imaging the metal distribution of genetically modified or mutant plants on a tissue, cell or subcellular level can provide information about the function of the gene of interest. By the same token, the distribution of an element is the result of functional transporters, and can be used as the starting point for an ionomic search for the responsible genes.
The ability to determine metal concentrations in specific cells or tissues, combined with expression data from these same cells, was used to describe the function of CAX1 and CAX3 in vacuolar-specific accumulation of Ca in mesophyll cells compared to epidermal cells66. The same approach showed that two Mg transporters are related to Mg accumulation in mesophyll cells67. By using whole leaf transcriptomic analyses of natural accessions68, 69 and leaf ionomic profiles found in ionomics Atlas, Conn et al.9 pulled out candidates for metal accumulation variation9. In a similar way, SXRF can be used to combine metal abundance variation in specific sites within plan organs with transcriptomics data from natural accessions, which would be a step forward to clone genes involved in site-specific metal distribution.
Here we have provided methodological approaches, with examples, for using SXRF in functional characterization of metal-related genes. Complementary techniques, such as X-ray Absorption Spectroscopy and its use in oxidation state mapping70 can be used to collect maps that distinguish between the oxidation states of elements, which is particularly important for elements like As and Cr. This approach was used to explore the role of an arsenate reductase gene in the As-accumulating fern Pteris vittata71. As synchrotrons continue to adjust to the needs of the life science community, this kind of imaging will be highly informative.
The use of fluorescent tags to show gene expression, or to identify a particular organelle is common practice in molecular genetics. Synchrotron beamlines frequently have fluorescence microscopy capabilities, although they have not been used extensively to show co-localization between gene expression and elemental distribution patterns. There is great potential to combine fluorescent tagging to either confirm organelle or membrane identity in elemental imaging, or to learn more about the spatial co-localization between gene expression and elemental distribution.
Acknowledgements
The authors wish to thank Antonio Lanzirotti (UCARS, University of Chicago) for helpful comments on this manuscript. This work was supported by grants from the National Institute of Environmental Health Sciences, (Superfund Research Program P42 ES007373-17), Department of Energy award DEFG02-06ER15809, the Children’s Environmental Health and Disease Prevention Center (P20 ES018175 and RD-83459901) and Center of Biomedical Research Excellence (COBRE) (P20 GM104416) to TP. We thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) for supporting FKR with a research scholarship. Portions of this work were performed at: (1) Beamline X26A, National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. X26A is supported by the Department of Energy (DOE) - Geosciences (DE-FG02-92ER14244 to The University of Chicago - CARS). Use of the NSLS was supported by DOE under Contract No. DE-AC02-98CH10886; (2) Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH. (3) Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Bibliography
- 1.Salt DE, Baxter I, Lahner B. Annual Review of Plant Biology. 2008;59:709–733. doi: 10.1146/annurev.arplant.59.032607.092942. [DOI] [PubMed] [Google Scholar]
- 2.Shi J, Wang H, Schellin K, Li B, Faller M, Stoop JM, Meeley RB, Ertl DS, Ranch JP, Glassman K. Nature Biotechnol. 2007;25:930–937. doi: 10.1038/nbt1322. [DOI] [PubMed] [Google Scholar]
- 3.Jung K-H, Jeon J-S, Gynheung A. J Pl Biol. 2011;54:65–80. [Google Scholar]
- 4.Gepts P, Beavis WD, Brummer EC, Shoemaker RC, Stalker HT, Weeden NF, Young ND. Plant Physiol. 2005;137:1228–1235. doi: 10.1104/pp.105.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donner E, Punshon T, Guerinot M, Lombi E. Analytical and Bioanalytical Chemistry. 2012:1–12. doi: 10.1007/s00216-011-5624-9. ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombi E, de Jonge MD, Donner E, Kopittke PM, Howard DL, Kirkham R, Ryan CG, Paterson D. PLoS ONE. 2011;6:e20626. doi: 10.1371/journal.pone.0020626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombi E, Scheckel KG, Kempson I. Environ Exp Bot. 2011;72:3–17. [Google Scholar]
- 8.Paunesku T, Vogt S, Maser J, Lai B, Woloschak G. J Cell Biochem. 2006;99:1489–1502. doi: 10.1002/jcb.21047. [DOI] [PubMed] [Google Scholar]
- 9.Conn SJ, Berninger P, Broadley MR, Gilliham M. New Phytol. 2012;193:859–866. doi: 10.1111/j.1469-8137.2011.03977.x. [DOI] [PubMed] [Google Scholar]
- 10.Hirschi KD. Nutrient biofortification of food crops. In: Cousins R, Bier D, Bowman B, Dean L, editors. Annual Reviews of Nutrition. Vol. 29. Annual Reviews, Inc.; Palo Alto, CA.: 2009. pp. 401–421. [DOI] [PubMed] [Google Scholar]
- 11.Murgia I, Arosio P, Tarantino D, Soave C. Trends Plant Sci. 2012;17:47–55. doi: 10.1016/j.tplants.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Nestel P, Bouis HE, M JV, Pfeiffer W. Journal of Nutrition. 2006;136:1064–1067. doi: 10.1093/jn/136.4.1064. [DOI] [PubMed] [Google Scholar]
- 13.Waters BM, Sankaran RP. Plant Science. 2011;180:562–574. doi: 10.1016/j.plantsci.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 14.White PJ, Broadley MR. Trends Plant Sci. 2005;10:586–593. doi: 10.1016/j.tplants.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhao F-J, McGrath SP. Curr. Opin. Plant Biol. 2009;12:373–380. doi: 10.1016/j.pbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Broadley MR, White PJ, Bryson RJ, Meacham MC, Bowen HC, Johnson SE, Hawkesford MJ, McGrath SP, Zhao FJ, Breward N, Harriman M, Tucker M. Proc Nutr Soc. 2006;65:169–181. doi: 10.1079/pns2006490. [DOI] [PubMed] [Google Scholar]
- 17.UNSCN . UNSCo Nutrition. Geneva: 2010. Sixth report on the world nutrition situation. [Google Scholar]
- 18.Grotto D, Zied E. Nutr. Clin. Pract. 2010;25:603–612. doi: 10.1177/0884533610386234. [DOI] [PubMed] [Google Scholar]
- 19.Meharg AA, Rahman M. Environ. Sci. Technol. 2003;37:229–234. doi: 10.1021/es0259842. [DOI] [PubMed] [Google Scholar]
- 20.Williams PN, Price AH, Raab A, Hossain MA, Feldmann J, Meharg AA. Environ. Sci. Technol. 2005;39:5531–5540. doi: 10.1021/es0502324. [DOI] [PubMed] [Google Scholar]
- 21.Mitani N, Chiba Y, Yamaji N, Ma JF. Plant Cell. 2009;21:2133–2142. doi: 10.1105/tpc.109.067884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB. Pl Mol Biol. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- 23.Zhu YG, Smolders E. J Exp Bot. 2000;51:1635–1645. doi: 10.1093/jexbot/51.351.1635. [DOI] [PubMed] [Google Scholar]
- 24.Mackova M, Dowling DN, Macek T. Phytoremediation and Rhizoremediation. Springer; New York: 2006. [Google Scholar]
- 25.Baker AJM. New Phytol. 1987;106(suppl.):93–111. [Google Scholar]
- 26.Hanikenne M, Nouet C. Curr. Opin. Plant Biol. 2011;14:252–259. doi: 10.1016/j.pbi.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Krämer U. Plant Biol. 2010;61:517–534. doi: 10.1146/annurev-arplant-042809-112156. [DOI] [PubMed] [Google Scholar]
- 28.Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR. Plant Cell. 2003;15:1131–1142. doi: 10.1105/tpc.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanquar V, Lelievre F, Bolte S, Hames C, Alcon C, Neumann D, Vansuyt G, Curie C, A. S, Kramer U, Barbier-Brygoo H, Thomine S. EMBO. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanquar V, Schnell Ramos M, Lelievre F, Barbier-Brygoo H, Krieger-Liszkay A, Kramer U, Thomine S. Plant Physiol. 2010;152:1986–1999. doi: 10.1104/pp.109.150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirschi KD, Korenkov VD, Wiganowski NL, Wagner GJ. Plant Physiol. 2000;124:125–133. doi: 10.1104/pp.124.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheoran V, Sheoran AS, Poonia P. Crit Rev Environ Sci Technol. 2010;41:168–214. [Google Scholar]
- 33.Seth CS, Remans T, Keunen E, Jozefczak M, Gielen H, Opdenakker K, Weyens N, Vangronsveld J, Cuypers A. Plant Cell Environ. 2012;35:334–346. doi: 10.1111/j.1365-3040.2011.02338.x. [DOI] [PubMed] [Google Scholar]
- 34.Sinclair SA, Sherson SM, Jarvis R, Camakaris J, Cobbett CS. New Phytol. 2007;174:39–45. doi: 10.1111/j.1469-8137.2007.02030.x. [DOI] [PubMed] [Google Scholar]
- 35.Miller EW, Zeng L, Domaille DW, Chang CJ. Nature Protocols. 2006;1:824–827. doi: 10.1038/nprot.2006.140. [DOI] [PubMed] [Google Scholar]
- 36.Dodani SC, Leary SC, Cobine PA, Winge DR, Change CJ. J Am Chem Soc. 2011;133:8606–8616. doi: 10.1021/ja2004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarret G. Plant Physiol. 2006;141:1021–1034. doi: 10.1104/pp.106.082743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawachi M, Kobae Y, Mimura T, Maeshima M. The Journal of Biological Chemistry. 2008;283:8374–8383. doi: 10.1074/jbc.M707646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanzirotti A, Tappero R, Schulze DG. Developments in Soil Science. Vol. 34. Elsevier B.V.; 2010. Practical application of synchrotron-based hard x-ray microprobes in soil sciences; pp. 27–72. [Google Scholar]
- 40.Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. Plant Physiol. 2007;143:600–611. doi: 10.1104/pp.106.092528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SA, Punshon T, Lanzirotti A, Liangtao L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- 42.Roschzttardtz H, Conéjéro G, Curie C, Mari S. Plant Signal Behavior. 2010;5:56–57. doi: 10.4161/psb.5.1.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Kramer U. Nature. 2008;453:391–U344. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- 45.Johnson AT, Kyriacou B, Callahan DL, Carruthers L, Stangoulis J. PLoS ONE. 2011;6:e24476. doi: 10.1371/journal.pone.0024476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Adv Bioinf. 20082008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R, Domrachev M, Lash AE. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterhouse PM, Helliwell CA. Nat Rev Genet. 2003;4:29–38. doi: 10.1038/nrg982. [DOI] [PubMed] [Google Scholar]
- 49.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punshon T, Hirschi KD, Lanzirotti A, Lai B, Guerinot ML. Plant Physiol. 2012;158:352–362. doi: 10.1104/pp.111.184812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Vogt S, Maser J, Jacobsen C. Journal De Physique Iv. 2003;104:617–622. [Google Scholar]
- 53.Fukuda N, Hokura A, Kitajima N, Terada Y, Saito H, Abe T, Nakai I. J Anal Atomic Spectr. 2008;23:1068–1075. [Google Scholar]
- 54.Webb SM. The MicroAnalysis Toolkit: X-ray fluorescence Image Processing Software. The 10th International Conference on X-ray microscopy; AIP Conference Proceedings; Chicago, IL (USA). 2011. pp. 196–199. [Google Scholar]
- 55.Criss JW. Naval Research Laboratory. Washington, D.C.: 1977. NRLXRF, A FORTRAN program for X-ray fluorescence analysis. [Google Scholar]
- 56.Ryan CG, Etschman BE, Vogt S, Masser J, Harland CL, Legnini D. Microscopy & Microanalysis. 2005;11:428–429. [Google Scholar]
- 57.Solé VA, Papillon E, Cotte M, Walter P, Susini J. Spectrochimica Acta Part B. 2007;62:63–68. [Google Scholar]
- 58.Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE. PLoS Genetics. 2012:8. doi: 10.1371/journal.pgen.1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML. The Plant Cell. 2009;21:3326–3338. doi: 10.1105/tpc.109.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baxter I, Muthukumar B, Park HC, Buchner P, Lahner B, Danku J, Zhao K, Lee J, Hawkesford MJ, Guerinot ML, Salt DE. PLoS Genetics. 2008:4. doi: 10.1371/journal.pgen.1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marks MD, Betancur L, Gilding E, Chen F, Bauer S, Wenger JP, Dixon RA, Haigler CH. Plant J. 2008;56:483–492. doi: 10.1111/j.1365-313X.2008.03611.x. [DOI] [PubMed] [Google Scholar]
- 62.Ager FJ, Ynsa MD, Dominguez-Solis JR, Lopez-Martin MC, Gotor C, Romero LC. Nuclear Instruments & Methods in Physics Research Section B-Beam Interactions with Materials and Journal of food protectionAtoms. 2003;210:401–406. Nuclear Instruments & Methods in Physics Research Section B-Beam Interactions with Materials and Atoms. [Google Scholar]
- 63.Sarret G, Willems G, Isaure M-P, Marcus MA, Fakra SC, Frerot H, Pairis S, Geoffroy N, Manceau A, Saumitou-Laprade P. New Phytol. 2009;184:581–595. doi: 10.1111/j.1469-8137.2009.02996.x. [DOI] [PubMed] [Google Scholar]
- 64.Marks MD, Wenger JP, Gilding E, Jilk R, Dixon RA. Molecular Plant. 2009;2:803–822. doi: 10.1093/mp/ssp037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srivastava AC, Palanichelvam K, Ma J, Steele J, Blancaflor EB, Tang Y. BioEnergy Research. 2010;3:278–294. [Google Scholar]
- 66.Conn SJ, Matthew Giliham, Athman A, Schreiber AW, Baumann U, Moller I, Cheng N-H, Stancombe MA, Hirschi KD, Webb AAR, Burton R, Kaiser BN, Tyerman SD, Leigh RA. Plant Cell. 2011 2011 Jan 21; doi: 10.1105/tpc.109.072769. Advance Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conn SJ, Conn V, Tyerman SD, Kaiser BN, Leigh RA, Gilliham M. New Phytol. 2011;190:583–594. doi: 10.1111/j.1469-8137.2010.03619.x. [DOI] [PubMed] [Google Scholar]
- 68.Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. PLoS Genetics. 2005;1:109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gan XC, Stegle O, Behr J, Steffen JG, Drewe P, Hildebrand KL, Lyngsoe R, Schultheiss SJ, Osborne EJ, Sreedharan VT, Kahles A, Bohnert R, Jean G, Derwent P, Kersey P, Belfield EJ, Harberd NP, Kemen E, Toomajian C, Kover PX, Clark RM, Ratsch G, Mott R. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schulze DG, Mccaybuis T, Sutton SR, Huber DM. Phytopathology. 1995;85:990–994. [Google Scholar]
- 71.Ellis DR, Gumaelius L, Indriolo E, Pickering IJ, Banks JA, Salt DE. Plant Physiol. 2006;141:1544–1554. doi: 10.1104/pp.106.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]